Abstract
Fecal microbiota transplantation (FMT) targeting gut microbiota has recently been successfully applied to ulcerative colitis. However, only a subset of patients responds to FMT and there is a pressing need for biomarkers of responsiveness. Fungi (the mycobiota) represent a highly immunologically reactive component of the gut microbiota. We analyzed samples from a large randomized controlled trial of FMT for ulcerative colitis (UC). High Candida abundance pre-FMT was associated with a clinical response, while decreased Candida abundance post-FMT was indicative of ameliorated disease severity. High pre-FMT Candida was associated with increased bacterial diversity post-FMT and the presence of genera linked to FMT responsiveness. Although we detected elevated anti-Candida antibodies in placebo recipients, this increase was abrogated in FMT recipients. Our data suggest that FMT might reduce Candida to contain pro-inflammatory immunity during intestinal disease and highlight the utility of mycobiota–focused approaches to identify FMT responders prior to therapy initiation.
Introduction
For decades, fecal microbiota transplantation (FMT) from heterologous donors has been successfully used to eradicate the causative toxin-producing bacterium in patients with intestinal Clostridioides difficile infections (CDI)(Quraishi et al., 2017; Tvede and Rask-Madsen, 1989; van Nood et al., 2013). This success has sparked interest in extending its application to other diseases affecting the gastrointestinal tract, such as inflammatory bowel disease (IBD)(Allegretti et al., 2019; Paramsothy et al., 2017; Weingarden and Vaughn, 2017), graft versus host disease (GVHD)(Kakihana et al., 2016), metabolic syndrome(Vrieze et al., 2012), and immune checkpoint inhibitor associated colitis (ICIAC)(Wang et al., 2018). In contrast to CDI, where FMT successfully treats >90% of cases (Quraishi et al., 2017; van Nood et al., 2013), FMT application in IBD faces significant challenges due to complex disease etiology and the absence of a clear microbial target. We and others have shown that FMT promotes symptoms resolution (also termed clinical remission) and mucosal healing in ulcerative colitis (UC), a major form of IBD(Costello et al., 2019; Fuentes et al., 2017; Imdad et al., 2018; Moayyedi et al., 2015; Paramsothy et al., 2017). However, the benefits were limited to a subset of patients (Paramsothy et al., 2017). This heterogeneity highlights the urgent need to identify mechanisms and define predictive markers to forecast FMT treatment response prior to the initiation of therapy.
In a large multi-center, double-blind, randomized, placebo-controlled FMT trial in UC (the FOCUS study), steroid-free clinical remission was achieved in 44% (18 of 41) of patients receiving multidonor intensive heterologous FMT versus 20% (8 of 40) of those receiving placebo (Paramsothy et al., 2017). FMT induced bacterial changes that related to the treatment outcome(Paramsothy et al., 2017; Paramsothy et al., 2019), such as increased bacterial diversity when compared to pre-FMT baseline and better engraftment of donor bacteria when compared to non-responders, consistent with other clinical studies(Costello et al., 2019; Fuentes et al., 2017; Imdad et al., 2018; Moayyedi et al., 2015). However, as in other trials for FMT in UC, we could not identify specific factors that influenced the observed bacterial engraftment and treatment response.
In addition to bacteria, FMT transfers all constituents of the intestinal microbiota including viruses and fungi(Zuo et al., 2018). Likewise, FMT treatment does not selectively target intestinal bacteria but it likely affects and is affected by the various members of the microbiota. In CDI, where the intestinal niche is severely affected by C. difficile domination, an overgrowth of Candida species has been associated with a negative outcome of FMT(Zuo et al., 2018), a finding that has been replicated in mouse models (Li et al., 2019; Panpetch et al., 2019; Zuo et al., 2018). Experimental studies, however, suggest that gut colonization with Candida can exert contrasting effects on intestinal inflammation and that this modulation is strongly dependent on the host’s genetic predisposition and gut microbiota composition (Jawhara et al., 2008; Leonardi et al., 2018; Li et al., 2019; Sovran et al., 2018). In hosts with an intact microbiota, targeting the healthy gut fungal communities can worsen the outcome of experimental colitis and lung disease(Li et al., 2018; Wheeler et al., 2016), suggesting that specific members of the gut mycobiota can play a protective function in the intestine. These data suggest that members of the mycobiota might be important players in the response to FMT.
Results
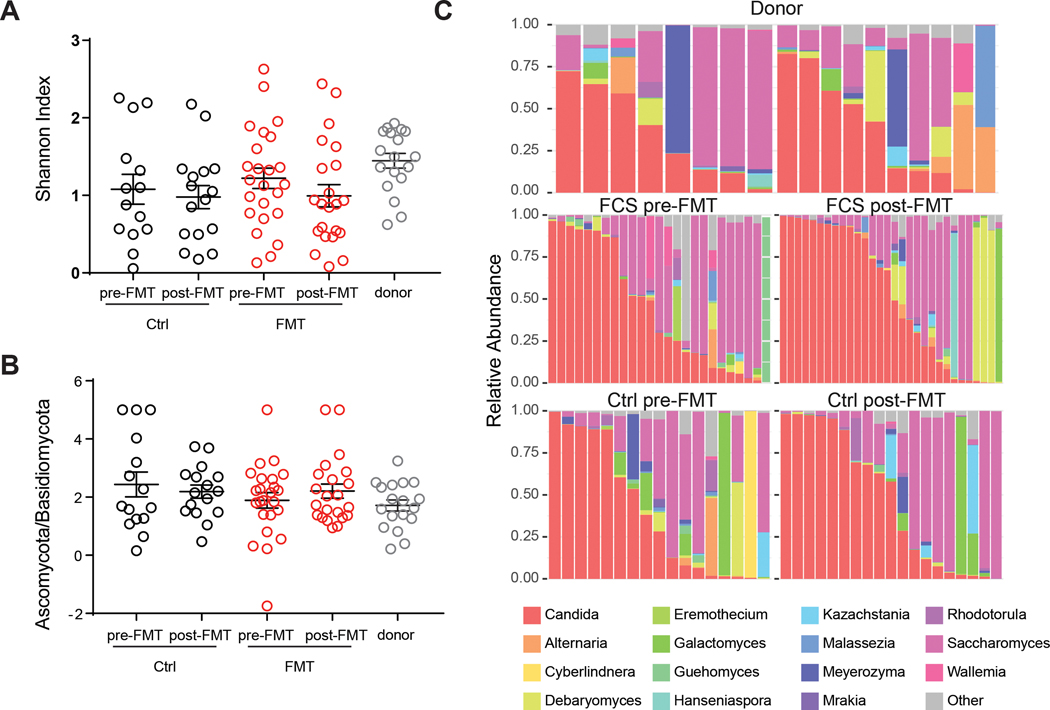
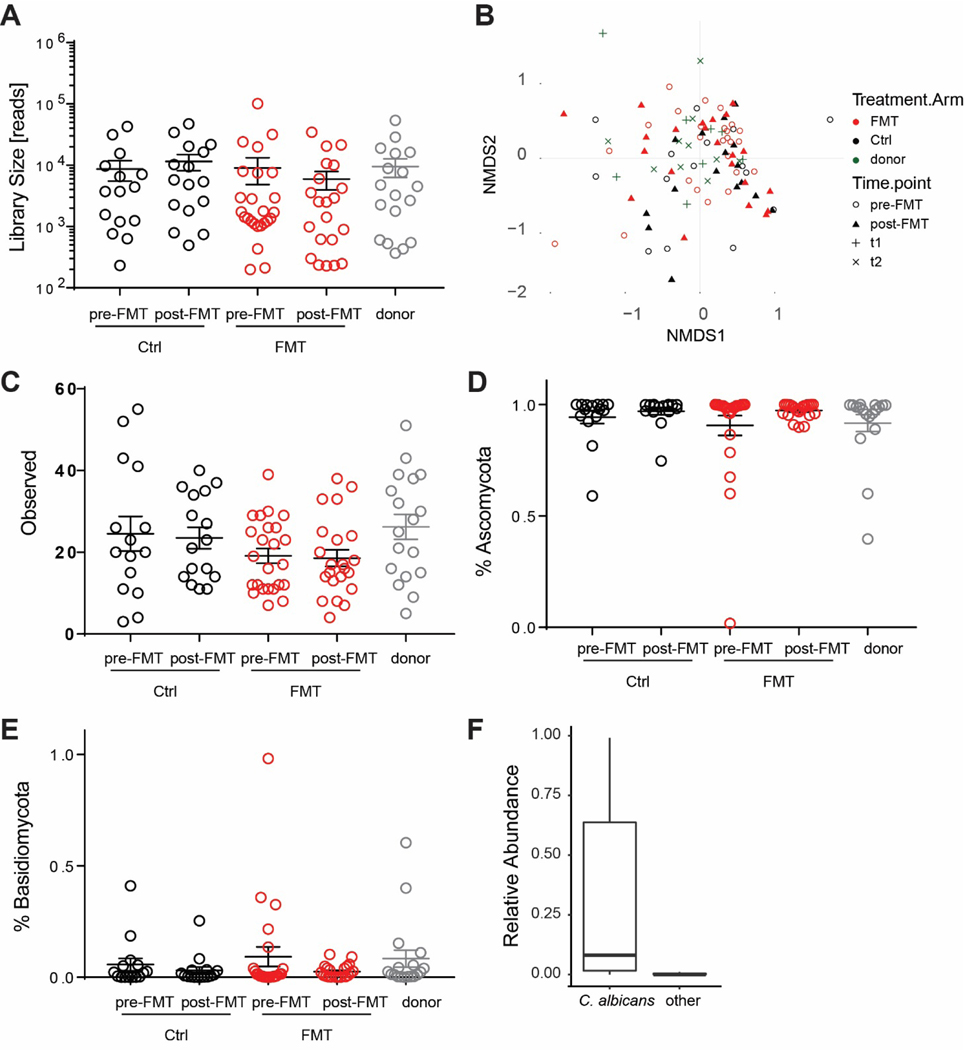
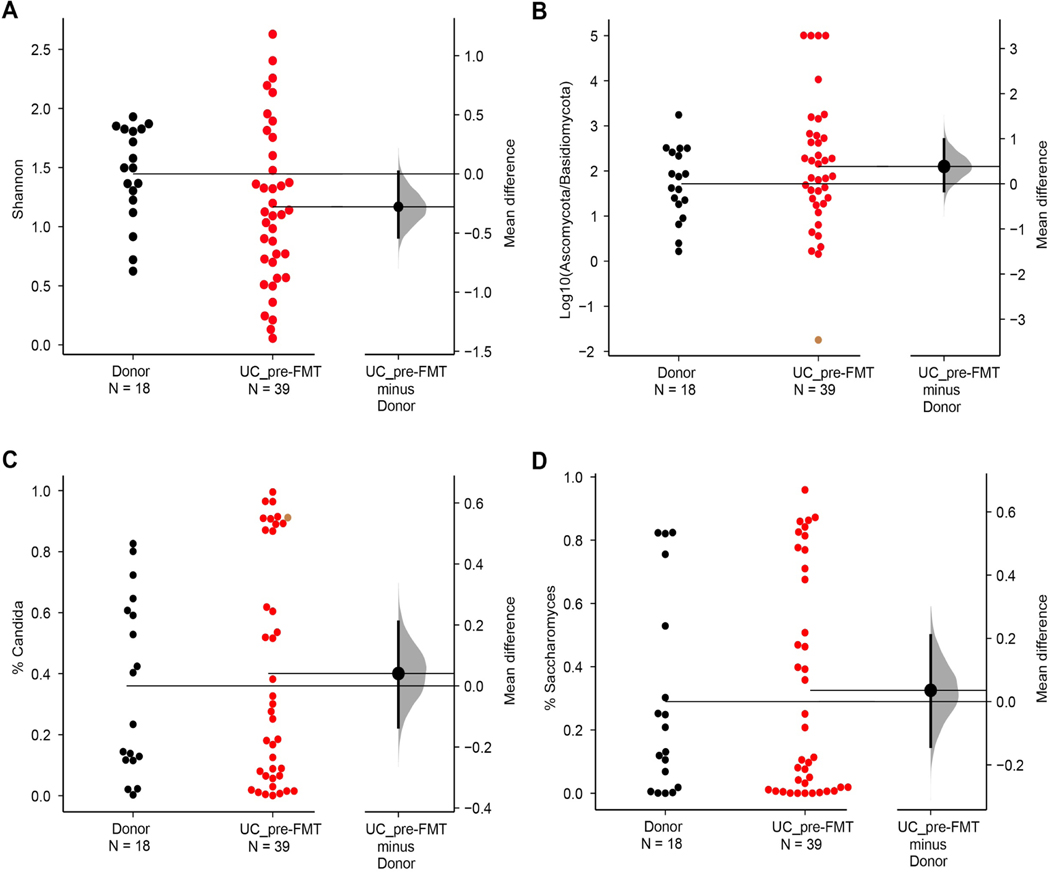
To investigate the dynamics of the intestinal mycobiota in UC patients undergoing FMT we used an Internal Transcribed Spacer (ITS)-1 based barcoding approach to deep sequence the ITS-1 regions of fungal rDNA in fecal samples from the FOCUS study(Paramsothy et al., 2017). Specifically, ITS sequencing was performed on 129 fecal samples collected from FMT donors and UC patients pre-FMT and following 8 weeks of intensive FMT or placebo treatment (post-FMT). ITS sequencing resulted in a uniform library size distribution across the analyzed samples (Extended Data Fig. 1A, Table 1). The mycobiota was generally dominated by the Ascomycota phylum in all groups (Extended Data Fig. 1 D–E). In line with previously published cohorts(Chehoud et al., 2015a; Hoarau et al., 2016; Sokol et al., 2017), we observed a trend towards lower fungal alpha-diversity and increased Ascomycota/Basidiomycota ratio in feces from UC patients pre-FMT when compared to healthy donors (Fig 1A, B and Extended Data Fig. 1C, 2A, B). However, these differences did not reach statistical significance, possibly due the mild-moderate nature of UC in our patients’ cohort.
Table 1:
Summary of the Patients characteristics pre-FMT f or the samples included in the study.
| FMT | Placebo | ||
|---|---|---|---|
| Count | 24 | 15 | |
|
| |||
| Age | 39.6±2.5 | 40.5±3.2 | |
|
| |||
| Gender | Female | 11 (45.83%) | 6 (40%) |
| Male | 13 (54.17%) | 9 (60%) | |
|
| |||
| Ethnicity | Arabic / Middle Eastern | 2 (8.33%) | 0 (0%) |
| Asian | 2 (8.33%) | 0 (0%) | |
| Caucasian | 18 (75%) | 12 (80%) | |
| Indian Subcontinental | 0 (0%) | 1 (6.67%) | |
| Mediterranean | 2 (8.33%) | 2 (13.33%) | |
|
| |||
| Smoking Status | Non-smoking | 12 (50%) | 7 (46.67%) |
| Ex-smoker | 8 (33.33%) | 8 (53.33%) | |
| Occasional | 4 (16.67%) | 0 (0%) | |
|
| |||
| Duration | 8.5±1.4 | 8.8±2.4 | |
|
| |||
| UCEIS | 4.2±0.2 | 4.4±0.4 | |
|
| |||
| IBDQ | 137.1±7.1 | 126.4±7.9 | |
|
| |||
| Total Mayo | 7.4±0.3 | 7.3±0.4 | |
|
| |||
| Bleed Mayo | 1.6±0.2 | 1.6±0.1 | |
|
| |||
| Stool Mayo | 1.9±0.1 | 1.8±0.2 | |
|
| |||
| PGA Mayo | 1.7±0.1 | 1.6±0.1 | |
|
| |||
| Endoscopic Mayo | 2.3±0.1 | 2.3±0.2 | |
|
| |||
| UCEIS | 4.2±0.2 | 4.4±0.4 | |
|
| |||
| ITS library size | 9089±4251 | 8704.7±3098 | |
Figure 1. Mycobiota composition in the FOCUS cohort.
A. Alpha diversity measured by observed OTU and Shannon Index for donor, fecal transplantation recipient (FMT), and placebo recipient (CTRL) pre- and post-FMT. B. Ratio between Ascomycota and Basidiomycota are shown for donor (grey), fecal transplantation recipient (FMT, red), and placebo recipient (CTRL. black) at pre- and post-FMT. C. Stack plot of relative abundances at the genus level for the 15 genera with the highest average abundance are shown. Dots represent individual patients, line and bars represent mean +/− SEM.
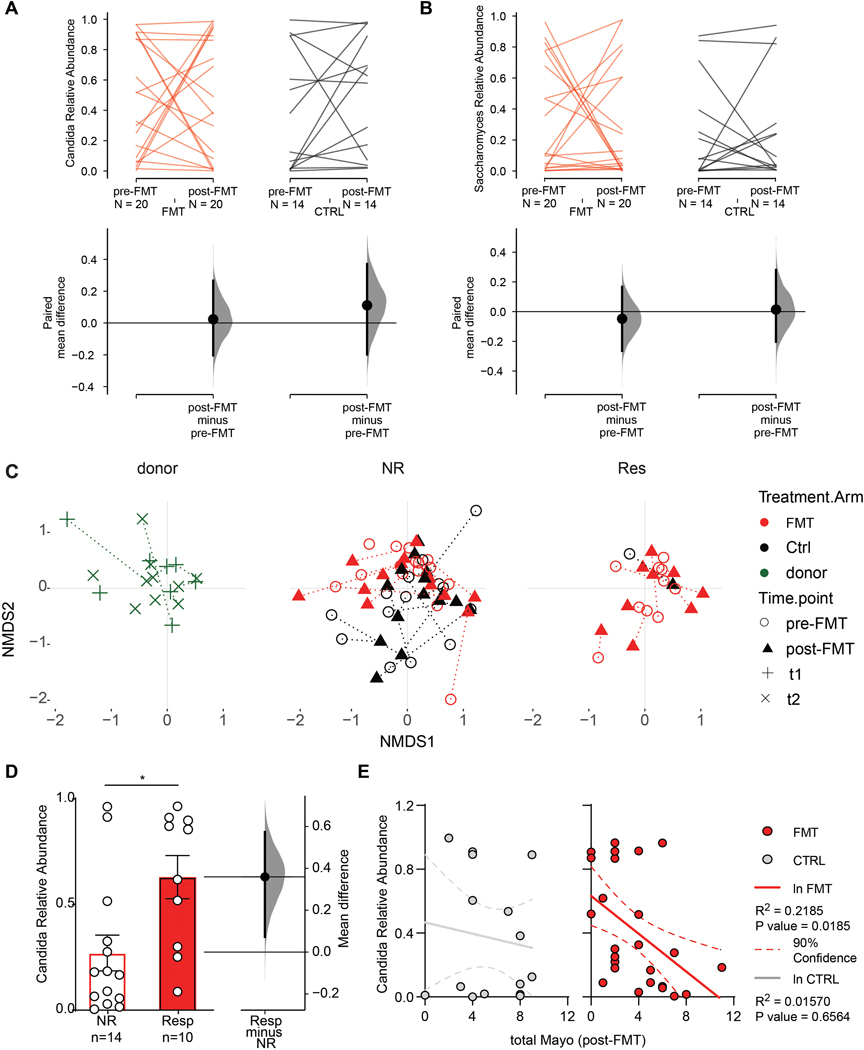
Although the bacterial composition changed post-FMT by both increased observed OTUs and diversity relative to baseline(Paramsothy et al., 2019), the gut mycobiota diversity, the abundance of the two major fungal phyla and observed OTUs all remained unaltered, suggesting that FMT did not affect the overall structure of gut fungal communities in our UC cohort. A deeper analysis of the mycobiota taxonomic composition revealed 15 major fungal genera with Candida and Saccharomyces being the most abundant in both healthy donors and UC patients (Fig 1C, Extended Data Fig. 2 C, D). Within the Candida genus, Candida albicans was the most abundant and prevalent species (Extended Data Fig. 1F). Both Candida and Saccharomyces showed a similar relative abundance between donors and recipients pre-FMT (Extended Data Fig. 2C, D). In contrast to previously reported post-FMT mycobiota outcomes in a CDI cohort (Zuo et al., 2018), multi-donor intensive FMT did not affect the relative abundance of Candida and Saccharomyces post-FMT in the UC recipients (Fig 2 A, B), consistent with major differences between FMT outcomes in these intestinal diseases of different etiology.
Figure 2.
A. Relative abundance of Candida in fecal transplantation recipient (FMT, red), and placebo recipient (CTRL. black) at pre- and post-FMT. The paired mean difference between FMT pre- and post-FMT is 0.0234 [95.0%CI −0.203, 0.266]; P value>0.1 (Wilcoxon test). The paired mean difference between CTRL pre- and post-FMT is 0.111 [95.0%CI −0.198, 0.371]; pValue>0.1 (Wilcoxon test). B. Relative abundance of Saccharomyces in the FMT and CTRL group at pre- and post-FMT. The paired mean difference between FMT pre- and post-FMT is −0.048 [95.0%CI −0.264, 0.167]; pValue>0.1 (Wilcoxon test). The paired mean difference between CTRL pre- and post-FMT is 0.0136 [95.0%CI −0.203, 0.28]; pValue>0.1 (Wilcoxon test). C. Non metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarities of the mycobiota community at different time points in the donor, Endoscopic non-responders (NR) and Endoscopic responders (Res). D. Pre-FMT Relative abundance of Candida among FMT recipient experiencing clinical response (Resp) and non-responders (NR). Two-sided P value, Mann-Whitney T test, * pValue= 0.0185. E. Correlation between the pre-FMT relative abundance of Candida and the total Mayo Index post-FMT in placebo (CTRL) and FMT recipient (FMT). Lines represent linear regression for the placebo and FMT groups. Lower and upper hinges represent the 90% confidence bands. A,B,E. Cumming estimation plot of the paired mean difference, raw data is plotted on the upper panel; each set of paired observations is connected by a line. Lower panel: each paired mean difference is plotted as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. Effect size [CI width lower bound; upper bound]
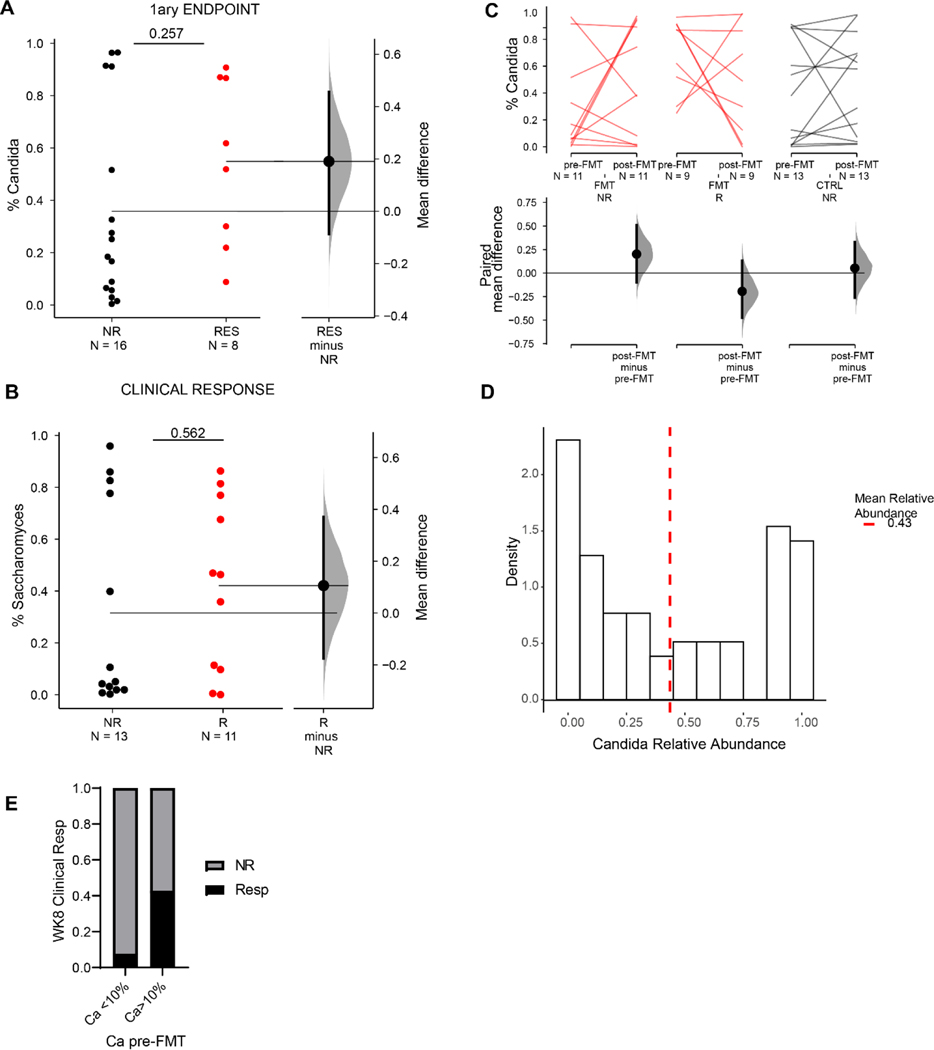
As the pre-existing intestinal landscape can dramatically affect the outcome of FMT(Ericsson et al., 2017; Zuo et al., 2018), we next assessed the composition of the gut mycobiota pre-FMT and analyzed correlations with the clinical outcomes post-FMT therapy. While FMT responders and non-responders did not cluster separately based on overall taxonomic composition (Fig 2C), clinical FMT response was associated with significantly higher levels of Candida in pre-FMT samples compared to samples from FMT non-responder recipients (Fig 2D).
These findings were corroborated by a significant negative correlation between the pre-FMT relative abundance of Candida and the total Mayo Score post-FMT in the FMT recipients but not in the placebo controls (Fig 2 E). Similarly, higher levels of Candida were observed in endoscopic responders with respect to non-responders (Extended Data Fig. 4A, B) and in patients achieving the primary endpoint (Extended Data Fig. 4A), although the difference did not reach statistical significance.
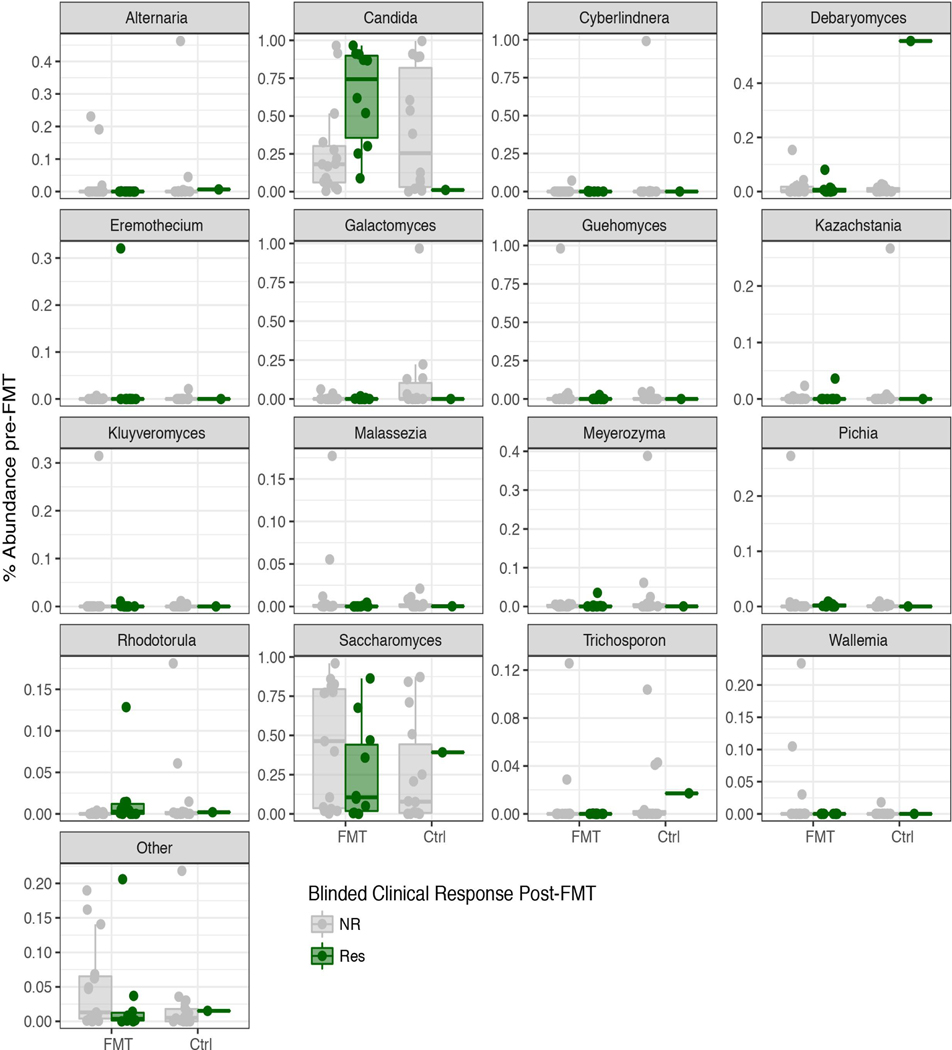
The pre-FMT difference between responders and non-responders was specific to Candida species as Saccharomyces and other less abundant genera were similar between the two groups (Extended Data Fig. 3, & 4C). Altogether, these findings suggest that preexisting intestinal Candida colonization is associated with positive clinical outcome of FMT.
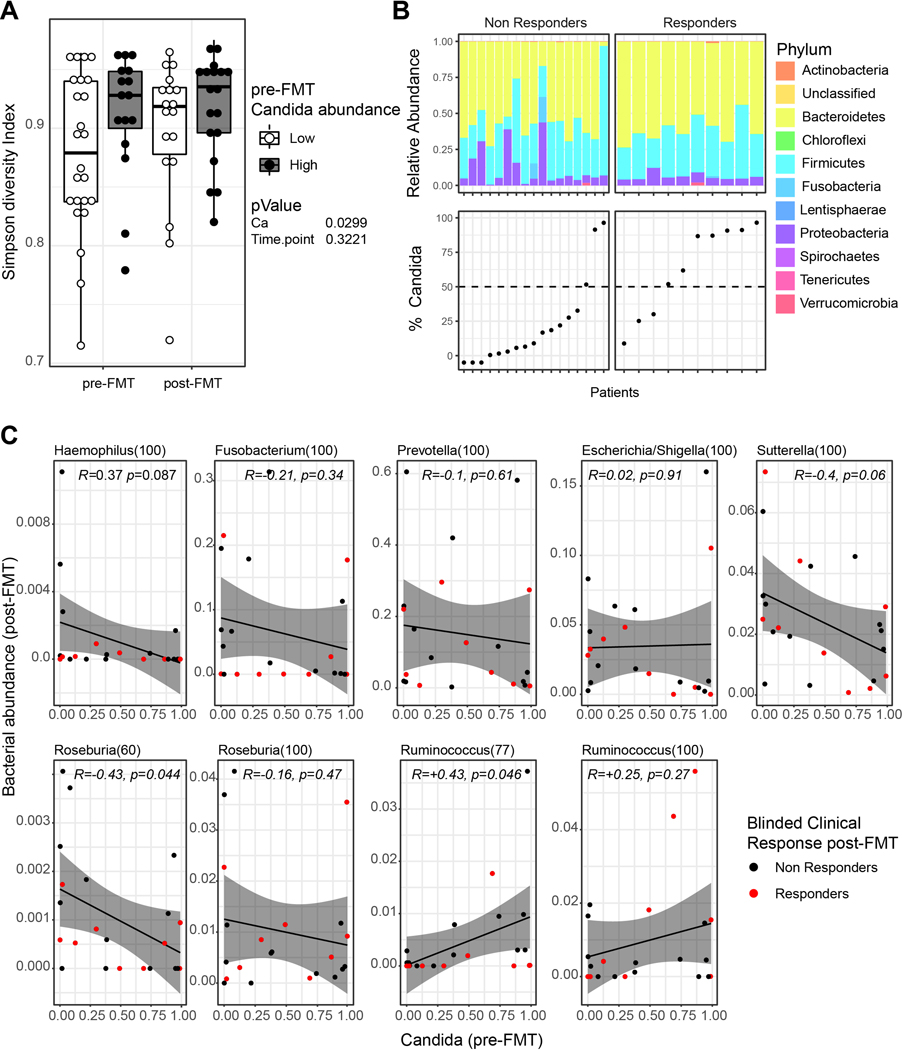
Candida species presence and expansion has been consistently observed in IBD patients (Chehoud et al., 2015b; Davidson et al., 2019; Hoarau et al., 2016; Limon et al., 2019; Sokol et al., 2017) and can negatively affect disease outcome in genetically susceptible host during experimental colitis(Jawhara et al., 2008; Leonardi et al., 2018). On the other hand, environmental competition and bacteria-dependent induction of a host response against intestinal Candida (Fan et al., 2015) have both been shown to eradicate this opportunistic fungus in mice. We thus hypothesized that in clinical responders, FMT might reduce the intestinal burden of Candida, thus attenuating its pro-inflammatory effects. Alternatively, Candida might directly affect bacterial expansion or engraftment and thus influence the outcome of FMT, as previously shown for specific bacteria (Paramsothy et al., 2019). To investigate the possible interaction between Candida species and the bacterial microbiota, we analyzed the bacterial composition of the trial participants stratifying pre-FMT subjects by Candida species abundance (Fig 3A, B, Extended Data Fig. 4D, 4E). Notably, UC patients with high levels of Candida pre-FMT, had significantly higher bacterial alpha diversity that persisted at week 8 post-treatment irrespective of FMT treatment (Fig 3A,B), suggesting that Candida might contribute to or associate with the establishment of a niche more permissive to FMT engraftment.
Figure 3. Changes in bacterial communities and association with fecal Candida levels.
A. Simpson diversity index of 16s rRNA batches in patients at pre- and post-FMT. Patients were grouped based on the relative abundance of Candida (Ca) reads among fungal ITS reads pre-FMT. Low and high samples were defined as samples with a Candida relative abundance smaller and higher than the average Candida relative abundance (see also Figure S 4D). Two way ANOVA. Lower and upper hinges correspond to the 25th and 75th percentiles, dots represent individual samples. B. Upper panel: Relative abundance of bacterial phyla among 16s rRNA reads in FMT recipients pre-FMT. Lower panel: Relative Candida abundance in FMT recipients pre-FMT (showed as % of total ITS reads). C. Pearson correlation between bacterial genera in FMT recipients post-FMT and Candida abundance pre-FMT; grey band represent 95% confidence interval. Dots represent individual samples.
We next assessed whether bacteria found to be associated with FMT outcome would correlate with the initial abundance of Candida. We have previously reported that, Haemophilus, Fusobacterium, Prevotella, and Sutterella genera correlated with a negative FMT outcome (Paramsothy et al., 2019). Among these genera, both Haemophilus and Sutterella negatively correlated with the abundance of Candida (Pearson correlation, Fig. 3C). In contrast, among the genera previously reported to positively correlate with FMT outcome, we found that Roseburia negatively correlated with Candida abundance, whereas Ruminococcus displayed a significant positive correlation with Candida abundance. Our results suggest the existence of complex trans-kingdom relationship where Candida might be associated with or directly affect the engraftment of particular bacterial strains.
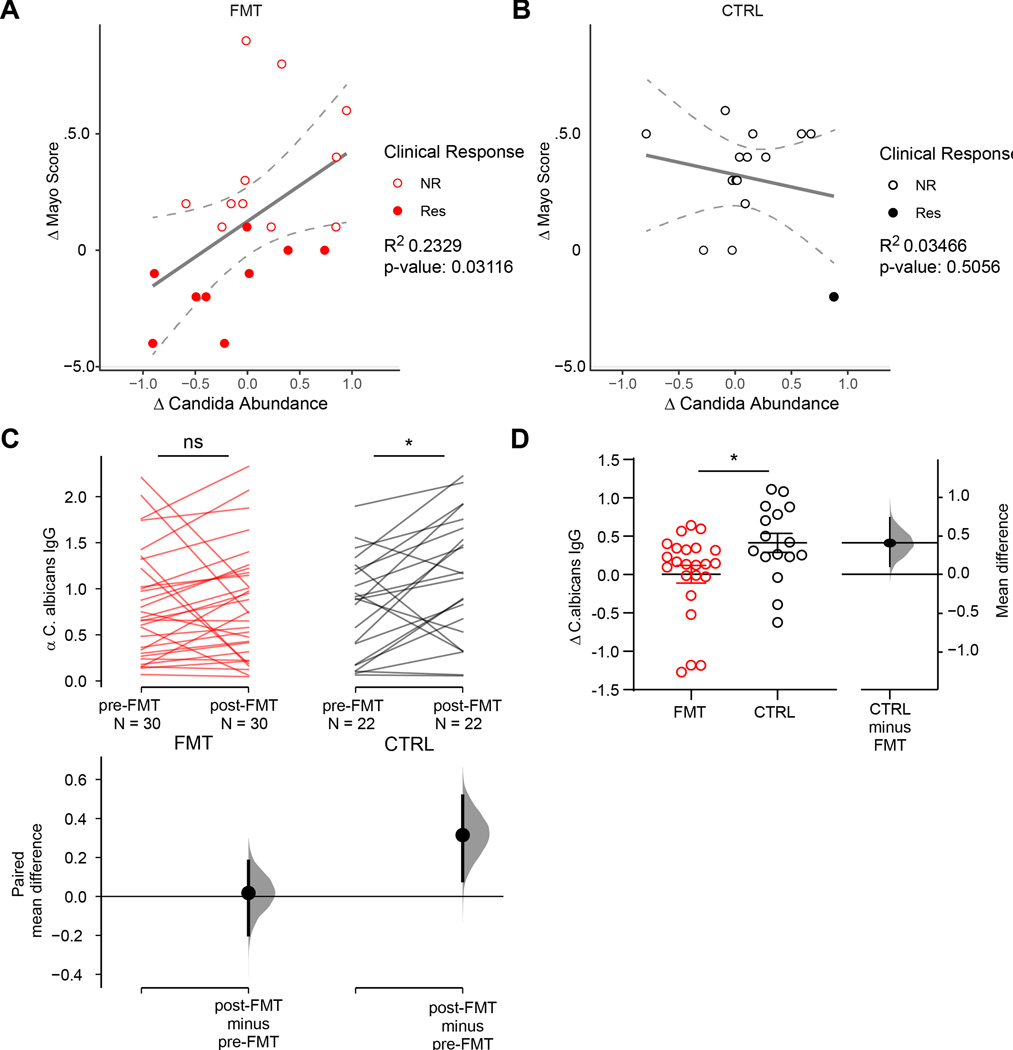
Candida spp. have been associated with intestinal inflammation and experimental studies have shown that several Candida species can worsen the outcome of intestinal inflammation in mouse models (Hoarau et al., 2016; Iliev et al., 2012; Jawhara et al., 2008; Leonardi et al., 2018; Sovran et al., 2018). We thus assessed the changes in Candida abundance following treatment in FMT and placebo recipients. In FMT recipients the reduction in Candida abundance correlated positively with a reduction of endoscopic and clinical disease severity as indicated by change in Mayo score (Fig 4A). This correlation was only observed in FMT recipients and not in placebo controls (Fig 4B), suggesting that a reduction in disease severity does not cause the reduction of Candida abundance. These result support the hypothesis that FMT might act at least partially by reducing the abundance of Candida in the gut. Candida species have the potential to induce strong pro-inflammatory immune responses during intestinal disease (Hoarau et al., 2016; Iliev et al., 2012; Jawhara et al., 2008; Leonardi et al., 2018; Sovran et al., 2018). In our cohort, Candida albicans was the most prevalent and abundant Candida species (Extended Data Fig. 1F). We thus assessed the effect of FMT on the induction of immune response against Candida by assessing the levels anti-C. albicans IgG antibodies in the patents’ serum. Over 8 weeks, placebo recipients experienced a significant increase in anti-Candida IgG responses (Fig 4C–D). In contrast, anti-C. albicans IgG titers were stable in FMT recipients (Fig 4C–D) during the same period. Collectively, these data suggest that FMT might act by reducing Candida abundance and containing pro-inflammatory immune responses induced by gut-fungi during intestinal inflammation.
Figure 4. A reduction in Candida abundance and associated immune responses correlate with clinical and endoscopic response in FMT patients.
A. Correlation between the change in relative abundance of Candida following FMT and the change in total Mayo Index following FMT in FMT recipient (FMT). Lines represent linear regression for the placebo and FMT groups. Lower and upper hinges represent the 90% confidence bands. B. Correlation between the change in relative abundance of Candida following FMT and the change in total Mayo Index following FMT in placebo (CTRL) patients. Lines represent linear regression for the placebo and FMT groups. Lower and upper hinges represent the 90% confidence bands. C. The paired mean difference for the anti C. albicans IgG in FMT and control patients before and after FMT shown as a Cumming estimation plot. The raw data is plotted on the upper axes; each paired set of observations is connected by a line. On the lower axes, each paired mean difference is plotted as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The paired mean difference between pre- and post-FMT in FMT recipients is 0.0175 [95.0%CI −0.198, 0.181], pValue= 0.141 (Wilcoxon test). The paired mean difference between pre- and post-FMT in Placebo recipients (CTRL) is 0.315 [95.0%CI 0.0802, 0.516], pValue= 0.0186 (Wilcoxon test). D. Variation of anti C. albicans IgG in FMT and placebo (CTRL) recipients between pre- and post- treatment. The unpaired mean difference between FMT and Placebo is 0.41 [95.0%CI 0.109, 0.738], pValue= 0.0215 (Mann-Whitney test).
Discussion
The positive outcomes of several pilot- and larger-scale clinical studies highlight the increasing promise of FMT for the treatment of diseases with complex etiology such as IBD, IBS, immune checkpoint inhibitor-associated colitis, metabolic, liver and neurological diseases(Allegretti et al., 2019; Imdad et al., 2018; Kakihana et al., 2016; Kang et al., 2017; Paramsothy et al., 2017; Vrieze et al., 2012; Wang et al., 2018; Weingarden and Vaughn, 2017). Our mycobiota analysis of participants in the largest, double blind, placebo-controlled FMT trial in UC to date, revealed a key role of trans-kingdom interactions between intestinal fungi and bacteria that positively affected the clinical outcome of FMT. Among the patients of the FOCUS cohort, we observed considerable heterogeneity in Candida abundance, reflecting the heterogeneity of IBD patient populations reported by previous studies (Chehoud et al., 2015a; Hoarau et al., 2016; Limon et al., 2019; Sokol et al., 2017). Nonetheless, preexisting high Candida abundance in UC patients prior to FMT treatment was associated with increased bacterial diversity and a better therapy outcome. Although the determination of the mechanistic basis for this phenomenon requires further studies, our data suggest that a reduction of intestinal Candida colonization contributes to the positive outcome of FMT on intestinal inflammation. While the exploration of mycobiota in FMT is still in its infancy(Zuo et al., 2018), the concept that gut mycobiota can influence intestinal health and bacteria engraftment has been explored by several studies(Li et al., 2018; Mason et al., 2012; Sovran et al., 2018; Wheeler et al., 2016). Fungi might affect the outcome of FMT in a context-dependent manner. Indeed, in CDI, where C. difficile dominates the intestinal niche, presence of Candida has been associated with a negative outcome of FMT (Zuo et al., 2018).
A growing number of studies support the existence of complex trans-kingdom interactions between fungi and bacteria (Mason et al., 2012; Sokol et al., 2017). In agreement with this notion, our data suggest that fungal and bacterial counterparts of the microbiota are interdependent, and that intestinal Candida colonization correlates with an increased diversity of the bacterial microbiota. This observation supports previous in vivo results demonstrating an increased recovery of bacterial diversity following antibiotic treatment in the presence of intestinal Candida (Mason et al., 2012). Analysis of the bacterial composition in UC patients from the FOCUS cohort, suggests that the presence of Candida might indicate a niche permissive to better bacterial engraftment. In contrast to CDI where FMT is effective in over 90% of the cases, in other diseases, such as IBD, response to FMT treatment appears to be highly heterogeneous and it is currently not possible to predict the clinical efficacy of FMT. Consequently, substantial effort is needed to identify factors that contribute to FMT treatment response that can be used as predictors prior to the initiation. Our findings suggest that mycobiota, host response to these organisms, and specific fungal species might harbor potential predictive value as to the outcome of microbiome-based therapies and should be further explored in future FMT trials.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents may be directed to and will be fulfilled by Lead Contact, Iliyan D. Iliev (iliev@med.cornell.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Patient samples were obtained from a previously reported double-blind, controlled trial of FMT in UC patients(Paramsothy et al., 2017; Paramsothy et al., 2019). Briefly, male and female patients aged 18–75 years were subjected to an initial colonoscopic infusion and then intensive multidonor FMT (32 patients) or placebo enemas (29 patients) 5 days/week for 8 weeks. Fourteen donors were used for the study, with random selection of 3–7 fixed donors for each of the 21 multidonor FMT batches.
Patients were selected based on clinically and endoscopically active UC, with a total Mayo score of 4–10(Paramsothy et al., 2017; Paramsothy et al., 2019). The Mayo endoscopy subscore was 1 or greater and physician’s global assessment subscore was 2 or less. Any disease extent was included except proctitis confined to the distal 5 cm. The following exclusion criteria were used: individuals with indeterminate colitis, major comorbid chronic disease, major food allergy, irritable bowel syndrome, or a history of bowel cancer, those who were pregnant, and patients who had previous gastrointestinal surgery apart from appendectomy more than 3 months before the study. Patients with gastrointestinal infections at study entry, including parasitic and C. difficile infections, were also excluded.
The following drugs were permitted as long as the dose was stable preceding enrolment: oral 5-aminosalicylates (stable dose for 4 weeks); thiopurines and methotrexate (on medication for ≥90 days and dose stable for 4 weeks); and oral prednisone (dose ≤20mg daily and stable for 2 weeks). During the study, patients were maintained on the same dose of 5-aminosalicylate, thiopurine, and methotrexate. For oral prednisone, a mandatory taper of up to 2·5 mg per week was performed, so that patients would be steroid-free by week 8. Rectal therapies were not permitted.
FMT infusions were constituted from the blended homogenized stool of 3–7 unrelated donors to increase microbial heterogeneity. Each patient received all of their FMT infusions from the same donor batch comprising fixed individual donors. This investigator-initiated study was sponsored by the University of New South Wales, approved by the St Vincent’s Hospital Sydney Human Research Ethics Committee (HREC/13/SVH/69), and registered with ClinicalTrials.gov (NCT01896635) and the Australian Therapeutic Goods Administration Clinical Trial Notification Scheme (2013/0523).
METHOD DETAILS
DNA isolation, fungal rDNA amplification, Illumina library generation and sequencing.
Fecal samples from donor and recipients were collected at various time points throughout the trial as reported previously(Paramsothy et al., 2017; Paramsothy et al., 2019). Samples from 24 FMT recipients and 19 placebo recipients were processed (Table 1). DNA for fungal sequencing was isolated from 50–80 mg material following lyticase treatment, bead beating, and processing using the Quick-DNA Fungal/Bacterial Kit (Zymo Research) as described(Li et al., 2018). Presence or absence of fungal DNA was validated in each sample by RT-qPCR. Fungal ITS1–2 regions were amplified using modified primers that include sample barcodes and sequencing adaptors(Li et al., 2018):
Primer sequences:
ITS1F CTTGGTCATTTAGAGGAAGTAA,
ITS2R GCTGCGTTCTTCATCGATGC
Forward overhang:
5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[locus-specific sequence]
Reverse overhang:
5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[locus-specific sequence]
Specifically, ITS amplicons were generated with 35 cycles using Invitrogen AccuPrime PCR reagents (Carlsbad, CA). Amplicons were then used in the second PCR reaction, using Illumina Nextera XT v2 (San Diego, CA) barcoded primers to uniquely index each sample and 2×300 paired end sequencing was performed on the Illumina MiSeq (Illumina, CA). DNA was amplified using the following PCR protocol: Initial denaturation at 94oC for 10 min, followed by 40 cycles of denaturation at 94oC for 30 s, annealing at 55oC for 30 s, and elongation at 72oC for 2 min, followed by an elongation step at 72oC for 30 min.
All libraries were subjected to quality control using qPCR, DNA 1000 Bioanalyzer (Agilent), and Qubit (Life Technologies) to validate and quantitate library construction prior to preparing a Paired End flow cell. Samples were randomly divided among flow cells to minimize sequencing bias. Clonal bridge amplification (Illumina) was performed using a cBot (Illumina) and 2×250 bp sequencing-by-synthesis was performed.
ITS1 fungal reads and 16S bacterial analysis
The generated raw FASTQ ITS1 sequencing data were filtered to enrich for high quality reads, removing the adapter sequence by cutadapt v1.4.1 or any reads that did not contain the proximal primer sequence(Kechin et al., 2017). Sequence reads were then quality-trimmed by truncating reads not having an average quality score of 20 over a 3 base pair sliding window and removing reads shorter than 100 bp(Leonardi et al., 2018). These reads were then aligned to Targeted Host Fungi (THF) ITS1 database, using BLAST v2.2.22 and the pick_otus.py pipeline in the QIIME v1.6 wrapper with an identity percentage ≥97% for OTU picking. The alignment results were subsequently tabulated across all reads, using the accession identifier of the ITS reference sequences as surrogate OTUs and using a Perl script(Tang et al., 2015). Among the analyzed samples, 23 were excluded due to insufficient quality size (ITS reads < 200). Bacterial reads where analyzed as previously described(Paramsothy et al., 2017). The R packages Phyloseq (1.26.1) and Vegan (2.5–5) were used for determining community properties such as Shannon index, Simpson index, Bray-Curtis index, NMDS scaling of Bray-Curtis dissimilarities, and relative abundance at various taxonomic levels. R version 3.5.0 was used.
Detection of anti-Candida antibodies in the human serum
The level of antibodies in the human serum that react with Candida antigens was determined by a sandwich enzyme-linked immunosorbent assay (ELISA). Briefly, to obtain bacterial or fungal lysates for ELISA the overnight Candida albicans SC5314 (ATCC MYA-2876) colonies were prepared through freeze and thawing disruption followed by 5 pulses of sonication(Leonardi et al., 2018). Candida lysates were used as the coating antigen in ELISA assay on high binding flat bottom plates (EIA/RIA 96-Well Plates, Corning). ELISA detection of IgG was carried out using Alk-Phosphatase coupled goat anti-Mouse IgG (Jackson Lab) and PNPP substrate (Life technologies). Samples were read at 405 nm on a microtiter plate reader (Menlo Park, CA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise indicated, statistics were computed using the dabestR package for Estimation Statistic in R(Ho et al., 2019). 5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated. Statistical details of experiments are reported in the figure legends. The P value(s) reported in the figure legends are the likelihood(s) of observing the effect size(s), if the null hypothesis of zero difference is true.
DATA AND CODE AVAILABILITY
The published article includes all datasets generated and analyzed during this study. Fecal ITS rDNA amplicon sequencing data were submitted to the Sequence Read Archive to the NCBI BioProject repository (BioProject ID PRJNA590898). Fecal 16S rDNA amplicon sequencing data were submitted as part of Paramsothy et al(Paramsothy et al., 2017) to the European Nucleotide Archive (accession numbers PRJEB26472, PRJEB26474, PRJEB26473, PRJEB20349 and PRJEB26357).
Extended Data
Extended Data Fig. 1: Mycobiota in the FOCUS cohort A, related to Figure 1.
Numbers of ITS reads recovered for the different patients’ samples at the indicated time-points. B. Non metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarities of the mycobiota community at different time points in the donor (green), fecal transplantation recipient (FMT, red), and placebo recipient (CTRL, black) pre- and post-FMT. C. Alpha diversity measured by observed OTU for the different patients’ samples at the indicated time-points. Relative abundance of Ascomycota (D), Basidiomycota (E), are shown for donor (green), fecal transplantation recipient (FMT, red), and placebo recipient (CTRL. black) pre- and post-FMT. (F) Relative abundance of C. albicans and non-C. albicans (other) Candida species across the analyzed samples. Dots represent individual patients, line and bars represent mean +/− SEM.
Extended Data Fig. 2: Mycobiota composition in UC patients and healthy donors pre-FMT, related to Figure 1.
A. Shannon diversity index, B. Log10 (Ascomycota/Basidiomycota) Ratio. C. Relative abundance of Candida reads among total ITS fungal reads. D. Relative abundance of Saccharomyces reads among total ITS fungal reads. Data are shown as Gardner-Altman estimation plot of the mean differences between groups. Dot represent individual patients. Mean difference is plotted on floating axes on the right as a bootstrap sampling distribution. The mean difference is depicted as a dot; the 95% confidence interval is indicated by the ends of the vertical error bar.
Extended Data Fig. 3: Relative abundance of major genera in the study cohort pre-FMT, Related to Figure 1.
Lower and upper hinges correspond to the 25th and 75th percentiles, dots represent individual samples. FMT: FMT recipients, CTRL: Placebo recipient. Colors denote the Blinded Clinical response post-FMT: NR: non responders, Res: Responders.
Extended Data Fig. 4: Candida and Saccharomyces relative abundance, related to Figure 1 and 2.
A. Relative abundance of Candida reads among total ITS fungal reads. The unpaired mean difference between FMT patients who met the primary endpoint at week 8 (RES) and patients who failed to meet the criteria (NR) is 0.19 [95.0%CI −0.0871, 0.456]. B. Relative abundance of Saccharomyces reads among total ITS fungal reads. The unpaired mean difference between NR and R is 0.106 [95.0%CI −0.176, 0.371]. Data in A, B, are shown as Gardner-Altman estimation plot of the mean differences between groups. Dot represent individual patients. Mean difference is plotted on a floating axes on the right as a bootstrap sampling distribution. The mean difference is depicted as a dot; the 95% confidence interval is indicated by the ends of the vertical error bar. C. Relative abundance of Candida in clinical non-responders (NR) and responders (R) of the FMT (red) and placebo (black) group pre- and post-FMT. The paired mean difference between pre- and post-FMT is 0.202 [95.0%CI −0.1, 0.51] in the FMT_NR group; −0.194 [95.0%CI −0.475, 0.131] in the FMT_R group; 0.0524 [95.0%CI −0.262, 0.33] in the Ctrl_NR group. pValue>0.1 (Wilcoxon test). Two-sided P value, Mann-Whitney T test is shown. D. Density distribution of Candida relative abundance in the analyzed patient samples. In Fig 3. Samples with Candida relative abundance smaller than the geometric mean (< 0.4347454) were defined as “low”. The remaining samples were defined as “high”. E. Ratio of clinical responders versus non-responders among FMT recipients post-FMT among patients with low Candida abundance (<10%) and high Candida abundance (>10%).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alkaline Phosphatase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson Immuno Research | Cat # 115-055-146 |
| Bacterial and fungal Strains | ||
| Candida albicans SC5314 | ATCC | MYA-2876 |
| Biological samples | ||
| Fecal samples from donor and UC FMT and placebo recipient | Paramsothy et al., 2017 | N/A |
| Serum samples from UC FMT and placebo recipient | Paramsothy et al., 2017 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fetal Bovine Serum | Corning | Cat# MT35016CV |
| Lyticase from Arthrobacter luteus | Sigma | Cat# L2524 |
| PNPP (p-nitrophenyl phosphate) | Sigma | Cat# 34045 |
| Sabouraud dextrose broth | VWR | Cat# 89406-400 |
| Critical Commercial Assays | ||
| Quick-DNA Fungal/Bacterial Kit | Zymo Research | Cat# D6007 |
| HighPrep™ PCR Clean-up System | Magbiogenomics | Cat# AC-60050 |
| Kapa BiosystemsSupplier Diversity Partner HIFI HOTSTART READYMIX 500RXN | Fisher Scientific | Cat# KK2602 |
| Nextera XT Index Kit v2 kit A Nextera XT Index Kit v2 Set B | Illumina | Cat# FC-131-200, FC-131-2002 |
| QIAEX II Gel Extraction Kit | Qiagen | Cat# 20021 |
| AccuPrime™ Taq DNA Polymerase, high fidelity | Thermofisher | Cat# 12346086 |
| Oligonucleotides | ||
| ITS1F: CTTGGTCATTTAGAGGAAGTAA, | Li et al., 2018 | N/A |
| ITS2R: GCTGCGTTCTTCATCGATGC | Li et al., 2018 | NA |
| Nextera Forward overhang: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[locus-specific sequence] | Illumina | N/A |
| Nextera Reverse overhang: 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[locus-specific sequence] | Illumina | NA |
| Deposited Data | ||
| 16S sequencing | Paramsothy et al., 2017 | European Nucleotide Archive, https://www.ebi.ac.uk/ena: PRJEB26472, PRJEB26474, PRJEB26473, PRJEB20349 and PRJEB26357 |
| ITS Sequencing | This paper | NCBI BioProject repository, https://www.ncbi.nlm.nih.gov/bioproject/: PRJNA590898 |
| Software and Algorithms | ||
| GraphPad Prim 8.3.0 (538) | GraphPad Software | N/A |
| RStudio Desktop 1.2.1335 | R Studio | https://rstudio.com/ |
| QIIME v1.6 | QIIME | www.qiime.org |
| R version 3.5.0 (2018-04-23) | R | www.r-project.org |
| Phyloseq v1.26.1 | Phyloseq | https://bioconductor.org/packages/release/bioc/html/phyloseq.html |
| Vegan v2.5-5 | Vegan | https://cran.r-project.org/web/packages/vegan/ |
| Estimation Stats DabestR v0.2.2 | Ho et al, 2019 | https://github.com/ACCLAB/dabestr |
Acknowledgements
Research in the Iliev laboratory is supported by US National Institutes of Health (DK113136 and AI137157), the Crohn’s and Colitis Foundation Senior Research Award, the Helmsley Charitable Trust, the Irma T. Hirschl Career Scientist Award, Pilot Project Funding from the Center for Advanced Digestive Care (CADC), and the JRI for Research in IBD. J.F.C. is supported by cutting-edge therapies for ulcerative colitis grant; H.M.M. is supported by Litwin IBD pioneer award from Crohn’s & Colitis Foundation; I.L. is supported by fellowships from the Crohn’s and Colitis Foundation.
Footnotes
Declaration of Interests
T.J.B. has a pecuniary interest in the Centre for Digestive Disease and holds patents in FMT treatment. The other authors declare no competing interests related to this study.
References
- Allegretti JR, Mullish BH, Kelly C, and Fischer M. (2019). The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. The Lancet 394, 420–431. [DOI] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, and Wu GD (2015a). Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 21, 1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, and Wu GD (2015b). Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis 21, 1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, et al. (2019). Effect of Fecal Microbiota Transplantation on 8-Week Remission in Patients With Ulcerative Colitis: A Randomized Clinical Trial. JAMA 321, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson K, Brancato A, Heetderks P, Mansour W, Matheis E, Nario M, Rajagopalan S, Underhill B, Wininger J, and Fox D. (2019). Outbreak of Electronic-Cigarette-Associated Acute Lipoid Pneumonia - North Carolina, July-August 2019. MMWR Morb Mortal Wkly Rep 68, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Personett AR, Turner G, Dorfmeyer RA, and Franklin CL (2017). Variable Colonization after Reciprocal Fecal Microbiota Transfer between Mice with Low and High Richness Microbiota. Front Microbiol 8, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, and Koh AY (2015). Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 21, 808–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S, Rossen NG, van der Spek MJ, Hartman JHA, Huuskonen L, Korpela K, Salojärvi J, Aalvink S, de Vos WM, D’Haens GR, et al. (2017). Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. The ISME Journal 11, 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Tumkaya T, Aryal S, Choi H, and Claridge-Chang A. (2019). Moving beyond P values: data analysis with estimation graphics. Nat Methods 16, 565–566. [DOI] [PubMed] [Google Scholar]
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, et al. (2016). Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 7, e01250–01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imdad A, Nicholson MR, Tanner-Smith EE, Zackular JP, Gomez-Duarte OG, Beaulieu DB, and Acra S. (2018). Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database of Systematic Reviews CD012774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhara S, Thuru X, Standaert-Vitse A, Jouault T, Mordon S, Sendid B, Desreumaux P, and Poulain D. (2008). Colonization of Mice by Candida albicans Is Promoted by Chemically Induced Colitis and Augments Inflammatory Responses through Galectin-3. J Infect Dis 197, 972–980. [DOI] [PubMed] [Google Scholar]
- Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, Mimura I, Morita H, Sugiyama D, Nishikawa H, et al. (2016). Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood 128, 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D-W, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, et al. (2017). Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechin A, Boyarskikh U, Kel A, and Filipenko M. (2017). cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J Comput Biol 24, 1138–1143. [DOI] [PubMed] [Google Scholar]
- Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, et al. (2018). CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, Kim Y, Kabata H, Artis D, Fiers WD, et al. (2018). Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host & Microbe 24, 847–856.e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XV, Leonardi I, and Iliev ID (2019). Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 50, 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. (2019). Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 25, 377–388 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason KL, Downward JRE, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, and Huffnagle GB (2012). Candida albicans and Bacterial Microbiota Interactions in the Cecum during Recolonization following Broad-Spectrum Antibiotic Therapy. Infection and Immunity 80, 3371–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, and Lee CH (2015). Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 149, 102–109.e106. [DOI] [PubMed] [Google Scholar]
- Panpetch W, Somboonna N, Palasuk M, Hiengrach P, Finkelman M, Tumwasorn S, and Leelahavanichkul A. (2019). Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLOS ONE 14, e0210798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, Bogaerde J.v.d., Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. (2017). Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. The Lancet 389, 1218–1228. [DOI] [PubMed] [Google Scholar]
- Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D, et al. (2019). Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis. Gastroenterology 156, 1440–1454.e1442. [DOI] [PubMed] [Google Scholar]
- Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, and Iqbal TH (2017). Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Alimentary Pharmacology & Therapeutics 46, 479–493. [DOI] [PubMed] [Google Scholar]
- Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovran B, Planchais J, Jegou S, Straube M, Lamas B, Natividad JM, Agus A, Dupraz L, Glodt J, Da Costa G, et al. (2018). Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 6, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Iliev ID, Brown J, Underhill DM, and Funari VA (2015). Mycobiome: Approaches to analysis of intestinal fungi. Journal of Immunological Methods 421, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvede M, and Rask-Madsen J. (1989). Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. The Lancet 333, 1156–1160. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al. (2013). Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. New England Journal of Medicine 368, 407–415. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Nood EV, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, Dallinga–Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. (2012). Transfer of Intestinal Microbiota From Lean Donors Increases Insulin Sensitivity in Individuals With Metabolic Syndrome. Gastroenterology 143, 913–916.e917. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, Jiang Z-D, Abu-Sbeih H, Sanchez CA, Chang C-C, et al. (2018). Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med 24, 1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden AR, and Vaughn BP (2017). Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 8, 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. (2016). Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 19, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, et al. (2018). Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat Commun 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets generated and analyzed during this study. Fecal ITS rDNA amplicon sequencing data were submitted to the Sequence Read Archive to the NCBI BioProject repository (BioProject ID PRJNA590898). Fecal 16S rDNA amplicon sequencing data were submitted as part of Paramsothy et al(Paramsothy et al., 2017) to the European Nucleotide Archive (accession numbers PRJEB26472, PRJEB26474, PRJEB26473, PRJEB20349 and PRJEB26357).