Abstract
Proteolytic activity is perturbed in tumors and their microenvironment, and proteases also affect cancer stem cells (CSCs). CSCs are the therapy-resistant subpopulation of cancer cells with tumor-initiating capacity that reside in specialized tumor microenvironment niches. In this review, we briefly summarize the significance of proteases in regulating CSC activities with a focus on brain tumor glioblastoma. A plethora of proteases and their inhibitors participate in CSC invasiveness and affect intercellular interactions, enhancing CSC immune, irradiation, and chemotherapy resilience. Apart from their role in degrading the extracellular matrix enabling CSC migration in and out of their niches, we review the ability of proteases to modulate CSC properties, which prevents their elimination. When designing protease-oriented therapies, the multifaceted roles of proteases should be thoroughly investigated.
Keywords: angiogenesis, cancer, cancer stem cells, epithelial-to-mesenchymal transition, glioblastoma, invasion, proteases, stemness, tissue niches, tumor heterogeneity, tumor immune infiltrate, tumor microenvironment
Introduction: Proteases and the Proteolytic System
To date, 884 known and putative protease genes have been annotated in the human genome (https://www.ebi.ac.uk/merops/). 1 According to the chemical mechanisms of catalysis, proteases are grouped into five major classes: serine proteases, cysteine proteases, threonine proteases, aspartic proteases, and metalloproteases. In addition, there are proteases of mixed and unknown catalytic types. Based on similarities in their primary amino acid sequences, proteases are grouped into families that are further combined into structural clans. 1
Metalloproteases are characterized by the presence of a catalytic metal ion in the active site of the enzyme and represent the largest protease group in humans. The family includes matrix metalloproteases (MMPs),2 –4 a disintegrin and metalloproteases (ADAMs), 5 and ADAMs with a thrombospondin motif (ADAMTSs).6 –8 In humans, 24 MMPs have been characterized, some of which are secreted, whereas others are anchored in the cell membrane. MMPs are best known for their role in remodeling the extracellular matrix (ECM).3,4 They degrade structural components of the ECM, including different types of native collagens, fibronectin, elastin, laminin, and proteoglycans, and proteolytically activate pro-MMPs. Their activity also results in the release and activation of matrix-bound growth factors and/or their precursors, for example, transforming growth factor-β (TGF-β) and vascular endothelial growth factor (VEGF). 9 Based on their protein structure and in relation to their substrate specificity, MMPs are divided into six groups: collagenases (MMP1, MMP8, MMP13), gelatinases (MMP2, MMP9), stromelysins (MMP3, MMP10, MMP11), matrylysins (MMP7, MMP26), membrane-type MMPs (MMP14, MMP15, MMP16, MMP17, MMP24), and other non-classified MMPs.10,11
Half of the 22 known ADAMs possess enzymatic activity. These transmembrane proteins shed the ectodomains of other transmembrane proteins, including growth factor and cytokine precursors, their receptors, and adhesion proteins.5,12 On the contrary, all 19 ADAMTSs are secreted enzymes involved in proteolytic processing of ECM proteoglycans, procollagens, and other substrates.6 –8
Another important group of proteases are cathepsins, that is, lysosomal proteases primarily responsible for intracellular protein catabolism.13,14 However, under certain conditions, cathepsins are released from lysosomes or even out of cells, where they perform other functions.14,15 According to the catalytic amino acid residue, cathepsins are divided into cysteine proteases (cathepsins B, C, F, H, K, L, O, S, V, W, and X), aspartic proteases (cathepsins D and E), and serine proteases (cathepsins A and G). 14
Proteases, their substrates, and endogenous protein inhibitors form a complex network called the degradome.16 –18 Proteolytic processing is tightly regulated and both directly and indirectly involved in virtually all biological processes, including cell proliferation, differentiation, death, and migration, and, importantly, in tissue remodeling.16,17,19,20
Among the numerous functions of proteases, there is evidence on their role in specialized anatomically delineated tissue microenvironments called niches that host and sustain certain cell types, such as stem cells (SCs).21 –23 Different cell types within SC niches provide a cocktail of cell-bound and secreted factors that, in concert with the niche ECM, shape the fate of resident SCs, either maintaining their quiescent state or inducing their proliferation and differentiation. 24 Among the best characterized SC niches is the bone marrow hematopoietic stem cell (HSC) niche,24,25 where proteolysis by MMPs23,26,27 and cathepsins27,28 has been demonstrated. Proteases regulate SC functions by reshaping the extracellular scaffold of the niche, degrading cell surface adhesion molecules, and/or regulating the bioavailability and activity of cytokines and growth factors such as stromal cell–derived factor 1α (SDF-1α), SC factor, TGF-β, and VEGF (reviewed in Tay et al., 24 Saw et al., 26 and Maurer et al. 27 ). SCs in the niches are mostly in a quiescent/dormant cellular state. 29 However, upon various endogenous cues, or cues from the microenvironment, several types of proteases are induced to release SCs from their niches. Secreted by SCs or from various niche-resident cells, such as neutrophil granulocytes, osteoblasts, and osteoclasts in the HSC niche, MMP8 30 and MMP931,32 have been reported to mobilize HSCs via cleavage of the cytokine, SDF-1α, thus preventing its binding to C-X-C chemokine receptor type 4 (CXCR4). The SDF-1α–CXCR4 axis and mobilization of HSCs are also affected by several cathepsins, including cathepsin X28,33 and cathepsin K. 34
Role of Proteases in Cancer
To maintain the fine tuning of complex proteolytic networks, proteolytic activity is tightly regulated at the levels of protease expression, activation, posttranslational modifications, and intercellular and intracellular trafficking. Furthermore, protease regulation is enhanced by the presence of selective endogenous protease inhibitors.19,35 –37 Dysregulated proteolysis underlies numerous pathologies, including cancer,38,39 where proteases are most commonly upregulated and act as protumorigenic factors.40 –42 However, some proteases are also involved in tumor suppression and are downregulated in cancer.43 –45 By taking part in various proteolytic cascades and networks, proteases are involved in several processes of tumor development and progression,39,46 –49 as summarized in Table 1. The primary function of proteases derived from either cancer or non-cancerous cells in the tumor microenvironment (TME)38,39,60 is not only ECM degradation, as originally anticipated, but also the modulation of other pericellular events, in particular the activity and bioavailability of growth factors, cytokines, cell surface receptors, and adhesion molecules, as well as the modulation of intracellular signaling pathways.39,49,50 Some proteases or their domains also possess non-enzymatic functions, 50 such as the inactive proform of cathepsin X 74 and several MMPs whose hemopexin domain is involved in protein–protein interactions. 23
Table 1.
Roles of Proteases in Cancer Development and Progression.
| Protease/Protease Group | Processes in Cancer | Mechanism of Action | References |
|---|---|---|---|
| MMPs | Cancer cell migration, invasion, EMT, proliferation, angiogenesis, inflammation, immune responses | ECM remodeling; processing of cytokines, growth factors, and their receptors | 42, 45, 50 –52 |
| ADAMs | Cancer cell adhesion, migration and proliferation, inflammation, immune responses | Sheddase activity releasing extracellular regions of growth factors, cytokines and their receptors, and adhesion and signaling molecules | 53 –57 |
| ADAMTSs | Cancer cell adhesion, migration and proliferation, angiogenesis | Processing of ECM proteoglycans | 7, 55, 58, 59 |
| Cathepsins | Cancer cell growth, migration, invasion, metastasis, apoptosis and therapy resistance, angiogenesis, immune responses | ECM remodeling; release and processing of growth factors and their receptors; functions, independent of the enzymatic activity; activation of intracellular signaling pathways | 36, 41, 47, 60 –66 |
| Kallikreins | Cancer cell growth, migration, invasion and chemoresistance, angiogenesis | ECM remodeling; processing of adhesion molecules, cytokines, growth factors, and cell-surface receptors; signaling via proteinase-activated receptors | 48, 67 –69 |
| Urokinase-type plasminogen activator | Cancer cell proliferation, adhesion, migration and invasion | Initiation of proteolytic cascade leading to ECM degradation, release, and activation of growth factors and cytokines | 70 –73 |
Abbreviations: MMPs, matrix metalloproteases; EMT, epithelial-to-mesenchymal-like transition; ECM, extracellular matrix; ADAMs, a disintegrin and metalloproteases; ADAMTSs, ADAMs with a thrombospondin motif.
Proteases in Cancer SCs and Their Niches
Cancer cells within a tumor are heterogeneous and occur in a variety of hierarchically organized and functionally distinct cell states. The modern concept of cancer stem cells (CSCs), which reside at the top of the tumor hierarchy or, alternatively, at the “bottom” of cancer evolution, was first introduced in leukemia 75 and later specified by the Eaves group. 76 The concept of a pool of CSCs with stemness characteristics has since been confirmed in many other malignancies, 77 for example, in breast, 78 colon,79,80 pancreatic, 81 and brain cancers, 82 including glioblastoma 83 and medulloblastoma. 84 CSCs, also called tumor-initiating cells, are a small subpopulation of cells within tumors that exhibit characteristics reminiscent of normal adult SCs. The key common characteristic of normal SCs and CSCs is asymmetric division (i.e., the ability to self-renew, yet differentiate via generation of progenies to mature, functionally differentiated cell types). In cancer, these mature cells comprise the bulk of the tumor mass, being initiated and maintained by CSCs, enabling fast tumor growth. 85 CSCs, like normal SCs, are mostly present in a so-called dormant state. In the TME, CSCs may undergo epithelial-to-mesenchymal-like transition (EMT) that allows their migration and, through a plethora of proteases, also invasion into the surrounding parenchyma and metastasis. 86
Apart from sharing the characteristic “stemness,” both normal SCs and CSCs reside in distinct microenvironments called niches. The niche milieu contains different cell types, including endothelial cells, immune cells, cancer-associated fibroblasts, and mesenchymal SCs.87 –89 In different cancer types, different types of niches have been identified. 88 They are best characterized in glioblastoma where perivascular, hypoxic, and invasive niches have been described. 90 A link between CSCs and the perivascular niche has also been reported in other brain tumors 91 and in several other cancers,92,93 for example, in colorectal cancer, 94 skin squamous cell carcinoma, 95 melanoma, 96 and head and neck squamous cell carcinoma. 97
Intercellular crosstalk and physical components of a niche either maintain the quiescent state of CSCs or induce their proliferation and differentiation, influence their metastatic potential, and play a role in protecting CSCs against the immune system.88 –90,98 Microenvi-ronmental conditions in the niche may also protect CSCs against therapeutic intervention. In medulloblastoma, CSCs display features imitating those of neural progenitor cells and SCs 84 ; they reside in perivascular niches and exhibit enhanced radioresistance compared with cells of the tumor bulk. 99 Similarly, slow-cycling perivascular tumor cells were shown to be therapy-resistant in a mouse model of glioma. 100 Due to their importance for disease progression and modulation of cancer-related processes such as inflammation and angiogenesis, stromal factors were proposed as molecular markers for diagnosis, prognosis, and treatment selection in prostate cancer. 101
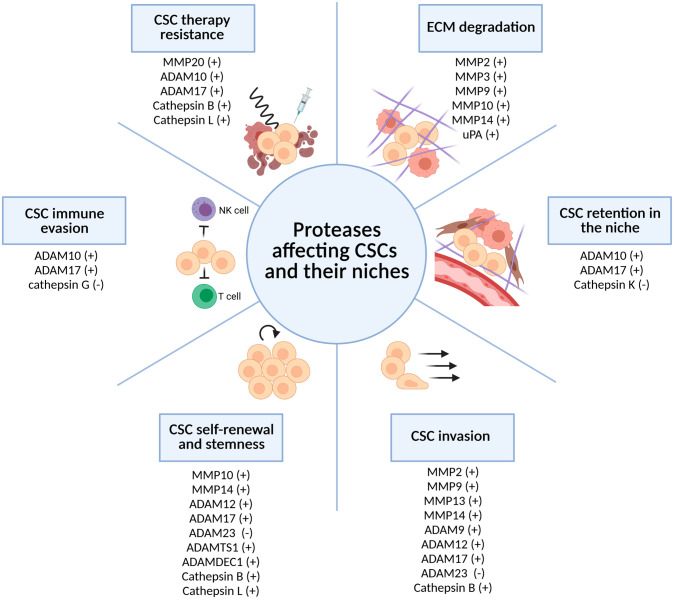
Proteases play an important role within the CSC niches. Expressed and/or released either by cancer (stem) cells or by other cells of the niche, some proteases, such as cathepsins, are activated by the hypoxic and acidified TME.102 –106 In the following sections, we briefly summarize the ways in which proteases affect CSCs and their niches (Fig. 1). Specifically, we focus on primary brain tumor glioblastoma and glioblastoma stem cells (GSCs) that are commonly identified by the expression of a selected set of biomarkers [CD133, CD44, CD15 (SSEA-1), CD70 (CD27 L), Nestin, Olig2, SOX2, ALDH1A3, S100A4, Nanog, and OCT4].107,108 We have localized GSCs in hypoxic periarteriolar niches109,110 that closely resemble bone marrow HSC niches. 111
Figure 1.
Proteases affect CSC functions and their niches. Proteases degrade niche ECM and influence several CSC properties, including their retention in the niche, invasion, self-renewal, stemness, immune evasion, and therapy resistance. Proteases either support (+) or inhibit (−) CSC-related processes. Created with BioRender.com. Abbreviations: CSC, cancer stem cell; ECM, extracellular matrix.
Proteases and the ECM
As mentioned previously, proteases were first investigated as key enzymes remodeling the ECM, which is increasingly recognized as a functional physical and biochemically active component of the niche, also with direct impact on the regulation of CSCs.112,113 Increased expression of several proteases in breast CSCs contributes to degradation of the ECM and invasion into their surroundings.114 –116 On the contrary, ECM components provide physical support and CSC anchorage, assuring their self-renewal.112,113,117 Furthermore, the ECM represents a reservoir of growth factor and cytokine precursors that are activated and released upon proteolysis and affect CSCs.112,113 Among these, TGF-β, 118 VEGF, 119 and insulin-like growth factors 120 can be highly protumorigenic in neoplasia. In addition, partial proteolytic degradation of certain ECM components generates bioactive peptide fragments (so-called matrikines) that are able to regulate cellular activities.121,122 Thus, besides their well-established roles in ECM degradation and direct promotion of cancer cell invasion, proteases also influence CSC self-renewal, EMT, chemoresistance, and immune evasion.116,123,124
Proteases and CSC Migration Out of Niches
Components of the ECM serve as anchoring sites for CSC adherence. CSCs interact with the ECM through several cell surface receptors, including integrins, discoidin domain receptors, and CD44, which maintain CSCs by inducing intracellular stem and proliferative signaling pathways. 112 Cathepsin K is an atypical papain-clan cysteine cathepsin, originally reported as a collagenolytic protease produced by osteoclasts, 125 which appears to be overexpressed in several cancers, particularly glioblastoma. 126 In addition to various types of native collagens, cathepsin K can also hydrolyze various other fibrillar proteins and proteoglycans. 125 In glioblastoma, cathepsin K, along with cathepsins B and X, is colocalized with GSC markers CD133 and nestin, and GSC niche markers osteopontin and SDF-1α and its receptor CXCR4.105,127 It was suggested that cathepsin K could induce GSCs to migrate out of their niches,104,109 and this notion was later supported by Hira and coworkers who demonstrated the ability of cathepsin K to cleave and inactivate SDF-1α in vitro. 128 This mechanism was first observed in the HSC niche where interactions between SDF-1α, secreted by osteoblasts and endothelial cells, and CXCR4, expressed by HSCs, retain HSCs,33,34 highlighting the similarity of the GSC niche and the physiological HSC niche.110,111 In addition, Siney et al. 129 suggested a possible role of ADAM17 and ADAM10 in the retention of GSCs in the tumorigenic niche, as inhibition of both proteases increased CSC migration and differentiation.
Proteases and CSC Invasion
In response to signals from the TME, CSCs can undergo the EMT and acquire migratory capacity to initiate the first steps in metastasis.86,130 CSCs also contribute to tumor spread via enhanced angiogenesis, that is, the formation of alternative vascular structures by vasculogenic mimicry, which is analogous to the mimicry of embryonic vasculogenesis by tumor cells first described by the Hendrix group in melanoma 131 and later in other cancers. 132 Vasculogenic mimicry requires activation of the transmembrane metalloproteases, MMP14 and MMP9, which support the proteolytic cascade in melanoma CSC invasion. 131 In breast cancer, CSCs line vasculogenic mimicry channels and synergize their formation in the perivascular niche. 133
Westhoff et al. 134 showed that invasion of GSCs is enhanced by activation of MMPs involved in fibronectin processing, thus forming routes facilitating GSC migration. The invasiveness of GSCs was shown to be supported by several metalloproteases, such as MMP2, MMP9, 135 and MMP13, 136 as well as ADAM9 and ADAM17.137,138 Invasion of GSCs is also directly supported by stromal cells proteases. Ye and coworkers have shown that tumor-associated microglia and macrophages enhance the invasiveness of GSCs via the release and activation of the TGF-β1 signaling pathway, leading to upregulation of MMP9, 139 whereas microglia-derived MMP14 activates GSC-derived MMP2. 140 The roles of urokinase-type plasminogen activator receptor (uPAR) and cathepsin B in GSCs were investigated by Alapati et al. 141 who showed that simultaneous downregulation of both proteins inhibited irradiation-induced integrin signaling to the cytoskeleton and to the nucleus via protein kinase C. In addition, the complex of mitogen-activated protein kinase kinase 1 and phospho-c-jun N-terminal kinase was translocated from the cytosol to the nucleus, resulting in migratory arrest of GSCs. 142
In breast CSCs, MMP14 is one of the most upregulated proteases, especially under hypoxic conditions that trigger its relocalization to the cell surface.143,144 This enzyme mediates the conversion of stationary CSCs into invasive CSCs—the mechanism thought to drive CSC metastasis. 144 MMP2,145 –147 MMP9,148,149 MMP14, 148 ADAM12, 150 and ADAM17 151 have also been shown to govern CSC invasion in other cancers. However, as emphasized above, proteases can also act as tumor suppressors. For example, in lung adenocarcinoma, ADAM23 suppresses migration and metastasis of CSCs by inhibiting integrin αvβ3 function. 152
Proteases and CSC Self-renewal and Stemness
CSC characteristics are maintained by a plethora of microenvironmental cues, including autocrine and paracrine growth factors (TGF-β, fibroblast growth factor, epidermal growth factor, VEGF), cytokines (interleukin [IL]-1β, IL-6, IL-8, tumor necrosis factor-α), and specific ligands that promote Hedgehog, Wnt, and Notch signaling pathways, leading to activation of NF-κB, JAK-STAT, GLI, β-catenin, LEF/TC, and NIC-CSL families of transcription factors.153 –156 Proteases play a crucial role as part of the signaling cascades in CSCs that sustain their self-renewal and stemness.
Metalloproteases
The metalloprotease ADAM17 assists in processing of Notch1 receptors and solubilization of the Notch ligands, Jagged-1 and Jagged-2. 157 In glioblastoma, ADAM17 has been identified as a mediator of stemness 158 and invasion of U87 GSCs 137 ; similar effects were proposed in colorectal CSCs 157 and in colorectal carcinoma CSC crosstalk with endothelial cells. 94 ADAM17-mediated activation of Notch1 was also detected in liver CSCs where the enzyme was activated by overexpression of inducible nitric oxide synthase. 159 Another protease secreted by GSCs, a disintegrin and metalloproteinase domain–like protein decysin 1, has been linked recently to the maintenance of GSCs, primarily via activation of an autocrine fibroblast growth factor signaling loop. 160 In addition, the claudin-low breast CSC phenotype was promoted by ADAM12, a protease induced during the EMT. 150 ADAMTS1 was recently associated with induction of stemness in uveal melanoma. 161 However, the tumor suppressor protease, ADAM23 (see above), was downregulated in the lung adenocarcinoma CSC subpopulation, which likely contributes to the stemness phenotype. 152
The maintenance of stemness of lung CSCs 162 and epithelial ovarian CSCs 163 was associated with MMP10, which inhibited the non-canonical Wnt signaling ligand, Wnt5a, and activated the canonical Wnt signaling pathway. MMP14 promoted the EMT and induced CSC properties in an oral squamous cell carcinoma cell line. 164 Consistent with these results, knockdown of MMP14 strongly affected CSC properties in breast carcinoma SCs. 143 In conclusion, metalloproteases of different classes affect multiple intracellular signaling pathways that are central to the maintenance of CSCs.
Cysteine and Serine Proteases
Cathepsin B and uPAR play important roles in regulating symmetric GSC division and self-renewal. Their expression correlates with the expression of the Hedgehog signaling components, SOX2 and BMI1, which are regulated by GLI factors, although the mechanisms have not been fully explained. 165 Wang et al. 166 demonstrated downregulation of the GSC marker, CD133, after knockdown of cathepsin L. We and others found that, due to excessive mRNA splicing in cancer cells, N-terminally truncated cathepsin L diffuses into the nucleus167,168 to proteolytically modify histones, thus affecting antiapoptotic gene expression and differentiation in cancer cells. 169
Proteases and SC Immune Evasion
The microenvironment around CSCs contains a broad spectrum of immune cells such as macrophages, myeloid-derived suppressor cells, natural killer (NK) cells, regulatory T-cells, cytotoxic T-lymphocytes (CTLs), and T-helper cells. 170 However, CSCs have evolved several ways to avoid their recognition and destruction by the immune system and to shape the TME into an immunosuppressive landscape. For example, CSCs can transform infiltrating macrophages from the M1 phenotype into the tumor-supportive M2 phenotype or tumor-associated macrophages.170,171 Among other factors, proteases and their endogenous inhibitors are involved in crosstalk between CSCs and immune cells in the TME.
An interesting example is NK cells, cytotoxic lymphocytes characterized by their ability to specifically recognize and eliminate target cells that lack the expression of major histocompatibility complex (MHC) class I molecules normally involved in cell surface presentation of antigenic peptides to CTLs. 172 GSCs have been shown to lack active serine cathepsin G, resulting in impaired cleavage of MHC class I molecules on these cells, thereby escaping recognition by NK cells.173,174 On the contrary, cysteine cathepsins normally contribute to NK cell cytotoxicity by proteolytically activating effector granzymes and perforin that are released by activated NK cells to induce target cell death. 175 In NK cells cocultured with oral squamous carcinoma CSCs, decreased levels of mature cathepsins C and H, and an increased level of their inhibitor, cystatin F, were observed, promoting the anergic state of NK cells.176,177 It has been proposed that CSCs or other stromal cells secrete cystatin F, which when internalized lowers the cytotoxic potential of NK and other cytotoxic cells in the TME. 123 Consistent with this suggestion, uptake of extracellular cystatin F by CTLs resulted in decreased activities of cathepsins C, H, and L, leading to impaired activation of granzymes A and B, and consequently lowered T-cell cytotoxicity. 178 High levels of proteinase inhibitor 9, a potent inhibitor of granzyme B, were detected in breast CSCs, most likely providing another means of CSC immune escape. 179 CSCs have also evolved mechanisms to evade γδ T-cells, a distinct subpopulation of T-cells that differ from the more commonly considered αβ T-cells in terms of their antigen recognition, activation, and effector functions. 180 In CSCs, increased ADAM10 and ADAM17 expression has been associated with increased shedding of the cell surface MHC class I polypeptide-related sequence A, which has been proposed as the main mechanism underlying CSC resistance to γδ T-cell cytotoxicity. 181
Proteases and CSC Therapy Resistance
Aside from residing in a quiescent state, being by itself protective against drugs that target exposed DNA during mitosis, CSCs also exhibit high expression of multidrug resistance proteins and enzymatic DNA damage repair mechanisms. Safeguarded in the shelter of their niche, these cells often represent the main reason for treatment failure and cancer recurrence.182 –184
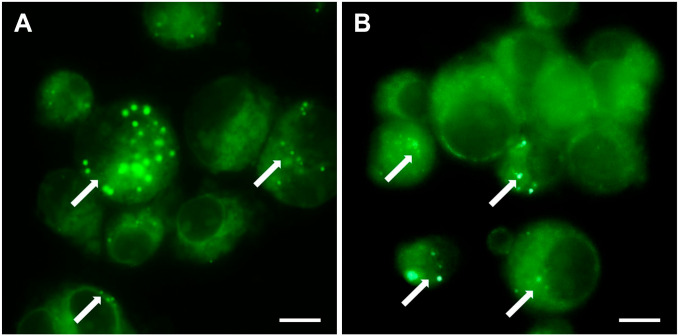
As mentioned previously, active cathepsin B has been detected in GSC niches. 105 Using fluorogenic metabolic mapping, we detected cathepsin B activity in both non-irradiated and irradiated GSC NCH644 cells (Fig. 2). Expression of this enzyme was upregulated upon irradiation of glioblastoma U87 stem-like cells. 187 Apart from the role of cathepsin B and uPAR in maintaining the stemness characteristics of GSCs described above, 165 highly expressed cathepsin B and uPAR also protected these cells from irradiation-induced DNA damage, and this effect was reversed by silencing of the two genes. 187 Similarly, radiation resistance mediated by cathepsin L has been shown. 166 The well-known sheddase, ADAM10, marks CSCs with active Notch signaling that mediates chemoresistance. Targeted inhibition of active ADAM10 inhibited Notch activity and tumor growth in mouse models, particularly regrowth following chemotherapy. This suggests targeted inhibition of active ADAM10 is a potential therapy for ADAM10-dependent tumor development and drug resistance. 188 Colorectal CSC chemoresistance has been linked to another sheddase, ADAM17. 157 In oral carcinoma CSCs, overexpression of MMP20 supported stemness and was proposed to reduce the sensitivity of this cell population to chemotherapeutic agents. 189 Overall, these studies confirm the involvement of proteases in the therapeutic resistance of CSCs and support their therapeutic potential.
Figure 2.
Metabolic mapping of cathepsin B activity. Mapping was done in non-irradiated (A) and irradiated (2 Gy) (B) NCH644 cells, a glioblastoma stem cell line. Enzymatic activity was detected as green fluorescent dots (marked by white arrows). The method is based on the coupling of NSA with 4MbNA, which is cleaved from a protease-specific substrate. Enzymatic release of 4MbNA and its coupling with NSA result in the formation of a fluorescent green product. Background fluorescence occurs due to the nonspecific binding of NSA to protein NH2 groups. The method was adapted from Van Noorden et al. 185 Cells were grown as floating spheres in supplemented NB medium as described by Podergajs et al. 186 Intact spheroids were irradiated with 2 Gy. Four hours after irradiation, the spheroids were washed with Ca2+- and Mg2+-free Hank’s balanced salt solution and with 100 mM sodium phosphate buffer (pH 6.0). Subsequently, spheroids were incubated in 100 mM phosphate buffer (pH 6.0) containing 1 mM dithiothreitol, 1.3 mM EDTA, 2.7 mM l-cysteine, 1 mM NSA, and 1 mg/ml of a specific substrate of cathepsin B (Z-Ala-Arg-Arg-4MbNA). After 30 min of incubation at room temperature, the cell suspension was transferred to a microscopy slide and covered with a coverslip. Images were taken using a FITC filter on an Eclipse Ti inverted microscope using NIS-Elements imaging software AR 4.13.04 and an Andor camera. Scale bar = 10 μm. Abbreviations: NSA, 2-hydroxy-5-nitrobenzaldehyde; 4MbNA, 4-methoxy-b-naphthylamide; NB, Neurobasal.
Proteases as Therapeutic Targets in Cancer
In anticancer therapies, targeting proteases has long been a promising therapeutic tool to counteract invasiveness of cancer cells. Selective targeting of CSCs in their specific niches has recently attracted much attention in the field of cancer therapy,190,191 and the inclusion of proteases as therapeutic targets seems to be an option. However, several attempts at protease inhibition have failed to deliver the desired outcomes. To date, only three proteasome inhibitors have been approved in cancer therapy for the treatment of multiple myeloma (Table 2).15,35,212 Reasons for the treatment failures with other protease inhibitors might, first, be due to inappropriate clinical trial designs, for example, including cancer patients with advanced disease where a hypermutated state was able to overcome single target therapy. Second, the lack of sufficient selectivity and specificity of the protease inhibitors may have led to undesired off-target effects by disturbing the normal physiological functions of these proteases in adjacent tissue or systemically. Finally, due to protease redundancy, the efficiency of selective inhibitors may hinder their effectiveness.15,40,212,213
Table 2.
Protease Inhibitors and Protease-cleavable Conjugates in Cancer Clinical Trials.
| Drug | Mechanism | Cancer Type | Clinical Study Identifier (Study Phase) | Clinical Study Recruitment Status | Results |
|---|---|---|---|---|---|
| Bortezomib | Proteasome inhibition | Multiple myeloma | NCT00048230 (phase 3) | Completed | Bortezomib is superior to high-dose dexamethasone for the treatment of relapsed multiple myeloma, 192 FDA-approved in 2003 |
| NCT00111319 (phase 3) | Completed | Bortezomib plus melphalan-prednisone is superior to melphalan-prednisone alone in patients with newly diagnosed myeloma 193 | |||
| Waldenström’s macroglobulinemia | NCT01788020 (phase 3) | Active, not recruiting | No results posted | ||
| Carfilzomib | Proteasome inhibition | Multiple myeloma | NCT00511238 (phase 2) | Completed | Clinically significant responses in heavily pretreated patients with relapsed and refractory multiple myeloma, 194 FDA-approved in 2012 |
| NCT01080391 (phase 3) | Completed | Addition of carfilzomib to lenalidomide and dexamethasone significantly improves progression-free survival in patients with relapsed multiple myeloma 195 | |||
| NCT01568866 (phase 3) | Completed | Carfilzomib extends overall survival in the relapsed setting over bortezomib 196 | |||
| Ixazomib | Proteasome inhibition | Multiple myeloma | NCT01564537 (phase 3) | Active, not recruiting | Addition of ixazomib to a regimen of lenalidomide and dexamethasone significantly prolongs progression-free survival, 197 FDA-approved in 2015 |
| Doxycycline | MMP inhibition | Relapsed non-Hodgkin lymphomas | NCT02086591 (phase 2) | Terminated (lack of accrual) | No results posted |
| Head and neck squamous cell carcinoma | NCT03076281 (phase 2) | Active, not recruiting | No results posted | ||
| Metastatic breast cancer with bone metastases | NCT01847976 (phase 2) | Completed | No significant effects 198 | ||
| Incyclinide | MMP inhibition | HIV-related Kaposi’s sarcoma | NCT00020683 (phase 2) | Terminated (termination of drug supply) | Active and well tolerated 199 |
| High-grade gliomas | NCT00004147 (phase 1, 2) | Completed | Poor single-agent activity 200 | ||
| Marimastat | MMP inhibition | Small-cell lung cancer | NCT00003011 (phase 3) | Completed | No improvement in survival 201 |
| Non–small-cell lung cancer | NCT00002911 (phase 3) | Completed | No results posted | ||
| Breast cancer | NCT00003010 (phase 3) | Completed | No improvement in survival 202 | ||
| Prinomastat | MMP (MMPs 2, 3, 9, 13, 14) inhibition | Non–small-cell lung cancer | NCT00004199 (phase 3) | Completed | No improvement in survival 203 |
| Prostate cancer | NCT00003343 (phase 3) | Completed | No results posted | ||
| Glioblastoma | NCT00004200 (phase 2) | Completed | No results posted | ||
| Rebimastat | MMP inhibition | Prostate cancer | NCT00040755 (phase 2) | Completed | Well tolerated, but limited efficacy 204 |
| NCT00039104 (phase 2) | Completed | No results posted | |||
| Lung cancer | NCT00006229 (phase 2,3) | Completed | No results posted | ||
| HIV-related Kaposi’s sarcoma | NCT00024024 (phase 1,2) | Completed | Inadequate efficacy 205 | ||
| Andecaliximab | MMP9 inhibition | Recurrent glioblastoma | NCT03631836 (phase 1) | Not yet recruiting | No results posted |
| Gastric or gastroesophageal junction adenocarcinoma | NCT02864381 (phase 2) | Completed | No improvement in survival 206 | ||
| NCT02545504 (phase 3) | Completed | No improvement in survival 207 | |||
| Upamostat | Serine proteases (uPA) inhibition | Metastatic breast cancer | NCT00615940 (phase 2) | Completed | Improved response rate 208 |
| Pancreatic cancer | NCT00499265 (phase 2) | Completed | No improvement in survival 209 | ||
| BXCL701 | Dipeptidyl peptidase 8/9 inhibition | Melanoma | NCT00083239 (phase 2) | Completed | No results posted |
| NCT00083252 (phase 2) | Completed | No results posted | |||
| Prostate cancer | NCT03910660 (phases 1 and 2) | Recruiting | No results posted | ||
| Advanced solid cancers | NCT04171219 (phase 2) | Recruiting | No results posted | ||
| Kidney cancer | NCT00489710 (phase 2) | Withdrawn (terminated for safety reasons) | No results posted | ||
| Non–small-cell lung cancer | NCT00243204 (phase 3) | Terminated (FDA hold May 2007) | No results posted | ||
| Adenocarcinoma of the pancreas | NCT00116389 (phase 2) | terminated (FDA hold May 2007) | No results posted | ||
| Chronic lymphocytic leukemia | NCT00086203 (phase 2) | Completed | No results posted | ||
| Rovalpituzumab tesirine | Protease-cleavable antibody-drug conjugate | Small-cell lung cancer | NCT03061812 (phase 3) | Completed | Inferior overall survival 210 |
| NCT03033511 (phase 3) | Terminated (independent data monitoring committee recommendation) | No improvement in survival 211 |
Abbreviations: FDA, Food and Drug Administration; MMPs, matrix metalloproteases; uPA, urokinase-type plasminogen activator.
Despite the initial clinical disappointments, novel approaches have emerged in which proteases are not used as treatment targets, but instead as therapeutic triggers (Table 2). These technologies enable the delivery of anticancer therapeutics directly to the tumor site where their activation or release is mediated by proteases (e.g., MMPs, cathepsins, or uPA) within the TME.213 –215
Based on current knowledge in the field, targeting or exploiting proteases in the CSC niches may contribute to the elimination of quiescent CSCs. This, arguably, represents the bottleneck in successful tumor eradication. Several examples of such approaches have already shown promising results,190,216,217 providing hope for future cancer treatments. Nevertheless, deliberate target selection may be the key to success. On one hand, proteases bolster CSC proliferation and invasion, whereas, on the other hand, they can also induce CSC detachment and mobilization out of the niche by modulating cytokines.104,109,128 This may lead to their differentiation and higher therapeutic sensitivity. However, CSC mobilization may not necessarily present the optimal therapeutic intervention because it may lead to enhanced aggressiveness of CSCs expressing hybrid phenotypes during the EMT.130,218 Hence, continued efforts are needed to better characterize the heterogeneous and phenotypically distinct CSC pool, and the interplay of these cells with the TME in the niches. When designing CSC niche protease-oriented therapeutic applications, the multifaceted roles of the proteases should be thoroughly examined and modeled in experimental animals in terms of the complex and dynamic TME, before human studies.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: BB and AH conceptualized the research. AH, MN, BM, and BB wrote the original draft of the manuscript. AH, TLT, and BB revised and edited the manuscript. All authors have read and approved the final version of the manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Slovenian Research Agency (grant program P1-0245, grant project J3-2526, post-doctoral project Z3-1870) and by the European Program of Cross-Border Cooperation for Slovenia-Italy Interreg TRANS-GLIOMA.
Contributor Information
Anamarija Habič, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; The Jožef Stefan International Postgraduate School, Ljubljana, Slovenia.
Metka Novak, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Bernarda Majc, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; The Jožef Stefan International Postgraduate School, Ljubljana, Slovenia.
Tamara Lah Turnšek, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; The Jožef Stefan International Postgraduate School, Ljubljana, Slovenia; Faculty of Chemistry and Chemical Technology, University of Ljubljana, Ljubljana, Slovenia.
Barbara Breznik, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Literature Cited
- 1. Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46(D1):D624–32. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yong VW, Power C, Forsyth P, Edwards DR. Metalloproteinases in biology and pathology of the nervous system. Nat Rev Neurosci. 2001;2(7):502–11. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41(2):271–90. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mittal R, Patel AP, Debs LH, Nguyen D, Patel K, Grati M, Mittal J, Yan D, Chapagain P, Liu XZ. Intricate functions of matrix metalloproteinases in physiological and pathological conditions. J Cell Physiol. 2016; 231(12):2599–621. doi: 10.1002/jcp.25430. [DOI] [PubMed] [Google Scholar]
- 5. Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008; 29(5):258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015; 16(1):113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cal S, López-Otín C. ADAMTS proteases and cancer. Matrix Biol. 2015;44–6:77–85. doi: 10.1016/j.matbio.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 8. Apte SS. ADAMTS proteins: concepts, challenges, and prospects. Methods Mol Biol. 2020;2043:1–12. doi: 10.1007/978-1-4939-9698-8_1. [DOI] [PubMed] [Google Scholar]
- 9. Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004; 16(5):558–64. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(Suppl. 1):177–83. doi: 10.3109/14756366.2016.1161620. [DOI] [PubMed] [Google Scholar]
- 11. Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69(3):562–73. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 12. Mullooly M, McGowan PM, Crown J, Duffy MJ. The ADAMs family of proteases as targets for the treatment of cancer. Cancer Biol Ther. 2016;17(8):870–80. doi: 10.1080/15384047.2016.1177684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The ins and outs of cathepsins: physiological function and role in disease management. Cells. 2020;9(7):1679. doi: 10.3390/cells9071679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5(9): 785–99. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 16. López-Otín C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol. 2002;3(7):509–19. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 17. López-Otín C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem. 2008;283(45):30433–7. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quesada V, Ordóñez GR, Sánchez LM, Puente XS, López-Otín C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucleic Acids Res. 2009;37(Database issue):D239–43. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turk B, Turk D, Turk V. Protease signalling: the cutting edge. EMBO J. 2012;31(7):1630–43. doi: 10.1038/emboj.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bond JS. Proteases: history, discovery, and roles in health and disease. J Biol Chem. 2019;294(5):1643–51. doi: 10.1074/jbc.TM118.004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 22. Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kessenbrock K, Wang CY, Werb Z. Matrix metalloproteinases in stem cell regulation and cancer. Matrix Biol. 2015;44–6:184–90. doi: 10.1016/j.matbio.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tay J, Levesque JP, Winkler IG. Cellular players of hematopoietic stem cell mobilization in the bone marrow niche. Int J Hematol. 2017;105(2):129–40. doi: 10.1007/s12185-016-2162-4. [DOI] [PubMed] [Google Scholar]
- 25. Man Y, Yao X, Yang T, Wang Y. Hematopoietic stem cell niche during homeostasis, malignancy, and bone marrow transplantation. Front Cell Dev Biol. 2021;9:621214. doi: 10.3389/fcell.2021.621214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saw S, Weiss A, Khokha R, Waterhouse PD. Metalloproteases: on the watch in the hematopoietic niche. Trends Immunol. 2019;40(11):1053–70. doi: 10.1016/j.it.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 27. Maurer A, Klein G, Staudt ND. Assessment of proteolytic activities in the bone marrow microenvironment. Methods Mol Biol. 2019;2017:149–63. doi: 10.1007/978-1-4939-9574-5_12. [DOI] [PubMed] [Google Scholar]
- 28. Staudt ND, Aicher WK, Kalbacher H, Stevanovic S, Carmona AK, Bogyo M, Klein G. Cathepsin X is secreted by human osteoblasts, digests CXCL-12 and impairs adhesion of hematopoietic stem and progenitor cells to osteoblasts. Haematologica. 2010;95(9):1452–60. doi: 10.3324/haematol.2009.018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo M, Li JF, Yang Q, Zhang K, Wang ZW, Zheng S, Zhou JJ. Stem cell quiescence and its clinical relevance. World J Stem Cells. 2020;12(11):1307–26. doi: 10.4252/wjsc.v12.i11.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steinl C, Essl M, Schreiber TD, Geiger K, Prokop L, Stevanović S, Pötz O, Abele H, Wessels JT, Aicher WK, Klein G. Release of matrix metalloproteinase-8 during physiological trafficking and induced mobilization of human hematopoietic stem cells. Stem Cells Dev. 2013;22(9):1307–18. doi: 10.1089/scd.2012.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jin F, Zhai Q, Qiu L, Meng H, Zou D, Wang Y, Li Q, Yu Z, Han J, Li Q, Zhou B. Degradation of BM SDF-1 by MMP-9: the role in G-CSF-induced hematopoietic stem/progenitor cell mobilization. Bone Marrow Transplant. 2008;42(9):581–8. doi: 10.1038/bmt.2008.222. [DOI] [PubMed] [Google Scholar]
- 32. Theodore LN, Hagedorn EJ, Cortes M, Natsuhara K, Liu SY, Perlin JR, Yang S, Daily ML, Zon LI, North TE. Distinct roles for matrix metalloproteinases 2 and 9 in embryonic hematopoietic stem cell emergence, migration, and niche colonization. Stem Cell Reports. 2017;8(5):1226–41. doi: 10.1016/j.stemcr.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Staudt ND, Maurer A, Spring B, Kalbacher H, Aicher WK, Klein G. Processing of CXCL12 by different osteoblast-secreted cathepsins. Stem Cells Dev. 2012;21(11):1924–35. doi: 10.1089/scd.2011.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, Elson A, Lapidot T. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12(6):657–64. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 35. Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–92. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 36. Lah TT, Obermajer N, Alonso MBD, Kos J. Cysteine cathepsins and cystatins as cancer biomarkers. In: Edwards D, Høyer-Hansen G, Blasi F, Sloane BF. editors. The cancer degradome: proteases and cancer biology. New York: Springer; 2008. p. 587–625. doi: 10.1007/978-0-387-69057-5_29. [DOI] [Google Scholar]
- 37. Breznik B, Mitrović AT, Lah T, Kos J. Cystatins in cancer progression: more than just cathepsin inhibitors. Biochimie. 2019;166:233–50. doi: 10.1016/j.biochi.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 38. Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21(4):228–37. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vizovisek M, Ristanovic D, Menghini S, Christiansen MG, Schuerle S. The tumor proteolytic landscape: a challenging frontier in cancer diagnosis and therapy. Int J Mol Sci. 2021;22(5):2514. doi: 10.3390/ijms22052514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lah TT, Durán Alonso MB, Van Noorden CJ. Antiprotease therapy in cancer: hot or not? Expert Opin Biol Ther. 2006;6(3):257–79. doi: 10.1517/14712598.6.3.257. [DOI] [PubMed] [Google Scholar]
- 41. Rudzińska M, Parodi A, Soond SM, Vinarov AZ, Korolev DO, Morozov AO, Daglioglu C, Tutar Y, Zamyatnin AA., Jr. The role of cysteine cathepsins in cancer progression and drug resistance. Int J Mol Sci. 2019;20(14):3602. doi: 10.3390/ijms20143602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy R, Morad G, Jedinak A, Moses MA. Metalloproteinases and their roles in human cancer. Anat Rec (Hoboken). 2020;303(6):1557–72. doi: 10.1002/ar.24188. [DOI] [PubMed] [Google Scholar]
- 43. López-Otín C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7(10):800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 44. López-Otín C, Palavalli LH, Samuels Y. Protective roles of matrix metalloproteinases: from mouse models to human cancer. Cell Cycle. 2009;8(22):3657–62. doi: 10.4161/cc.8.22.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noël A, Gutiérrez-Fernández A, Sounni NE, Behrendt N, Maquoi E, Lund IK, Cal S, Hoyer-Hansen G, López-Otín C. New and paradoxical roles of matrix metalloproteinases in the tumor microenvironment. Front Pharmacol. 2012;3:140. doi: 10.3389/fphar.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 47. Levicar N, Strojnik T, Kos J, Dewey RA, Pilkington GJ, Lah TT. Lysosomal enzymes, cathepsins in brain tumour invasion. J Neurooncol. 2002;58(1):21–32. doi: 10.1023/a:1015892911420. [DOI] [PubMed] [Google Scholar]
- 48. Filippou PS, Karagiannis GS, Musrap N, Diamandis EP. Kallikrein-related peptidases (KLKs) and the hallmarks of cancer. Crit Rev Clin Lab Sci. 2016;53(4):277–91. doi: 10.3109/10408363.2016.1154643. [DOI] [PubMed] [Google Scholar]
- 49. Breznik B, Motaln H, Lah Turnšek T. Proteases and cytokines as mediators of interactions between cancer and stromal cells in tumours. Biol Chem. 2017;398(7):709–19. doi: 10.1515/hsz-2016-0283. [DOI] [PubMed] [Google Scholar]
- 50. Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Turunen SP, Tatti-Bugaeva O, Lehti K. Membrane-type matrix metalloproteases as diverse effectors of cancer progression. Biochim Biophys Acta Mol Cell Res. 2017;1864(11, Pt. A):1974–88. doi: 10.1016/j.bbamcr.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 52. Quintero-Fabián S, Arreola R, Becerril-Villanueva E, Torres-Romero JC, Arana-Argáez V, Lara-Riegos J, Ramírez-Camacho MA, Alvarez-Sánchez ME. Role of matrix metalloproteinases in angiogenesis and cancer. Front Oncol. 2019;9:1370. doi: 10.3389/fonc.2019.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98(5):621–8. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15(4):1140–4. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 55. Turner SL, Blair-Zajdel ME, Bunning RA. ADAMs and ADAMTSs in cancer. Br J Biomed Sci. 2009;66(2):117–28. doi: 10.1080/09674845.2009.11730257. [DOI] [PubMed] [Google Scholar]
- 56. Moss ML, Minond D. Recent advances in ADAM17 research: a promising target for cancer and inflammation. Mediators Inflamm. 2017;2017:9673537. doi: 10.1155/2017/9673537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schumacher N, Rose-John S. ADAM17 activity and IL-6 trans-signaling in inflammation and cancer. Cancers (Basel). 2019;11(11):1736. doi: 10.3390/cancers11111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sun Y, Huang J, Yang Z. The roles of ADAMTS in angiogenesis and cancer. Tumour Biol. 2015;36(6):4039–51. doi: 10.1007/s13277-015-3461-8. [DOI] [PubMed] [Google Scholar]
- 59. Binder MJ, McCoombe S, Williams ED, McCulloch DR, Ward AC. The extracellular matrix in cancer progression: role of hyalectan proteoglycans and ADAMTS enzymes. Cancer Lett. 2017;385:55–64. doi: 10.1016/j.canlet.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 60. Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6(10):764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 61. Khaket TP, Kwon TK, Kang SC. Cathepsins: potent regulators in carcinogenesis. Pharmacol Ther. 2019;198:1–19. doi: 10.1016/j.pharmthera.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 62. Tan GJ, Peng ZK, Lu JP, Tang FQ. Cathepsins mediate tumor metastasis. World J Biol Chem. 2013;4(4):91–101. doi: 10.4331/wjbc.v4.i4.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kos J, Werle B, Lah T, Brunner N. Cysteine proteinases and their inhibitors in extracellular fluids: markers for diagnosis and prognosis in cancer. Int J Biol Markers. 2000;15(1):84–9. [DOI] [PubMed] [Google Scholar]
- 64. Zhang L, Wang H, Xu J. Cathepsin S as a cancer target. Neoplasma. 2015;62(1):16–26. doi: 10.4149/neo_2015_003. [DOI] [PubMed] [Google Scholar]
- 65. Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prébois C, Rochefort H, Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237(2):167–79. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 66. Benes P, Vetvicka V, Fusek M. Cathepsin D: many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kryza T, Silva ML, Loessner D, Heuzé-Vourc’h N, Clements JA. The kallikrein-related peptidase family: dysregulation and functions during cancer progression. Biochimie. 2016;122:283–99. doi: 10.1016/j.biochi.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 68. Kontos CK, Scorilas A. Kallikrein-related peptidases (KLKs): a gene family of novel cancer biomarkers. Clin Chem Lab Med. 2012;50(11):1877–91. doi: 10.1515/cclm-2012-0247. [DOI] [PubMed] [Google Scholar]
- 69. Oikonomopoulou K, Diamandis EP, Hollenberg MD. Kallikrein-related peptidases: proteolysis and signaling in cancer, the new frontier. Biol Chem. 2010;391(4): 299–310. doi: 10.1515/BC.2010.038. [DOI] [PubMed] [Google Scholar]
- 70. Mahmood N, Mihalcioiu C, Rabbani SA. Multifaceted role of the urokinase-type plasminogen activator (uPA) and its receptor (uPAR): diagnostic, prognostic, and therapeutic applications. Front Oncol. 2018;8:24. doi: 10.3389/fonc.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mekkawy AH, Pourgholami MH, Morris DL. Involvement of urokinase-type plasminogen activator system in cancer: an overview. Med Res Rev. 2014;34(5):918–56. doi: 10.1002/med.21308. [DOI] [PubMed] [Google Scholar]
- 72. McMahon B, Kwaan HC. The plasminogen activator system and cancer. Pathophysiol Haemost Thromb. 2008;36(3–4):184–94. doi: 10.1159/000175156. [DOI] [PubMed] [Google Scholar]
- 73. Santibanez JF, Krstic J. Transforming growth factor-beta and urokinase type plasminogen interplay in cancer. Curr Protein Pept Sci. 2018;19(12):1155–63. doi: 10.2174/1389203718666171030103801. [DOI] [PubMed] [Google Scholar]
- 74. Kos J, Vižin T, Fonović UP, Pišlar A. Intracellular signaling by cathepsin X: molecular mechanisms and diagnostic and therapeutic opportunities in cancer. Semin Cancer Biol. 2015;31:76–83. doi: 10.1016/j.semcancer.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 75. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 76. Valent P, Bonnet D, De Maria R, Lapidot T, Copland M, Melo JV, Chomienne C, Ishikawa F, Schuringa JJ, Stassi G, Huntly B, Herrmann H, Soulier J, Roesch A, Schuurhuis GJ, Wöhrer S, Arock M, Zuber J, Cerny-Reiterer S, Johnsen HE, Andreeff M, Eaves C. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12(11):767–75. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 77. Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauß A, Blaudszun AR, Yevsa T, Fricke S, Kossatz-Boehlert U. Cancer stem cells-origins and biomarkers: perspectives for targeted personalized therapies. Front Immunol. 2020;11:1280. doi: 10.3389/fimmu.2020.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 80. O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 81. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 82. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 83. Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 84. Bahmad HF, Poppiti RJ. Medulloblastoma cancer stem cells: molecular signatures and therapeutic targets. J Clin Pathol. 2020;73(5):243–9. doi: 10.1136/jclinpath-2019-206246. [DOI] [PubMed] [Google Scholar]
- 85. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 86. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 87. Borovski T, De Sousa E, Melo F, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71(3):634–9. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 88. Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24(1):41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Oshimori N, Guo Y, Taniguchi S. An emerging role for cellular crosstalk in the cancer stem cell niche microenvironment. J Pathol. 2021;254(4):384–94. doi: 10.1002/path.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1(4):252–65. doi: 10.1016/j.trecan.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 92. Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10(2):138–46. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ping YF, Zhang X, Bian XW. Cancer stem cells and their vascular niche: do they benefit from each other? Cancer Lett. 2016;380(2):561–7. doi: 10.1016/j.canlet.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 94. Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, Maru DM, Hawke DH, Rak J, Mani SA, Zweidler-McKay P, Ellis LM. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23(2):171–85. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Beck B, Driessens G, Goossens S, Youssef KK, Kuchnio A, Caauwe A, Sotiropoulou PA, Loges S, Lapouge G, Candi A, Mascre G, Drogat B, Dekoninck S, Haigh JJ, Carmeliet P, Blanpain C. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478(7369):399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- 96. Lai CY, Schwartz BE, Hsu MY. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer Res. 2012;72(19):5111–8. doi: 10.1158/0008-5472.CAN-12-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Krishnamurthy S, Dong Z, Vodopyanov D, Imai A, Helman JI, Prince ME, Wicha MS, Nör JE. Endothelial cell-initiated signaling promotes the survival and self-renewal of cancer stem cells. Cancer Res. 2010;70(23):9969–78. doi: 10.1158/0008-5472.CAN-10-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16(3):225–38. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22(4):436–48. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jung E, Osswald M, Ratliff M, Dogan H, Xie R, Weil S, Hoffmann DC, Kurz FT, Kessler T, Heiland S, von Deimling A, Sahm F, Wick W, Winkler F. Tumor cell plasticity, heterogeneity, and resistance in crucial microenvironmental niches in glioma. Nat Commun. 2021;12(1):1014. doi: 10.1038/s41467-021-21117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bahmad HF, Jalloul M, Azar J, Moubarak MM, Samad TA, Mukherji D, Al-Sayegh M, Abou-Kheir W. Tumor microenvironment in prostate cancer: toward identification of novel molecular biomarkers for diagnosis, prognosis, and therapy development. Front Genet. 2021;12:652747. doi: 10.3389/fgene.2021.652747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66(10):5216–23. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 103. Rankin EB, Nam JM, Giaccia AJ. Hypoxia: signaling the metastatic cascade. Trends Cancer. 2016;2(6):295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Verbovšek U, Van Noorden CJ, Lah TT. Complexity of cancer protease biology: cathepsin K expression and function in cancer progression. Semin Cancer Biol. 2015;35:71–84. doi: 10.1016/j.semcancer.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 105. Breznik B, Limbaeck Stokin C, Kos J, Khurshed M, Hira VVV, Bošnjak R, Lah TT, Van Noorden CJF. Cysteine cathepsins B, X and K expression in peri-arteriolar glioblastoma stem cell niches. J Mol Histol. 2018;49(5):481–97. doi: 10.1007/s10735-018-9787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vander Linden C, Corbet C. Therapeutic targeting of cancer stem cells: integrating and exploiting the acidic niche. Front Oncol. 2019;9:159. doi: 10.3389/fonc.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hassn Mesrati M, Behrooz AB, Y Abuhamad A, Syahir A. Understanding glioblastoma biomarkers: knocking a mountain with a hammer. Cells. 2020;9(5):1236. doi: 10.3390/cells9051236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–12. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hira VV, Ploegmakers KJ, Grevers F, Verbovšek U, Silvestre-Roig C, Aronica E, Tigchelaar W, Turnšek TL, Molenaar RJ, Van Noorden CJ. CD133+ and nestin+ glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J Histochem Cytochem. 2015;63(7):481–93. doi: 10.1369/0022155415581689. [DOI] [PubMed] [Google Scholar]
- 110. Hira VVV, Wormer JR, Kakar H, Breznik B, van der Swaan B, Hulsbos R, Tigchelaar W, Tonar Z, Khurshed M, Molenaar RJ, Van Noorden CJF. Periarteriolar glioblastoma stem cell niches express bone marrow hematopoietic stem cell niche proteins. J Histochem Cytochem. 2018;66(3):155–73. doi: 10.1369/0022155417749174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hira VVV, Breznik B, Vittori M, Loncq de Jong A, Mlakar J, Oostra RJ, Khurshed M, Molenaar RJ, Lah T, Van Noorden CJF. Similarities between stem cell niches in glioblastoma and bone marrow: rays of hope for novel treatment strategies. J Histochem Cytochem. 2020;68(1):33–57. doi: 10.1369/0022155419878416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Nallanthighal S, Heiserman JP, Cheon DJ. The role of the extracellular matrix in cancer stemness. Front Cell Dev Biol. 2019;7:86. doi: 10.3389/fcell.2019.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kesh K, Gupta VK, Durden B, Garrido V, Mateo-Victoriano B, Lavania SP, Banerjee S. Therapy resistance, cancer stem cells and ECM in cancer: the matrix reloaded. Cancers (Basel). 2020;12(10):3067. doi: 10.3390/cancers12103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Raja AM, Xu S, Zhuo S, Tai DC, Sun W, So PT, Welsch RE, Chen CS, Yu H. Differential remodeling of extracellular matrices by breast cancer initiating cells. J Biophotonics. 2015;8(10):804–15. doi: 10.1002/jbio.201400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hillebrand LE, Bengsch F, Hochrein J, Hülsdünker J, Bender J, Follo M, Busch H, Boerries M, Reinheckel T. Proteolysis—a characteristic of tumor-initiating cells in murine metastatic breast cancer. Oncotarget. 2016;7(36):58244–60. doi: 10.18632/oncotarget.11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hillebrand LE, Reinheckel T. Impact of proteolysis on cancer stem cell functions. Biochimie. 2019;166:214–22. doi: 10.1016/j.biochi.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 117. Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–74. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14(2):163–76. doi: 10.1101/gad.14.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Vempati P, Popel AS, Mac Gabhann F. Extracellular regulation of VEGF: isoforms, proteolysis, and vascular patterning. Cytokine Growth Factor Rev. 2014;25(1):1–19. doi: 10.1016/j.cytogfr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Brahmkhatri VP, Prasanna C, Atreya HS. Insulin-like growth factor system in cancer: novel targeted therapies. Biomed Res Int. 2015;2015:538019. doi: 10.1155/2015/538019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Monboisse JC, Oudart JB, Ramont L, Brassart-Pasco S, Maquart FX. Matrikines from basement membrane collagens: a new anti-cancer strategy. Biochim Biophys Acta. 2014;1840(8):2589–98. doi: 10.1016/j.bbagen.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 122. Papadas A, Arauz G, Cicala A, Wiesner J, Asimakopoulos F. Versican and versican-matrikines in cancer progression, inflammation, and immunity. J Histochem Cytochem. 2020;68(12):871–85. doi: 10.1369/0022155420937098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Pišlar A, Jewett A, Kos J. Cysteine cathepsins: their biological and molecular significance in cancer stem cells. Semin Cancer Biol. 2018;53:168–77. doi: 10.1016/j.semcancer.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 124. Mitschke J, Burk UC, Reinheckel T. The role of proteases in epithelial-to-mesenchymal cell transitions in cancer. Cancer Metastasis Rev. 2019;38(3):431–44. doi: 10.1007/s10555-019-09808-2. [DOI] [PubMed] [Google Scholar]
- 125. Novinec M, Lenarčič B. Cathepsin K: a unique collagenolytic cysteine peptidase. Biol Chem. 2013;394(9):1163–79. doi: 10.1515/hsz-2013-0134. [DOI] [PubMed] [Google Scholar]
- 126. Verbovšek U, Motaln H, Rotter A, Atai NA, Gruden K, Van Noorden CJ, Lah TT. Expression analysis of all protease genes reveals cathepsin K to be overexpressed in glioblastoma. PLoS ONE. 2014;9(10):e111819. doi: 10.1371/journal.pone.0111819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Breznik B, Limback C, Porcnik A, Blejec A, Krajnc MK, Bosnjak R, Kos J, Van Noorden CJF, Lah TT. Localization patterns of cathepsins K and X and their predictive value in glioblastoma. Radiol Oncol. 2018;52(4):433–42. doi: 10.2478/raon-2018-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hira VV, Verbovšek U, Breznik B, Srdič M, Novinec M, Kakar H, Wormer J, der Swaan BV, Lenarčič B, Juliano L, Mehta S, Van Noorden CJ, Lah TT. Cathepsin K cleavage of SDF-1α inhibits its chemotactic activity towards glioblastoma stem-like cells. Biochim Biophys Acta Mol Cell Res. 2017;1864(3):594–603. doi: 10.1016/j.bbamcr.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 129. Siney EJ, Holden A, Casselden E, Bulstrode H, Thomas GJ, Willaime-Morawek S. Metalloproteinases ADAM10 and ADAM17 mediate migration and differentiation in glioblastoma sphere-forming cells. Mol Neurobiol. 2017;54(5):3893–905. doi: 10.1007/s12035-016-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Celià-Terrassa T, Jolly MK. Cancer stem cells and epithelial-to-mesenchymal transition in cancer metastasis. Cold Spring Harb Perspect Med. 2020;10(7):a036905. doi: 10.1101/cshperspect.a036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Seftor RE, Seftor EA, Koshikawa N, Meltzer PS, Gardner LM, Bilban M, Stetler-Stevenson WG, Quaranta V, Hendrix MJ. Cooperative interactions of laminin 5 gamma2 chain, matrix metalloproteinase-2, and membrane type-1-matrix/metalloproteinase are required for mimicry of embryonic vasculogenesis by aggressive melanoma. Cancer Res. 2001;61(17):6322–7. [PubMed] [Google Scholar]
- 132. Wang SS, Gao XL, Liu X, Gao SY, Fan YL, Jiang YP, Ma XR, Jiang J, Feng H, Chen QM, Tang YJ, Tang YL, Liang XH. CD133+ cancer stem-like cells promote migration and invasion of salivary adenoid cystic carcinoma by inducing vasculogenic mimicry formation. Oncotarget. 2016;7(20):29051–62. doi: 10.18632/oncotarget.8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sun H, Yao N, Cheng S, Li L, Liu S, Yang Z, Shang G, Zhang D, Yao Z. Cancer stem-like cells directly participate in vasculogenic mimicry channels in triple-negative breast cancer. Cancer Biol Med. 2019;16(2):299–311. doi: 10.20892/j.issn.2095-3941.2018.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Westhoff MA, Zhou S, Nonnenmacher L, Karpel-Massler G, Jennewein C, Schneider M, Halatsch ME, Carragher NO, Baumann B, Krause A, Simmet T, Bachem MG, Wirtz CR, Debatin KM. Inhibition of NF-κB signaling ablates the invasive phenotype of glioblastoma. Mol Cancer Res. 2013;11(12):1611–23. doi: 10.1158/1541-7786.MCR-13-0435-T. [DOI] [PubMed] [Google Scholar]
- 135. Wang F, Zhang P, Yang L, Yu X, Ye X, Yang J, Qian C, Zhang X, Cui YH, Bian XW. Activation of toll-like receptor 2 promotes invasion by upregulating MMPs in glioma stem cells. Am J Transl Res. 2015;7(3):607–15. [PMC free article] [PubMed] [Google Scholar]
- 136. Inoue A, Takahashi H, Harada H, Kohno S, Ohue S, Kobayashi K, Yano H, Tanaka J, Ohnishi T. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int J Oncol. 2010;37(5):1121–31. doi: 10.3892/ijo_00000764. [DOI] [PubMed] [Google Scholar]
- 137. Chen X, Chen L, Chen J, Hu W, Gao H, Xie B, Wang X, Yin Z, Li S, Wang X. ADAM17 promotes U87 glioblastoma stem cell migration and invasion. Brain Res. 2013;1538:151–8. doi: 10.1016/j.brainres.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 138. Sarkar S, Zemp FJ, Senger D, Robbins SM, Yong VW. ADAM-9 is a novel mediator of tenascin-C-stimulated invasiveness of brain tumor-initiating cells. Neuro Oncol. 2015;17(8):1095–105. doi: 10.1093/neuonc/nou362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ye XZ, Xu SL, Xin YH, Yu SC, Ping YF, Chen L, Xiao HL, Wang B, Yi L, Wang QL, Jiang XF, Yang L, Zhang P, Qian C, Cui YH, Zhang X, Bian XW. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-β1 signaling pathway. J Immunol. 2012;189(1):444–53. doi: 10.4049/jimmunol.1103248. [DOI] [PubMed] [Google Scholar]
- 140. Markovic DS, Vinnakota K, Chirasani S, Synowitz M, Raguet H, Stock K, Sliwa M, Lehmann S, Kälin R, van Rooijen N, Holmbeck K, Heppner FL, Kiwit J, Matyash V, Lehnardt S, Kaminska B, Glass R, Kettenmann H. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc Natl Acad Sci U S A. 2009;106(30):12530–5. doi: 10.1073/pnas.0804273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Alapati K, Gopinath S, Malla RR, Dasari VR, Rao JS. uPAR and cathepsin B knockdown inhibits radiation-induced PKC integrated integrin signaling to the cytoskeleton of glioma-initiating cells. Int J Oncol. 2012;41(2):599–610. doi: 10.3892/ijo.2012.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Alapati K, Kesanakurti D, Rao JS, Dasari VR. uPAR and cathepsin B-mediated compartmentalization of JNK regulates the migration of glioma-initiating cells. Stem Cell Res. 2014;12(3):716–29. doi: 10.1016/j.scr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hillebrand LE, Wickberg SM, Gomez-Auli A, Follo M, Maurer J, Busch H, Boerries M, Reinheckel T. MMP14 empowers tumor-initiating breast cancer cells under hypoxic nutrient-depleted conditions. FASEB J. 2019;33(3):4124–40. doi: 10.1096/fj.201801127R. [DOI] [PubMed] [Google Scholar]
- 144. Li J, Zucker S, Pulkoski-Gross A, Kuscu C, Karaayvaz M, Ju J, Yao H, Song E, Cao J. Conversion of stationary to invasive tumor initiating cells (TICs): role of hypoxia in membrane type 1-matrix metalloproteinase (MT1-MMP) trafficking. PLoS ONE. 2012;7(6):e38403. doi: 10.1371/journal.pone.0038403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Xin YH, Bian BS, Yang XJ, Cui W, Cui HJ, Cui YH, Zhang X, Xu C, Bian XW. POU5F1 enhances the invasiveness of cancer stem-like cells in lung adenocarcinoma by upregulation of MMP-2 expression. PLoS ONE. 2013;8(12):e83373. doi: 10.1371/journal.pone.0083373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Chavali PL, Saini RK, Zhai Q, Vizlin-Hodzic D, Venkatabalasubramanian S, Hayashi A, Johansson E, Zeng ZJ, Mohlin S, Påhlman S, Hansford L, Kaplan DR, Funa K. TLX activates MMP-2, promotes self-renewal of tumor spheres in neuroblastoma and correlates with poor patient survival. Cell Death Dis. 2014;5(10):e1502. doi: 10.1038/cddis.2014.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. You N, Tan Y, Zhou L, Huang X, Wang W, Wang L, Wu K, Mi N, Li J, Zheng L. Tg737 acts as a key driver of invasion and migration in liver cancer stem cells and correlates with poor prognosis in patients with hepatocellular carcinoma. Exp Cell Res. 2017;358(2):217–26. doi: 10.1016/j.yexcr.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 148. Annabi B, Rojas-Sutterlin S, Laflamme C, Lachambre MP, Rolland Y, Sartelet H, Béliveau R. Tumor environment dictates medulloblastoma cancer stem cell expression and invasive phenotype. Mol Cancer Res. 2008;6(6):907–16. doi: 10.1158/1541-7786.MCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 149. Long H, Xie R, Xiang T, Zhao Z, Lin S, Liang Z, Chen Z, Zhu B. Autocrine CCL5 signaling promotes invasion and migration of CD133+ ovarian cancer stem-like cells via NF-κB-mediated MMP-9 upregulation. Stem Cells. 2012;30(10):2309–19. doi: 10.1002/stem.1194. [DOI] [PubMed] [Google Scholar]
- 150. Duhachek-Muggy S, Qi Y, Wise R, Alyahya L, Li H, Hodge J, Zolkiewska A. Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol Cancer. 2017;16(1):32. doi: 10.1186/s12943-017-0599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hong SW, Hur W, Choi JE, Kim JH, Hwang D, Yoon SK. Role of ADAM17 in invasion and migration of CD133-expressing liver cancer stem cells after irradiation. Oncotarget. 2016;7(17):23482–97. doi: 10.18632/oncotarget.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ota M, Mochizuki S, Shimoda M, Abe H, Miyamae Y, Ishii K, Kimura H, Okada Y. ADAM23 is downregulated in side population and suppresses lung metastasis of lung carcinoma cells. Cancer Sci. 2016;107(4):433–43. doi: 10.1111/cas.12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore). 2016;95(1, Suppl. 1):S8–19. doi: 10.1097/MD.0000000000004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Cabarcas SM, Mathews LA, Farrar WL. The cancer stem cell niche—there goes the neighborhood? Int J Cancer. 2011;129(10):2315–27. doi: 10.1002/ijc.26312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. López de Andrés J, Griñán-Lisón C, Jiménez G, Marchal JA. Cancer stem cell secretome in the tumor microenvironment: a key point for an effective personalized cancer treatment. J Hematol Oncol. 2020;13(1):136. doi: 10.1186/s13045-020-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5(1):8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wang R, Ye X, Bhattacharya R, Boulbes DR, Fan F, Xia L, Ellis LM. A disintegrin and metalloproteinase domain 17 regulates colorectal cancer stem cells and chemosensitivity via Notch1 signaling. Stem Cells Transl Med. 2016;5(3):331–8. doi: 10.5966/sctm.2015-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Chen X, Chen L, Zhang R, Yi Y, Ma Y, Yan K, Jiang X, Wang X. ADAM17 regulates self-renewal and differentiation of U87 glioblastoma stem cells. Neurosci Lett. 2013;537:44–9. doi: 10.1016/j.neulet.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 159. Wang R, Li Y, Tsung A, Huang H, Du Q, Yang M, Deng M, Xiong S, Wang X, Zhang L, Geller DA, Cheng B, Billiar TR. iNOS promotes CD24+CD133+ liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc Natl Acad Sci USA. 2018;115(43):E10127–36. doi: 10.1073/pnas.1722100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Jimenez-Pascual A, Hale JS, Kordowski A, Pugh J, Silver DJ, Bayik D, Roversi G, Alban TJ, Rao S, Chen R, McIntyre TM, Colombo G, Taraboletti G, Holmberg KO, Forsberg-Nilsson K, Lathia JD, Siebzehnrubl FA. ADAMDEC1 maintains a growth factor signaling loop in cancer stem cells. Cancer Discov. 2019;9(11):1574–89. doi: 10.1158/2159-8290.CD-18-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Peris-Torres C, Plaza-Calonge MDC, López-Domínguez R, Domínguez-García S, Barrientos-Durán A, Carmona-Sáez P, Rodríguez-Manzaneque JC. Extracellular protease ADAMTS1 is required at early stages of human uveal melanoma development by inducing stemness and endothelial-like features on tumor cells. Cancers (Basel). 2020;12(4):801. doi: 10.3390/cancers12040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Justilien V, Regala RP, Tseng IC, Walsh MP, Batra J, Radisky ES, Murray NR, Fields AP. Matrix metalloproteinase-10 is required for lung cancer stem cell maintenance, tumor initiation and metastatic potential. PLoS ONE. 2012;7(4):e35040. doi: 10.1371/journal.pone.0035040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Mariya T, Hirohashi Y, Torigoe T, Tabuchi Y, Asano T, Saijo H, Kuroda T, Yasuda K, Mizuuchi M, Saito T, Sato N. Matrix metalloproteinase-10 regulates stemness of ovarian cancer stem-like cells by activation of canonical Wnt signaling and can be a target of chemotherapy-resistant ovarian cancer. Oncotarget. 2016;7(18):26806–22. doi: 10.18632/oncotarget.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yang CC, Zhu LF, Xu XH, Ning TY, Ye JH, Liu LK. Membrane type 1 matrix metalloproteinase induces an epithelial to mesenchymal transition and cancer stem cell-like properties in SCC9 cells. BMC Cancer. 2013;13:171. doi: 10.1186/1471-2407-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Gopinath S, Malla R, Alapati K, Gorantla B, Gujrati M, Dinh DH, Rao JS. Cathepsin B and uPAR regulate self-renewal of glioma-initiating cells through GLI-regulated Sox2 and Bmi1 expression. Carcinogenesis. 2013;34(3):550–9. doi: 10.1093/carcin/bgs375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wang W, Long L, Wang L, Tan C, Fei X, Chen L, Huang Q, Liang Z. Knockdown of Cathepsin L promotes radiosensitivity of glioma stem cells both in vivo and in vitro. Cancer Lett. 2016;371(2):274–84. doi: 10.1016/j.canlet.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 167. Kenig S, Frangež R, Pucer A, Lah T. Inhibition of cathepsin L lowers the apoptotic threshold of glioblastoma cells by up-regulating p53 and transcription of caspases 3 and 7. Apoptosis. 2011;16(7):671–82. doi: 10.1007/s10495-011-0600-6. [DOI] [PubMed] [Google Scholar]
- 168. Goulet B, Sansregret L, Leduy L, Bogyo M, Weber E, Chauhan SS, Nepveu A. Increased expression and activity of nuclear cathepsin L in cancer cells suggests a novel mechanism of cell transformation. Mol Cancer Res. 2007;5(9):899–907. doi: 10.1158/1541-7786.MCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 169. Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135(2):284–94. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Vahidian F, Duijf PHG, Safarzadeh E, Derakhshani A, Baghbanzadeh A, Baradaran B. Interactions between cancer stem cells, immune system and some environmental components: friends or foes? Immunol Lett. 2019;208:19–29. doi: 10.1016/j.imlet.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 171. Müller L, Tunger A, Plesca I, Wehner R, Temme A, Westphal D, Meier F, Bachmann M, Schmitz M. Bidirectional crosstalk between cancer stem cells and immune cell subsets. Front Immunol. 2020;11:140. doi: 10.3389/fimmu.2020.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kärre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9(5):477–80. doi: 10.1038/ni0508-477. [DOI] [PubMed] [Google Scholar]
- 173. Palesch D, Wagner J, Meid A, Molenda N, Sienczyk M, Burkhardt J, Münch J, Prokop L, Stevanovic S, Westhoff MA, Halatsch ME, Wirtz CR, Zimecki M, Burster T. Cathepsin G-mediated proteolytic degradation of MHC class I molecules to facilitate immune detection of human glioblastoma cells. Cancer Immunol Immunother. 2016;65(3):283–91. doi: 10.1007/s00262-016-1798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Bischof J, Westhoff MA, Wagner JE, Halatsch ME, Trentmann S, Knippschild U, Wirtz CR, Burster T. Cancer stem cells: the potential role of autophagy, proteolysis, and cathepsins in glioblastoma stem cells. Tumour Biol. Epub 2017 Mar. doi: 10.1177/1010428317692227. [DOI] [PubMed] [Google Scholar]
- 175. Perišić Nanut M, Sabotič J, Jewett A, Kos J. Cysteine cathepsins as regulators of the cytotoxicity of NK and T cells. Front Immunol. 2014;5:616. doi: 10.3389/fimmu.2014.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Magister Š, Tseng HC, Bui VT, Kos J, Jewett A. Regulation of split anergy in natural killer cells by inhibition of cathepsins C and H and cystatin F. Oncotarget. 2015;6(26):22310–27. doi: 10.18632/oncotarget.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Perišić Nanut M, Sabotič J, Švajger U, Jewett A, Kos J. Cystatin F affects natural killer cell cytotoxicity. Front Immunol. 2017;8:1459. doi: 10.3389/fimmu.2017.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Prunk M, Perišić Nanut M, Jakoš T, Sabotič J, Švajger U, Kos J. Extracellular cystatin F is internalised by cytotoxic T lymphocytes and decreases their cytotoxicity. Cancers (Basel). 2020;12(12):3660. doi: 10.3390/cancers12123660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Lauricella M, Carlisi D, Giuliano M, Calvaruso G, Cernigliaro C, Vento R, D’Anneo A. The analysis of estrogen receptor-positive breast cancer stem-like cells unveils a high expression of the serpin proteinase inhibitor PI-9: possible regulatory mechanisms. Int J Oncol. 2016;49(1):352–60. doi: 10.3892/ijo.2016.3495. [DOI] [PubMed] [Google Scholar]
- 180. Chien YH, Meyer C, Bonneville M. γδ T cells: first line of defense and beyond. Annu Rev Immunol. 2014;32:121–55. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 181. Dutta I, Dieters-Castator D, Papatzimas JW, Medina A, Schueler J, Derksen DJ, Lajoie G, Postovit LM, Siegers GM. ADAM protease inhibition overcomes resistance of breast cancer stem-like cells to γδ T cell immunotherapy. Cancer Lett. 2021;496:156–68. doi: 10.1016/j.canlet.2020.10.013. [DOI] [PubMed] [Google Scholar]
- 182. Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T. Drug resistance driven by cancer stem cells and their niche. Int J Mol Sci. 2017;18(12):2574. doi: 10.3390/ijms18122574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Marzagalli M, Fontana F, Raimondi M, Limonta P. Cancer stem cells-key players in tumor relapse. Cancers (Basel). 2021;13(3):376. doi: 10.3390/cancers13030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Van Noorden CJF, Vogels IMC, Everts V, Beertsen W. Localization of cathepsin B activity in fibroblasts and chondrocytes by continuous monitoring of the formation of a final fluorescent reaction product using 5-nitrosalicylaldehyde. Histochem J. 1987;19(9):483–7. doi: 10.1007/bf01675418. [DOI] [PubMed] [Google Scholar]
- 186. Podergajs N, Brekka N, Radlwimmer B, Herold-Mende C, Talasila KM, Tiemann K, Rajcevic U, Lah TT, Bjerkvig R, Miletic H. Expansive growth of two glioblastoma stem-like cell lines is mediated by bFGF and not by EGF. Radiol Oncol. 2013;47(4):330–7. doi: 10.2478/raon-2013-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Malla RR, Gopinath S, Alapati K, Gorantla B, Gondi CS, Rao JS. uPAR and cathepsin B inhibition enhanced radiation-induced apoptosis in gliomainitiating cells. Neuro Oncol. 2012;14(6):745–60. doi: 10.1093/neuonc/nos088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Atapattu L, Saha N, Chheang C, Eissman MF, Xu K, Vail ME, Hii L, Llerena C, Liu Z, Horvay K, Abud HE, Kusebauch U, Moritz RL, Ding BS, Cao Z, Rafii S, Ernst M, Scott AM, Nikolov DB, Lackmann M, Janes PW. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J Exp Med. 2016;213(9):1741–57. doi: 10.1084/jem.20151095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Nikitakis NG, Gkouveris I, Aseervatham J, Barahona K, Ogbureke KUE. DSPP-MMP20 gene silencing downregulates cancer stem cell markers in human oral cancer cells. Cell Mol Biol Lett. 2018;23:30. doi: 10.1186/s11658-018-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhao Y, Dong Q, Li J, Zhang K, Qin J, Zhao J, Sun Q, Wang Z, Wartmann T, Jauch KW, Nelson PJ, Qin L, Bruns C. Targeting cancer stem cells and their niche: perspectives for future therapeutic targets and strategies. Semin Cancer Biol. 2018;53:139–55. doi: 10.1016/j.semcancer.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 191. Ray SK, Mukherjee S. Cancer stem cells: current status and therapeutic implications in cancer therapy—a new paradigm. Curr Stem Cell Res Ther. Epub 2021 Feb. doi: 10.2174/1574888X16666210203105800. [DOI] [PubMed] [Google Scholar]
- 192. Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, Reece D, San-Miguel JF, Bladé J, Boccadoro M, Cavenagh J, Dalton WS, Boral AL, Esseltine DL, Porter JB, Schenkein D, Anderson KC; Assessment of Proteasome Inhibition for Extending Remissions Investigators. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med. 2005;352(24):2487–98. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 193. San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M, Spicka I, Petrucci MT, Palumbo A, Samoilova OS, Dmoszynska A, Abdulkadyrov KM, Schots R, Jiang B, Mateos MV, Anderson KC, Esseltine DL, Liu K, Cakana A, van de Velde H, Richardson PG; VISTA Trial Investigators. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359(9):906–17. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 194. Siegel DS, Martin T, Wang M, Vij R, Jakubowiak AJ, Lonial S, Trudel S, Kukreti V, Bahlis N, Alsina M, Chanan-Khan A, Buadi F, Reu FJ, Somlo G, Zonder J, Song K, Stewart AK, Stadtmauer E, Kunkel L, Wear S, Wong AF, Orlowski RZ, Jagannath S. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 2012;120(14):2817–25. doi: 10.1182/blood-2012-05-425934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Stewart AK, Rajkumar SV, Dimopoulos MA, Masszi T, Špička I, Oriol A, Hájek R, Rosiñol L, Siegel DS, Mihaylov GG, Goranova-Marinova V, Rajnics P, Suvorov A, Niesvizky R, Jakubowiak AJ, San-Miguel JF, Ludwig H, Wang M, Maisnar V, Minarik J, Bensinger WI, Mateos MV, Ben-Yehuda D, Kukreti V, Zojwalla N, Tonda ME, Yang X, Xing B, Moreau P, Palumbo A; ASPIRE Investigators. Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. N Engl J Med. 2015;372(2):142–52. doi: 10.1056/NEJMoa1411321. [DOI] [PubMed] [Google Scholar]
- 196. Dimopoulos MA, Goldschmidt H, Niesvizky R, Joshua D, Chng WJ, Oriol A, Orlowski RZ, Ludwig H, Facon T, Hajek R, Weisel K, Hungria V, Minuk L, Feng S, Zahlten-Kumeli A, Kimball AS, Moreau P. Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): an interim overall survival analysis of an open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18(10):1327–37. doi: 10.1016/S1470-2045(17)30578-8. [DOI] [PubMed] [Google Scholar]
- 197. Moreau P, Masszi T, Grzasko N, Bahlis NJ, Hansson M, Pour L, Sandhu I, Ganly P, Baker BW, Jackson SR, Stoppa AM, Simpson DR, Gimsing P, Palumbo A, Garderet L, Cavo M, Kumar S, Touzeau C, Buadi FK, Laubach JP, Berg DT, Lin J, Di Bacco A, Hui AM, van de Velde H, Richardson PG; TOURMALINE-MM1 Study Group. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med. 2016;374(17):1621–34. doi: 10.1056/NEJMoa1516282. [DOI] [PubMed] [Google Scholar]
- 198. Addison CL, Simos D, Wang Z, Pond G, Smith S, Robertson S, Mazzarello S, Singh G, Vandermeer L, Fernandes R, Iyengar A, Verma S, Clemons M. A phase 2 trial exploring the clinical and correlative effects of combining doxycycline with bone-targeted therapy in patients with metastatic breast cancer. J Bone Oncol. 2016;5(4):173–9. doi: 10.1016/j.jbo.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Dezube BJ, Krown SE, Lee JY, Bauer KS, Aboulafia DM. Randomized phase II trial of matrix metalloproteinase inhibitor COL-3 in AIDS-related Kaposi’s sarcoma: an AIDS malignancy consortium study. J Clin Oncol. 2006;24(9):1389–94. doi: 10.1200/JCO.2005.04.2614. [DOI] [PubMed] [Google Scholar]
- 200. Rudek MA, New P, Mikkelsen T, Phuphanich S, Alavi JB, Nabors LB, Piantadosi S, Fisher JD, Grossman SA. Phase I and pharmacokinetic study of COL-3 in patients with recurrent high-grade gliomas. J Neurooncol. 2011;105(2):375–81. doi: 10.1007/s11060-011-0602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Shepherd FA, Giaccone G, Seymour L, Debruyne C, Bezjak A, Hirsh V, Smylie M, Rubin S, Martins H, Lamont A, Krzakowski M, Sadura A, Zee B. Prospective, randomized, double-blind, placebo-controlled trial of marimastat after response to first-line chemotherapy in patients with small-cell lung cancer: a trial of the National Cancer Institute of Canada-Clinical Trials Group and the European Organization for Research and Treatment of Cancer. J Clin Oncol. 2002;20(22):4434–9. doi: 10.1200/JCO.2002.02.108. [DOI] [PubMed] [Google Scholar]
- 202. Sparano JA, Bernardo P, Stephenson P, Gradishar WJ, Ingle JN, Zucker S, Davidson NE. Randomized phase III trial of marimastat versus placebo in patients with metastatic breast cancer who have responding or stable disease after first-line chemotherapy: Eastern Cooperative Oncology Group trial E2196. J Clin Oncol. 2004;22(23):4683–90. doi: 10.1200/JCO.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 203. Bissett D, O’Byrne KJ, von Pawel J, Gatzemeier U, Price A, Nicolson M, Mercier R, Mazabel E, Penning C, Zhang MH, Collier MA, Shepherd FA. Phase III study of matrix metalloproteinase inhibitor prinomastat in non-small-cell lung cancer. J Clin Oncol. 2005;23(4):842–9. doi: 10.1200/JCO.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 204. Lara PN, Jr, Stadler WM, Longmate J, Quinn DI, Wexler J, Van Loan M, Twardowski P, Gumerlock PH, Vogelzang NJ, Vokes EE, Lenz HJ, Doroshow JH, Gandara DR. A randomized phase II trial of the matrix metalloproteinase inhibitor BMS-275291 in hormone-refractory prostate cancer patients with bone metastases. Clin Cancer Res. 2006;12(5):1556–63. doi: 10.1158/1078-0432.CCR-05-2074. [DOI] [PubMed] [Google Scholar]
- 205. Brinker BT, Krown SE, Lee JY, Cesarman E, Chadburn A, Kaplan LD, Henry DH, Von Roenn JH. Phase 1/2 trial of BMS-275291 in patients with human immunodeficiency virus-related Kaposi sarcoma: a multicenter trial of the AIDS Malignancy Consortium. Cancer. 2008;112(5):1083–8. doi: 10.1002/cncr.23108. [DOI] [PubMed] [Google Scholar]
- 206. Shah MA, Metges JP, Cunningham D, Shiu KK, Wyrwicz L, Thai D, Brachmann C, Bhargava P, Catenacci DVT, Wainberg ZA. A phase II, open-label, randomized study to evaluate the efficacy and safety of andecaliximab combined with nivolumab versus nivolumab alone in subjects with unresectable or recurrent gastric or gastroesophageal junction adenocarcinoma. J Clin Oncol. 2019;37(Suppl. 4):75. doi: 10.1200/JCO.2019.37.4_suppl.75. [DOI] [Google Scholar]
- 207. Shah MA, Bodoky G, Starodub A, Cunningham D, Yip D, Wainberg ZA, Bendell J, Thai D, He J, Bhargava P, Ajani JA. Phase III study to evaluate efficacy and safety of andecaliximab with mFOLFOX6 as first-line treatment in patients with advanced gastric or GEJ adenocarcinoma (GAMMA-1). J Clin Oncol. 2021;39(9):990–1000. doi: 10.1200/JCO.20.02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Goldstein LJ, Oliveira CT, Heinrich B, Stemmer SM, Mala C, Kastner S, Bevan P, Richters L, Schmalfeldt B, Harbeck N. A randomized double-blind phase II study of the combination of oral WX-671 plus capecitabine versus capecitabine monotherapy in first-line HER2-negative metastatic breast cancer (MBC). J Clin Oncol. 2013;31(Suppl. 15):508. [Google Scholar]
- 209. Heinemann V, Ebert MP, Laubender RP, Bevan P, Mala C, Boeck S. Phase II randomised proof-of-concept study of the urokinase inhibitor upamostat (WX-671) in combination with gemcitabine compared with gemcitabine alone in patients with non-resectable, locally advanced pancreatic cancer. Br J Cancer. 2013;108(4):766–70. doi: 10.1038/bjc.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Blackhall F, Jao K, Greillier L, Cho BC, Penkov K, Reguart N, Majem M, Nackaerts K, Syrigos K, Hansen K, Schuette W, Cetnar J, Cappuzzo F, Okamoto I, Erman M, Langer SW, Kato T, Groen H, Sun Z, Luo Y, Tanwani P, Caffrey L, Komarnitsky P, Reinmuth N. Efficacy and safety of rovalpituzumab tesirine compared with topotecan as second-line therapy in DLL3-high SCLC: results from the phase 3 TAHOE study. J Thorac Oncol. Epub 2021 Feb. doi: 10.1016/j.jtho.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 211. Johnson ML, Zvirbule Z, Laktionov K, Helland A, Cho BC, Gutierrez V, Colinet B, Lena H, Wolf M, Gottfried M, Okamoto I, van der Leest C, Rich P, Hung JY, Appenzeller C, Sun Z, Maag D, Luo Y, Nickner C, Vajikova A, Komarnitsky P, Bar J. Rovalpituzumab tesirine as a maintenance therapy after first-line platinum-based chemotherapy in patients with extensive-stage-SCLC: results from the phase 3 MERU study. J Thorac Oncol. Epub 2021 Apr. doi: 10.1016/j.jtho.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 212. Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhibitors in cancer therapy: turning past failures into future successes. Mol Cancer Ther. 2018;17(6):1147–55. doi: 10.1158/1535-7163.MCT-17-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Dudani JS, Warren AD, Bhatia SN. Harnessing protease activity to improve cancer care. Annu Rev Cancer Biol. 2018;2:353–76. doi: 10.1146/annurev-cancerbio-030617-050549. [DOI] [Google Scholar]
- 214. Anderson CF, Cui H. Protease-sensitive nanomaterials for cancer therapeutics and imaging. Ind Eng Chem Res. 2017;56(20):5761–77. doi: 10.1021/acs.iecr.7b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Raeeszadeh-Sarmazdeh M, Do LD, Hritz BG. Metalloproteinases and their inhibitors: potential for the development of new therapeutics. Cells. 2020;9(5):1313. doi: 10.3390/cells9051313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Mamaeva V, Niemi R, Beck M, Özliseli E, Desai D, Landor S, Gronroos T, Kronqvist P, Pettersen IK, McCormack E, Rosenholm JM, Linden M, Sahlgren C. Inhibiting notch activity in breast cancer stem cells by glucose functionalized nanoparticles carrying γ-secretase inhibitors. Mol Ther. 2016;24(5):926–36. doi: 10.1038/mt.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Mohanty S, Chen Z, Li K, Morais GR, Klockow J, Yerneni K, Pisani L, Chin FT, Mitra S, Cheshier S, Chang E, Gambhir SS, Rao J, Loadman PM, Falconer RA, Daldrup-Link HE. A novel theranostic strategy for MMP-14-expressing glioblastomas impacts survival. Mol Cancer Ther. 2017;16(9):1909–21. doi: 10.1158/1535-7163.MCT-17-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, De Clercq S, Minguijón E, Balsat C, Sokolow Y, Dubois C, De Cock F, Scozzaro S, Sopena F, Lanas A, D’Haene N, Salmon I, Marine JC, Voet T, Sotiropoulou PA, Blanpain C. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–8. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]