Abstract
The neural stem cell niche of the ventricular–subventricular zone supports the persistence of stem and progenitor cells in the mature brain. This niche has many notable cytoarchitectural features that affect the activity of stem cells and may also support the survival and growth of invading tumor cells. Histochemical studies of the niche have revealed many proteins that, in combination, can help to reveal stem-like cells in the normal or cancer context, although many caveats persist in the quest to consistently identify these cells in the human brain. Here, we explore the complex relationship between the persistent proliferative capacity of the neural stem cell niche and the malignant proliferation of brain tumors, with a special focus on histochemical identification of stem cells and stem-like tumor cells and an eye toward the potential application of high-dimensional imaging approaches to the field.
Keywords: angiogenesis, glioblastoma, glioma stem cell, image analysis, mass cytometry, multiplex imaging, neural stem cell, SEZ, stemness, SVZ, tumor heterogeneity, tumor immune infiltrate, tumor microenvironment, V-SVZ
Introduction
The ventricular–subventricular zone (V-SVZ) is one of two locations which retains detectable stem/progenitor cell capacity after birth in the brains of many mammals. The ability to generate new neurons and glia in vitro and in vivo, while offering the potential of cell replacement, has also led to intense focus on the potential relationships of stem cell niches such as V-SVZ to malignant growth in cancer. In human brain and brain tumors, the identification of stem-like cells frequently relies on careful histochemical characterization, as classic lineage tracing approaches used in model organisms are rarely feasible. Here, we review challenges and successes in using histochemical and tissue culture approaches to study this niche in mouse and human, with a special focus on the use of antibody-based approaches in normal and tumor-bearing brain. Finally, we discuss the plethora of multidimensional, single-cell approaches now emerging for tissue imaging and sequence analysis, and the analytical tools for revealing known and novel cell phenotypes within these data.
Histochemical and Cytochemical Approaches to Identifying Neural Stem Cells
In mammals, postnatal neurogenesis primarily occurs in two germinative regions: the V-SVZ and the subgranular zone (SGZ) within the hippocampus. The V-SVZ is the larger of these two regions—in the mouse, neurogenic capacity is found in the medial, lateral, and subcallosal faces of the lateral ventricles, whereas studies in human have largely focused on the lateral walls of the lateral ventricles. 1 The stem cells of the V-SVZ are derived from a subset of prenatal stem/progenitor cells termed radial glia. During embryonic development, these radial glia have cell bodies that are close to the ventricles, an apical process that contacts the cerebrospinal fluid (CSF) within the ventricles, and a long radial process that extends toward the outer (pial) surface of the brain. 2 While this radial process gradually retracts during the emergence of the postnatal V-SVZ, many postnatal stem cells retain both the apical contact with the CSF and a basal process that contacts the vasculature underlying this niche, thus maintaining a radial structure.3–5 The cytoarchitecture of the adult V-SVZ has been described in rodent and human using both light microscopy and electron microscopy approaches; indeed, identification of V-SVZ cell types using electron microscopy was essential to early studies defining lineage progressions within this niche.6,7 Surprisingly, neural stem cells in both the V-SVZ and the SGZ share many features with mature astrocytes, including expression of multiple marker proteins (discussed below), ultrastructural characteristics, and functional features such as contact of the cellular endfoot with the vasculature. In addition to stem cell–blood vessel contact, the architecture of the V-SVZ is also distinguished by several additional features: (1) the collection of transit-amplifying cells near the vasculature, 4 (2) the presence of specialized brain-resident immune cells (microglia) with different phenotypes than those in other brain regions,8–10 (3) the ensheathing of chains of newly born neurons by the processes of stem cells,11,12 (4) the arrangement of ependymal cells lining the ventricle into “pinwheels” that surround the apical contact of neural stem cells with the CSF,3,13 and (5) the infiltration of subdomains of the niche by processes from multiple populations of neurons.14,15
When activated, quiescent V-SVZ neural stem cells (also termed type B-cells) can generate rapidly dividing intermediate progenitor cells (type C-cells), which in turn produce maturing progeny: neuroblasts and oligodendrocyte precursors. In rodents, the majority of V-SVZ stem cells generate neuroblasts that migrate along a structure termed the rostral migratory stream and then enter the olfactory bulb, where they have long been known to differentiate and integrate as interneurons.11,16–20 Specific subregions of the V-SVZ also generate oligodendrocytes, which contribute to white matter structures such as the corpus callosum.21–25
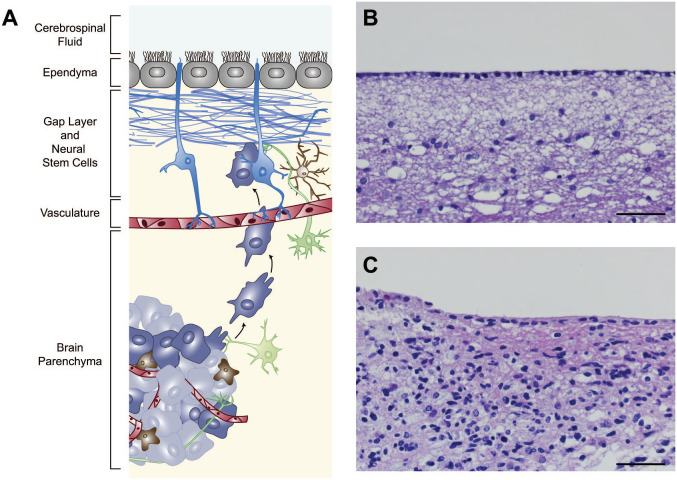
Although studies in the rodent have been highly informative in studying core pathways regulating the organization, persistence, and activation of stem cells within the V-SVZ niche, 26 some key differences exist between mouse and human, which are relevant to studies of the normal and cancerous brain. As explored in the discussion of other stem cell niches below, the extent of olfactory bulb neurogenesis in human brain has been subject to conflicting reports.27,28 One feature that is readily observable in histological sections (Fig. 1) is the presence of a “gap layer” in the human V-SVZ which is not seen in the mouse. 29 In adult brain, this layer is filled with astrocytic processes but is relatively devoid of nuclei. This distinctive organization emerges in early postnatal human brain, coinciding with a marked decline in the detection of proliferating cells and immature neurons. 30 These neuroblasts initially fill a space between the ependymal cells lining the ventricle and the neural stem cells, and the gap emerges as neuroblast production declines. This decline in proliferation differs from the young adult mouse brain, where proliferating cells are abundant, as well as older (greater than one year old) rodents, where proliferation is decreased but still detectable.31,32
Figure 1.
The V-SVZ in schematic and in tumor histology. The mature human V-SVZ is found in the lateral walls of the lateral ventricles and is shown in a cross-sectional schematic (A) and in histological section (B). The neural stem cells that persist in this niche (shown in blue, A) have contact with the cerebrospinal fluid (at top) and the underlying vasculature (shown in red). Processes of neurons (green) and immune cells including brain-resident microglia (brown) are also found in this region, as well as the multiciliated ependymal cells (gray) which line the ventricles. The human V-SVZ is also distinguished by a gap layer (blue, bracket in A) that is largely devoid of nuclei (visible in B). Brain tumor cells (purple and gray) can be found invading this niche and migrate toward it in mouse models. In tissue from a glioblastoma patient with radiographic contact with the ventricles, abnormal cells can be seen in this region (C). Scale bars for B and C = 50 µm. Abbreviation: V-SVZ, ventricular–subventricular zone.
At the anatomic level, although the bulk of V-SVZ-derived neurons in the mouse migrate to and populate the olfactory bulbs, other migration patterns have been identified in human. Studies of pediatric brain described a medial migratory stream (MMS), which branches from the proximal rostral migratory stream to supply the ventromedial prefrontal cortex. 30 These clustered cells coexpressed proteins that interact with the cytoskeleton or adhesion complexes and are typical of neuroblasts. A comparable MMS has not been identified in other vertebrates.
Examinations of the anterior forebrain of children younger than 3 months of age 33 further described regions of high cell density adjacent to the anterior body of the lateral ventricle and within the neighboring subcortical white matter, forming a distinct arching structure (“Arc”) in sagittal sections. These migrating neurons target an extensive region of the anterior forebrain, including the cingulate gyrus and prefrontal cortex, where they differentiate into interneurons. Similar to other V-SVZ-derived populations, these young neurons appear to migrate during a specific early postnatal period and are not seen in mature brain. However, functional studies of label-retaining cells and cells cultured in vitro suggest that some level of latent proliferative potential persists in human V-SVZ. These functional assays, and their application in the V-SVZ and SGZ niches, are further discussed below.
Functional Features of Neural Stem Cells and Their Study Across Species
Functional features of neural stem cells that have been studied in both normal and cancer contexts include (1) infrequent entry into the cell cycle, resulting in resistance to antimitotic treatment and retention of labels diluted by dividing cells; (2) long-term in vitro propagation and self-renewal in vivo; and (3) multilineage differentiation—the generation of multiple mature cell types. Each of these features was used in early research as the identification of neural stem cells grew increasingly specific, progressing from tissues to subregions to precise subcategories of cells. More recently, histochemical studies of both V-SVZ and SGZ have highlighted both new approaches and accompanying challenges in validation of these approaches in human tissue specimens.
At a time when the possibility of postnatal neurogenesis was doubted by many, Altman and Das34,35 made a pioneering report of postnatal and adult neurogenesis by injection of radiolabeled thymidine in rats to acutely label cells in S phase at different postnatal ages. Labeling was present within neurogenic niches, including a small number of labeled cells in adult animals. These early findings indicated that dividing populations of cells persisted in the adult mammalian brain, and the use of nucleoside analogs or radiolabeled thymidine to detect DNA replication is a method that continues to be widely used.32,36 However, it should be noted that this method, like others, can be susceptible to false-positive signals (discussed in Breunig et al. 37 ).
In the mouse, a series of studies using infusion of the antimitotic and chemotherapeutic Ara-C (cytosine arabinoside, also called cytarabine) into the cerebrospinal fluid subsequently demonstrated that a subpopulation of cells within the V-SVZ, while resistant to the effects of Ara-C, was able to enter the cell cycle and produce transit-amplifying cells, neuroblasts, and, ultimately, mature neurons and oligodendrocytes after the drug was discontinued. 38 Furthermore, the astrocyte-like type B-cells identified using this approach were shown to be slow-cycling and label-retaining using pulse-chase administration paradigms which labeled S-phase cells.
In 1998, similar approaches revealed potential conservation of properties of stem cells in the human by studying postmortem brain tissue from cancer patients who had been treated with the thymidine analog and S-phase DNA marker bromodeoxyuridine (BrdU) for diagnostic purposes. 39 Using immunofluorescent detection of BrdU, potential S-phase cells were found in the hippocampal dentate gyrus and SGZ.
Other rare human populations that provided a label-retention assay view into postnatal neuron birth in the V-SVZ include cancer patients receiving iododeoxyuridine (IdU) for radiosensitization and people whose lives spanned the beginning of nuclear bomb testing–based atmospheric 14C exposure.36,40,41 Examination of these samples found infrequent IdU-labeled neuronal cells expressing NeuN, MAP2, and calretinin in both the caudate nucleus and the putamen regions of the striatum, as well as in the hippocampal dentate gyrus, indicating that cells which incorporated IdU gave rise to mature neurons. This group also isolated neuronal nuclei from the lateral ventricle wall and striatum of adult humans, and analyzed the 14C content of genomic DNA. Using a mathematical model based on the 14C content expected in non-dividing cells and dividing cells, differing amounts were found in nuclei in the striatum adjoining the V-SVZ, suggesting that some neurons in this region were generated postnatally. However, given their reliance on a specific population (humans alive during the 1950s, when 14C levels were elevated), these experiments cannot be readily or widely repeated. A second concern is that the method of nuclear isolation, via prospective fluorescence-activated cell sorting, prohibited the simultaneous collection of anatomic or cytoarchitectural information beyond the initial dissection of the regions analyzed.
Progress in tissue culture assays for quantifying postnatal neurogenic capacity advanced in the 1990s with the isolation of a multipotent population from the adult mouse periventricular region and striatum, which could be induced to proliferate on a non-adhesive substrate with multiple specialized culture mediums.42,43 The proliferating cells formed spheres and could be passaged as single cells to form new spheres, suggesting a self-renewing population was contained within these cultures. Following transfer to a poly-ornithine-coated surface, adherent spheres differentiated to form neurons and astrocytes, demonstrating multipotency. Human neural stem/progenitor cells cultured under these conditions also showed the capacity for integration and multilineage differentiation when engrafted in rodent brain in vivo. This feature has been demonstrated for human brain cells by multiple groups.44–46
Since the initial generation of neurosphere cultures, these assays have been widely used as a proxy for self-renewal and multipotency in both normal and cancer populations. However, a note of caution has been sounded by multiple studies in the rodent V-SVZ, which have demonstrated that standard sphere culture conditions (using both epidermal growth factor and basic fibroblast growth factor) enrich for spheres that are primarily derived from transit-amplifying cells and a small population of activated stem cells.13,47 By contrast, culture conditions containing other factors identified from the cerebrospinal fluid and choroid plexus have more successfully elicited sphere growth from the long-lived, quiescent neural stem cell population. 48
Similarly, as the ability of researchers to follow the activity of small populations and, in some cases, single stem/progenitor cells has advanced, previously unappreciated levels of heterogeneity have been found within the V-SVZ niche.49,50 Type B-cells develop “positional identity” during embryonic development, with the dorsoventral patterning of radial glia, the progenitors of the developing brain, persisting into the mature niche.51–56 Within the adult brain, the precise position of a cell within the dorsal or ventral V-SVZ predicts the type of glia or olfactory bulb neurons it will generate.25,57 Thus, although bulk sphere cultures of stem cells or stem-like cancer cells may suggest multipotency, at the per-cell level cells may be more limited in the types of progeny they produce.
Histochemical Features of Neural Stem Cells
Strategies such as BrdU/IdU labeling and 14C dating are not readily available for most studies of human neural progenitors, and consequently immunohistochemical markers of cells with stem-like or immature properties are widely used to identify these cells. However, many factors can alter the quality of such staining, leading to controversy in the field regarding the extent of ongoing neural stem cell activity in human brain and the presence of stem-like subpopulations within human brain tumors. In these as in other histochemical studies, proper use of multiple relevant positive and negative controls for key antibodies and other reagents is essential to interpreting these findings. Since the functional and structural identification of neural stem cells in mouse and human, a large number of proteins have been found to be expressed on stem and progenitor cells within the V-SVZ. Many of these proteins are dynamically expressed and not perfectly exclusive to the stem cell population, meaning that combinations of markers (typically, three to six, as in Mich et al. 58 and Codega et al. 13 ) are often used to isolate and study these cells. Below, a subset of widely used markers are discussed (a more extensive review of marker proteins is available at Rushing and Ihrie 55 ).
Nestin is an intermediate filament protein often used to mark proliferating neuroepithelial cells. It was originally identified by the monoclonal antibody Rat 401 which transiently stained mitotic regions of the rat embryo neural tube.59,60 The distribution of Nestin immunoreactive cells in the neural tube and their proliferative capacity during the period of neurogenesis were found to be characteristic of neural stem cells. 59 Subsequent work has used animal models with reporters or enzymes driven by the Nestin promoter to mark and manipulate these cells. However, Nestin may also be expressed by some mature cells, and some subpopulations of neural stem cells lack Nestin expression. 48 Similarly, the intermediate filament protein vimentin distinguishes stem cells in the developing and mature human and mouse V-SVZ, but can be expressed by other cell types outside the brain as well as parenchymal astrocytes. 61
Expression of the intermediate filament protein glial fibrillary acidic protein (GFAP) and the glutamate-aspartate transporter (GLAST) is also characteristic of V-SVZ stem cells,38,62 reflective of their astrocyte-like properties. Within the V-SVZ, GFAP or GLAST expression distinguishes B-cells from their transit-amplifying and neuroblast progeny. However, GFAP and GLAST are widely expressed by astrocytes throughout the brain, and thus must typically be coupled with additional markers and/or physical dissection of the region of interest (ROI) to be used in stem cell characterization. Specific isoforms of GFAP, such as GFAPδ, have been proposed to be more specific to the stem/progenitor lineage but may also be present in immature neurons. 63
Doublecortin (DCX) can be used to label neuroblasts of the subventricular zone and the hippocampal dentate gyrus. DCX is a brain-specific microtubule-associated protein which acts as a microtubule stabilizer in the setting of migration. 64 Its expression has been carefully described in the adult mouse hippocampus and V-SVZ, where it marks lineage-determined progenitor cells through immature postmitotic neurons.65,66 Although it is not expressed in the stem population, expression of DCX is often cited as evidence that production of neurons is ongoing,30,67 especially when it is present in the same region as proteins which are thought to distinguish cycling cells, such as Ki67, Mcm2, and PCNA. 68 However, an important caveat, further discussed below, is that the fixation of samples and the conditions under which staining is performed can result in false-positive or nonspecific labeling for several of these antigens.
Musashi is an RNA-binding protein that specifies cell fate by translational regulation and is present in the developing mouse central nervous system, especially in mitotically active cells.69–71 In cultured progenitors, its expression overlaps with Nestin expression, and these progenitors are capable of generating neurons and glia. Low levels of Musashi1 expression have been detected in human brain tissue and gliomas by RT-PCR, as well as neurospheres derived from human embryonic brains.69,72
Sox2 is a highly conserved transcription factor expressed in cell types including the totipotent cell lineage, but which also marks stem cells of the central nervous system. Sox2 expression was reported in the neural plate and telencephalic ventricular zone using transgenic mice. 73 Similarly, clonal analysis showed that multipotential neural stem cells are present within the Sox2-positive embryonic population. 74 Constitutive expression of Sox2 was sufficient to maintain neural progenitor characteristics in vivo, whereas Sox2 inhibition causes cell cycle exit, loss of progenitor marker expression, and differentiation. More recently, regulation of the translation of Sox2 mRNA has been suggested to be a key regulator of lineage progression in the V-SVZ. 75 However, like many of the markers discussed here, this protein has also been reported to be expressed by mature cells outside the stem cell niche. 76
Also known as LeX or SSEA-1, CD15 (leukocyte cluster of differentiation 15) is a carbohydrate expressed by embryonic stem cells and in adult brain regions, including the pial surface, corpus callosum, the SGZ of the hippocampal dentate gyrus, and a subset of SVZ cells. 77 Under neurosphere growth-promoting conditions, the CD15-positive cell fraction from mouse brain was capable of generating multipotent and self-renewing spheres, in contrast to the CD15-negative population which only rarely made spheres.
CD133 (Prominin) is a five-transmembrane surface glycoprotein originally described as a marker for hematopoietic stem and progenitor cells 78 and as associating with a microvillus-associated structure in epithelial cells including brain ependymal cells. 79 It was identified in a monoclonal antibody screen as a prospective marker that can enrich for sphere-forming cells in dissociated brain (PMID: 11121071) and has been widely used to enrich for stem-like populations in the V-SVZ and other brain regions.80–82 More recent high-resolution studies have confirmed that it is expressed by both ependymal cells, which lack stem cell activity, and type B stem cells within the V-SVZ. 13
Although the emergence of these and other markers has informed both studies of human neurogenesis and the detection of stem-like populations within human brain tumors, reliable staining for these proteins can be strongly influenced by tissue quality and preparation, an important consideration when working with intraoperative, biopsy, or postmortem samples of brain tissue. For example, within the adult human hippocampus, immunohistochemical detection of subsets of stem cell markers has produced contradictory evidence. Findings that adult neurogenesis may drop to undetectable levels were based on a lack of Ki67+ proliferating cells as well as a lack of DCX+ and PSA-NCAM+ neurons and a lack of adult dentate gyrus Nestin+/Ki67+ cells.83,84 Within the same study, DCX expression was found in cells that had already developed typical dendrites and axons of granule cell neurons, suggesting that these neurons continue to express DCX as they differentiate. On the contrary, the existence of PSA-NCAM+ DCX+ cells, presumed to be immature, in the adult human hippocampus has also been demonstrated.85,86 These differences have in part been suggested to be due to variation in reagent quality and usage, tissue fixation, postmortem interval, and the pathological state of the tissue collected (discussed in depth in Moreno-Jimenez et al. 87 and Sorrells et al. 88 ).
In addition, some cross-species variation in marker expression may exist. In another example, an analysis of CD133 expression in the human brain found expression by embryonic neural stem cells, by an intermediate radial glial/ependymal cell type in the early postnatal stage, and by ependymal cells in the adult brain, but not for neurogenic astrocytes in the adult subventricular zone. 89 In these rare and difficult to collect samples, higher dimensional cell phenotyping may also help shed light on this controversy by allowing simultaneous examination of many markers, as discussed in section “V-SVZ–Glioma Interactions.” 90
Stem-like Cells in Gliomas
As one of two sites which harbors latent proliferative potential, the V-SVZ has been a major focus of study for those seeking to understand the origins of adult brain tumors. The vast majority of rodent models of these tumors are generated through the genetic modification of stem/progenitor cells from either embryonic brain or adult V-SVZ, and these cells efficiently form tumors when transduced with constructs replicating core mutational features of human brain tumors. Furthermore, genetic analyses of V-SVZ cells located adjacent to resected human brain tumors showed that some mutations found within the tumor were also detectable in V-SVZ cells, arguing that the V-SVZ was the site of origin. 91 However, studies of genetically manipulated mature astrocytes indicate that these cells can also form tumors, albeit with decreased efficiency. 92 Furthermore, elegant tracking of mutant cell outgrowth using mouse models suggests that in some cases although stem cells may sustain an initial oncogenic mutational event, aberrant outgrowth does not occur until the cell in question differentiates further and reaches a transit-amplifying or precursor state. 93 Thus, although it is likely that many adult brain tumors in humans originate in the V-SVZ, the possibility remains that some brain tumors originate elsewhere but share some molecular features with the populations of this niche.
In adults, the large majority of malignant tumors originating in the brain are aggressive World Health Organization grade IV glioblastomas, which fall within the larger class of grade II to grade IV gliomas. 94 Glioblastomas almost inevitably recur following standard treatment. The nature of this treatment—maximally achievable resection of the tumor mass, followed by radiation and treatment with alkylating chemotherapy temozolomide—and the apparent resistance of a subpopulation of tumor cells to this approach suggested that a population of stem-like cells might exist within tumors. The cancer stem cell theory (reviewed in Lauko et al. 95 and Zong et al. 96 ) initially proposed that within tumors such as glioblastoma, there exists a population of cells that are self-renewing and generate the diverse populations of abnormal cells found within a tumor. A subset of these cells, like normal neural stem cells, may divide infrequently, and thus be resistant to many chemotherapies that target rapidly dividing cells.
Since the framing of the cancer stem cell model, multiple studies have used a variety of lineage-tracing, prospective-sorting, and lineage-reconstruction strategies to identify and follow stem-like subpopulations of cells in gliomas. Consequently, an array of proteins that are enriched in the stem cells of the V-SVZ and SGZ have been used to prospectively identify and sort such cells from within human brain tumors. CD133 (Prominin-1) was the first reported cell-surface antigen to enrich for the tumor-propagating, stem-like fraction. 97 Subsequent work has used the cell-surface proteins CD44, CD15, CD49f, PDGFRβ, and L1CAM, among others, to prospectively identify these cells. 96 Furthermore, SOX2, Nestin, the oligodendrocyte and peripheral nervous system transcription factor SOX10, and other intracellular factors have been proposed as markers of stem-like states within tumors. Many of these factors are enriched in tumor-propagating fractions of cells derived from tissue, as in the normal niche there is no single marker that reliably identifies a cancer stem cell state. Similarly, the formation of spheres using neurosphere culture conditions in combination with limiting dilution analyses has been used to estimate the fraction of stem-like cells within intraoperative samples from brain tumors. 98 Neurosphere culture approaches have allowed propagation of “glioma stem cell” lines that have been invaluable in modeling tumor growth and resistance to therapy. 99 However, as in the normal niche, there are multiple protocols and approaches for sphere culture and tumor cell propagation, which likely enrich for differing populations of stem and progenitor-like cells.
In longitudinal studies of tumor initiation and growth, the identification and tracing of stem-like subsets are complicated by the likelihood that multiple stem-like populations may emerge, differentiate, and de-differentiate over time—that is, that plasticity exists within the malignant cell population. As high-dimensional approaches begin to be applied to these tumors, a high degree of heterogeneity has been observed, with multiple markers and combinations of markers found across patient cohorts and individual subregions of tumors.95,100,101 Intriguingly, recent studies of a small set of primary glioblastoma samples revealed a stem-like population that most closely resembles outer radial glial cells—a progenitor cell type that is typically seen only during embryonic brain development. 102 These outer radial glia-like cells resembled embryonic progenitors both in their transcript expression and in their pattern of division, exhibiting mitotic somal translocation (movement of the cell body just prior to division), a feature that is stereotypic of these progenitors. Functional features such as these, which resemble those of early progenitors, may in part explain the well-known invasive and migratory behavior of glioblastoma tumors. Cells with these stem/progenitor phenotypes may also be well adapted to exploit the environment of the V-SVZ, which is rich in factors that can drive cell quiescence, self-renewal, and immune suppression (reviewed in Sinnaeve et al. 103 ).
V-SVZ–Glioma Interactions
Beyond the potential origins and constituent cells of glioblastomas, the environment of the V-SVZ has recently emerged as a potential regulator of cancer cell spread and aggression. Many of the factors which are enriched in the cerebrospinal fluid or the niche itself have been proposed to attract glioblastoma cells, support their growth, or alter immune cell activity, and in turn glioma cell invasion has been noted to affect V-SVZ cell proliferation.104–106 Systematic review of the effects of tumor contact with the lateral ventricles on MRI (and, by extension, with the V-SVZ niche) has demonstrated that this contact, and not contact with the SGZ, is an independent predictor of shorter progression-free and overall survival, underscoring the importance of understanding these interactions when working to improve glioblastoma treatment.107–109 Studies of V-SVZ-contacting and V-SVZ-distant tumors using MRI imaging and bulk transcriptomics have not reliably identified broad differences in the profiles of these two classes, arguing that a finer dissection of areas proximal to the V-SVZ, or an approach that enables quantification of cellular-level phenotypes and spatial relationship, may be needed to understand the biological effects of this contact.
The environment of the V-SVZ has been proposed as a source of multiple trophic signals, resulting in preferential recruitment of glioma cells to this region rather than other locations within the brain in studies using intracranial engraftment of human tumor cells.110,111 Case studies of human brain tumor cases at autopsy have reported detection of abnormal cells within the V-SVZ, distant from the primary tumor mass, supporting the idea that this niche is attractive and supportive for glioma cells.112,113
Studies in the mouse have also indicated that the vascular plexus underlying the V-SVZ is supportive of both stem cell persistence and of transit-amplifying cell proliferation.3–5,114 Furthermore, testing of barrier function via systemic administration of high-molecular-weight dyes has indicated that the vasculature in this region is more permeable than in other brain regions, 5 suggesting that this environment may provide blood-borne factors to invading tumor cells more readily than other regions, providing a natural harbor that enhances therapy resistance and the survival of cells that are distant from the main tumor mass. Such features may help to explain why an increased percentage of recurrent tumors, versus primary, display contact with the lateral ventricles/V-SVZ.107,108 Similarly, the innervation of the V-SVZ may provide support to glioblastoma cells, although how such support may compare with the supportive neuron–glioma interactions demonstrated in other brain regions, or interaction with the vascularized niche of the SGZ, is not yet clear. Emerging vascularized organoid models (reviewed in Matsui et al. 115 ) may provide a platform to systematically test these interactions.
To date, high-dimensional characterization of either the tumor mass or tumor-associated immune, neural, and glial cells has largely relied on dissociated tissue, removing the ability to quantify spatial relationships and cytoarchitecture. 101 However, recent advances in the development of complex culture models and platforms to produce high-dimensional imaging cytometry suggest that such analyses are now achievable. The design and rapid adoption in the field of organoid culture models, which produce “mini-brains” recapitulating several key architectural features of the developing brain, have led to multiple approaches adapting this method to study brain tumor biology. Organoid approaches have been used both to generate “host” tissues for studying glioblastoma cell invasion and persistence and to propagate “tumoroids” that contain multiple spatially distinct populations and may support the survival of tumor-infiltrating immune cells.116,117 As these systems advance, it will be of interest to explore whether they enrich for specific stem- and progenitor-like features in the cancer lineage cells. Comparison of these systems, which are typically used to recapitulate the embryonic stages of human brain growth, with the environment of the postnatal and maturing V-SVZ will also be informative to understanding how this niche may support brain tumor growth in adults.
Emerging New Histochemical and Imaging Approaches and Their Potential Impact on the Field
Conventional imaging approaches such as immunohistochemistry (IHC) and immunofluorescence (IF) have been widely used in the identification and characterization of neural stem cells in the V-SVZ and stem-like cells within brain tumors. However, these approaches are often limited in the number of markers that can be labeled and measured within a single sample. This can result in discordant identification strategies or a loss of specificity in population identification. In addition, to more fully parse the potential interactions between V-SVZ-constituent cells, cancer lineage cells, vasculature, and immune cells, simultaneous staining and analysis for markers of many cell types and cell states will be a valuable tool. Recent developments in the field of highly multiplexed, antibody-based imaging such as multiplex IHC/IF, imaging mass cytometry (IMC)/multiplexed ion beam imaging (MIBI), and CODEX (Fig. 2) present the opportunity to circumvent these limitations and simultaneously measure a wide array of markers, allowing for a deeper phenotyping of protein expression within the V-SVZ. Beyond immunohistochemical labeling, the proliferation of spatially resolved transcriptomics, metabolomics, high-resolution mass spectrometry, and approaches combining these methods are also likely to reveal new features of this niche.118–120 Ultimately, the ability to simultaneously measure and combine many parameters, and to subsequently identify those which are most essential to revealing populations or responses of interest, has the potential to lead to new diagnostic tools or biomarkers in these tumors. The development and proliferation of highly multiplexed techniques have also necessitated data analysis tools and platforms designed to manipulate datasets of considerable size and high dimensionality.
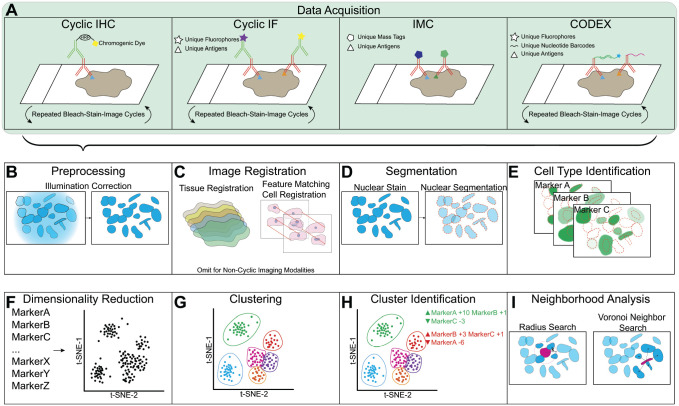
Figure 2.
Highly multiplexed imaging data collection and analysis pipelines. A selection of multiplexed imaging approaches is illustrated in brief (A). In cyclic IHC, primary antibodies for a protein of interest are detected by a secondary antibody conjugated to horseradish peroxidase, which catalyzes a reaction depositing a chromogenic dye which can then be visualized through light microscopy in rounds of bleaching, staining, and imaging. In Cyclic IF, secondary antibodies are detected using fluorophores, followed by bleaching, staining, and imaging. In IMC or MIBI, primary antibodies are directly conjugated to unique mass tags, and data are collected simultaneously. In CODEX, primary antibodies conjugated to unique nucleotide barcodes are stained simultaneously and are detected cyclically using complementary barcodes conjugated to fluorophores. After data collection, imaging data must be processed to quantify features of interest (B–E). Preprocessing steps include illumination correction (B), background subtraction, bleedthrough correction, and autofluorescence correction. Image registration often begins at the level of the gross tissue and ideally is followed by a cell–cell registration (C). Nuclear segmentation identifies single cells (D). Expression of proteins of interest and their localization can then be quantified per detected cell (E). Once an expression dataset has been produced, specific techniques are necessary to analyze high-dimensional imaging data (F–I). Dimensionality reduction tools such as t-SNE and UMAP can make high-dimensional relationships between cells easier to visualize (F). Clustering tools such as FlowSOM and SPADE identify populations within the dataset (G). Phenotypic characteristics of identified clusters can be determined using marker enrichment modeling (MEM) or expert-guided median expression analysis (H). Local niche characteristics can be discovered by identifying neighbor cells either by distance from the index cell or by selecting all first-order neighbors (I). Abbreviations: IHC, immunohistochemistry; IF, immunofluorescence; IMC, imaging mass cytometry; MIBI, multiplexed ion beam imaging.
As highlighted by the controversies over detection of neurogenesis discussed above, in all antibody-based detection assays, adequate controls are essential to ensure that the target of interest is selectively and uniquely detected by the protocol. In a highly multiplexed context, this need becomes ever greater as the complexity of the dataset grows. It is crucial to test candidate antibodies for multiplexing on known positive and negative control samples processed and prepared using the same conditions as the multiplex staining protocol. In addition, cyclic staining protocols necessitate testing and validation to ensure that late round antibody–antigen binding is not affected by the repeated staining and stripping procedures. 121
Multiplexed immunohistochemistry or immunofluorescence is perhaps the most straightforward technique for collecting high-dimensional imaging data, as it is built on traditional IHC and IF. These techniques use iterative cycles of antibody staining, imaging, antibody stripping, or fluorophore bleaching and restaining of a single sample to achieve relatively high-dimensional measurements.122–124 The resulting images are typically computationally registered using a nuclear marker that is maintained throughout the imaging cycles. Multiplex IF was recently used to measure biomarkers or signaling proteins in tumor tissue sections from pediatric glioblastoma and astrocytoma samples, demonstrating the feasibility of this approach.125,126
IMC builds on mass cytometry techniques in which single cells labeled with unique mass-tagged antibodies are measured using a time-of-flight mass spectrometry (TOF-MS). However, instead of measuring cells in suspension, IMC rasters an ROI within a slide-mounted tissue section stained with a cocktail of mass-tagged antibodies using laser ablation. The resulting plumes of particles are then measured by TOF-MS. IMC allows for routine simultaneous measurement of 32 markers. In the brain, IMC has been used to study myeloid and astrocytic cell phenotypes in multiple sclerosis (MS) lesions.8,127,128 MIBI is similarly able to detect mass-tagged antibodies; in addition, the use of specialized slides and a dedicated imaging chamber allow simultaneous detection of isotopic variants of lower mass elements including those naturally present in tissues, providing additional channels of information for imaging analysis. 129 CODEX imaging deploys oligonucleotide barcodes conjugated to antibodies that are stained simultaneously but are imaged cyclically. Sets of fluorescently labeled oligonucleotides complementary to sets of barcoded antibodies are administered, imaged, stripped, and repeated to measure up to 58 unique barcoded antibodies.130,131 The resulting images are computationally registered and analyzed similarly to cyclic IF or IHC datasets, but have the advantage of having been stained together, removing variation in staining conditions, reagents, and time.
A throughline in all highly multiplexed imaging techniques is the need for specialized data analysis tools and expertise after data collection. Broadly, these tools can be categorized into commercial and open source. Commercial options include HALO, developed by Indica Labs, and Oncotopix, developed by Visiopharm. These options have the advantages of wide adoption and support, but require costly license agreements. Open source options also exist, such as histoCAT, which was developed for IMC analysis, QuPath, CellProfiler, and Steinbock (github.com/BodenmillerGroup/steinbock).132–134 These options are free to use and include powerful analysis capabilities, but often require some degree of user expertise to access their full potential.
An analysis pipeline for multiplex imaging data from any source will typically include a similar series of steps. Before any analysis begins, quality control of the images is paramount to screen for clear staining failures and artifacts. Preprocessing of the images is often necessary, and the specific algorithms used are dependent on the imaging technique used (Fig. 2B). For cyclic imaging techniques such as multiplex IF/IHC and CODEX, registration of images between many rounds of staining is necessary (Fig. 2C). A single-cell segmentation is often performed using a strong ubiquitous specific nuclear stain (Fig. 2D). Next, expression of proteins of interest is measured by quantification of signal overlapping each segmented cell (Fig. 2E). The expression dataset can be followed by expert or automatic thresholding to identify marker “high/positive” cells or populations exhibiting known combinations of marker expression. Simple thresholding and gating are often insufficient to fully analyze high-dimensional imaging data; thus, a series of tools have been developed to deal with such datasets. Tools such as t-SNE or UMAP can be used to reduce the dimensionality of the expression data while maintaining the relationships between objects in high-dimensional space (Fig. 2F).135,136 Clustering tools such as FlowSOM, and SPADE can be used to group cells of similar high-dimensional phenotype for further analysis (Fig. 2G).137,138 Once clusters of cell populations of interest are identified, it is useful to determine the phenotypic features of those populations. Statistical descriptors such as MEM produce labels for populations that represent the relative positive or negative enrichment of markers in the population compared with the rest of the sample. 139 Alternatively, expert knowledge can be used to infer cell identity from median expression data.140,141
A major opportunity presented by a highly multiplexed imaging study of NSC and GSC niches, beyond better identification and phenotyping of these cells, is the exploration of the neighborhood of cells that surround individual NSC or GSC cells (Fig. 2I). The multiplex imaging cytometry analysis toolbox histoCAT performs a “neighborhood” analysis by splitting the observed cells into clusters of similar phenotype in high-dimensional space and comparing the observed frequency of interaction between cells of different clusters compared with a computationally randomized distribution of the cells. Cells within a user-defined threshold distance from the index cell are defined as interacting. The fraction of ROIs in which there is a significant interaction or avoidance between cell types is then reported. 134 Such an analysis on a high-dimensional imaging dataset could reveal cell phenotypes that specifically interact with NSCs or GSCs, or alternately immune cell subpopulations that are specifically excluded from the NSC neighborhood. In another approach, an i-niche around each cell in the high-dimensional imaging dataset was defined by the collection of cell types in the ring of first-tier neighbors surrounding each central index cell. These i-niches are then clustered to find types of i-niches that exist in the tissue. The distribution of these i-niches can then be compared across conditions or defined tissue regions. 130 In the NSC/GSC context, such an analysis could identify specific niche patterns around NSCs or GSCs whose presence or absence correlates with other biological features.
Collectively, the advent of high-dimensional imaging, coupled with the ever-expanding amount of information available on the V-SVZ and glioblastoma, offers exciting opportunities for dissecting the complex relationships between the many cell constituents of this niche. Ultimately, a better understanding of how contact with the CSF, vasculature, and stem cells impacts glioma cell survival and resistance may inform the development of treatments that inhibit migration to this region, as well as modulating the effects of the V-SVZ on cancer cells and immune infiltrate.
Acknowledgments
We apologize to our colleagues whose work could not be discussed in depth within space limitations. The authors thank Dr. Justine Sinnaeve for assistance in generating Fig. 1, Dr. Akshitkumar Mistry for assistance in collecting the postmortem tissues depicted in Fig. 1, the three reviewers of the manuscript for their careful recommendations, and the members of the Ihrie and Irish labs at Vanderbilt for thoughtful discussions and figure review.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All authors contributed to the literature review and manuscript preparation.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research in the Ihrie lab is supported by the National Institutes of Health (R01NS096238 and R01NS118580 to R.A.I.), DOD Idea Development Award W81XWH-16-1-0171, and a gift from the Michael David Greene Brain Cancer Fund at the Vanderbilt-Ingram Cancer Center.
Contributor Information
Asa A. Brockman, Department of Cell and Developmental Biology, Vanderbilt University School of Medicine, Nashville, Tennessee
Bret C. Mobley, Departments of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, Tennessee
Rebecca A. Ihrie, Department of Cell and Developmental Biology, Vanderbilt University School of Medicine, Nashville, Tennessee; Departments of Neurological Surgery, Vanderbilt University Medical Center, Nashville, Tennessee.
Literature Cited
- 1. Paredes MF, Sorrells SF, Garcia-Verdugo JM, Alvarez-Buylla A. Brain size and limits to adult neurogenesis. J Comp Neurol. 2016;524:646–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. In Cell Stem Cell. 2008;3:265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. In Cell Stem Cell. 2008;3:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. In Cell Stem Cell. 2008;3:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Regeneration of a germinal layer in the adult mammalian brain. Procnatlacadsciusa. 1999;96:11619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottcher C, van der Poel M, Fernandez-Zapata C, Schlickeiser S, Leman JKH, Hsiao CC, Mizee MR, Adelia Vincenten MCJ, Kunkel D, Huitinga I, Hamann J, Priller J. Single-cell mass cytometry reveals complex myeloid cell composition in active lesions of progressive multiple sclerosis. Acta Neuropathol Commun. 2020;8:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goings GE, Kozlowski DA, Szele FG. Differential activation of microglia in neurogenic versus non-neurogenic regions of the forebrain. Glia. 2006;54:329–42. [DOI] [PubMed] [Google Scholar]
- 10. Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci. 2014;34:2231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–81. [DOI] [PubMed] [Google Scholar]
- 12. Sawamoto K, Hirota Y, Alfaro-Cervello C, Soriano-Navarro M, He X, Hayakawa-Yano Y, Yamada M, Hikishima K, Tabata H, Iwanami A, Nakajima K, Toyama Y, Itoh T, Alvarez-Buylla A, Garcia-Verdugo JM, Okano H. Cellular composition and organization of the subventricular zone and rostral migratory stream in the adult and neonatal common marmoset brain. J Comp Neurol. 2011;519:690–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paul A, Chaker Z, Doetsch F. Hypothalamic regulation of regionally distinct adult neural stem cells and neurogenesis. Science. 2017;356:1383–6. [DOI] [PubMed] [Google Scholar]
- 15. Tong CK, Chen J, Cebrian-Silla A, Mirzadeh Z, Obernier K, Guinto CD, Tecott LH, Garcia-Verdugo JM, Kriegstein A, Alvarez-Buylla A. Axonal control of the adult neural stem cell niche. Cell Stem Cell. 2014;14:500–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–8. [DOI] [PubMed] [Google Scholar]
- 17. Luskin MB. Neuroblasts of the postnatal mammalian forebrain: their phenotype and fate. J Neurobiol. 1998;36:221–33. [PubMed] [Google Scholar]
- 18. Luskin MB, Zigova T, Soteres BJ, Stewart RR. Neuronal progenitor cells derived from the anterior subventricular zone of the neonatal rat forebrain continue to proliferate in vitro and express a neuronal phenotype. Mol Cell Neurosci. 1997;8:351–66. [DOI] [PubMed] [Google Scholar]
- 19. Pencea V, Bingaman KD, Freedman LJ, Luskin MB. Neurogenesis in the subventricular zone and rostral migratory stream of the neonatal and adult primate forebrain. Exp Neurol. 2001;172:1–16. [DOI] [PubMed] [Google Scholar]
- 20. Zigova T, Pencea V, Betarbet R, Wiegand SJ, Alexander C, Bakay RA, Luskin MB. Neuronal progenitor cells of the neonatal subventricular zone differentiate and disperse following transplantation into the adult rat striatum. Cell Transplant. 1998;7:137–56. [DOI] [PubMed] [Google Scholar]
- 21. Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Radecki DZ, Messling HM, Haggerty-Skeans JR, Bhamidipati SK, Clawson ED, Overman CA, Thatcher MM, Salzer JL, Samanta J. Relative levels of Gli1 and Gli2 determine the response of ventral neural stem cells to demyelination. Stem Cell Reports. 2020;15:1047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samanta J, Grund EM, Silva HM, Lafaille JJ, Fishell G, Salzer JL. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature. 2015;526:448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seri B, Herrera DG, Gritti A, Ferron S, Collado L, Vescovi A, Garcia-Verdugo JM, Alvarez-Buylla A. Composition and organization of the SCZ: a large germinal layer containing neural stem cells in the adult mammalian brain. Cereb Cortex. 2006;16(suppl 1):i103–11. [DOI] [PubMed] [Google Scholar]
- 25. Tong CK, Fuentealba LC, Shah JK, Lindquist RA, Ihrie RA, Guinto CD, Rodas-Rodriguez JL, Alvarez-Buylla A. A dorsal SHH-dependent domain in the V-SVZ produces large numbers of oligodendroglial lineage cells in the postnatal brain. Stem Cell Reports. 2015;5:461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Obernier K, Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146(4):dev156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Curtis MA, Kam M, Nannmark U, Anderson MF, Axell MZ, Wikkelso C, Holtas S, van Roon-Mom WM, Bjork-Eriksson T, Nordborg C, Frisen J, Dragunow M, Faull RL, Eriksson PS. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315:1243–49. [DOI] [PubMed] [Google Scholar]
- 28. Sanai N, Berger MS, Garcia-Verdugo JM, Alvarez-Buylla A. Comment on “human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension.” Science. 2007;318:393. [DOI] [PubMed] [Google Scholar]
- 29. Sanai N, Tramontin AD, Quinones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Manuel-Garcia Verdugo J, Berger MS, Alvarez-Buylla A. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–44. [DOI] [PubMed] [Google Scholar]
- 30. Sanai N, Nguyen T, Ihrie RA, Mirzadeh Z, Tsai HH, Wong M, Gupta N, Berger MS, Huang E, Garcia-Verdugo JM, Rowitch DH, Alvarez-Buylla A. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 2011;478:382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Obernier K, Cebrian-Silla A, Thomson M, Parraguez JI, Anderson R, Guinto C, Rodas Rodriguez J, Garcia-Verdugo JM, Alvarez-Buylla A. Adult neurogenesis is sustained by symmetric self-renewal and differentiation. Cell Stem Cell. 2018;22:221–34.e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ponti G, Obernier K, Guinto C, Jose L, Bonfanti L, Alvarez-Buylla A. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc Natl Acad Sci U S A. 2013;110:E1045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paredes MF, James D, Gil-Perotin S, Kim H, Cotter JA, Ng C, Sandoval K, Rowitch DH, Xu D, McQuillen PS, Garcia-Verdugo JM, Huang EJ, Alvarez-Buylla A. Extensive migration of young neurons into the infant human frontal lobe. Science. 2016;354:aaf7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altman J. Autoradiographic investigation of cell proliferation in the brains of rats and cats. Anat Rec. 1963;145:573–91. [DOI] [PubMed] [Google Scholar]
- 35. Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–36. [DOI] [PubMed] [Google Scholar]
- 36. Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisen J. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156:1072–83. [DOI] [PubMed] [Google Scholar]
- 37. Breunig JJ, Arellano JI, Macklis JD, Rakic P. Everything that glitters isn’t gold: a critical review of postnatal neural precursor analyses. Cell Stem Cell. 2007;1:612–27. [DOI] [PubMed] [Google Scholar]
- 38. Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–16. [DOI] [PubMed] [Google Scholar]
- 39. Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–17. [DOI] [PubMed] [Google Scholar]
- 40. Bergmann O, Liebl J, Bernard S, Alkass K, Yeung MS, Steier P, Kutschera W, Johnson L, Landen M, Druid H, Spalding KL, Frisen J. The age of olfactory bulb neurons in humans. Neuron. 2012;74:634–9. [DOI] [PubMed] [Google Scholar]
- 41. Yeung MS, Zdunek S, Bergmann O, Bernard S, Salehpour M, Alkass K, Perl S, Tisdale J, Possnert G, Brundin L, Druid H, Frisen J. Dynamics of oligodendrocyte generation and myelination in the human brain. Cell. 2014;159:766–74. [DOI] [PubMed] [Google Scholar]
- 42. Carpenter MK, Cui X, Hu ZY, Jackson J, Sherman S, Seiger A, Wahlberg LU. In vitro expansion of a multipotent population of human neural progenitor cells. Exp Neurol. 1999;158:265–78. [DOI] [PubMed] [Google Scholar]
- 43. Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–10. [DOI] [PubMed] [Google Scholar]
- 44. Englund U, Fricker-Gates RA, Lundberg C, Bjorklund A, Wictorin K. Transplantation of human neural progenitor cells into the neonatal rat brain: extensive migration and differentiation with long-distance axonal projections. Exp Neurol. 2002;173:1–21. [DOI] [PubMed] [Google Scholar]
- 45. Flax JD, Aurora S, Yang C, Simonin C, Wills AM, Billinghurst LL, Jendoubi M, Sidman RL, Wolfe JH, Kim SU, Snyder EY. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat Biotechnol. 1998;16:1033–9. [DOI] [PubMed] [Google Scholar]
- 46. Fricker RA, Carpenter MK, Winkler C, Greco C, Gates MA, Bjorklund A. Site-specific migration and neuronal differentiation of human neural progenitor cells after transplantation in the adult rat brain. J Neurosci. 1999;19:5990–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A. 2009;106:6387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell. 2016;19:643–52. [DOI] [PubMed] [Google Scholar]
- 49. Borrett MJ, Innes BT, Jeong D, Tahmasian N, Storer MA, Bader GD, Kaplan DR, Miller FD. Single-cell profiling shows murine forebrain neural stem cells reacquire a developmental state when activated for adult neurogenesis. Cell Rep. 2020;32:108022. [DOI] [PubMed] [Google Scholar]
- 50. Mizrak D, Levitin HM, Delgado AC, Crotet V, Yuan J, Chaker Z, Silva-Vargas V, Sims PA, Doetsch F. Single-cell analysis of regional differences in adult V-SVZ neural stem cell lineages. Cell Rep. 2019;26:394–406.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delgado RN, Mansky B, Ahanger SH, Lu C, Andersen RE, Dou Y, Alvarez-Buylla A, Lim DA. Maintenance of neural stem cell positional identity by mixed-lineage leukemia 1. Science. 2020;368:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kohwi M, Osumi N, Rubenstein JL, Alvarez-Buylla A. Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J Neurosci. 2005;25:6997–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kohwi M, Petryniak MA, Long JE, Ekker M, Obata K, Yanagawa Y, Rubenstein JL, Alvarez-Buylla A. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J Neurosci. 2007;27:6878–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Merkle FT, Fuentealba LC, Sanders TA, Magno L, Kessaris N, Alvarez-Buylla A. Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat Neurosci. 2014;17:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rushing G, Ihrie RA. Neural stem cell heterogeneity through time and space in the ventricular-subventricular zone. Front Biol (Beijing). 2016;11:261–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Young KM, Fogarty M, Kessaris N, Richardson WD. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J Neurosci. 2007;27:8286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, Morrison SJ. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife. 2014;3:e02669s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Frederiksen K, McKay RD. Proliferation and differentiation of rat neuroepithelial precursor cells in vivo. J Neurosci. 1988;8:1144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hockfield S, McKay RD. Identification of major cell classes in the developing mammalian nervous system. J Neurosci. 1985;5:3310–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O’Leary LA, Mechawar N. Implication of cerebral astrocytes in major depression: a review of fine neuroanatomical evidence in humans. Glia. Epub 2021 March. doi: 10.1002/glia.23994. [DOI] [PubMed] [Google Scholar]
- 62. Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. [DOI] [PubMed] [Google Scholar]
- 63. Middeldorp J, Boer K, Sluijs JA, De Filippis L, Encha-Razavi F, Vescovi AL, Swaab DF, Aronica E, Hol EM. GFAPdelta in radial glia and subventricular zone progenitors in the developing human cortex. Development. 2010;137:313–21. [DOI] [PubMed] [Google Scholar]
- 64. Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–71. [DOI] [PubMed] [Google Scholar]
- 65. Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, Alvarez-Buylla A. Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron. 2011;71:250–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27:447–52. [DOI] [PubMed] [Google Scholar]
- 67. Wang C, Liu F, Liu YY, Zhao CH, You Y, Wang L, Zhang J, Wei B, Ma T, Zhang Q, Zhang Y, Chen R, Song H, Yang Z. Identification and characterization of neuroblasts in the subventricular zone and rostral migratory stream of the adult human brain. Cell Res. 2011;21:1534–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Knoth R, Singec I, Ditter M, Pantazis G, Capetian P, Meyer RP, Horvat V, Volk B, Kempermann G. Murine features of neurogenesis in the human hippocampus across the lifespan from 0 to 100 years. PLoS ONE. 2010;5:e8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Devneurosci. 2000;22:139–53. [DOI] [PubMed] [Google Scholar]
- 70. Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesigi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–42. [DOI] [PubMed] [Google Scholar]
- 71. Sakakibara S, Nakamura Y, Satoh H, Okano H. RNA-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. [DOI] [PubMed] [Google Scholar]
- 73. Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S, Nicolis SK. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–382. [DOI] [PubMed] [Google Scholar]
- 74. Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–65. [DOI] [PubMed] [Google Scholar]
- 75. Baser A, Skabkin M, Kleber S, Dang Y, Gulculer Balta GS, Kalamakis G, Gopferich M, Ibanez DC, Schefzik R, Lopez AS, Bobadilla EL, Schultz C, Fischer B, Martin-Villalba A. Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature. 2019;566:100–4. [DOI] [PubMed] [Google Scholar]
- 76. Foglio B, Rossini L, Garbelli R, Regondi MC, Mercurio S, Bertacchi M, Avagliano L, Bulfamante G, Coras R, Maiorana A, Nicolis S, Studer M, Frassoni C. Dynamic expression of NR2F1 and SOX2 in developing and adult human cortex: comparison with cortical malformations. Brain Struct Funct. 2021;226:1303–22. [DOI] [PubMed] [Google Scholar]
- 77. Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–75. [DOI] [PubMed] [Google Scholar]
- 78. Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–12. [PubMed] [Google Scholar]
- 79. Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci U S A. 1997;94:12425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Corti S, Nizzardo M, Nardini M, Donadoni C, Locatelli F, Papadimitriou D, Salani S, Del Bo R, Ghezzi S, Strazzer S, Bresolin N, Comi GP. Isolation and characterization of murine neural stem/progenitor cells based on Prominin-1 expression. Exp Neurol. 2007;205:547–62. [DOI] [PubMed] [Google Scholar]
- 81. Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8:723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97:14720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cipriani S, Ferrer I, Aronica E, Kovacs GG, Verney C, Nardelli J, Khung S, Delezoide AL, Milenkovic I, Rasika S, Manivet P, Benifla JL, Deriot N, Gressens P, Adle-Biassette H. Hippocampal radial glial subtypes and their neurogenic potential in human fetuses and healthy and Alzheimer’s disease adults. Cereb Cortex. 2018;28:2458–78. [DOI] [PubMed] [Google Scholar]
- 84. Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, Kelley KW, James D, Mayer S, Chang J, Auguste KI, Chang EF, Gutierrez AJ, Kriegstein AR, Mathern GW, Oldham MC, Huang EJ, Garcia-Verdugo JM, Yang Z, Alvarez-Buylla A. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature. 2018;555:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, Rosoklija GB, Stankov A, Arango V, Dwork AJ, Hen R, Mann JJ. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22:589–99.e585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Moreno-Jimenez EP, Flor-Garcia M, Terreros-Roncal J, Rabano A, Cafini F, Pallas-Bazarra N, Avila J, Llorens-Martin M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med. 2019;25:554–60. [DOI] [PubMed] [Google Scholar]
- 87. Moreno-Jimenez EP, Terreros-Roncal J, Flor-Garcia M, Rabano A, Llorens-Martin M. Evidences for adult hippocampal neurogenesis in humans. J Neurosci. 2021;41:2541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sorrells SF, Paredes MF, Zhang Z, Kang G, Pastor-Alonso O, Biagiotti S, Page CE, Sandoval K, Knox A, Connolly A, Huang EJ, Garcia-Verdugo JM, Oldham MC, Yang Z, Alvarez-Buylla A. Positive controls in adults and children support that very few, if any, new neurons are born in the adult human hippocampus. J Neurosci. 2021;41:2554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pfenninger CV, Roschupkina T, Hertwig F, Kottwitz D, Englund E, Bengzon J, Jacobsen SE, Nuber UA. CD133 is not present on neurogenic astrocytes in the adult subventricular zone, but on embryonic neural stem cells, ependymal cells, and glioblastoma cells. Cancer Res. 2007;67:5727–36. [DOI] [PubMed] [Google Scholar]
- 90. Kempermann G, Gage FH, Aigner L, Song H, Curtis MA, Thuret S, Kuhn HG, Jessberger S, Frankland PW, Cameron HA, Gould E, Hen R, Abrous DN, Toni N, Schinder AF, Zhao X, Lucassen PJ, Frisen J. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell. 2018;23:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um JY, Kim WK, Lee JK, Park J, Kim EH, Lee JH, Lee JH, Chung WS, Ju YS, Park SH, Chang JH, Kang SG, Lee JH. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–7. [DOI] [PubMed] [Google Scholar]
- 92. Marumoto T, Tashiro A, Friedmann-Morvinski D, Scadeng M, Soda Y, Gage FH, Verma IM. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 2017;19:v1–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lauko A, Lo A, Ahluwalia MS, Lathia JD. Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin Cancer Biol. 2021;S1044–579X(21):00049–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zong H, Parada LF, Baker SJ. Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb Perspect Biol. 2015;7:a020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. [DOI] [PubMed] [Google Scholar]
- 98. Kim Y, Kim E, Wu Q, Guryanova O, Hitomi M, Lathia JD, Serwanski D, Sloan AE, Weil RJ, Lee J, Nishiyama A, Bao S, Hjelmeland AB, Rich JN. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26:1247–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lan X, Jorg DJ, Cavalli FMG, Richards LM, Nguyen LV, Vanner RJ, Guilhamon P, Lee L, Kushida MM, Pellacani D, Park NI, Coutinho FJ, Whetstone H, Selvadurai HJ, Che C, Luu B, Carles A, Moksa M, Rastegar N, Head R, Dolma S, Prinos P, Cusimano MD, Das S, Bernstein M, Arrowsmith CH, Mungall AJ, Moore RA, Ma Y, Gallo M, Lupien M, Pugh TJ, Taylor MD, Hirst M, Eaves CJ, Simons BD, Dirks PB. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Galdieri L, Jash A, Malkova O, Mao DD, DeSouza P, Chu YE, Salter A, Campian JL, Naegle KM, Brennan CW, Wakimoto H, Oh ST, Kim AH, Chheda MG. Defining phenotypic and functional heterogeneity of glioblastoma stem cells by mass cytometry. JCI Insight. 2021; e128456:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Leelatian N, Sinnaeve J, Mistry AM, Barone SM, Brockman AA, Diggins KE, Greenplate AR, Weaver KD, Thompson RC, Chambless LB, Mobley BC, Ihrie RA, Irish JM. Unsupervised machine learning reveals risk stratifying glioblastoma tumor cells. eLife. 2020; e56879:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bhaduri A, Di Lullo E, Jung D, Muller S, Crouch EE, Espinosa CS, Ozawa T, Alvarado B, Spatazza J, Cadwell CR, Wilkins G, Velmeshev D, Liu SJ, Malatesta M, Andrews MG, Mostajo-Radji MA, Huang EJ, Nowakowski TJ, Lim DA, Diaz A, Raleigh DR, Kriegstein AR. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26:48–63.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sinnaeve J, Mobley BC, Ihrie RA. Space invaders: brain tumor exploitation of the stem cell niche. Am J Pathol. 2018;188:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Carrano A, Zarco N, Phillipps J, Lara-Velazquez M, Suarez-Meade P, Norton ES, Chaichana KL, Quinones-Hinojosa A, Asmann YW, Guerrero-Cazares H. Human cerebrospinal fluid modulates pathways promoting glioblastoma malignancy. Front Oncol. 2021;11:624145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lara-Velazquez M, Zarco N, Carrano A, Phillipps J, Norton ES, Schiapparelli P, Alkharboosh R, Rincon-Torroella J, Jeanneret S, Corona T, Segovia J, Jentoft ME, Chaichana KL, Asmann YW, Quinones-Hinojosa A, Guerrero-Cazares H. Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of Glioblastoma. Neuro Oncol. 2021;23:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu Q, Sanai N, Jin WN, La Cava A, Van Kaer L, Shi FD. Neural stem cells sustain natural killer cells that dictate recovery from brain inflammation. Nat Neurosci. 2016;19:243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mistry AM, Dewan MC, White-Dzuro GA, Brinson PR, Weaver KD, Thompson RC, Ihrie RA, Chambless LB. Decreased survival in glioblastomas is specific to contact with the ventricular-subventricular zone, not subgranular zone or corpus callosum. J Neurooncol. 2017;132:341–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mistry AM, Hale AT, Chambless LB, Weaver KD, Thompson RC, Ihrie RA. Influence of glioblastoma contact with the lateral ventricle on survival: a meta-analysis. J Neurooncol. 2017;131:125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Mistry AM, Wooten DJ, Davis LT, Mobley BC, Quaranta V, Ihrie RA. Ventricular-subventricular zone contact by glioblastoma is not associated with molecular signatures in bulk tumor data. Sci Rep. 2019;9:1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Goffart N, Lombard A, Lallemand F, Kroonen J, Nassen J, Di Valentin E, Berendsen S, Dedobbeleer M, Willems E, Robe P, Bours V, Martin D, Martinive P, Maquet P, Rogister B. CXCL12 mediates glioblastoma resistance to radiotherapy in the subventricular zone. Neuro Oncol. 2017;19:66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Qin EY, Cooper DD, Abbott KL, Lennon J, Nagaraja S, Mackay A, Jones C, Vogel H, Jackson PK, Monje M. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell. 2017;170:845–59.e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Caretti V, Bugiani M, Freret M, Schellen P, Jansen M, van Vuurden D, Kaspers G, Fisher PG, Hulleman E, Wesseling P, Vogel H, Monje M. Subventricular spread of diffuse intrinsic pontine glioma. Acta Neuropathol. 2014;128:605–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tamura M, Ohye C, Nakazato Y. Pathological anatomy of autopsy brain with malignant glioma. Neurol Med Chir (Tokyo). 1993;33:77–80. [DOI] [PubMed] [Google Scholar]
- 114. Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Matsui TK, Tsuru Y, Hasegawa K, Kuwako KI. Vascularization of human brain organoids. Stem Cells. Epub 2021 Mar. doi: 10.1002/stem.3368. [DOI] [PubMed] [Google Scholar]
- 116. Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, Thokala R, Sheikh S, Saxena D, Prokop S, Liu DA, Qian X, Petrov D, Lucas T, Chen HI, Dorsey JF, Christian KM, Binder ZA, Nasrallah M, Brem S, O’Rourke DM, Ming GL, Song H. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, Schenkein E, Singhania R, Cirigliano S, Magdeldin T, Lin Y, Nanjangud G, Chadalavada K, Pisapia D, Liston C, Fine HA. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26:3203–11.e3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Burgess DJ. Spatial transcriptomics coming of age. Nat Rev Genet. 2019;20:317. [DOI] [PubMed] [Google Scholar]
- 119. Rayaprolu S, Higginbotham L, Bagchi P, Watson CM, Zhang T, Levey AI, Rangaraju S, Seyfried NT. Systems-based proteomics to resolve the biology of Alzheimer’s disease beyond amyloid and tau. Neuropsychopharmacology. 2021;46:98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang LB, Karpova A, Gritsenko MA, Kyle JE, Cao S, Li Y, Rykunov D, Colaprico A, Rothstein JH, Hong R, Stathias V, Cornwell M, Petralia F, Wu Y, Reva B, Krug K, Pugliese P, Kawaler E, Olsen LK, Liang WW, Song X, Dou Y, Wendl MC, Caravan W, Liu W, Cui Zhou D, Ji J, Tsai CF, Petyuk VA, Moon J, Ma W, Chu RK, Weitz KK, Moore RJ, Monroe ME, Zhao R, Yang X, Yoo S, Krek A, Demopoulos A, Zhu H, Wyczalkowski MA, McMichael JF, Henderson BL, Lindgren CM, Boekweg H, Lu S, Baral J, Yao L, Stratton KG, Bramer LM, Zink E, Couvillion SP, Bloodsworth KJ, Satpathy S, Sieh W, Boca SM, Schurer S, Chen F, Wiznerowicz M, Ketchum KA, Boja ES, Kinsinger CR, Robles AI, Hiltke T, Thiagarajan M, Nesvizhskii AI, Zhang B, Mani DR, Ceccarelli M, Chen XS, Cottingham SL, Li QK, Kim AH, Fenyo D, Ruggles KV, Rodriguez H, Mesri M, Payne SH, Resnick AC, Wang P, Smith RD, Iavarone A, Chheda MG, Barnholtz-Sloan JS, Rodland KD, Liu T, Ding L, Clinical Proteomic Tumor Analysis Consortium. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–28.e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget MA, Bernatchez C, Haymaker C, Wistuba II, Rodriguez-Canales J. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep. 2017;7:13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem. 2009;57:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lin JR, Fallahi-Sichani M, Sorger PK. Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 2015;6:8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. [DOI] [PubMed] [Google Scholar]
- 125. Bernstock JD, Vicario N, Rong L, Valdes PA, Choi BD, Chen JA, DiToro D, Osorio DS, Kachurak K, Gessler F, Johnston JM, Jr, Atkinson TP, Whitley RJ, Bag AK, Gillespie GY, Markert JM, Maric D, Friedman GK. A novel in situ multiplex immunofluorescence panel for the assessment of tumor immunopathology and response to virotherapy in pediatric glioblastoma reveals a role for checkpoint protein inhibition. Oncoimmunology. 2019;8:e1678921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Geben LC, Mobley BC, Brockman AA, Pastakia D, Naftel R, Ihrie RA, Esbenshade AJ. Sustained response to erlotinib and rapamycin in a patient with pediatric anaplastic oligodendroglioma. Pediatr Blood Cancer. 2020;68:e28750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Park C, Ponath G, Levine-Ritterman M, Bull E, Swanson EC, De Jager PL, Segal BM, Pitt D. The landscape of myeloid and astrocyte phenotypes in acute multiple sclerosis lesions. Acta Neuropathol Commun. 2019;7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ramaglia V, Sheikh-Mohamed S, Legg K, Park C, Rojas OL, Zandee S, Fu F, Ornatsky O, Swanson EC, Pitt D, Prat A, McKee TD, Gommerman JL. Multiplexed imaging of immune cells in staged multiple sclerosis lesions by mass cytometry. eLife. 2019;8:e48051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, Nolan GP. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–81.e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kennedy-Darling J, Bhate SS, Hickey JW, Black S, Barlow GL, Vazquez G, Venkataraaman VG, Samusik N, Goltsev Y, Schurch CM, Nolan GP. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur J Immunol. 2021;51:1262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, Sabatini DM. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106:1826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, Giesen C, Catena R, Varga Z, Bodenmiller B. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14:873–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 136. Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–45. [DOI] [PubMed] [Google Scholar]
- 139. Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nat Methods. 2017;14:275–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Czech E, Aksoy BA, Aksoy P, Hammerbacher J. Cytokit: a single-cell analysis toolkit for high dimensional fluorescent microscopy imaging. BMC Bioinformatics. 2019;20:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Scheeder C, Heigwer F, Boutros M. Machine learning and image-based profiling in drug discovery. Curr Opin Syst Biol. 2018;10:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]