Abstract
Obesity increases mortality risk because of metabolic sequelae such as type 2 diabetes and cardiovascular disease1. Thermogenesis by adipocytes can counter obesity and metabolic diseases2,3. In thermogenic fat, creatine liberates a molar excess of mitochondrial ADP to drive thermogenic respiration, hypothesized to be caused by a phosphorylation cycle4. However, the proteins that control this futile creatine cycle are unknown. Here we show that creatine kinase B (CKB) is indispensable for futile creatine cycling-based thermogenesis by trafficking to mitochondria through the utilization of an internal mitochondrial targeting sequence. CKB abundance is powerfully induced by thermogenic stimuli in both mouse and human adipocytes. Adipocyte-selective inactivation of Ckb in mice diminishes thermogenic capacity, increases the predisposition to obesity, and disrupts glucose homeostasis. Thus, CKB is a key effector of the futile creatine cycle.
The obesity epidemic is a major public health concern1. Thermogenesis by brown and beige adipocytes can combat obesity and cardiometabolic diseases2,5-7. Adipocyte-selective creatine depletion impairs thermogenesis and promotes diet-induced obesity8,9, recapitulating the higher fat accretion and lower energy expenditure that occurs with germline deletion of creatine transport (Slc6a8)10 and biosynthesis (Gatm)11 genes, respectively. Creatine kinases catalyze the reversible phosphoryl transfer from ATP to creatine, and comprise four discrete genes in mammals exhibiting tissue-selective expression and varied subcellular localization12,13. Mitochondrial creatine kinases are positioned to phosphorylate creatine from ATP derived from oxidative phosphorylation. This activity liberates ADP in proportion to the amount of accessible creatine such that creatine and ATP exhibit a strict 1:1 stoichiometric relationship with phosphocreatine (PCr) and ADP. However, through the futile creatine cycle, creatine promotes the liberation of a molar excess of ADP to drive thermogenic respiration in fat cells4. Two proteins are posited to regulate futile creatine cycling. First, a creatine kinase catalyzes the phosphoryl transfer from mitochondrial ATP to creatine to simultaneously generate PCr and liberate ADP. Second, a phosphatase that hydrolyzes PCr will regenerate creatine to allow creatine kinase to engage another round of ADP liberation and futile cycling. However, the proteins that control the two steps of this thermogenic cycle have not been defined.
Creatine kinase activity in brown fat
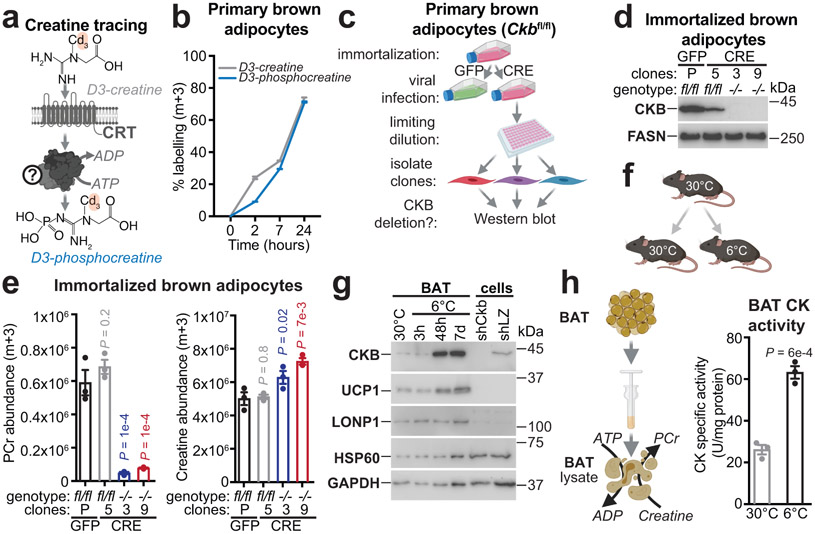
We applied stable isotope tracing by liquid chromatography coupled to mass spectrometry (LC-MS) to investigate creatine kinase activity in brown adipocytes (Fig. 1a). Supplementation of these cells with labeled m+3 deuterated creatine (D3-creatine) led to its accumulation and subsequent appearance of the labeled m+3 isotopologue of PCr (D3-phosphocreatine) (Fig. 1b). D3-phosphocreatine was undetectable in cell media, suggesting that it was synthesized intracellularly (Extended Data Fig. 1a). Thus, brown adipocytes exhibit robust creatine kinase activity.
Fig. 1: Creatine kinase activity in BAT is dependent on CKB.
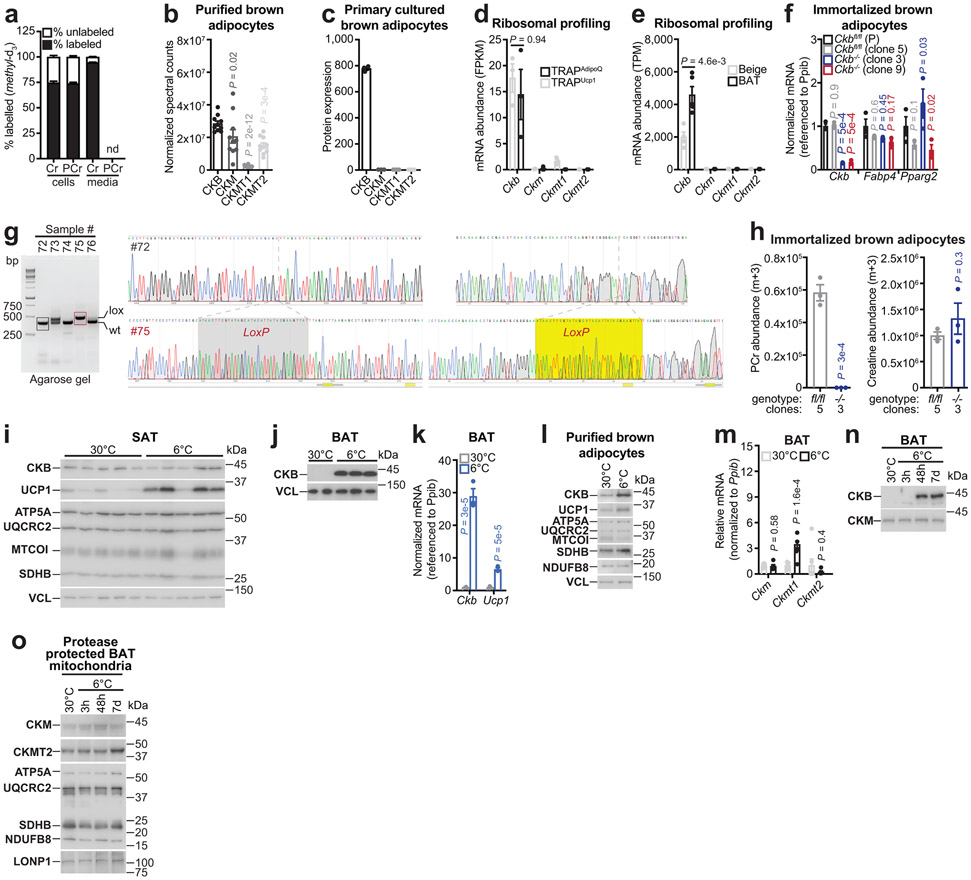
a, Schematic of creatine methyl-D3 (D3-creatine) supplementation of brown adipocytes to test phosphocreatine methyl-d3 (D3-phosphocreatine) synthesis. CRT, creatine transporter. b, Percent-labeling (m+3) of intracellular D3-creatine and D3-phosphocreatine, using LC-MS (n = 3 per time point). c, Cartoon of limiting dilution. d, Western blot of immortalized brown adipocyte clones (parental cell genotype: Ckbfl/fl) following infection with Gfp (P) or with Cre recombinase. Limiting dilution allowed generation of clones (3 and 9) that were devoid of CKB expression (Ckb−/−). e, LC-MS analysis of labeled (m+3) PCr (e, left) and creatine (e, right) levels after labeling for 24 hours (n = 3 per group). f, Cartoon of cold exposure. g, Western blot of BAT from wild-type (C57BL6/N, 6-8 weeks old) male mice, reared at 22°C, housed at 30°C for 7 days and compared to single-housing at 6°C. Primary brown adipocytes (cells) infected with shRNA targeting LacZ (shLZ) or Ckb (shCkb) was used to demonstrate specificity of CKB antibody. h, Cartoon of creatine kinase activity from BAT lysates (h, left). Creatine kinase activity (PCr synthesis) from BAT extracts of wild-type (C57BL6/N, 6-8 weeks old) female mice, reared at 22°C, housed at 30°C for 5 days and then subjected to single-housing at 30°C or 6°C for 48 hours (n = 3 per group) (h, right). For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. e, one-way ANOVA (Fisher’s LSD); h, two-tailed Student’s t-tests. Cartoons were created with biorender.com.
Using quantitative proteomics, we found creatine kinase B (CKB) to be the most abundant creatine kinase isoenzyme in purified brown adipocytes (Extended Data Fig. 1b) and the only isoenzyme detected in cultured primary brown adipocytes (Extended Data Fig. 1c). Mining of two independent ribosomal profiling studies14,15 revealed Ckb to be the major isoform expressed in brown/beige adipocytes (Extended Data Fig. 1d, e). To test the hypothesis that CKB is the dominant creatine kinase in murine brown adipocytes, we constructed mice with loxP sites flanking exon 3 of Ckb (Ckbfl/fl) (Extended Data Fig. 1g). We immortalized Ckbfl/fl brown preadipocytes, infected them with Cre recombinase-expressing adenovirus (or Gfp adenovirus as control), and isolated clones that were CKB-positive (Ckbfl/fl) or CKB-negative (Ckb−/−) (Fig. 1c, d). Ckb inactivation did not affect adipocyte differentiation (Fig. 1d, Extended Data Fig. 1f). Using D3-creatine isotope tracing, we found that the synthesis of D3-phosphocreatine was blocked (85%), while D3-creatine levels were increased (37-56%), in Ckb−/− compared to Ckbfl/fl adipocytes (Fig. 1e, Extended Data Fig. 1h). Thus, CKB constitutes the major creatine kinase activity in brown adipocytes.
CKB expression is regulated by cAMP
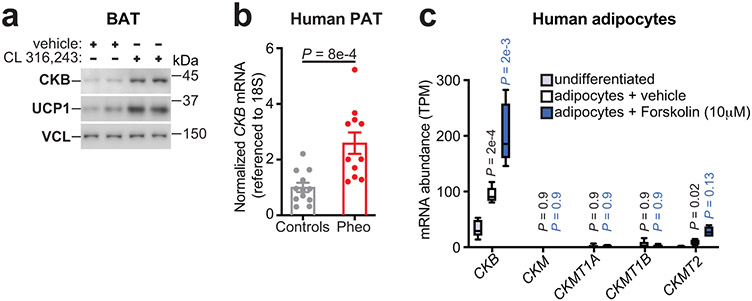
To investigate the physiological regulation of CKB expression in fat, we exposed mice to environmental cold (6°C) or 30°C (Fig. 1f). Ckb mRNA was dramatically elevated (~30-fold) in cold-exposed brown adipose tissue (BAT) (Extended Data Fig. 1k). Similarly, CKB protein abundance was selectively increased in bulk BAT and purified brown adipocytes following cold exposure (Fig. 1g, Extended Data Fig. 1i, j, l). Notably, 6°C exposure increased (~3-fold) creatine kinase activity in BAT compared to 30°C exposure (Fig. 1h). In BAT, Ckm and Ckmt2 mRNA and their protein products were not cold-induced (Extended Data Fig. 1m-o). Ckmt1 mRNA was moderately cold-inducible (Extended Data Fig. 1m), although the expression levels were low (Extended Data Fig. 1b-e). Much of the adipose thermogenic pathway is under cAMP regulation, typically downstream of β3-adrenergic signaling. Pharmacological β3-adrenoreceptor stimulation elicited a profound increase in CKB protein in BAT (Fig. 2a) Similarly, in human adipose tissue, CKB abundance was higher in perirenal BAT of patients with catecholamine-secreting pheochromocytomas, compared to non-tumor-bearing control subjects (Fig. 2b). Furthermore, using publicly available RNA sequencing data from cultured thermogenic human adipocytes16, we discovered that CKB mRNA levels were higher than all the other creatine kinase isoforms (Fig. 2c) Furthermore, direct activation of adenylyl cyclase in these cells significantly increased CKB levels (Fig. 2c). Thus, control of CKB abundance in both mouse and human adipocytes appears to be downstream of cAMP-mediated thermogenic signaling.
Fig. 2: CKB expression is regulated by cAMP signaling.
a, Western blot from male mice injected i.p. with CL 316,243 (1 mg/kg) or vehicle once daily for 48 hours at 30°C. b, RT-qPCR from human perirenal BAT (pheo: n = 11; controls: n = 12). c, Creatine kinase isoform expression from publicly available RNA-seq16 of human cultured thermogenic adipocytes. Statistical comparisons are between undifferentiated and vehicle-treated adipocytes, and between vehicle- and forskolin-treated adipocytes (n = 5 per group). Box plots were generated in Graphpad Prism 8. Boxes stretch from the 25th to the 75th percentile; black horizontal line: median. For gel source data, see Supplementary Figure 1. Mice used were wild-type (C57BL6/N) male (6-8 weeks old). All mice were reared at 22°C and housed at 30°C for 5 days before treatments. Data are presented as mean ± s.e.m. of biologically independent samples. b, c, two-tailed Student’s t-tests.
CKB regulates thermogenesis
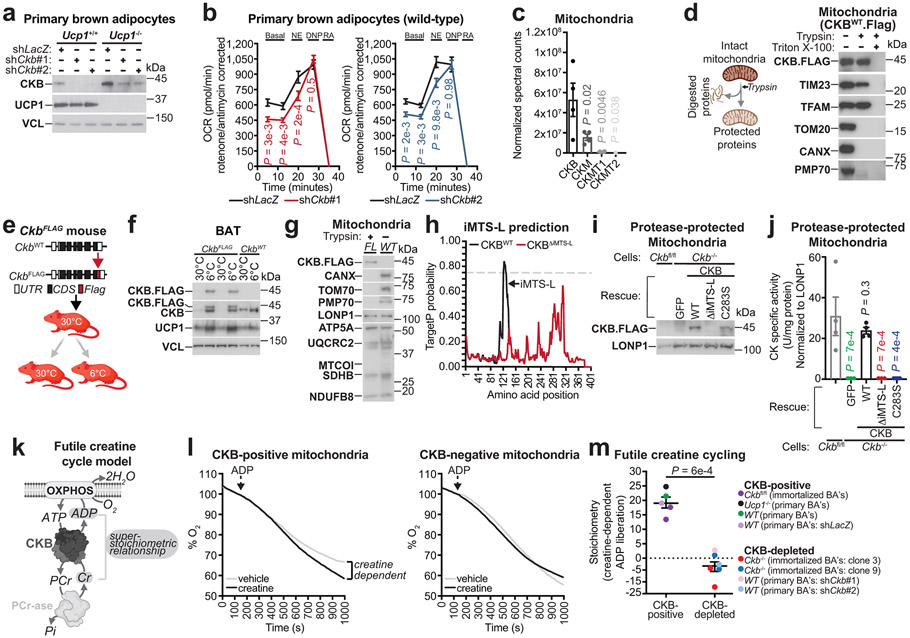
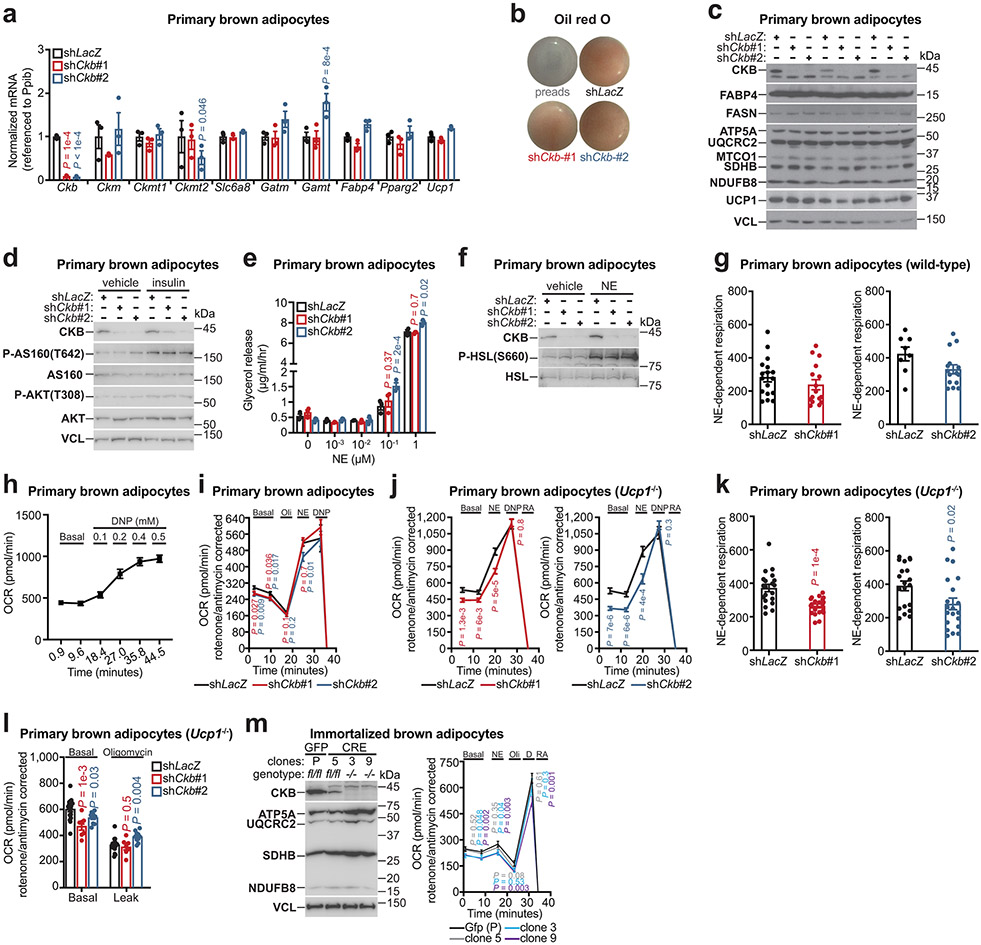
The regulation of CKB expression by thermogenic signals in mouse and human adipocytes prompted us to explore the degree to which CKB controls adipocyte energy expenditure. We silenced Ckb in primary mouse brown adipocytes with two distinct short hairpin RNAs (shRNAs) (Fig. 3a). Ckb silencing did not affect the mRNA expression levels of other creatine kinase isoforms (Ckm, Ckmt1, Ckmt2), the creatine transporter (Slc6a8), creatine biosynthetic genes (Gatm and Gamt), adipogenic markers (Fabp4 and Pparg2), or Ucp1 (Extended Data Fig. 3a). Ckb silencing did not influence adipocyte differentiation, mitochondrial abundance, insulin signaling, or lipolysis (Extended Data Fig. 3b-f). However, silencing of Ckb in wild-type or Ucp1−/− primary brown adipocytes decreased basal oxygen consumption (principally controlled by ATP turnover) and this reduction was maintained following norepinephrine stimulation (Fig. 3b, Extended Data Fig. 3g, j-l). Notably, CKB depletion had no effect on 2,4-dinitrophenol (DNP)-mediated uncoupling (Fig. 3b, Extended Data Fig. 3h-j). Moreover, inhibition of mitochondrial ATP synthesis eliminated the respiration differences between Ckb-depleted and control cells (Extended Data Fig. 3i, l), and silencing of Ckb had no discernable effect on norepinephrine-dependent leak respiration (Extended Data Fig. 3i). Akin to shRNA-mediated knockdown, Ckb−/− immortalized brown adipocyte clones exhibited decreased basal- and norepinephrine-dependent respiration with no effect on DNP-mediated uncoupling (Extended Data Fig. 3m) or mitochondrial abundance (Extended Data Fig. 3m). In aggregate, these data suggest that CKB regulates mitochondrial ATP turnover, not leak respiration.
Fig. 3: CKB is targeted to mitochondria and regulates futile creatine cycling.
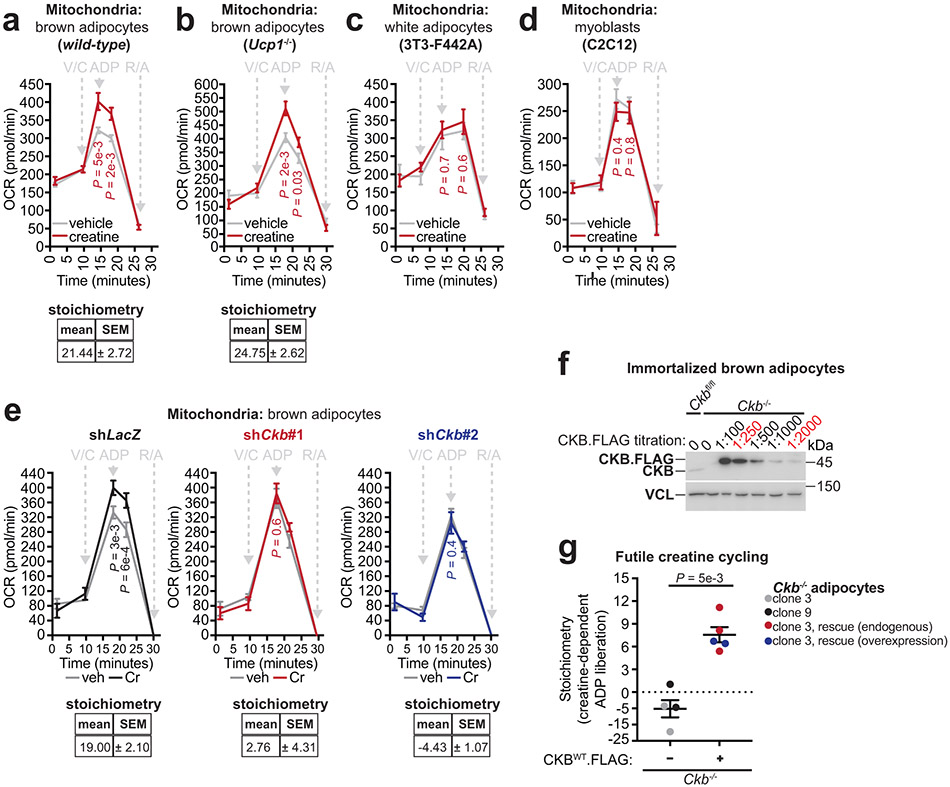
a, Western blot of primary brown adipocytes. b, Oxygen consumption rate (OCR). Left panel: shLacZ, n = 17; shCkb#1, n = 16. Right panel: shLacZ, n = 8; shCkb#2, n = 15. NE (Norepinephrine); DNP (2,4-dinitrophenol); RA (Rotenone and antimycin A). c, Quantitative proteomics of primary brown adipocyte mitochondria (n = 4). d, Cartoon of protease protection assay and Western blot of mitochondrial extracts with and without protease treatment. e, Cartoon of Flag insertion into the Ckb locus (CkbFLAG mice) and cold exposure experiment. UTR (untranslated region); CDS (coding sequence). f, Western blot from BAT of male CkbFLAG or wild-type mice (6-8 weeks of age), reared at 22°C, housed at 30°C for 5 days, then single-housed at 30°C or 6°C for 48 hours. g, Western blot of protease-protected BAT mitochondria from CkbFLAG mice (FL) or non-protease-treated wild-type (WT) mice housed at 22°C. h, TargetP probability plotted along the CKB amino acid sequence. i, Western blot of protease-treated mitochondria. j, Creatine kinase activity from protease-treated mitochondrial extracts. Ckbfl/fl (n =4), Ckb−/− brown adipocytes infected with FLAG-tagged GFP (n = 3), CKBWT (n = 4), CKBΔiMTS-L (n = 4), or CKBC283S (n = 3). k, Futile creatine cycle model. OXPHOS (oxidative phosphorylation); Pi (inorganic phosphate); PCr-ase (protein catalyzing PCr hydrolysis). l, Representative oxygen consumption trace. ADP (70 nmoles) addition is indicated by the arrow. Change in % O2 is relative to ADP (set to 100%). m, Super-stoichiometric action of creatine on ADP-dependent respiration (CKB-positive: n = 5; CKB-negative: n = 6). For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. b, multiple two-tailed Student’s t-tests; c, j, one-way ANOVA (Fisher’s LSD); m, two-tailed Student’s t-test. Cartoons were created with biorender.com.
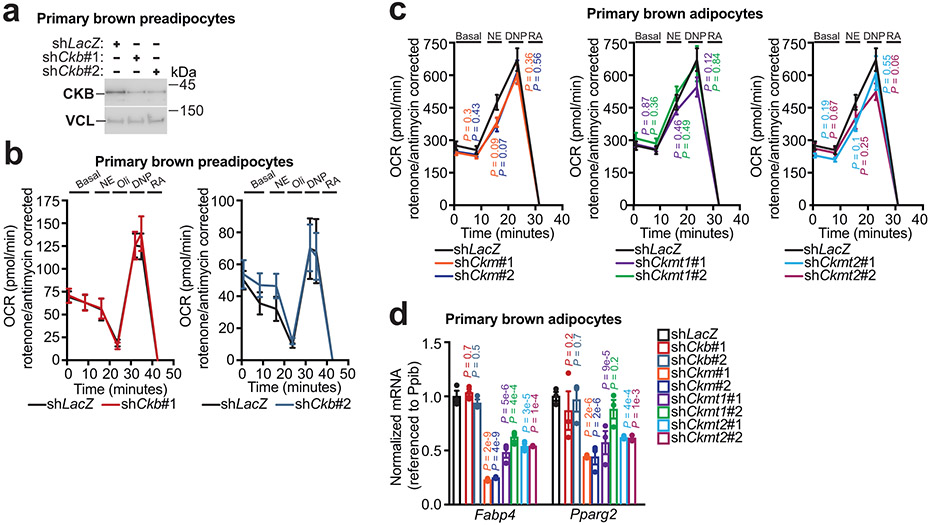
To determine if the decreased respiration following CKB loss was specific to brown adipocytes or was an effect on cellular metabolism more generally, we silenced Ckb in preadipocytes (Extended Data Fig. 4a). However, Ckb silencing did not alter respiration in these non-thermogenic cells (Extended Data Fig. 4b). Next, we evaluated if brown adipocyte respiration is regulated by the other creatine kinase isoenzymes. However, knockdown attempts of Ckm, Ckmt1, and Ckmt2 did not significantly alter respiration (Extended Data Fig. 4c), and any mild respiratory reduction could be explained by compromised differentiation, based on Fabp4 and Pparg2 mRNA abundance (Extended Data Fig. 4d). Thus, brown adipocyte respiration is selectively regulated by CKB with respect to other creatine kinase isoenzymes.
CKB is partly targeted to mitochondria
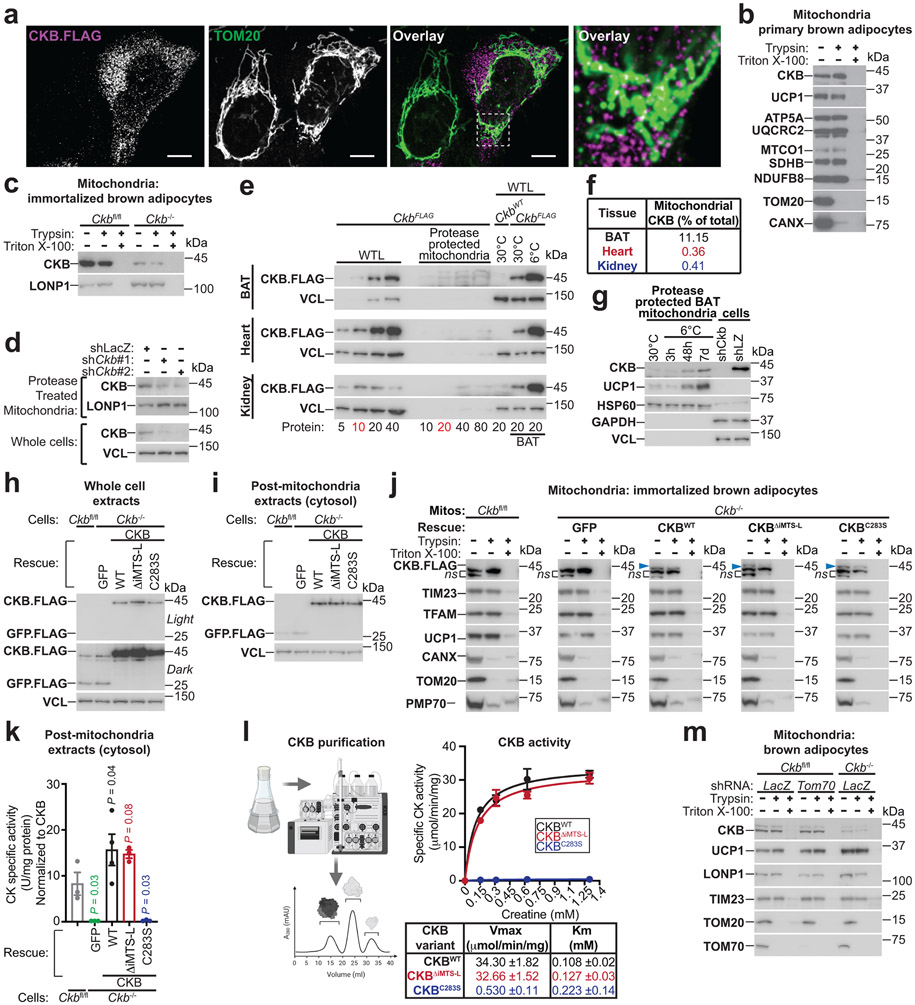
The futile creatine cycling model predicts that mitochondrial creatine kinase phosphorylates creatine from mitochondrial-derived ATP. However, the annotation of CKB as a non-mitochondrial protein appeared inconsistent with this model. Remarkably, using quantitative proteomics, we identified CKB as the most abundant creatine kinase isoenzyme in mitochondria of primary brown adipocytes (Fig. 3c). In addition, carboxy-terminus flag-tagged CKB (CKB.FLAG) co-localized with TOM20 (an import receptor located on the outer mitochondrial membrane), based on immunofluorescence (Extended Data Fig. 5a). Next, we exposed brown adipocyte mitochondria to protease treatment; shielding from proteolysis is a well-established method of demonstrating mitochondrial internalization of multi-localized proteins17,18. Consistent with our proteomics data, both exogenous CKB.FLAG and endogenous CKB was spared from proteolysis, whereas TOM20, CANX (an integral endoplasmic reticulum membrane protein), and PMP70 (a peroxisomal membrane transporter) were degraded (Fig. 3d, Extended Data Fig. 5b-d). To investigate if CKB was targeted to mitochondria in vivo, we constructed knock-in mice with a Flag epitope tag inserted at the carboxy-terminus of the open reading frame of the Ckb locus (CkbFLAG mice) (Fig. 3e). Exposing CkbFLAG mice to environmental cold (Fig. 3e) demonstrated that CKB.FLAG was markedly cold-inducible (Fig. 3f). CKB.FLAG was internalized into mitochondria in vivo, as determined by its protection from proteolysis, whilst CANX, TOM70 and PMP70 were completely degraded (Fig. 3g). In BAT, ~10% of the total CKB pool was internalized by mitochondria, a substantially greater (~30-fold) amount than other CKB-expressing tissues (Extended Data Fig. 5e, f). In addition, BAT mitochondrial CKB levels increased following cold exposure (Extended Data Fig. 5g).
Analysis of the primary CKB amino acid sequence revealed that it contains a strong (0.8 TargetP score) putative internal mitochondrial-targeting signal-like (iMTS-L) sequence (Fig. 3h)19. iMTS-L sequences harbor a high frequency of arginine, and low occurrence of glutamate amino acids19. Mutation of four arginine residues, each to glutamatic acid, within the CKB iMTS-L, was predicted to abolish this putative signal (Fig. 3h). To test this hypothesis directly, we expressed flag-tagged green fluorescent protein (GFP), wild-type CKB (CKBWT), CKB lacking the supposed iMTS-L (CKBΔiMTS-L), or CKB with defective enzymatic activity (CKBC283S)20 in Ckb−/− brown adipocytes. All CKB variants displayed equal expression (Extended Data Fig. 5h, i). CKBWT and CKBC283S were both internalized into mitochondria; however, the import of CKBΔiMTS-L was impeded as demonstrated by its proteolysis (Fig. 3i, Extended Data Fig. 5j). Protease-treated mitochondria from control Ckbfl/fl brown adipocytes exhibited creatine kinase activity, which was nonexistent in Ckb−/− mitochondria (Fig. 3j). Notably, creatine kinase activity was re-established in Ckb−/− adipocyte mitochondria following CKBWT, but not CKBΔiMTS-L, expression (Fig. 3j), supporting the hypothesis that the iMTS-L is required for mitochondrial targeting of CKB. The loss of mitochondrial import and inability to re-establish creatine kinase activity by CKBΔiMTS-L was not caused by misfolding or aggregation, because cytosolic extracts expressing CKBΔiMTS-L displayed similar creatine kinase activity as CKBWT-expressing samples (Extended Data Fig. 5k) and bacteria-purified CKBWT and CKBΔiMTS-L showed comparable Michaelis constants for creatine phosphorylation (Extended Data Fig. 5l). As expected20,21, CKBC283S did not rescue creatine kinase activity in mitochondrial (Fig. 3j) or cytosolic extracts (Extended Data Fig. 5k), and purified CKBC283S protein had a dramatic reduction (98%) in specific activity and an increased Kmcreatine compared to CKBWT and CKBΔiMTS-L (Extended Data Fig. 5l). The utilization of an iMTS-L for mitochondrial import can depend on TOM70 engagement19. However, silencing of Tom70 did not alter CKB levels in brown adipocyte mitochondria (Extended Data Fig. 5m). Collectively, these data demonstrate that CKB is the mitochondrial creatine kinase in brown adipocytes, and that its targeting to the organelle is controlled by recognition of a cis-regulatory iMTS-L element.
CKB regulates futile creatine cycling
The mitochondrial localization of CKB is consistent with its possible role in the futile creatine cycle. We added ADP to mitochondria respiring in an oxygen electrode cell and calculated the stoichiometry of creatine-dependent ADP liberation (measured as oxygen consumption), based on the established P/O coupling ratio22. Our futile creatine cycling model predicts that addition of a given amount of creatine will drive the release of a super-stoichiometric quantity of ADP, resulting in a surplus of oxygen consumption under ADP-limiting conditions (Fig. 3k). Consistent with this hypothesis, creatine liberated an excess of ADP from wild-type (stoichiometry: 21.44, ±2.72) and Ucp1−/− (stoichiometry: 24.75, ±2.62) primary brown adipocyte mitochondria (Extended Data Fig. 6a, b). In contrast, mitochondria from 3T3-F442A white adipocytes and C2C12 myoblasts did not display creatine-dependent super-stoichiometric respiration (Extended Data Fig. 6c, d), suggesting that futile creatine cycling is a unique feature of thermogenic adipocytes.
Next, we investigated if CKB is required for futile creatine cycling. Mitochondria from CKB-expressing (stoichiometry: 19.25, ±1.89), but not Ckb-depleted (stoichiometry: −5.97, ±3.35), brown adipocytes displayed futile creatine cycling (Fig. 3l, m, Extended Data Fig. 6e). Furthermore, futile creatine cycling was rescued to a similar extent in Ckb−/− mitochondria whether CKBWT expression was restored to endogenous levels or overexpressed 5-fold (Extended Data Fig. 6f, g). Thus, Ckb is required and is not limiting for futile creatine cycling-based thermogenesis.
Adipocyte CKB promotes thermogenesis
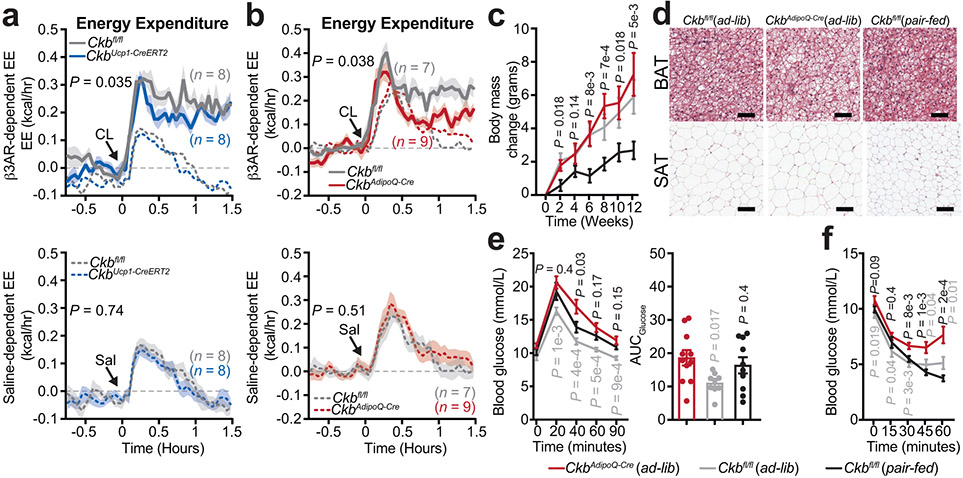
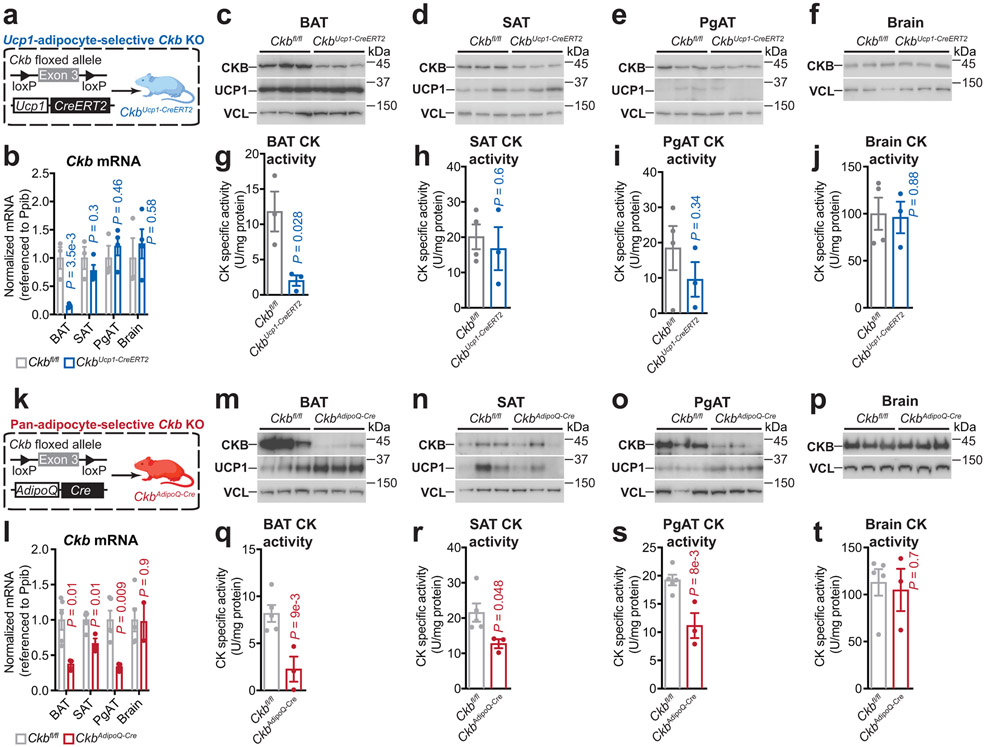
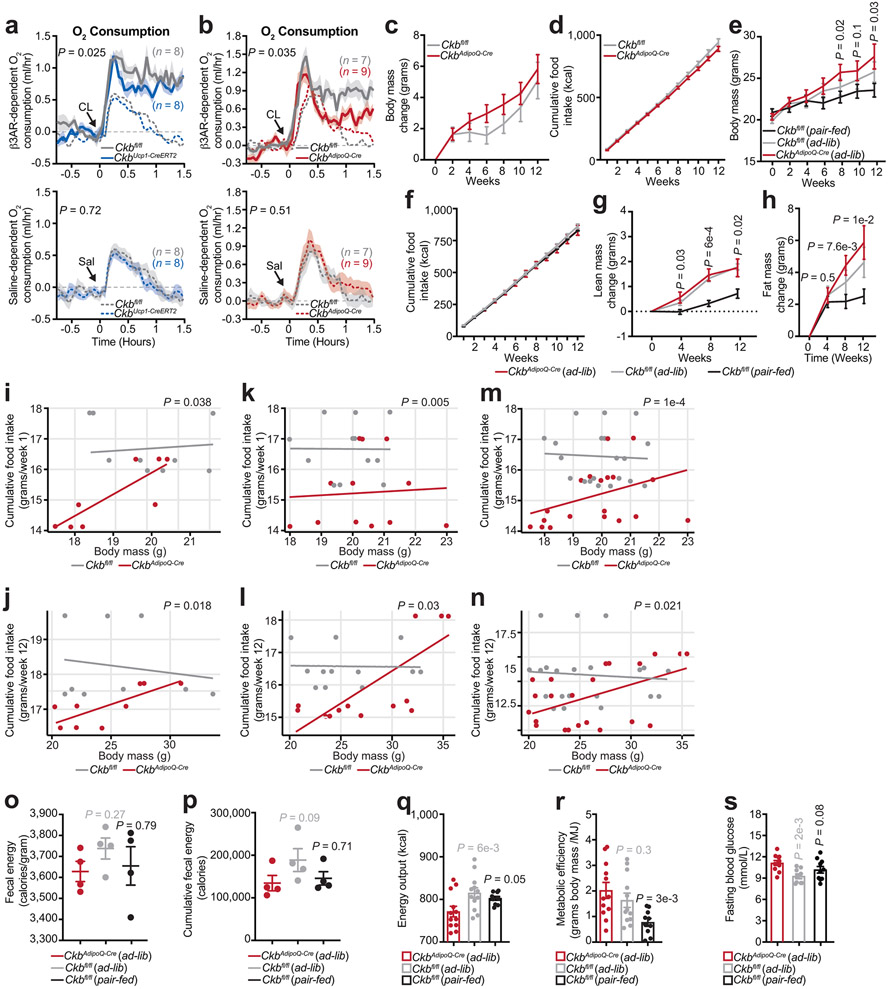
We used an inducible model to inactivate Ckb in Ucp1+ adipocytes (CkbUcp1-CreERT2 mice) (Extended Data Fig. 7a-j). Strikingly, the rise in β3-adrenoreceptor-elicited whole body energy expenditure and O2 consumption was markedly blunted (21%) in CkbUcp1-CreERT2 compared to Ckbfl/fl mice (Fig. 4a, Extended Data Fig. 8a). Thus, CKB regulates thermogenesis in Ucp1+ adipocytes. We also generated a pan-adipocyte-specific loss-of-function model of Ckb (CkbAdipoQ-Cre mice) (Extended Data Fig. 7k-t). Importantly, like CkbUcp1-CreERT2 mice, CkbAdipoQ-Cre mice displayed a profound reduction (45%) of β3-adrenoreceptor-dependent energy expenditure and O2 consumption compared to Ckbfl/fl mice (Fig. 4b, Extended Data Fig. 8b). Thus, adipocyte CKB regulates thermogenesis in vivo.
Fig. 4: Fat-selective deletion of Ckb decreases energy expenditure and predisposes mice to obesity.
a, CL-dependent (a, top) and saline-dependent (a, bottom) energy expenditure (EE) of male mice at 30°C (n = 8 per group). b, CL-dependent (b, top) and saline-dependent (b, bottom) EE of Ckbfl/fl (n = 7) and CkbAdipoQ-Cre (n = 9) male mice at 30°C. c, Body weight during high-fat feeding of female mice at 22°C (CkbAdipoQ-Cre(ad-lib): n = 12; Ckbfl/fl (ad-lib): n = 12; Ckbfl/fl (pair-fed): n = 10). d, Representative histological images of BAT and SAT from female mice following 16 weeks high fat feeding. Scale bar (black line), 50 μm for BAT and 125 μm for SAT. e, glucose tolerance test (e, left) and area under the curve (AUC) of glucose tolerance test (e, right) following 12 weeks of high-fat feeding of female mice at 22°C (CkbAdipoQ-Cre(ad-lib): n = 11; Ckbfl/fl (ad-lib): n = 9; Ckbfl/fl (pair-fed): n = 10). f, insulin tolerance test following 16 weeks of high-fat feeding of female mice at 22°C (CkbAdipoQ-Cre(ad-lib): n = 12; Ckbfl/fl (ad-lib): n = 11; Ckbfl/fl (pair-fed): n = 10). P values indicate comparison to CkbAdipoQ-Cre(ad-lib) mice. Data are presented as mean ± s.e.m. of biologically independent samples. a-c, e(left), f, two-way ANOVA (Fisher’s LSD); e(right), one-way ANOVA (Fisher’s LSD).
CkbAdipoQ-Cre mice tended to gain more weight than Ckbfl/fl mice, despite consuming less food (Extended Data Fig. 8c, d, i, j, m, n), but gained significantly more weight (primarily fat) compared to pair-fed Ckbfl/fl mice (Fig. 4c, Extended Data Fig. 8f-h), with no change in energy absorption (Extended Data Fig. 8o, p). Application of the energy balance method23 revealed that CkbAdipoQ-Cre mice displayed lower total energy expenditure compared to Ckbfl/fl controls (Extended Data Fig. 8q), and the metabolic efficiency of CkbAdipoQ-Cre mice was markedly greater than pair-fed Ckbfl/fl mice (Extended Data Fig. 8r). Ckb-deficient BAT and SAT accumulated more lipid than Ckbfl/fl controls (Fig. 4d). Loss of adipocyte CKB also elevated fasting blood glucose levels (Extended Data Fig. 8s), impaired glucose tolerance, (Fig. 4e) and reduced insulin sensitivity (Fig. 4f) following high fat feeding. These findings indicate that adipocyte CKB promotes metabolic health.
Discussion:
We reveal a targeting signal that is compulsory for CKB import into mitochondria, rendering it a key protein of the futile creatine cycle. In the human and mouse adipose samples that we have examined, CKB levels are regulated by canonical thermogenic cAMP signaling. Fat-selective inactivation of Ckb in mice impairs thermogenic capacity and promotes weight gain upon high fat feeding. Unlike most cell types that express dedicated cytosolic and mitochondrial creatine kinase isoenzymes, brown adipocytes orchestrate creatine kinase activity by subcellular trafficking of a single isoenzyme. The fraction of mitochondrial CKB in relation to the total cellular CKB pool is greater in brown adipocytes compared to other tissues we have examined. However, expression of a single creatine kinase isoenzyme can dominate in some non-thermogenic cell types24, suggesting that mitochondrial targeting of CKB may be of general implication. Moreover, because translation of Ckb from the canonical open reading frame is sufficient to target the protein to mitochondria, we speculate that mitochondrial CKB only partly explains the unique capacity of thermogenic adipocytes to elicit futile creatine cycling. As we have shown, the dynamic nature of CKB expression in thermogenic adipocytes in response to thermogenic stimuli is the most of any other creatine kinase isoenzyme. In addition, CKB shows the highest Vmax and lowest Km values for all substrates involved in the creatine kinase phosphoryl transfer reaction which would facilitate creatine kinase activity at low substrate concentrations21,25. Linking CKB to futile creatine cycling may ensure that the cycle can be triggered despite lower rates of mitochondrial ATP synthesis in brown adipocytes relative to other cell types. The nature of acute regulation of futile creatine cycling through enzymatic regulation by allosteric modulation or covalent posttranslational modifications on CKB or other proteins in this pathway merits investigation.
Methods:
Primary interscapular brown stromal vascular cell preparation
Interscapular brown stromal vascular cells (SVC) were obtained from 2-6 day old pups. BAT was dissected, washed in PBS, minced, and digested for 45 minutes at 37°C in PBS containing 1.5 mg ml−1 collagenase B (Roche), 123 mM NaCl, 5 mM KCl, 1.3 mM CaCl2, 5 mM glucose, 100 mM HEPES, and 4 % essentially fatty acid-free BSA. Tissue suspension was filtered through a 100 μm cell strainer and centrifuged at 600 g for 5 min to pellet the SVF. The cell pellet was resuspended in adipocyte culture medium (DMEM/F12 with GlutaMAX (1:1, Invitrogen), 10 % FBS and 0.1 % PenStrep), filtered through a 40 μm cell strainer, centrifuged at 600 g for 5 min, resuspended and plated in adipocyte culture medium.
Brown adipocyte differentiation
Brown preadipocytes were grown to post-confluency and differentiated with adipogenic cocktail (1 μM rosiglitazone, 0.5 mM IBMX, 5 μM dexamethasone, 0.114 μg ml−1 insulin, 1 nM T3, and 125 μM Indomethacin). Cells were re-fed every 48 hours with maintenance cocktail (1 μM rosiglitazone and 0.5 μg ml−1 insulin). Adipocytes were fully differentiated 6 days after initial exposure to adipogenic cocktail.
Metabolite analyses of brown adipocytes by mass spectrometry
Metabolites were profiled at the Rosalind and Morris Goodman Cancer Research Centre Metabolomics Core Facility. Specifically, brown preadipocytes were seeded in 6 cm plates, grown to post-confluency (~ 1x106 cells) and differentiated. 6 days post-differentiation, adipocytes were supplemented with 150 μM D3-creatine (Cambridge Isotope Laboratories, DLM-1302) in maintenance cocktail containing dialyzed FBS for 24 hours. 1X106 adipocytes were lysed in 350 μl of methanol/HPLC-grade water (50:50 (v/v)), 0.22 ml of ice-cold acetonitrile (ACN) (at −20°C or colder) and six ceramic beads (1.4 mm diameter). The mixture was homogenized in a bead beater (Qiagen TissueLyser) for 2 min at 30 Hz. Next, 0.6 ml of ice-cold dichloromethane and 0.3 ml of ice-cold HPLC-grade water were added. The samples were vortexed for 1 min, incubated on ice for 10 min and centrifuged at 1,500 g for 10 min at 1°C. Water-soluble metabolites in the upper polar phase were collected and dried using a chilled vacuum centrifuge operating at a sample temperature of 4°C (Labconco). Samples were resuspended in 50 μl of HPLC-grade water before LC–MS analysis. Metabolites were profiled at the Rosalind and Morris Goodman Cancer Research Centre Metabolomics Core Facility by LC-MS using an Agilent 6530 QTOF equipped with a 1290 UPLC (Agilent Santa Clara, CA). Compounds were chromatographically separated using a Scherzo SM-C18 column 3 μm, 3.0×150mm (Imtakt Corp, Japan). The gradient started at 100% mobile phase A (5 mM ammonium acetate in water) with a 5 min gradient to 100% B (200 mM ammonium acetate in 80% water / 20% ACN) at a flow rate of 0.4 ml/min. This was followed by a 5 min hold time at 100% mobile phase B and a subsequent re-equilibration time (6 min) before next injection. Eluting compounds were ionized using an Agilent dual jet stream electrospray ionization source operating in positive mode. The source was set with the following parameters: Gas temperature of 325°C, drying gas of 9 l/min, sheath gas temperature 400°C, Sheath gas flow of 12 l/min, nebulizer pressure of 45 psi capillary voltage of 4000 V, nozzle voltage of 500 V. Ion optics were set with a fragmentor of 140 V, skimmer of 65 V. To maintain the instrument resolution reference masses, 121.05083 from purine and 922.009798 from HP0921, were infused using the second nebulizer. Data were collected in scan mode (100 to 1600 m/z) and MS/MS mode using a collision energy of 10, 20 and 30 V. Accurate peak mass and retention times were compared to authentic standards and MS/MS. Data were collected on matched samples that were treated with unlabeled-creatine to separate deuterium incorporation from interfering ions. Stable isotope tracer data were analyzed using MassHunter Profinder Software B.08.00 (Agilent Technologies), which includes matrix correction, or data were analyzed using area under the curve from MassHunter Quant (Agilent Technologies).
Immortalization of brown adipocytes
Stromal vascular cells from BAT were infected with retrovirus (pBabe-Neo largeT; Addgene #1780), passaged 4 times and frozen as immortalized stocks. Parental cell lines were tested empirically for differentiation capacity.
Limiting dilution
Immortalized Ckbfl/fl preadipocytes were infected with adenovirus expressing green fluorescent protein (Gfp) or cre recombinase (1:500 dilution) for 12 hours. Gfp-infected cells were kept as a pooled population. Cre recombinase-infected cells were subjected to limiting dilution. Single clones were selected and screened for CKB expression by Western blotting. Several distinct clones were null for CKB (clones 3 and 9), whereas others still retained CKB expression (clone 5).
Western blotting
Samples were prepared in Lysis Buffer (50 mM Tris, pH 7.4, 500 mM NaCl, 1% NP40, 20% glycerol, 5 mM EDTA and 1 mM phenylmethylsulphonyl fluoride (PMSF), supplemented with a cocktail of Roche protease inhibitors. The homogenates were centrifuged at 16,000 g for 10 minutes at 4°C, and the supernatants were used for subsequent analyses. Protein concentration was determined using the bicinchoninic acid assay (Pierce). Quantity of protein lysate to use for each antibody was determined empirically. Protein lysates were denatured in Laemmli buffer (60 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 0.05 % bromophenol blue, 0.7 M β-mercaptoethanol), resolved by 4-12% NuPAGE Bis-Tris SDS–PAGE (Invitrogen) and transferred to a polyvinylidene difluoride (PVDF) membrane. Primary antibodies were diluted in TBS containing 0.05% Tween (TBS-T), 5% BSA and 0.02% NaN3. Membranes were incubated overnight with primary antibodies at 4°C. For secondary antibody incubation, anti-rabbit or anti-mouse HRP (Promega) was diluted at 1:10,000 (v/v) in TBS-T containing 5% milk. Results were visualized with enhanced chemiluminescence (ECL) Western blotting substrates (Bio-rad). Uncropped and unprocessed scans of blots are found in Supplementary Figure 1. Dilutions for antibodies were as follows: HSP60 (Abcam; Cat. No. ab46798; dilution: 1:10,000); GAPDH (Abcam; Cat. No. ab8245; clone 6C5; dilution: 1:20,000); VCL (Cell Signaling; Cat. No. 13901; clone E1E9V; dilution: 1:5,000); TIM23 (BD Biosciences; Cat. No. 611222; clone 32; dilution: 1:200); CANX (Abcam; Cat. No. ab22595; dilution: 1:4,000); PMP70 (Abcam; Cat. No. ab3421; dilution: 1:500); P-AS160(T642) (Cell Signaling; Cat. No. 8730; clone D8E4; dilution: 1:1,000); AS160 (Cell Signaling; Cat. No. 2670; clone C69A7; dilution: 1:1,000); P-AKT(T308) (New England Biolabs; Cat. No. 9275; dilution: 1:1,000); AKT (Cell Signaling; Cat. No. 4691; clone C67E7; dilution: 1:1,000); P-HSL(S660) (Cell Signaling; Cat. No. 4126; dilution: 1:1,000); HSL (Cell Signaling; Cat. No. 4107; dilution: 1:1,000); FLAG M2 (Sigma; Cat. No. F1804; clone M2; dilution: 1:1,000); FLAG-HRP (Sigma; Cat. No. A8592; clone M2; dilution: 1:2,000); CKB (Abclonal; Cat. No. ab12631; 1:1,000); FASN (Abcam; Cat. No. ab128856; clone EPR7465; dilution: 1:1,000); UCP1 (Abcam; Cat. No. ab10983; dilution: 1:2,000); TOM20 (Santa Cruz; Cat. No. sc-11415; clone FL-145; dilution: 1:4,000); LONP1 (Abcam; Cat. No. ab103809; dilution: 1:1,500); TFAM (Abcam; Cat. No. ab131607; dilution: 1:1,000); TOTAL OXPHOS (Abcam; Cat. No. ab110413; dilution: 1:2,000); FABP4 (Cell Signaling; Cat. No. 3544; clone D25B3; dilution: 1:1,000); TOM70 (Santa Cruz; sc-390545; clone A-8; dilution: 1:1,000); Anti-rabbit (Promega; Cat. No. W4011; dilution: 1:10,000); Anti-mouse (Promega; Cat. No. W4021; dilution: 1:10,000).
Animals
Mouse experiments were performed according to procedures approved by the Animal Resource Centre at McGill University and complied with guidelines set by the Canadian Council of Animal Care. Unless otherwise specified, mice were housed at 22°C under a 12-h light/dark cycle and given free access to food and water. Wild-type C57BL/6N mice were purchased from Charles River (strain code: 027). Ckbfl/fl mice were generated by Applied StemCell using CRISPR/Cas9 gene editing. Ucp1-CreERT2 mice26, were bred to Ckbfl/fl mice to generate experimental groups (Ckbfl/fl and CkbUcp1-CreERT2). Adiponectin-Cre mice (B6;FVB-Tg(AdipoQ-Cre)1Evdr/J, stock 028020), maintained on a C57BL/6J background, were bred to Ckbfl/fl mice to generate experimental groups (Ckbfl/fl and CkbAdipoQ-Cre). CkbFlag mice were generated by CRISPR/Cas9 gene editing at the McGill Integrated Core for Animal Modeling (MICAM). Mouse experiments used age-matched littermates and were conducted at the temperature indicated in the figure legend. For energy balance studies, mice were housed in groups of three. For cold exposure experiments, mice were singly housed.
Inducible deletion of Ckb
Mice were reared at 22°C until 7 week of age, then injected with tamoxifen (75mg/kg) for 5 days and allowed to recover for 4 days. Mice were then placed in metabolic cages at 30°C with chow diet for 1 week until CL injection.
Genotyping
Ucp1 alleles: Primers to detect wild-type (Ucp1+/+), heterozygous (Ucp1−/+) and knockout (Ucp1−/−) alleles in Ucp1 were multiplexed: Ucp1 common F, 5’-GGG GTA GTA TGC AAG AGA GGT G-3’; Ucp1 WT R, 5’-CCT TTA TAC CTA ATG GTA CTG GAA GC-3’; Ucp1 KO R, 5’-CCT ACC CGC TTC CAT TGC TCA-3’. Ckb alleles: Primers to detect WT (Ckb+/+) and floxed (Ckbfl/+ and Ckbfl/fl) alleles: Ckb F, 5’-AGG TGG TGG CTA GAG TGA GC-3’; Ckb R, 5’-CAA GGA TCC CAC TGC TCT TC-3’. CkbFLAG alleles: Primers to detect CkbFLAG alleles: CkbFLAG F, 5’-AGA AGG TGC AGG TGT TGT CC-3’; CkbFLAG R, 5’-GTC CCT AGG AAA GGA GAA GCA A-3’.
Gene expression analysis (RT-qPCR)
Total RNA was extracted from frozen tissue using QIAzol (Qiagen), purified with RNeasy Mini spin columns (QIAGEN) and reverse transcribed using a High- Capacity cDNA Reverse Transcription kit (Applied Biosystems). The resultant cDNA was analyzed by RT–qPCR. Briefly, 20 ng cDNA and 150 nmol of each primer were mixed with GoTaq qPCR Master Mix (Promega). Reactions were performed in a 384-well format using a CFX384 Real-time PCR system (Bio-rad). Normalized mRNA expression was calculated using the ΔΔCt method, using Ppib mRNA as the reference gene. CFX Maestro 2017 was used for data collection. Primer sequences used for RT-qPCR of mouse samples are as follows: Adipoq (Fwd: TGT TCC TCT TAA TCC TGC CCA; Rev: CCA ACC TGC ACA AGT TCC CTT); Ckb (Fwd: GCC TCA CTC AGA TCG AAA CTC; Rev: GGC ATG TGA GGA TGT AGC CC); Ckm (Fwd: CCT CTA TAT AAC CCA GGG GCA CA; Rev: AGT GTC TGT CTG TGC TGT GGA); Ckmt1 (Fwd: TGA GGA GAC CTA TGA GGT ATT TGC; Rev: TCA TCA AAG TAG CCA GAA CGG A); Ckmt2 (Fwd: CCA GTG CCT TCT CAA AGT TGC; Rev: AGT CCG CAC TTG GGG GAA AGA G); Fabp4 (Fwd: AAG GTG AAG AGC ATC ATA ACC CT; Rev: TCA CGC CTT TCA TAA CAC ATT CC); Gamt (Fwd: GCA GCC ACA TAA GGT TGT TCC; Rev: CTC TTC AGA CAG CGG GTA CG); Gatm (Fwd: ATG CCT GTG TCG CAC CAT TC; Rev: TTG CAC ATC TCT TCG ACC TCA); Pparg2 (Fwd: TGC CTA TGA GCA CTT CAC AAG AAA T; Rev: CGA AGT TGG TGG GCC AGA A); Ppib (Fwd: GGA GAT GGC ACA GGA GGA A; Rev: GCC CGT AGT GCT TCA GCT T); Slc6a8 (Fwd: GTG TGG AGA TCT TCC GCC AT; Rev: CCC GTG GAG AGC CTC AAT AC); Ucp1 (Fwd: AAG CTG TGC GAT GTC CAT GT; Rev: AAG CCA CAA ACC CTT TGA AAA). Primer sequences used for RT-qPCR of human samples are as follows: CKB (Fwd: TTC TCA GAG GTG GAG CTG GT; Rev: AGG CAT GAG GTC GTC GAT); 18S rRNA (Fwd: ACC GCA GCT AGG AAT AAT GGA; Rev: GCC TCA GTT CCG AAA ACC A).
Creatine kinase activity
A coupled reaction (pyruvate kinase and lactate dehydrogenase) was used to determine creatine kinase activity (PCr synthesis). Absorbance at 340nM was measured to determine ATP turnover rate using a VarioSKAN plate reader in kinetic mode. The assay was performed at 25°C with the following conditions: 5 mM ATP, 4 mM PEP, 20 mM MgCl2, 100 mM KCl, 0.45 mM NADH, 50 mM Tris buffer pH 9, 10 mM creatine. For Km determination creatine concentration was varied between 0-10 mM. Approximately 0.5 μg purified protein was used to determine creatine kinase activity from bacteria-purified protein. Approximately 100 μg of protein was used to determine creatine kinase activity from whole cell, post-mitochondrial supernatant, or mitochondrial extracts.
Human perirenal fat qPCR
CKB mRNA levels were determined in human perirenal fat biopsies as described previously27. CKB expression levels were normalized to 18S rRNA. Norepinephrine levels were determined in plasma collected at the same time as the fat biopsies as previously described27.
Mitochondrial isolation
Two 15 cm dishes of differentiated adipocytes were scraped gently in 6 ml of hypotonic buffer (20 mM Hepes, pH7.8; 5 mM KCl; 1.5 mM MgCl2; 1 mg/ml fatty acid-free BSA), and placed on ice for 10 minutes. Cells were homogenized on ice in a tight-fitting dounce homogenizer with 25 strokes, and rapidly equilibrated to a 1X MSH buffer (210 mM mannitol; 70 mM sucrose; 20 mM Hepes, pH7.8) using a 2.5X MSH stock. Homogenate was spun at 8,500 g for 10 min at 4°C and fat layer was disgarded. Pellet was resuspended in MSH buffer and spun at 600 g for 5 min at 4°C to pellet nuclei and cell debris. The post-nuclear supernatant was spun at 8,500 g for 10 min at 4°C to pellet mitochondria. Mitochondria were resuspended in MSH buffer and protein was quantified using the bicinchoninic acid method.
Mitochondrial protease protection
400 μg mitochondria were treated with 5 μg trypsin in MSH buffer (210 mM mannitol; 70 mM sucrose; 20 mM Hepes, pH7.8) in a final volume of 200 μl for 20 minutes at room temperature rotating end over end. 25 μl FBS was spiked into the reaction to inhibit trypsin and samples were spun at 10,000 g for 2 min at 4°C to pellet mitochondria, and the supernatant was removed. Mitochondria were washed in MSH buffer twice by rotating the tube with pellet and spinning at 10,000 g for 2 min at 4°C to pellet mitochondria to the other side of the tube. Mitochondrial pellets were lysed in adipocyte lysis buffer (50 mM Tris, pH7.5; 500 mM NaCl; 1% IGEPAL; 20% glycerol; 2 mM EDTA), supplemented with protease and phosphatase inhibitor cocktail tablets (Roche) to a final concentration of 5 μg/μl. Samples were stored at −80°C until further analysis by Western blotting.
Purification of mature brown adipocytes
Interscapular BAT was digested in a Krebs-Ringer Bicarbonate Modified buffer (KRBMB: 135 mM NaCl; 5 mM KCl; 1 mM CaCl2; 1 mM MgCl2; 0.4 mM K2HPO4; 25 mM NaHCO3; 20 mM Hepes; 10 mM glucose; 4% fatty acid-free BSA), supplemented with 2mg/ml collagenase B (Worthington) and 1 mg/ml soybean trypsin inhibitor (Worthington). BAT was minced and placed in KRBMB digestion buffer with continuous shaking at 37°C for 45 minutes. The tissue suspension was filtered through a 100 μm cell strainer and spun 500 rpm for 5 minutes at room temperature. The infranatant was removed, and adipocytes were washed twice with 20 ml KRBMB and spun 500 rpm for 5 minutes at room temperature. Adipocytes were left for 30 minutes at room temperature on the bench in between washes and spins for 30 minutes to allow adipocytes to float. After final spin, the infranatant was removed and cells were snap-frozen in liquid nitrogen until downstream analysis.
Construct design and site-directed mutagenesis
The mouse Ckb open reading frame was cloned from a BAT cDNA library. Ckb cDNA was cloned into the pENTR Directional TOPO (pENTR™/D-TOPO®) cloning kit following the manufacturer’s instructions (Invitrogen; K2400-20). Mutagenesis primers (nucleotides that cause mutation are bolded) used to construct CKBC283S.Flag and CKBΔiMTS-L.Flag were as follows: CKBC283S.Flag: Fwd, 5’- CTG GGC TAC ATC CTC ACA AGC CCA TCC AAC CTG GGC AC -3’; Rev, 5’- GTG CCC AGG TTG GAT GGG CTT GTG AGG ATG TAG CCC AG -3’; CKBΔiMTS-L.Flag: Fwd, 5’- CTA CGT GCT GAG CTC GGA AGT GGA GAC AGG CGA GAG CAT CGA GGG CTT CTG TCT CCC CCC -3’; Rev, 5’- GGG GGG AGA CAG AAG CCC TCG ATG CTC TCG CCT GTC TCC ACT TCC GAG CTC AGC ACG TAG -3’.
CKB purification from bacteria
The mouse Ckb open reading frame with a flag epitope on the c-terminus (CKB.Flag) was cloned from a BAT cDNA library into pet6xHN plasmid. The reverse primer contained the flag epitope tag and flexible linker, 5’-TTA CTT GTC ATC GTC GTC CTT GTA ATC CTG AGA ACC AGG TGC GGT GTC AGA CTT CTG GGC CGG C-3’. Plasmids were transformed into E. coli, [BL-21 (DE3)pLysS cells], and 100 ml cultures were grown at 37°C to an OD600 of 0.6-0.9. IPTG (final concentration 1mM) was added to induce protein expression for 5 hours after which pellets were collected by centrifugation at 2,300 g for 15 min at 4°C. Cell pellets were frozen at −80°C. The following day, cell pellet was resuspended in PN buffer (50 mM NaPO4, pH 7.4; 300 mM NaCl) and cells were lysed by sonication. Lysate was clarified through a 0.2 mm filter, and subsequently batch bound by end over end rotation with 50 μl FLAG-M2 beads (Sigma, M8823) for 2 hours. Beads were washed with 1ml of TBS (50 mM Tris, pH 7.4; 150 mM NaCl) three times. The sample was eluted with a 3X FLAG Peptide (Sigma, F4799), diluted in Protein storage buffer (TBS supplemented with 20% glycerol), and flash frozen in liquid nitrogen and stored at −80°C. Purity and yield of purification was determined by coomassie blue staining using BSA as a standard.
Adenoviral shRNA cloning, construction, and use
Complementary DNA oligos for shRNA generation were annealed and cloned into the pENTRTM/U6 entry vector according to the manufacturer’s instructions (Invitrogen; K4944). Sequences for shRNA targeting were as follows: shCkb-#1 sense: CAC CGC GAG GAG AGT TAC GAC GTA TTC GAA AAT ACG TCG TAA CTC TCC TCG; antisense: AAA ACG AGG AGA GTT ACG ACG TAT TTT CGA ATA CGT CGT AAC TCT CCT CGC; shCkb-#2 sense: CAC CGC ACA ATG ACA ATA AGA CTT TCG AAA AAG TCT TAT TGT CAT TGT GC; antisense: AAA AGC ACA ATG ACA ATA AGA CTT TTT CGA AAG TCT TAT TGT CAT TGT GC; shLacZ sense: CAC CGC TAC ACA AAT CAG CGA TTT CGA AAA ATC GCT GAT TTG TGT AG, antisense: AAA ACT ACA CAA ATC AGC GAT TTT TCG AAA TCG CTG ATT TGT GTA GC. Cloned shRNA’s were shuttled into the pAD/BLOCK-iT™-DEST Gateway® vector and transfected into 293A cells. Crude adenovirus was generated according to the manufacturer’s instructions (Invitrogen; K4941). Crude adenovirus was amplified by infecting 293A cells, and iterative freeze thawing until ten 15 cm dishes were attained. Glycerol was added to a final concentration of 20% to the final adenovirus amplification extract, which was stored in aliquots at −80°C. Primary adipocytes were transduced with adenovirus (1/20 dilution) in the evenings of day 1 and day 3 of adipocyte differentiation, with medium replacement the following morning each time.
Adenoviral overexpression cloning, construction, and use
The mouse Ckb open reading frame was cloned from a BAT cDNA library. Ckb cDNA was cloned into the pENTR Directional TOPO (pENTR™/D-TOPO®) cloning kit following the manufacturer’s instructions (Invitrogen; K2400-20). Cloned cDNA’s were shuttled into the pAD/CMV/V5-DEST Gateway® vector and transfected into 293A cells. Crude adenovirus was generated according to the manufacturer’s instructions (Invitrogen; V493-20). Crude adenovirus was amplified by infecting 293A cells, and iterative freeze thawing until ten 15 cm dishes were attained. Glycerol was added to a final concentration of 20% to the final adenovirus amplification extract, which was stored in aliquots at −80°C. Primary adipocytes were transduced with adenovirus to reach endogenous levels (determined empirically for each preparation) in the evening of day 3 of adipocyte differentiation, with medium replacement the following morning.
Lipid staining by Oil Red O
Prior to staining, adipocyte culture media was discarded. Cells were rinsed twice with PBS, and fixed in 4% paraformaldehyde for 30 minutes. Cells were washed with PBS three times, incubated for 5 minutes with 60% (vol/vol) isopropanol, and then incubated for 15 minutes at room temperature with Oil Red O solution. Next, stained cells were rinsed with 60% isopropanol, and three times with PBS.
Glycerol release
Primary brown preadipocytes (40,000 cells per well) were plated in a 48-well dish. 24 hours after plating brown preadipocytes were differentiated with adipogenic cocktail (1 μM rosiglitazone, 0.5 mM IBMX, 5 μM dexamethasone, 0.114 μg ml−1 insulin, 1 nM T3, and 125 μM Indomethacin). Cells were re-fed every 48 hours with maintenance cocktail (1 μM rosiglitazone and 0.5 μg ml−1 insulin). Adipocytes were fully differentiated 6 days after initial exposure to adipogenic cocktail. On day 6 post-differentiation, maintenance cocktail was replaced with 300 μl of DMEM/F-12 (ThermoFisher Scientific) supplemented with 2% fatty acid-free BSA, and different doses of NE for 1 hour. Adipocytes were washed twice with warm D-PBS and lysed with lysis solution (0.3 N NaOH, 0.1% SDS) for three hours at room temperature. Glycerol content of the samples was determined using Free glycerol reagent (Sigma) and Glycerol Standard Solution (Sigma) at an absorbance of 540 after incubating sample and standards with free glycerol reagent for 15 minutes at room temperature in the dark.
Immunofluorescence
U-2 OS cells (ATCC; Cat. No. HTB-96) were seeded on glass coverslips in 24-well format and transfected with Ckb.Flag cDNA. Cells were fixed with 5% paraformaldehyde (PFA) in PBS at 37°C for 15 min. After washing with PBS (3x), the cells were quenched with 50 mM NH4Cl/PBS for 10 min at room temperature. 0.1% Triton X-100/PBS (v/v) was added for 10 min at room temperature to permeabilize the cells. Blocking was performed with 5% FBS/PBS for 10 min at room temperature. Next, cells were incubated overnight with primary antibodies at a dilution of 1:1,000: FLAG M2 (Sigma; Cat. No. F1804) and TOM20 (Santa Cruz; Cat. No. sc-11415). Cells were washed with PBS (3X) and then incubated with fluorescent-conjugated secondary antibodies: Alexa Fluor Plus 488 (ThermoFisher; Cat. No. A32723) and Alexa Fluor Plus 647 (ThermoFisher; Cat. No. A32733) at 1:2,000 dilution for 1 hour. Dako fluorescence mounting medium (Agilent; Cat. No. S3023) was used to mount the coverslips. Images were obtained with spinning confocal microscopy (Olympus IX81 with Andor/Yokogawa spinning disk system (CSU-X), sCMOS camera and 100× or 60× objective lenses (NA1.4)). Adobe Photoshop CS6 was used to apply 1 pixel Gaussian blur to the images.
Plate-based adipocyte respirometry
Preadipocytes were plated at 6,000 cells per well of a XF96 cell culture microplate (Agilent), and differentiation was induced 4 hours later. On day 6 post-differentiation, adipocytes were washed once and maintained for ~45 minutes in unbuffered DMEM supplemented with 1 mM pyruvate. A Seahorse XF analyzer was used for respirometry analysis, where the respirometry protocol was set to a mix:wait:measure ratio of 4:0:2. Norepinephrine (NE) (0.1 μM for wild-type adipocytes and for 1 μM Ucp1−/− adipocytes) was used to stimulate respiration. Oligomycin (2.5 μM) was used to inhibit mitochondrial ATP synthesis. 2,4-dinitrophenol (DNP) was added to a final concentration of 0.5 mM, which was based on titration experiments of investigating maximal respiration. A mix of rotenone and antimycine A (3 μM each) was used to inhibit mitochondrial respiration. Wave, v.2.4 software was used for data collection.
Primary inguinal stromal vascular cell preparation
Inguinal SVC’s were obtained from 3-5 week old mice. Inguinal fat was dissected, washed in PBS, minced, and digested for 45 minutes at 37°C in PBS containing 10 mg ml−1 collagenase D (Roche), 3 U ml−1 dispase II (Roche), and 10 mM CaCl2. Tissue suspension was filtered through a 100 μm cell strainer and centrifuged at 600 g for 5 min to pellet the SVF. The cell pellet was resuspended in adipocyte culture medium (DMEM/F12 with GlutaMAX (1:1, Invitrogen), 10 % FBS and 0.1 % PenStrep), filtered through a 40 μm cell strainer, centrifuged at 600 g for 5 min, resuspended and plated in adipocyte culture medium.
Prediction of iMTS-L
iMTS-L signals were predicted as previously described19. Multiple CKB sequences were generated by sequentially removing single amino acids from the N-terminus and submitted all sequences to TargetP28 with options set to non-plant organism and without cut-off and cleavage-site prediction. The mTP scores were plotted against the corresponding amino acid position. A threshold of 0.75 was used to define the presence of an iMTS-L.
Estimation of mitochondrial CKB across tissues
Whole tissue lyasates and purified mitochondria were prepared from BAT, heart, and kidney of CkbFLAG mice. Whole tissue and mitochondrial lysates were loaded on the same polyacrylamide gel and were used for quantification of CKB.FLAG protein by densitometry after Western blotting using FLAG antibody. Non-specific signal of FLAG antibody was determined based on any signal found in tissue from wild-type mice (loaded on the same gel). BAT from 30°C- and 6°C-exposed mice was loaded on each gel to confirm proper annotation of CKB.FLAG. Following densitometry-mediated quantification of CKB.FLAG abundance (10 μg loaded for whole tissue lysate or 20 μg loaded for mitochondria), the total amount of CKB.FLAG was determined by multiplying this signal by the total amount of protein isolated in the respective extract (whole tissue or mitochondria). Contamination of mitochondria with non-mitochondrial proteins was determined based on VCL densitometry. The percentage of mitochondrial CKB.FLAG was calculated by dividing the total mitochondrial pool by the total pool in whole tissue lysates. The VCL pool was subtracted from the CKB.FLAG pool to obtain the mitochondrial CKB pool, but did not alter the results much due to the high quality of mitochondrial purification from these tissues.
Calculation of ADP:creatine stoichiometry (futile creatine cycling) using plate-based respirometry
The extracellular flux analyzer, XFe24 (Agilent) was used to determine futile creatine cycling. To this end, 5,550 pmoles of creatine (10 μM creatine in a volume of 0.56 ml) were added to 30 μg mitochondria in respiration buffer (125 mM sucrose, 20 mM K-TES, pH7.2, 2mM MgCl2, 1 mM EDTA, 4 mM KH2PO4, 4% fatty acid-free BSA, 10 mM pyruvate, 5 mM malate, 1 mM GDP). According to the details provided by the manufacturer, we assumed oxygen consumption was measured in a micro chamber volume of 2 - 7 μl. We used the most conservative estimate (7 μl) of micro chamber volume (which would underestimate the sub-stoichiometric action of creatine). A microchamber volume of 7 μl would contain 70 pmoles creatine. Total pmoles of oxygen consumed was measured over this time, based on the oxygen consumption rate (measured as pmoles/min). The excess of oxygen consumed by creatine supplementation was used to determine the sub-stoichiometric action of creatine by quantifying ADP liberation based on a P/O ratio of 2.7. Wave, v.2.4 software was used for data collection.
Calculation of ADP:creatine stoichiometry (futile creatine cycling) using an oxygen electrode
A Clark type electrode (Rank Bros) was used to determine futile creatine cycling with an oxygen electrode. To this end, 7 nmoles of creatine (10 μM creatine in a volume of 0.7 ml) were added to 0.25 mg mitochondria in respiration buffer (125 mM sucrose, 20 mM K-TES, pH7.2, 2mM MgCl2, 1 mM EDTA, 4 mM KH2PO4, 4% fatty acid-free BSA, 10 mM pyruvate, 5 mM malate, 1 mM GDP). The medium (0.7 ml) contained 0.25 mg of brown adipocyte mitochondria; 70 nmoles of ADP was added to initiate ADP-dependent respiration. Pilot experiments were conducted to determine the length of time and ADP concentration required to reach state 4 respiration (100 μM). State 4 respiration was defined as the respiratory rate, following ADP addition, that closely matched the respiratory rate prior to ADP addition. Once these parameters were established, total nmoles of oxygen consumed was measured over this time, based on the oxygen consumption rate (measured as nmoles/min). The excess of oxygen consumed by creatine supplementation was used to determine the super-stoichiometric action of creatine by quantifying ADP liberation based on a P/O ratio of 2.7. The data (in Fig. 3m) represent the moles of liberated ADP (measured as the creatine-dependent change in ADP-dependent O2 consumption) over moles of added creatine. Rank Brothers Dual Digital model 20: Picolog 6 data logging software was used for data collection.
Indirect calorimetry
Mice were placed, single-housed, in metabolic cages (Sable Systems International, Promethion high-definition behavioral phenotyping system) housed in thermal cabinets set to 30°C with a 12-hour light and 12-hour dark schedule. The following morning at Zeitgeber (ZT) ZT3, mice were injected with vehicle (saline) and placed back in the metabolic cages to monitor the stress response to i.p. injection. The next morning (at ZT3), the same volume of the β3-adrenoreceptor agonist, CL 316,243, was administered i.p. at 1mg/kg, and mice were placed back in the metabolic cages. Saline and Cl 316,243 responses were followed every 3 minutes for 90 minutes. Mass-dependent variables (VO2, VCO2, and energy expenditure) were not normalized to body weight. Oxygen consumption (VO2), carbon dioxide expiration (VCO2), energy expenditure (kcal), respiratory exchange ratio, and total physical movement (meters; measured by infrared beam breaks) were recorded every 3 min using Sable Systems data acquisition software (IM-3 v.20.0.3). Data were parsed/analyzed using Sable Systems International MacroInterpreter software (v.2.41) using One-Click Macro (v2.37).
Determination of free-living whole-body total energy expenditure
The energy balance method (law of energy conservation) was used to delineate energy output in mice. This method accounts for cumulative caloric intake and changes in lean and fat mass throughout the dietary intervention, as described previously29. Mouse body composition (fat mass and lean mass) was determined before, and every two weeks following dietary intervention until completion of the study. Caloric intake was concluded from the energy density of the diet (5.21 kcal/g). Energy density of accumulated fat mass in mice was 9.4 kcal/g and fat-free mass was 1.8 kcal/g. To solve for energy output, the following formula was used: Energy Output (kcal) = Energy Intake − (change in kcal Fat mass) + (change in kcal Lean mass).
Body composition determination on live mice
Body composition was examined with an Echo MRI-100 body composition analyzer (EchoMRI LLC).
Metabolic efficiency
Metabolic efficiency was calculated as the fraction of assimilated food energy that is stored somatically, and expressed as grams of body mass gained over megajoules (MJ) food eaten.
Histological analysis
Tissues were harvested, placed in tissue clamps in 4% paraformaldehyde (PFA) overnight at 4°C. The following day, samples were rinsed three times in PBS and stored in 70% ethanol at 22°C. Tissue fragments were embedded in paraffin, sectioned and mounted on glass slides at the Goodman Cancer Research Centre Histology Core Facility. Sections were stained with hematoxylin and eosin and scanned using the Aperio ScanScope XT at 20x magnification, creating an .svs image file per slide. For each slide, a snapshot has been taken and based on the image captured, a single region of interest (ROI) was determined. A subset of these ROI’s required manual adjustment to correct the blank areas.
Intraperitoneal glucose tolerance test
Mice were fasted for 6 hours (ZT0 – ZT6). Glucose (2 g/kg lean mass) was administered i.p., and blood glucose levels were measured from the tail without use of restraint at 0, 20, 40, 60, 90, and 120 minutes using a glucometer (ONETOUCH Ultramini). Basal glucose levels were subtracted before calculating the area under the curve (AUC). Glucose was administered based on lean mass, not body weight; lean mass was not different between groups.
Intraperitoneal insulin tolerance test
Mice were fasted for 2 hours (ZT4 – ZT6). Insulin (1 U/kg lean mass) was administered i.p., and blood glucose levels were measured from the tail without use of restraint at 0, 15, 30, 45, and 60 minutes using a glucometer (ONETOUCH Ultramini). Insulin was administered based on lean mass, not body weight; lean mass was not different between groups.
Oxygen bomb calorimetry of feces
Calorimetry was conducted using a Parr 6725/6772 Semimicro Calorimeter and 1109A oxygen bomb. During dietary intervention, fecal samples from mice were collected over a 24 hour period. Samples were baked at 60°C for 48 hours to remove water content. Fecal samples were ignited and the energy content was measured as heat of combustion (calories/g).
Statistical analyses
Statistical analyses were performed with Graphpad Prism 8. Data analysis was obtained with Microsoft office Excel 2016. Data were expressed as mean ±s.e.m. Unpaired two-tailed Student’s t-test for pairwise comparison, one-way ANOVA and two-way ANOVA for multiple comparisons involving two independent variables, F-test for linear regression, and Analysis of Co-Variance (ANCOVA) using body weight as covariate where appropriate for in vivo metabolic analyses were used to calculate P values to determine statistical differences. Significance was considered as P < 0.05. Mice were randomly assigned to treatment groups for in vivo studies. n values represent independent biological replicates for isolated mitochondria, cultured cell experiments or individual mice for in vivo experiments. Specific details for n value are noted in each figure legend.
Statistics and reproducibility
Sample sizes for each experiment are reported in the figure legends. The numbers of experimental repetitions were as follows: Fig. 1d, 3 times; Fig. 1g, 4 times; Fig. 2a, 3 times; Fig. 3a, 7 times; Fig. 3d, 3 times; Fig. 3f, g, i, 2 times; Fig. 4a, b, 4 times; Fig. 4d, 2 times; Extended Data Fig. 1g, 1 time; Extended Data Fig. 1i, 5 times; Extended Data Fig. 1j, 4 times; Extended Data Fig. 1l, 2 times; Extended Data Fig. 1n, o, 4 times; Extended Data Fig. 2c, d, f, 7 times; Extended Data Fig. 2m, 3 times; Extended Data Fig. 3a, 3 times; Extended Data Fig. 4a-c, 3 times; Extended Data Fig. 4d, e, g-j, 2 times; Extended Data Fig. 4m, 4 times; Extended Data Fig. 5f, 1 time; Extended Data Fig. 6c-f , 3 times; Extended Data Fig. 6m-p, 3 times.
Extended Data
Extended Data Fig. 1: CKB is the primary creatine kinase isoenzyme in brown adipocytes.
a, percent labeling (m+3) of deuterated creatine (D3-creatine) and phosphocreatine (D3-phosphocreatine) in brown adipocytes and media (n = 3). b, Quantitative proteomics (n = 10). c, Quantitative proteomics (n = 3). d, Ribosomal profiling of AdipoQ+ and Ucp1+ adipocytes14 (n = 3). e, Ribosomal profiling from BAT (n = 5) and beige fat (n = 4)15. f, RT-qPCR (n = 3). g, Genotyping and sequencing. h, LC-MS after 1 hour labeling with deuterated (m+3) creatine (n = 3). i-j, Western blot from male (i) or female (j) mice, single-housed at 30°C or 6°C for 48 hours. k, RT-qPCR from mice treated as i (n = 3). l, Western blot as i. m, RT-qPCR as k. 30°C (n = 7) or 6°C (n = 5). n, Western blot of BAT. o, Western blot of protease-protected mitochondria from mice treated as n. For gel source data, see Supplementary Figure 1. Mice used were wild-type (C57BL6/N) male (6-8 weeks old) or female (20 weeks old) mice. All mice were reared at 22°C and housed at 30°C for 5 days before treatments. Data are presented as mean ± s.e.m. of biologically independent samples. b, one-way ANOVA (Fisher’s LSD); d-f, two-way ANOVA (Fisher’s LSD); h, k, m, two-tailed Student’s t-tests.
Extended Data Fig. 2: Ckb silencing impairs brown adipocyte respiration.
a, RT-qPCR of primary brown adipocytes (n = 3 per group). b, Oil red O staining of primary brown adipocytes. Preads (preadipocytes). c-d, Western blot examining the effect of Ckb silencing on mitochondrial abundance and adipocyte differentiation (c) and insulin signaling (d). e, Glycerol release assay (n = 3 per group). f, Western blot examining the effect of Ckb silencing on lipolytic signaling. g, NE-dependent OCR, obtained by subtracting basal from NE-induced respiration (from Fig. 3b). Left panel: shLacZ, n = 17; shCkb#1, n = 16. Right panel: shLacZ, n = 8; shCkb#2, n = 15. h, DNP-dependent OCR (n = 33). i, OCR following Ckb silencing (shLacZ, n = 31; shCkb#1, n = 27, shCkb#2, n = 27). Oli (oligomycin); RA (Rotenone and antimycin A). j, OCR of Ucp1−/− primary brown adipocytes (n = 20 per group). k, NE-dependent OCR, obtained by subtracting basal from NE-induced respiration from j (n = 20 per group). l, OCR of Ucp1−/− primary brown adipocytes (shLacZ, n = 17 shCkb#1, n = 7 or shCkb#2, n = 10). m, Western blot (m, left) and OCR (m, right) of immortalized brown adipocytes. Gfp (P): n = 21; clone 5: n = 20, clone 3: n = 19, clone 9: n = 23). For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. a, e, two-way ANOVA (Dunnett’s multiple comparisons test); g, i-l, m(right), multiple two-tailed Student’s t-tests.
Extended Data Fig. 3: Ckb silencing selectively impairs brown adipocyte respiration.
a, Western blot of primary brown preadipocytes following Ckb silencing. b, OCR of primary brown preadipocytes following Ckb silencing. Left panel: shLacZ, n = 10; shCkb#1, n = 10. Right panel: shLacZ, n = 10; shCkb#2, n = 10. c, OCR of primary brown adipocytes with silencing of Ckmt2 (shCkmt2#1: n = 8; shCkmt2#2: n = 11), Ckm (shCkm#1: n = 11; shCkm#2: n = 11), or Ckmt1 (shCkmt1#1: n = 23; shCkmt1#2: n = 12), compared to shLacZ (n = 23). d, RT-qPCR following silencing of creatine kinase isoforms (n = 3 per group). For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. b, c, multiple two-tailed Student’s t-tests; d, two-way ANOVA (Fisher’s LSD).
Extended Data Fig. 4: CKB is targeted to mitochondria by an internal mitochondrial targeting signal-like.
a, Confocal images of U2OS cells transfected with Ckb.Flag cDNA. Mitochondria were labeled with anti-TOM20 antibody (green); CKB.FLAG was labeled with anti-FLAG antibody (magenta). Scale bar (white line), 5 μm. b, Western blot of mitochondrial extracts with and without protease (10 μg trypsin) treatment. c, Western blot of mitochondrial extracts with and without protease treatment. d, Western blot of mitochondrial extracts following protease treatment and extracts from whole cells, following Ckb silencing. e, Western blot of whole tissue lysates (WTL) and protease-protected mitochondrial extracts from BAT, heart, and kidney of CkbFLAG mice housed at 22°C. CkbFLAG mice and wild type (CkbWT mice) exposed to 30°C or 6°C were used to confirm Flag antibody cross-reactivity with CKB.FLAG. f, Quantification of mitochondrial CKB from Western blots in e. g, Western blot of protease-protected BAT mitochondria from wild-type (C57BL6/N, 6-8 weeks old) male mice, reared at 22°C, housed at 30°C for 5 days and then subjected to single-housing at 30°C or 6°C. h, Western blot of whole cell extracts. i, Western blot of cytosol extracts. j, Western blot of mitochondrial extracts with and without protease treatment. Blue arrows: CKB.FLAG protein; ns: non-specific bands. k-l, Creatine kinase activity from Ckbfl/fl (n = 3), Ckb−/− brown adipocytes expressing FLAG-tagged GFP (n = 3), CKBWT (n = 4), CKBΔiMTS-L (n = 3), or CKBC283S (n = 3) (k) or bacteria-purified CKB.Flag variants (n = 3) (l). m, Western blots of control or Tom70-silenced mitochondrial extracts with and without protease treatment. For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. k, one-way ANOVA (Fisher’s LSD). Cartoons were created with biorender.com.
Extended Data Fig. 5: Mitochondrial CKB triggers futile creatine cycling selectively in thermogenic adipocytes.
a-d, Effect of creatine on the rate of respiration in mitochondria from wild-type brown adipocytes (n = 10 per group) (a), Ucp1−/− brown adipocytes (n = 14 per group) (b), 3T3-F442A white adipocytes (vehicle: n = 4; creatine: n = 5) (c), or C2C12 myoblasts (vehicle: n = 4; creatine: n = 5) (d). e, Effect of creatine on the rate of respiration in mitochondria from brown adipocytes infected with shLacZ (vehicle: n = 4; creatine: n = 5) (e, left), shCkb#1 (vehicle: n = 4; creatine: n = 5) (e, middle), or shCkb#2 (vehicle: n = 5; creatine: n = 5) (e, right). f, Western blot CKB abundance in Ckbfl/fl brown adipocytes compared to Ckb−/− brown adipocytes titrated with various amounts of FLAG-tagged CKBWT. g, Super-stoichiometric action of creatine on ADP-dependent respiration in Ckb−/− brown adipocytes without rescue (same data as Fig. 4m) (n = 4) compared to re-expression of FLAG-tagged CKBWT to endogenous levels (n = 3) or levels 5-fold above endogenous (n = 2). Endogenous re-expression and over-expression data were not different, and were thus pooled together (n = 5). Futile creatine cycling data represent the moles of liberated ADP (measured as the creatine-dependent change in ADP-dependent O2 consumption) over moles of added creatine. For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. a-e, multiple two-tailed Student’s t-tests; g, two-tailed Student’s t-test.
Extended Data Fig. 6: Selective reduction of creatine kinase protein and activity in adipose tissues of CkbUcp1-CreERT2 and CkbAduoiQ-Cre mice.
a, Cartoon of breeding strategy to generate inducible Ucp1-adipocyte-selective Ckb knockout mice (CkbUcp1-CreERT2). b, RT–qPCR of Ckb mRNA levels in various tissues of Ckbfl/fl (n = 3) and CkbUcp1-CreERT2 (n = 4) female mice housed at 22°C. c-f, Western blot of BAT (c), subcutaneous adipose tissue (SAT) (d), perigonadal adipose tissue (PgAT) (e), and brain (f) from Ckbfl/fl and CkbUcp1-CreERT2 male mice, housed at 22°C (n = 3 per group). g, Creatine kinase activity in BAT lysates from Ckbfl/fl (n = 3) and CkbUcp1-CreERT2 (n = 3) male mice, housed at 22°C. h-j, Creatine kinase activity in SAT (h), PgAT (i), and brain (j) lysates from Ckbfl/fl (n = 4) and CkbUcp1-CreERT2 (n = 3) male mice, housed at 22°C. k, Cartoon of breeding strategy to generate pan-adipocyte-selective Ckb knockout mice (CkbAdipoQ-Cre). l, RT–qPCR of Ckb mRNA levels in various tissues of Ckbfl/fl (n = 5) and CkbAdipoQ-Cre (n = 3) male mice housed at 22°C. m-p, Western blot from BAT (m), SAT (n), PgAT (o), and brain (p) from Ckbfl/fl and CkbAdipoQ-Cre male mice, housed at 22°C. q-t, Creatine kinase activity in BAT (q), SAT (r), PgAT (s), and brain (t) lysates from Ckbfl/fl (n = 5) and CkbAdipoQ-Cre (n = 3) male mice, housed at 22°C. For gel source data, see Supplementary Figure 1. Data are presented as mean ± s.e.m. of biologically independent samples. b, g-j, l, q-t, two-tailed Student’s t-tests. Cartoons were created with biorender.com.
Extended Data Fig. 7: Oxygen consumption, food intake and nutrient absorption.
a, CL-dependent (a, top) and saline-dependent (a, bottom) oxygen consumption of male mice at 30°C (n = 8 per group). b, CL-dependent (b, top) and saline-dependent (b, bottom) oxygen consumption of Ckbfl/fl (n = 7) and CkbAdipoQ-Cre (n = 9) male mice at 30°C. c-d, Body mass change (c) and cumulative food intake (d) of female mice (n = 9 per group). e-f, Body mass change (e) and cumulative food intake (f) of female mice. CkbAdipoQ-Cre(ad-lib): n = 12; Ckbfl/fl(ad-lib): n = 12 and Ckbfl/fl(pair-fed): n = 10. g-h, Lean (g) and fat (h) mass change of mice from e. i-j, ANCOVA of cumulative food intake derived from data in d, for the first (i) and last (j) week of HFD. k-l, ANCOVA of cumulative food intake derived from data in f, for the first (k) and last (l) week of HFD. m-n, ANCOVA of combined data from i and k for the first (m), and from j and l for the last (n) week of HFD. o-p, Fecal energy density (o) and output (p) (n = 4 per group). q-r, Energy output (q) and metabolic efficiency (r) of data from f-h. s, fasting blood glucose levels following a fast from ZT0-ZT6. CkbAdipoQ-Cre(ad-lib): n = 11; Ckbfl/fl (ad-lib): n = 9; Ckbfl/fl (pair-fed): n = 10. Data are presented as mean ± s.e.m. of biologically independent samples. P values on graphs containing 3 experimental groups are relative to CkbAdipoQ-Cre(ad-lib) mice. a-c, e, g, h, two-way ANOVA (Fisher’s LSD); i-n, two-sided analysis of co-variance (ANCOVA); o-s, one-way ANOVA (Fisher’s LSD).
Supplementary Material
Acknowledgements:
We thank Patrick Seale for helpful discussions. We acknowledge technical assistance from the McGill Goodman Cancer Research Centre (GCRC) Metabolomics core facility. We acknowledge technical assistance from the Histology Core Facility at the McGill/GCRC for assistance in embedding and processing tissue samples. We thank all members of the Kazak lab for critical reading of the manuscript.
Funding:
Supported by Canadian Institutes of Health Research grant (PJT-159529) and Canadian Foundation for Innovation John R. Evans Leaders Fund (37919) (to L.K.); Canderel and Charlotte and Leo Karassik fellowships (to J.F.R.); Canderel studentship (to M.F.H.); and the GCRC metabolomics core facility is funded by the Dr. John R and Clara M. Fraser Memorial Trust; the Terry Fox Foundation; the Quebec Breast Cancer Foundation and McGill University. L.K. is a Canada Research Chair in Adipocyte Biology.
Footnotes
Competing interests: The authors declare no competing interests.
Additional information: Supplementary information is available for this paper.
Data availability:
Please address correspondence and requests for materials to L.K. Source data are provided with this paper. All data is available in the main text or the supplementary materials.
References:
- 1.Ward ZJ et al. Projected U.S. State-Level Prevalence of Adult Obesity and Severe Obesity. N Engl J Med 381, 2440–2450, doi: 10.1056/NEJMsa1909301 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Bartelt A et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med 17, 200–205, doi: 10.1038/nm.2297 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Carpentier AC et al. Brown Adipose Tissue Energy Metabolism in Humans. Front Endocrinol (Lausanne) 9, 447, doi: 10.3389/fendo.2018.00447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazak L et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655, doi: 10.1016/j.cell.2015.09.035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challa TD et al. A Genetic Model to Study the Contribution of Brown and Brite Adipocytes to Metabolism. Cell Rep 30, 3424–3433 e3424, doi: 10.1016/j.celrep.2020.02.055 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Lowell BB et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366, 740–742, doi: 10.1038/366740a0 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Betz MJ & Enerback S Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nat Rev Endocrinol 14, 77–87, doi: 10.1038/nrendo.2017.132 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Kazak L et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab 26, 660–671 e663, doi: 10.1016/j.cmet.2017.08.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazak L et al. Ablation of adipocyte creatine transport impairs thermogenesis and causes diet-induced obesity. Nature Metabolism 1, doi: 10.1038/s42255-019-0035-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perna MK et al. Creatine transporter deficiency leads to increased whole body and cellular metabolism. Amino Acids 48, 2057–2065, doi: 10.1007/s00726-016-2291-3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corrigan JK et al. A big-data approach to understanding metabolic rate and response to obesity in laboratory mice. eLife 9, doi: 10.7554/eLife.53560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallimann T, Wyss M, Brdiczka D, Nicolay K & Eppenberger HM Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the 'phosphocreatine circuit' for cellular energy homeostasis. The Biochemical journal 281 (Pt 1), 21–40 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlattner U, Tokarska-Schlattner M & Wallimann T Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta 1762, 164–180, doi: 10.1016/j.bbadis.2005.09.004 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Long JZ et al. A smooth muscle-like origin for beige adipocytes. Cell Metab 19, 810–820, doi: 10.1016/j.cmet.2014.03.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roh HC et al. Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab 27, 1121–1137 e1125, doi: 10.1016/j.cmet.2018.03.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min SY et al. Diverse repertoire of human adipocyte subtypes develops from transcriptionally distinct mesenchymal progenitor cells. Proc Natl Acad Sci U S A 116, 17970–17979, doi: 10.1073/pnas.1906512116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazak L et al. Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res 41, 2354–2369, doi: 10.1093/nar/gks1347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazak L et al. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS One 8, e62340, doi: 10.1371/journal.pone.0062340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backes S et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. The Journal of cell biology 217, 1369–1382, doi: 10.1083/jcb.201708044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furter R, Furter-Graves EM & Wallimann T Creatine kinase: the reactive cysteine is required for synergism but is nonessential for catalysis. Biochemistry 32, 7022–7029, doi: 10.1021/bi00078a030 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Hornemann T, Rutishauser D & Wallimann T Why is creatine kinase a dimer? Evidence for cooperativity between the two subunits. Biochim Biophys Acta 1480, 365–373, doi: 10.1016/s0167-4838(00)00098-4 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Watt IN, Montgomery MG, Runswick MJ, Leslie AG & Walker JE Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci U S A 107, 16823–16827, doi: 10.1073/pnas.1011099107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J & Hall KD Estimating the continuous-time dynamics of energy and fat metabolism in mice. PLoS Comput Biol 5, e1000511, doi: 10.1371/journal.pcbi.1000511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe MT, Kim EH, Faull RL, Christie DL & Waldvogel HJ Dissociated expression of mitochondrial and cytosolic creatine kinases in the human brain: a new perspective on the role of creatine in brain energy metabolism. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 33, 1295–1306, doi: 10.1038/jcbfm.2013.84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eppenberger HM, Dawson DM & Kaplan NO The comparative enzymology of creatine kinases. I. Isolation and characterization from chicken and rabbit tissues. J Biol Chem 242, 204–209 (1967). [PubMed] [Google Scholar]
- 26.Rosenwald M, Perdikari A, Rulicke T & Wolfrum C Bi-directional interconversion of brite and white adipocytes. Nature cell biology 15, 659–667, doi: 10.1038/ncb2740 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Vergnes L et al. Adipocyte Browning and Higher Mitochondrial Function in Periadrenal But Not SC Fat in Pheochromocytoma. J Clin Endocrinol Metab 101, 4440–4448, doi: 10.1210/jc.2016-2670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelsson O, Brunak S, von Heijne G & Nielsen H Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols 2, 953–971, doi: 10.1038/nprot.2007.131 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Guo J & Hall KD Predicting changes of body weight, body fat, energy expenditure and metabolic fuel selection in C57BL/6 mice. PLoS One 6, e15961, doi: 10.1371/journal.pone.0015961 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Please address correspondence and requests for materials to L.K. Source data are provided with this paper. All data is available in the main text or the supplementary materials.