Abstract
Marine sponges often house small molecule producing symbionts extracellularly in their mesohyl, providing the host with means of chemical defense against predation and microbial infection. Here, we report an intriguing case of chemically mediated symbiosis between the renieramycin-containing sponge Haliclona sp. and its herein discovered renieramycin producing symbiont Candidatus Endohaliclona renieramycinifaciens. Remarkably, Ca. E. renieramycinifaciens has undergone extreme genome reduction where it has lost almost all necessary elements for free living while maintaining a complex, multi-copy-plasmid encoded biosynthetic gene cluster for renieramycin biosynthesis. In return, the sponge houses Ca. E. renieramycinifaciens in previously uncharacterized cellular reservoirs (chemobacteriocytes), where it can acquire nutrients from the host and avoid bacterial competition. This relationship is highly specific to a single clade of Haliclona sponges. Our study reveals intracellular symbionts as an understudied source for defense chemicals in the oldest living metazoans, and paves the way towards discovering similar systems in other marine sponges.
Marine sponges are among the richest metazoan hosts of microbial diversity. They are considered to be ancestral organisms, and are a good model system for studying the evolution of microbe-host interactions1,2. Having evolved mechanisms to be recognized as symbiotic partners instead of food3, or to avoid host detection altogether4, most sponge symbionts are housed in their mesohyl matrix. These symbionts include those that provide nutrition for the host and help with waste recycling5,6, provide structural elements such as calcium or phosphorus7,8, and produce potent molecules as defense chemicals against predators and infectious agents. The latter is best exemplified by the filamentous bacterial symbiont of Theonella sponges, Candidatus Entotheonella sp., which produces numerous complex metabolites with different biological activities9,10, and the cyanobacterial symbiont of Dysidea sponges, Hormoscilla spongeliae, which produces several halogenated toxins11. These two remarkable examples, and the fact that more than 8000 complex small molecules have been isolated from marine sponges suggest that microbiome-derived biosynthesis of sponge metabolites is a widespread phenomenon that is largely unexplored12.
Examples of such understudied sponge metabolites include the renieramycins: a group of tetrahydroisoquinoline quinones (THQs) originally isolated from marine sponges of the genus Haliclona (previously known as Reniera), and subsequently from Xestospongia and Neopetrosia sponges13–17. Approximately 30 renieramycins have been discovered to date, exerting a wide range of antimicrobial and cytotoxic activities that are relevant both ecologically and therapeutically13–16. These biological activities suggest that the renieramycins may have defensive or competitive allelopathic roles that benefit the sponge host, although further studies are needed to fully understand their ecological role18,19. From a biosynthetic point of view, much less is known about the renieramycins. The renieramycin core structure resembles that of the saframycin group of THQs, members of which are produced by disparate bacterial origins: free living terrestrial Myxobacteria and Actinobacteria (the saframycins)20–22, free living Proteobacteria (the safracins)23,24, and obligate symbionts of marine ascidians (the ecteinascidins, which include the recently approved anticancer drug ET743, or trabectedin)25–28. Intrigued by the widespread nature of this group of molecules, we set out to identify the source and molecular bases of renieramycin production in the most primitive metazoans: marine sponges.
RESULTS
Discovery of the renieramycin biosynthetic gene cluster
Because of the structural similarity between the renieramycins and other THQs, we hypothesized that they are biosynthesized by a bacterial symbiont of Haliclona sponges through a nonribosomal peptide synthetase (NRPS) pathway. To test this hypothesis, we studied four renieramycin containing Haliclona sponge samples from different years of collection and geographical locations in the Tropical Pacific Ocean: Ren-Pal-02 (Palau, 2002), Ren-PNG-07060, Ren-PNG-07113 (Papua New Guinea, 2007), and Ren-Bali-16–03 (Bali, 2016) (Fig. 1). First, we used chemical analysis, High Performance Liquid Chromatography coupled with High Resolution Tandem Mass Spectrometry (HPLC-HR-MS/MS), and Nuclear Magnetic Resonance (NMR) to verify the presence of renieramcyins in these sponges. In all samples, renieramycin E was the most prominent derivative, accompanied by the typical renieramycin degradation products renierone and N-formyl dehydrorenierone (Fig. 1c, Supplementary Fig. 1, and Supplementary Fig. 2). Second, we molecularly identified the four sponges (collected as Reniera or Haliclona based on their morphology) by amplifying and sequencing ~500 bps of their 18S rRNA gene. As expected, the four sequences were 99.9% identical to each other and to previously characterized Haliclona sponges.
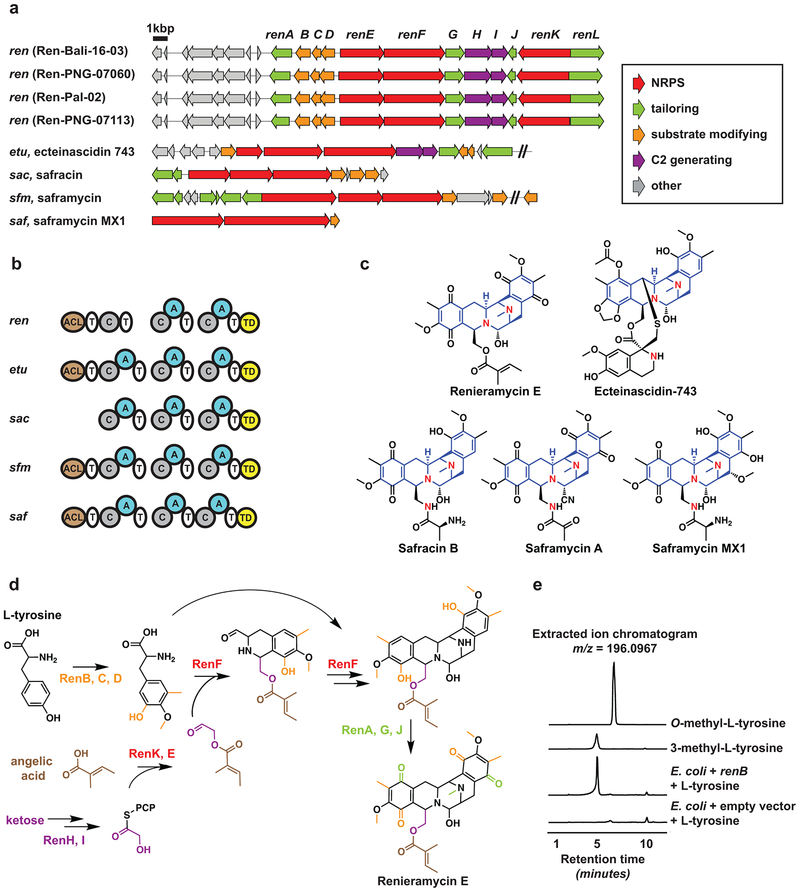
Figure 1. Chemistry of Haliclona sponges.
(a) Blue Haliclona sponges from Papua New Guinea (top) and Bali (bottom): sources of the renieramycins. (b) Chemical structures of renieramycin natural products previously reported from sponges13–16. Renieramycins A-J share the same core structure and vary only in five positions (R1-R5), which are indicated for each molecule below the drawn structure. (c) Extracted ion chromatogram of renieramycin E, m/z 549.2237 (M+H-H2O)+, the major renieramycin derivative from the four Haliclona sponge extracts reported in this study. Chemical analysis of the four sponges was repeated twice, and produced the same results.
Having confirmed the presence of renieramycins in our Haliclona sponges, we undertook an unbiased strategy towards identifying the renieramcyin biosynthetic gene cluster (BGC) in their microbiome. We isolated metagenomic DNA from the four sponges, and subjected them to deep metagenomic sequencing using Illumina (30–110 M paired-end reads each of 141 or 175 bps in length, See Methods and Supplementary Table 1). Several rounds of assembly yielded major scaffolds with lengths up to ~700 Kbps. To identify the renieramycin BGC, we took two approaches: a targeted one, where we used tBLASTn to search the final assemblies (scaffolds > 5 Kbps) of the four metagenomes for homologs of the saframycin Mx1 NRPSs SafA and SafB29, and an untargeted one, where we analyzed them using antiSMASH (a specialized tool for automatic identification of small molecule BGCs)30. Remarkably, this analysis revealed that the four assembled Haliclona metagenomes have very limited biosynthetic capacity, unlike previously studied sponge metagenomes10,31. Overall, antiSMASH detected a single BGC from each of Ren-Pal-02, Ren-PNG-07060, Ren-PNG-07113 and three BGCs from Ren-Bali-16–03 (Supplementary Fig. 3). Only one BGC was common to the four metagenomes: NRPS-1, which was found on a ~33 Kbps scaffold with >99% DNA sequence identity between the samples. Interestingly, the same BGC was also retrieved using the targeted saframycin Mx1-based search strategy, and encodes for a complete saframycin-like BGC: ren (Fig. 2a).
Figure 2. Renieramycin biosynthesis.
(a) Renieramycin BGCs (ren) discovered from the four Haliclona sponges in this study (top), and previously characterized THQ BGCs from other microorganisms (bottom). (b) Comparison of the nonribosomal peptide synthetase domain architecture between ren and related BGCs: ACL: Acyl-Coenzyme A Ligase, A: Adenylation, C: Condensation, T: Thiolation, TD: Terminal reductase. (c) Chemical structures of the products encoded by the BGCs in b, showing the common pentacyclic core of the molecules in blue. Note that renieramycin E is one amino acid shorter than the typical molecules in this class: safracins and saframycins (Nitrogen atoms of individual amino acids are shown in red), which agrees with ren missing the first A domain. (d) Proposed biosynthesis of renieramycin E based on characterized homologs from this study and previous ones. (e) Extracted ion chromatograms (HPLC-HR-MS) monitored at m/z = 196.0967 for the following samples (from top to bottom): an authentic standard of O-methyl-L-tyrosine, an authentic standard of 3-methyl-L-tyrosine, an organic extract generated from the supernatant of E. coli cells expressing renB and supplemented with L-tyrosine, an organic extract generated from the supernatant of E. coli cells harboring an empty vector and supplemented with L-tyrosine. This experiment was repeated three independent times, each in a triplicated setup, and produced the same results.
Analogous to previously characterized THQ BGCs, ren encodes all of the enzymes necessary for the formation of the pentacyclic core of the molecule as well as substrate-modifying and tailoring enzymes, allowing us to propose a biosynthetic scheme for the renieramycins (Fig. 2, Supplementary Table 2, and Supplementary Note). To unequivocally demonstrate that ren encodes for the renieramycins, we cloned and sought to heterologously express the entire ~25 Kbps BGC in Escherichia coli. Several attempts under different conditions and using additional genes (e.g., a phosphopantetheinyl transferase and an MbtH-like protein, see Methods) were however unsuccessful in establishing recombinant renieramycin production. We therefore turned into the characterization of a substrate modifying enzyme that is specific to THQ pathways. ren encodes three such enzymes: the methyltransferases RenB and RenC, and the peroxygenase RenD, homologs of which from other THQ pathways are responsible for converting L-tyrosine to the essential NRPS substrate 3-hydroxy-5-methyl-O-methyl-L-tyrosine32,33. A phylogenetic analysis of THQ methyltransferases placed RenB in a clade of L-tyrosine C(3)-methyltransferases, and RenC in a clade of L-tyrosine O-methyltransferase (Supplementary Fig. 4). To provide experimental support for this prediction, we cloned renB and heterologously expressed it in E. coli cultures supplemented with L-tyrosine. We then compared the organic extracts of culture supernatants from renB expression lines to those of E. coli harboring empty vectors using HPLC-HR-MS/MS. Indeed, in three triplicated experiments, only E. coli cultures expressing renB and not controls produced 3-methyl-L-tyrosine, as confirmed by comparison to an authentic standard (Fig. 2e). These results confirm the predicted function of RenB in providing a THQ specific substrate, and provide biochemical evidence for the involvement of ren in renieramycin biosynthesis.
Discovery of the renieramycin producer
After the discovery of ren, which is evidently of bacterial origin, we sought to identify the member of the microbiome whose genome harbors it, and is therefore responsible for renieramycin production. To achieve this goal, we revisited our metagenomic assemblies in an attempt to connect ren to a bacterial chromosome or a taxonomic marker. However, this task proved unsuccessful despite several iterations of assemblies. In all four metagenomes, our assemblies consistently produced ren on ~ 33 Kbps scaffold despite its high coverage (232–335 × coverage). We then wondered if ren exists on an extrachromosomal element. Indeed, analysis of paired-end reads spanning the ~33 Kbps scaffolds proved that they are joined end to end, and that ren exists on a small bacterial plasmid in all four metagenomes: p-ren (Fig. 3). Although very interesting from an evolutionary stand point, this fact presented us with a technical challenge to unequivocally associate ren to specific members of the Haliclona microbiome.
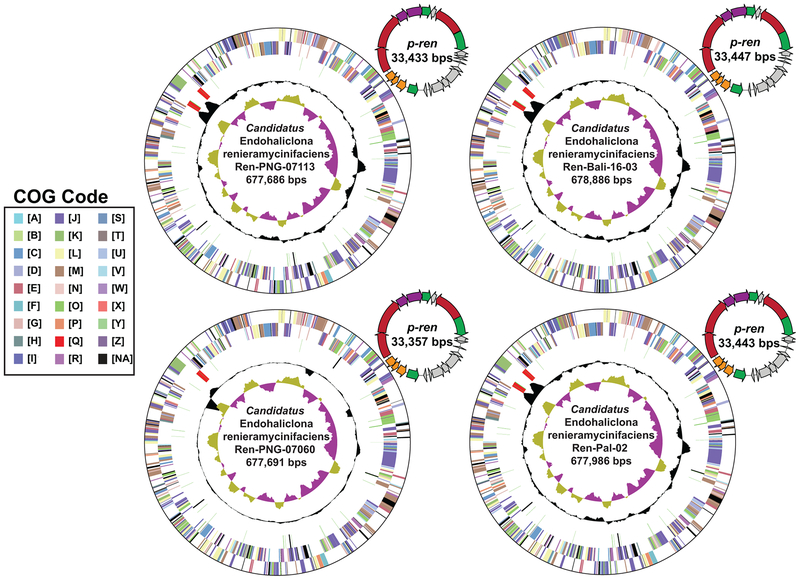
Figure 3. Candidatus Endohaliclona renieramycinifaciens genomes and plasmids.
Assembled circular chromosomes of Ca. E. renieramycinifaciens from the four sponge metagenomes and the corresponding renieramycin gene cluster containing plasmids, p-ren. Concentric rings (from outside to inside) indicate genes on the forward frame, genes on the reverse frame, RNAs, GC content and GC skew. Genes are classified according to general Cluster of Orthologous Groups (COG) categories in IMG (A: RNA processing and modification, B: Chromatin structure and dynamics, C: Energy production and conversion, D: Cell cycle control, cell division, chromosome partitioning, E: Amino acid transport and metabolism, F: Nucleotide transport and metabolism, G: Carbohydrate transport and metabolism, H: Coenzyme transport and metabolism, I: Lipid transport and metabolism, J: Translation, ribosomal structure and biogenesis, K: Transcription, L: Replication, recombination and repair, M: Cell wall/membrane/envelope biogenesis, N: Cell motility, O: Posttranslational modification, protein turnover, chaperones, P: Inorganic ion transport and metabolism, Q: Secondary metabolites biosynthesis, transport and catabolism, R: General function prediction only, S: Function unknown, T: Signal transduction mechanisms, U: Intracellular trafficking, secretion, and vesicular transport, V: Defense mechanisms, W: Extracellular structures, X: Mobilome, prophages, transposons, Y: Nuclear structure, Z: Cytoskeleton, NA: Not Assigned). The color code of genes in p-ren follows the same key in Fig. 2.
To identify the bacterium that harbors p-ren, we undertook two independent approaches: a computational one and an experimental one. First, based on the extreme sequence conservation between p-ren in the four metagenomes, we hypothesized that the genomes of the bacterium harboring it would also be conserved and present at a similar coverage. To test this hypothesis, we identified all scaffolds that are in common between the four metagenomes by mapping the reads from Ren-PNG-07060, Ren-Pal-02, and Ren-PNG-07113 to the assembled scaffolds of Ren-Bali-16–03. Only 43 of 6013 scaffolds (7 %) are shared between the Ren-Bali-16–03 metagenome and the other three. Satisfyingly, 40 of these scaffolds originated from the sponge genome, one corresponded to the sponge mitochondrion (a circularized 18,605 bps), one corresponded to p-ren, and only the final one appeared to be of an unknown prokaryotic origin (Ren-Bali-16–03-NODE_1, ~680 Kbps). Ren-Bali-16–03-NODE_1 had three notable features: a) its matching scaffolds in the four samples had an average pairwise nucleotide identity of 99.73 – 99.98 %, b) the coverage ratio of this scaffold to p-ren is 1:4.4, 1:3, 1:1.7, and 1:2.6 for Ren-PNG-07060, Ren-PNG-07113, Ren-Pal-02, and Ren-Bali-16–03, respectively (calculated by computing their RPKM values: number of mapped Reads Per Kbps per Million of sequenced reads), and c) its GC content is similar to that of p-ren (38% versus 34%, respectively). Remarkably, paired-end read analysis circularized Ren-Bali-16–03-NODE_1 and its corresponding scaffolds into complete bacterial chromosomes of an extremely small size (~680 Kbps) (Fig. 3). Taken together, these results show that only one bacterial chromosome is consistently found in p-ren containing samples at a sequence conservation level, coverage, and GC content similar to that of p-ren, motivating us to hypothesize that these two genetic elements are harbored by the same bacterial cell. We propose to name this bacterium Candidatus Endohaliclona renieramycinifaciens.
To test this hypothesis experimentally, we wondered whether the two elements can be physically co-localized. Inspired by elegant studies in Theonella sponges10, we gently homogenized fresh tissues of Ren-Bali-16–03 and subjected them to flow cytometry and cell sorting, guided only by particle size (Supplementary Fig. 5). Overall, we sorted the homogenate into 8 partitions (P3–P10), isolated DNA from each, and performed high-throughput 16S rRNA gene amplicon sequencing on all of them and metagenomic sequencing on a selected subset that had widely varying levels of Ca. E. renieramycinifaciens (P4, and P7–P10) (Fig. 4a and Supplementary Table 1). Surprisingly, relative abundance of the 16S rRNA gene sequence of Ca. E. renieramycinifaciens was most enriched in the last partition (P10), which contained the largest particle size of the sponge homogenate. This enrichment is corroborated in the metagenomic sequencing data, and corresponds to an increase in the coverage of both the Ca. E. renieramycinifaciens chromosome and p-ren (Fig. 4b, c and Supplementary Table 3). As expected, a covariance analysis between the coverage of the Ca. E. renieramycinifaciens chromosome and p-ren in the five partition metagenomes revealed a strong and positive correlation: Pearson correlation coefficient of 0.99, p-value = 4.08 × 10−07. This covariance is maintained at an average RPKM ratio of 1:2.4 (Ca. E. renieramycinifaciens chromosome: p-ren), which agrees with their ratio in the full Ren-Bali-16–03 metagenome (1:2.6). In addition, the overall compositional complexity of the partition metagenomes decreases as the particle size increases, as shown by GC content versus coverage binning analyses (Supplementary Fig. 6). These results not only provide strong evidence that the plasmid and chromosome are co-localized, but also that they are co-localized with sponge cells in the largest size particle of the entire homogenate.
Figure 4. p-ren and Ca. E. renieramycinifaciens co-localize with the largest sponge particles.
(a) A schematic representation of the flow cytometry experiment and subsequent analyses performed on Ren-Bali-16–03. (b) Relative abundance of Ca. E. renieramycinifaciens 16S rRNA gene sequence in all 8 flow partitions. Note that the relative abundance of Ca. E. renieramycinifaciens increases in later partitions containing larger particles (P9, P10). (c) Coverage (measured in RPKM: number of mapped Reads Per Kbps per Million of sequenced reads) of the Ca. E. renieramycinifaciens chromosome and p-ren in the five partitions analyzed by metagenomic sequencing. Note that the coverage of both genetic elements also increases in later partitions containing larger particles (P9, P10).
Intracellular localization of Ca. E. renieramycinifaciens
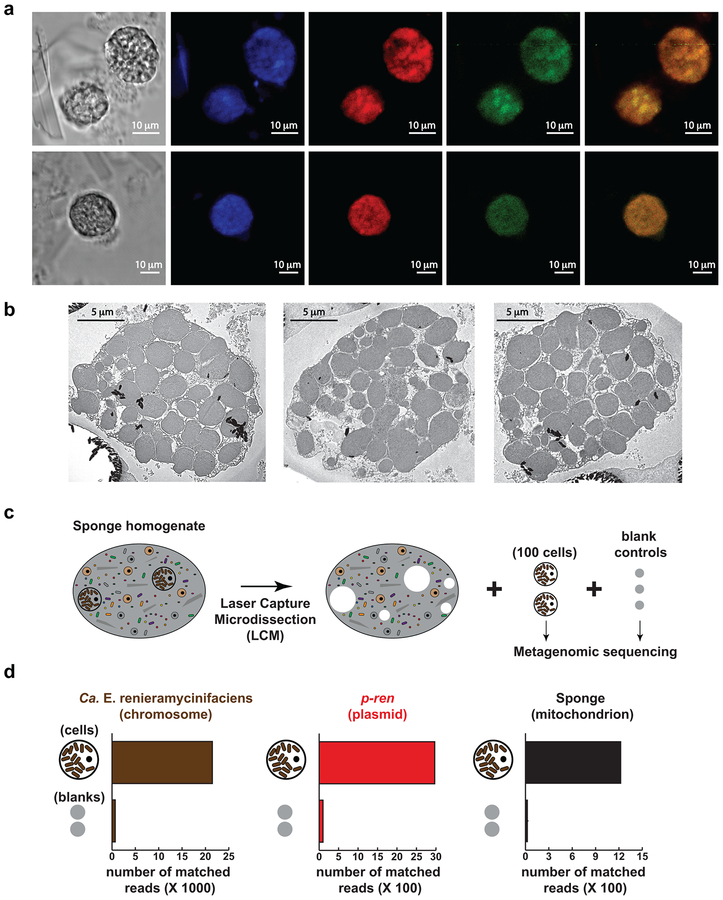
We reasoned that there are two main possibilities for the co-localization of Ca. E. renieramycinifaciens and p-ren in large particles: a) Ca. E. renieramycinifaciens cells are clumped in a colony-like shape that is not easily disrupted; or b) Ca. E. renieramycinifaciens cells exist intracellularly in an enlarged sponge cell (bacteriocyte). To differentiate these two possibilities, we performed Fluorescence In Situ Hybridization (FISH) on Ren-Bali-16–03 using both a mixture of general eubacterial probes (EU338 I, II, and III) and a Ca. E. renieramycinifaciens specific probe designed here (CE75), in addition to 4’,6-diamidino-2-phenylindole (DAPI) staining for DNA visualization34,35. The sponge tissue contains a heterogeneous population of cells, including typical sponge cells of 2–5 μm in diameter and noticeably large spherical, bacteriocyte-like cells of 15–25 μm in diameter, as well as siliceous sponge spicules (Supplementary Fig. 7).
Interestingly, the bacteriocyte-like cells, but not other sponge cells, appear packed with smaller DAPI-stained particles. Hybridization using universal eubacterial probes showed strong fluorescent signals from the bacteriocyte-like cells only, indicating that the smaller particles that fill them are indeed bacterial cells. Hybridization with a Ca. E. renieramycinifaciens-specific probe also showed localized signals in the bacteriocyte-like cells. Moreover, a composite image and co-localization microscopy analyses revealed that signals from the eubacterial and Ca. E. renieramycinifaciens-specific probes mostly superimpose in the bacteriocyte-like cells (Pearson correlation coefficient of 0.94) (Fig. 5a). While signals corresponding to the universal eubacterial probes can be observed elsewhere in the sponge tissue (other bacteria), Ca. E. renieramycinifaciens specific signals are exclusively detected in the bacteriocyte-like cells (Supplementary Fig. 7). Finally, Transmission Electron Microscopy (TEM) images of the bacteriocyte-like cells confirmed that they harbor what appears to be a single morphotype of bacterial cells (Fig. 5b). Altogether, these hybridization and microscopy experiments demonstrate that Ca. E. renieramycinifaciens is an intracellular symbiont that resides solely in large sponge bacteriocytes.
Figure 5. Localization of Ca. E. renieramycinifaciens in sponge chemobacteriocytes.
(a) Fluorescence In Situ Hybridization (FISH) of chemobacteriocytes in Ren-Bali-16–03. From left to right: bright field image, DAPI staining showing chemobacteriocytes packed with smaller cells, hybridization with the universal eubacterial probes, EU338 I, II, and III (red), showing that chemobacteriocytes harbor bacterial cells, hybridization with the Ca. E. renieramycinifaciens specific probe, CE75 (green), localizing Ca. E. renieramycinifaciens to chemobacteriocytes, composite of the green and red signals, showing their predominant co-localization. Scale bars indicate 10 μm. FISH experiments were performed six independent times, and produced the same results. (b) Transmission electron microscopy images of single Ren-Bali-16–03 chemobacteriocytes. Circular cuts on the resin result from hard spicules during thin slicing. Scale bars indicate 5 μm. Imaging experiments using transmission electron microscopy were performed two independent times, and produced the same results. (c) Schematic representation of the isolation of sponge chemobacteriocytes or blank controls by laser capture microdissection. (d) Quantification of reads that mapped to Ca. E. renieramycinifaciens chromosome (left), p-ren (middle), and sponge mitochondrion (right), showing a clear enrichment of target DNA in chemobacteriocytes (out of 850K paired-end reads in total) compared to the membrane background (out of 1M paired-end reads in total).
While the flow cytometry, cell sorting, and metagenomic binning analyses provided strong support for the co-localization of p-ren with the Ca. E. renieramycinifaciens chromosome, and in light of the microscopic revelation that Ca. E. renieramycinifaciens resides in specialized sponge bacteriocytes, we sought to perform one final experiment that definitively localizes p-ren to Ca. E. renieramycinifaciens. Aided by the easily recognizable spherical shape of the large sponge bacteriocytes, we used laser capture microdissection (LCM) to isolate 100 single cells of their kind from diluted Ren-Bali-16–03 sponge homogenates. We then isolated and sequenced DNA from the captured cells, and analyzed their metagenomes in comparison to DNA isolated from captured background membranes as a control. Finally, we quantified the number of reads that mapped to the Ca. E. renieramycinifaciens chromosome, p-ren, and the sponge mitochondrion from both datasets. As expected, reads mapping to all three genetic elements were overwhelmingly enriched in the sequencing dataset originating from the sponge bacteriocytes compared to the control dataset (Fig. 5c, d). These results unequivocally show that Ca. E. renieramycinifaciens harbors p-ren, and further localize the production of the renieramycins to the sponge bacteriocytes. We therefore propose the term “chemobacteriocytes” for describing sponge cells where chemical production of defense molecules by an intracellular bacterial symbiont takes place.
Extreme reduction of the Ca. E. renieramycinifaciens genome
Based on a phylogenetic tree constructed from the 16S rRNA gene, Ca. E. renieramycinifaciens sequences derived from this study and three closely related ones (~99% identical to Ca. E. renieramycinifaciens over ~800 bps) obtained previously from Australian Haliclona sp. form a distinct clade within the class Gammaproteobacteria, with members from the families Coxiellaceae and Legionellaceae (order Legionellales) as the nearest neighbors (Supplementary Fig. 8)36. To gain more insights into their biochemical and metabolic capabilities, we annotated the Ca. E. renieramycinifaciens genomes using the Integrated Microbial Genome platform (IMG, http://img.jgi.doe.gov) and compared their encoded functions to that of Coxiella HT99, the closest free-living relative with a sequenced genome (89% 16S rDNA sequence identity) (Supplementary Table 4)37.
The four Ca. E. renieramycinifaciens genomes are syntenic and share an average nucleotide sequence identity of 99.98%. Moreover, an overview of all metabolic functions using TIGRFAM pathways shows that the four genomes shared the same abundance profiles of genes in all pathways, indicating that they are mostly identical. Consistent with its intracellular lifestyle, the Ca. E. renieramycinifaciens genome shows several hallmarks of reduction: a) it encodes all genes for DNA replication, but no genes for DNA repair, recombination, or transposable and mobile elements, b) central metabolic pathways (i.e., glycolysis, pentose-phosphate, and TCA cycle) are incomplete, c) except for a partial biosynthetic pathway for lysine, it lacks all pathways for amino acid, cofactor, prosthetic group, nucleoside, and nucleotide biosynthesis, and d) genes for cell surface structures, chemotaxis, toxin production and detoxification, and signal transduction are mostly absent. The loss of all of these functions further establishes Ca. E. renieramycinifaciens as an obligate long-term symbiont incapable of free living (Supplementary Table 4).
Reniermacyin biosynthesis requires several substrates and cofactors; remarkably, almost none of which seem to be produced by Ca. E. renieramycinifaciens. These include direct substrates, namely angelic acid and tyrosine, and cofactors needed for the enzymatic activity, such as (R)-4’-phosphopantothenate (PPT), S-adenosyl methionine (SAM) and nicotinamide adenine dinucleotide (NAD). Because of the exclusive intracellular nature of Ca. E. renieramycinifaciens, these substrates and cofactors are likely provided by the host cell, although other bacteria can also contribute if they occasionally co-inhabit the chemobacteriocyte. Taken together, renieramycin production represents a rare case of collaborative biosynthesis between the host and symbiont, where the host provides the substrates and cofactors, while the symbiont provides a complex enzymatic assembly line that is unattainable in animal biochemistry.
A specific symbiosis between Ca. E. renieramycinifaciens and a clade of Haliclona sponges
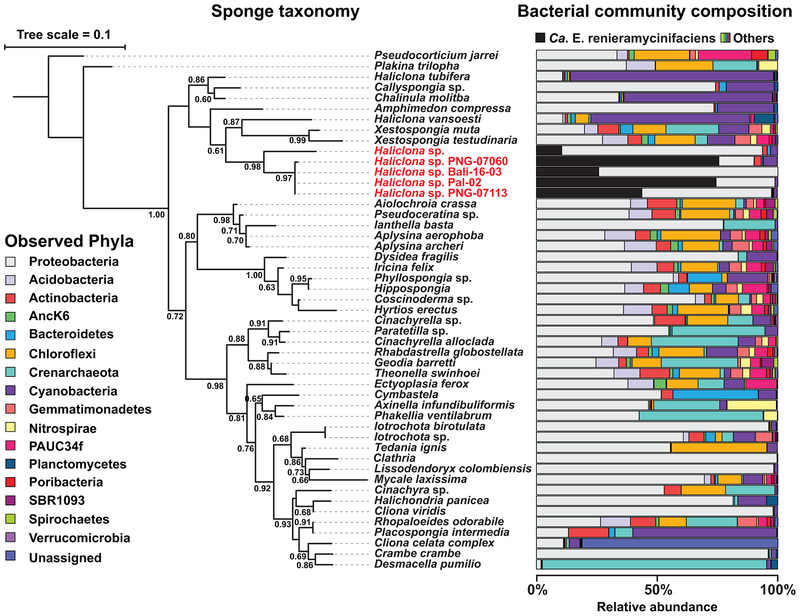
To gain insights into the distribution of Ca. E. renieramycinifaciens in the Ocean, we compared the bacterial community composition of our four Haliclona sponges with that of 1435 marine samples belonging to 1194 sponges, 37 sediment metagenomes, 195 seawater metagenomes, and 9 unclassified metagenomes from a Global Sponge Microbiome analysis (GSM) using high-throughput 16S rRNA gene sequencing (See Methods)1. The 1194 GSM sponge metagenomes belonged to 96 types of sponges, 89 of which have genus/species level assignmenst while 7 are unclassified (Supplementary Table 5). After data processing using the Qiime pipeline38, we searched all samples for OTUs (operational taxonomic units) matching Ca. E. renieramycinifaciens, and mapped their distribution to a phylogenetic tree of the host sponges that we constructed based on their 18S rRNA gene sequences. Overall, we discovered OTUs matching Ca. E. renieramycinifaciens exclusively in sponge samples, and not in the sea water or sediment samples included, supporting its obligate symbiotic lifestyle. Importantly, sequences matching Ca. E. renieramycinifaciens exist specifically in one clade of Haliclona sponges, namely the four from this study and 7 ‘blue Haliclonas’ from the Pacific Ocean (Fig. 6), and is absent in all other sponges including Haliclona individuals that belong to other clades. Finally, when present, the relative abundance of Ca. E. renieramycinifaciens in Haliclona sponges is unusually high, ranging from 10 to 75% of the total 16S rRNA gene sequences in a given sample (Fig. 6 and Supplementary Fig. 9). These results establish Ca. E. renieramycinifaciens as an intracellular symbiont of a specific clade of marine sponges, where it dominates the bacterial community and specializes in producing the renieramcyins.
Figure 6. Host specificity of Ca. E. renieramycinifaciens.
18S rRNA gene-based phylogenetic tree of representative sponges (left) and the 16S rRNA gene-based bacterial composition of their corresponding microbiomes (samples are from this study and the GSM). The phylogenetic tree was constructed using Fasttree-ML (Jukes-Cantor model), and local support values as fractions of 1000 resamples are shown. When sponge species from the GSM are represented by multiple individuals, their average composition is shown (see Methods and Supplementary Table 5). Ca. E. renieramycinifaciens (indicated in black) is specific to a single clade of Haliclona sponges (names shown in red), where it constitutes a major component of their bacterial microbiome. The bacterial composition of the rest of the microbiome is shown at the phylum level, following the color code on the left.
DISCUSSION
The Haliclona-Ca. E. renieramycinifaciens system described here differs from other examples of defensive symbiosis in marine sponges in several aspects10,11. First, unlike H. spongeliae and Ca. Entotheonella sp., Ca. E. renieramycinifaciens does not reside in the mesohyl or pinacoderm of the sponge, but lives instead in specialized sponge chemobacteriocytes. Second, Ca. E. renieramycinifaciens harbors an extremely reduced genome that is incapable of supporting a free-living state (in contrast, Ca. Entotheonella sp. harbors large, almost intact genomes)10,39,40. Finally, it is predominantly a one-molecule system, where ren is the only (or one of three) small molecule BGC recovered from several deeply sequenced metagenomes, and the renieramycins are the major molecules observed (in contrast, dozens of BGCs and molecules are usually recovered from Ca. Entotheonella-harboring sponges)10,39,40. Several of these features, however, are paralleled in cases of defensive symbioses of other host organisms41. Small molecule producing symbionts with reduced genomes and a known or presumed intracellular lifestyle have been reported from several marine ascidians, including the ecteinascidin producer Candidatus Endoecteinascidia frumentensis27,42,43. It is interesting that two distant bacteria, Ca. E. frumentensis and Ca. E. renieramycinifaciens (83% 16S rRNA gene sequence identity), which live an intracellular symbiotic lifestyle with two very distant marine animals (an ascidian and a sponge), base their symbioses on the production of very similar defensive molecules (the ecteinascidins and the renieramycins).
In insects, defensive symbionts with extremely reduced or evolutionarily degenerated genomes are relatively common41,44. For example, specialized cells of the psyllid Diaphorina citri contain an intracellular symbiont, Candidatus Profftella armature, which harbors an extremely reduced genome (~460 kbps) and produces the defensive molecule diaphorin45. Similarly, Lagria villosa beetles harbor a genome-reduced symbiont, Burkholderia gladioli Lv-StB, both in female accessory glands and on eggs, which produce the defensive molecule lagriamide41. It is remarkable that widely different symbionts, hosts, and molecules are involved in otherwise very similar defensive strategies.
Bacteriocytes have been previously observed in marine sponges, but the identity and function of the bacterial symbionts residing in them have been rarely studied46,47. The Ca. E. renieramycinifaciens-Haliclona system described here represents the second case where intracellular sponge symbionts have been identified7,48, and one where the symbiont genome has been sequenced. Hemimycale sponges harbor intracellular bacterial symbionts in specialized spherical cells termed calcibacteriocytes. In these cells, calcibacteria are cultivated in sponge vacuoles where the pH promotes nucleation of calcium carbonate in bacterial membranes, which are later exported to deposit on the sponge exoskeleton7,48. Similarly, Ca. E. renieramycinifaciens are cultivated in Haliclona sponge chemobacteriocytes, where they produce the reniermaycins. Altogether, it is remarkable that sponges – which have a very limited number of cell types – outsource essential structural and defensive roles to symbionts that are cultivated in specialized cells (calcibacteriocytes and chemobacteriocytes). Taking place in the oldest living metazoans, these examples represent an ancestral view of microbe-host interactions and highlight the importance of uncovering more cases of intracellular symbioses in marine sponges.
METHODS
Sponge sample processing and storage
Sponge samples were collected by SCUBA diving from different locations in the Pacific Ocean: Ren-PNG-07060, 5° 17’S. 150° 06’ E.; Ren-PNG-07113, 5° 17.5’S. 150° 06.1’ E; and Ren-Pal-02: 7° 30’ N. 134° 30’ E. For Palau and PNG samples, freshly collected samples were processed in the field for chemistry and DNA work. For chemistry work, a portion of each sponge sample was cut and frozen immediately. For DNA work, a portion was stored in RNALater solution (ThermoFisher Scientific, USA). Ren-Bali-16–03 was freshly collected in Bali from a commercial mariculture farm (Quality Marine, USA), shipped alive to the laboratory, and processed immediately upon arrival. For chemistry and DNA, Ren-Bali-16–03 was processed as described above; for microscopy, small portions of Ren-Bali-16–03 were first fixed in paraformaldehyde (4%), sequentially dehydrated in ethanol (30 %, 50%, and 70 %), and finally stored in 70% ethanol at −20 °C until use; and for flow cytometry, see below.
DNA extraction and sequencing
Metagenomic DNA was extracted from each RNALater preserved sponge sample using the Genomic Tip Kit (Qiagen, MD, USA) with some modifications. The sponge tissue (~1 cm3) was flash frozen in liquid nitrogen and homogenized using a sterile pestle in a conical tube. Buffer B1 (3.5 mL) containing Rnase A (0.2 mg mL−1) was then added and further homogenization was done. The solution was then treated with proteinase K (0.5 mg mL−1) and lysozyme (2.5 mg mL−1) for 5 hours at 37 °C while shaking gently. Buffer B2 (1.2 mL) was added and the solution was further incubated at 50 °C for 30 minutes. Samples were then centrifuged and the supernatant was treated according to the Genomic Tip protocol (Qiagen, MD, USA). Metagenomic DNA was mechanically sheared to an average size of ~500 bps, and Illumina sequencing libraries were prepared using Apollo 324™ NGS Library Prep System and the PrepX DNA library kit (Wafergen, CA) and sequenced on an Illumina HiSeq 2500 Rapid Flow cell as paired-end 2×175 bps reads for Ren-PNG-07113, Ren-Pal-02, and Ren-Bali-16–03, and 2×141 bps for Ren-PNG-07060 (Supplementary Table 1).
Metagenomic analysis, genome assembly and annotation
Raw Illumina reads were filtered as follows using PRINSEQ49. Reads below an average quality score of 30 and with more than two percent undetermined (N) bases were discarded, bases with quality scores below 30 on either end of a read were trimmed, and trimmed reads shorter than half of the original read length were discarded. Filtered reads (pairs and singletons) were assembled using SPAdes with default parameters50. The four chromosomes and four plasmids were circularized by either mapping corresponding paired-end, filtered reads to the edges of the SPAdes scaffolds using BLASTn, or by simply aligning the scaffold edges and then repeating the assembly in Geneious with manual inspection51. The general overview of all functions in the Ca. E. renieramycinifaciens and Coxiella HT99 genomes was obtained by comparison of all the TIGRFAM categories in IMG. Analysis of individual pathways was done by examining the COG, TIGRfam, and KEGG pathways for the presence or absence of genes in each pathway. The circular map was downloaded from IMG with standard COG categories as annotated37.
Identification of common scaffolds between metagenomes
The scaffolds from the SPAdes assembly of Ren-Bali-16–03 were filtered to a minimum length of 5000 bps. To identify the scaffolds that are in common between Ren-Bali-16–03 and the other three metagenomes, filtered reads from the three metagenomes were mapped to Ren-Bali-16–03 scaffolds > 5000 bps using Bowtie 2 (end-to-end alignment, fast mode, with minimum alignment score set as L −0.6, −0.3)52. The breadth coverage of each scaffold (% of scaffold length covered by reads) was then calculated using Samtools scripts, and scaffolds with less than 90% breadth coverage were discarded53. Scaffolds that were covered by reads from all three metagenomes using the above cutoff were counted as common to all metagenomes and then taxonomically assigned using BLASTx against the non-redundant protein database on NCBI (nr).
16S Amplicon sequencing and meta-analysis
The V4 region of the 16S rRNA gene (~250 bps) was PCR amplified and used to construct Illumina sequencing libraries following the previously published design and primers54. Libraries were pooled, sequenced on an Illumina HiSeq 2500 Rapid Flowcell as paired-end reads of 2×175 bps, along with 8 bps index reads, following the manufacturer’s protocol (Illumina, CA). Raw reads were de-multiplexed based on their index reads, and overlapping forward and reverse reads were merged using FLASH with a minimum overlap of 80, a maximum overlap of 100, and a maximum mismatch density of 0.255. The resulting FastQ sequences were preprocessed in Qiita (qiita.ucsd.edu) using split_libraries.py. The resulting Fasta sequences were trimmed to the first 100 nucleotides and the resulting file was downloaded for downstream processing in Qiime (qiime.org)38. To compare the bacterial composition of Haliclona sponges with the existing sponge microbiome data, we used a published dataset which we refer to here as GSM (Global Sponge Microbiome)1. The de-multiplexed, trimmed, Fasta sequences from GSM was downloaded from Qiita (study 10346). GSM and Haliclona sequences were then combined and submitted to subsampled open reference OTU picking using pick-open-reference-otus.py in Qiime using default parameters except for enabled reverse strand matching and the number of parallel jobs. OTUs were defined at 97% identity. The resulting OTU table was filtered to 0.005% minimum relative abundance and then split according to sample type (sponge metagenomes, seawater metagenomes and marine sediment metagenomes) for downstream analyses. To map the composition of each sample to the host sponges, the filtered OTU table for sponges was combined with the metadata which was downloaded from Qiita after the demultiplexing and trimming of sequences. Samples were then grouped according to host taxonomy (Supplementary Table 5) and the relative abundance of each OTU was calculated from the total abundance of OTUs in each host. A phylum level summary of the OTU table with Ca. E. renieramycinifaciens OTU as a group was constructed for mapping with the sponge phylogeny (described below).
Mapping of microbial composition to sponge phylogeny
To construct the phylogenetic tree of host sponges, the 18S rRNA gene sequences were amplified from the metagenomic DNA of the Haliclona sponges using 18S rRNA gene universal primers 18S-A1 (5’- AACCTGGTTGATCCTGCCAGT) and 18S-564R (5’- GGCACCAGACTTGCCCTC). The following thermocycler program was used: initial denaturation, 1 min, 98 °C; 34 cycles of 98 °C for 10 sec, 65 °C for 30 sec, 72 °C for 30 sec; and final extension of 5 min at 72 °C. Phusion high fidelity polymerase (New England Biolabs, USA) was used. The amplification products were gel purified and submitted for direct sequencing using both forward and reverse primers. For samples with sequences that returned as mixed, the gel purified bands were cloned into TOPO-TA (Invitrogen, USA) vectors using manufacturer protocols and individual clones were sequenced to obtain the host sponge 18S rDNA sequence. To construct the tree, available 18S sequences were downloaded from NCBI using the taxon ID (Supplementary Table 5). The sequences, including those of Haliclona sponges were aligned with 45 sponge 18S rRNA gene sequences from NCBI using MUSCLE56. The resulting alignment was trimmed in Geneious to include only sequence regions with good alignment coverage51. The trimmed alignment was subsequently refined by MUSCLE and uploaded to the CIPRES portal for phylogenetic tree construction using Fasttree-ML (Jukes-Cantor model)57. The phylum level composition of the sponges in the tree were then mapped in ITOL58.
Cell sorting by flow cytometry and processing of fractions
A piece (~1cm3) of fresh Ren-PNG-Bali-16–03 was cut and homogenized gently in calcium-magnesium-free (CMF) artificial seawater (2 mL) using a sterile tissue homogenizer. The resulting suspension was passed through a 70 μm filter and the filtrate was used for cell sorting on a FACSVantage SE w/DiVa cell sorter (BD Biosciences, San Jose, CA USA). Particles were observed on the forward scatter (FSC) and side scatter (SSC) parameters, representing increasing 488 nm laser light scatter due to particle size and granularity, respectively. Eight gates were created along the FSC axis and particles from each were collected into separate tubes, resulting in partitions with increasing particle size composition (Supplementary Fig. 5). Metagenomic DNA was extracted from each using the Masterpure complete DNA extraction kit following manufacturer’s protocol (Epicenter, Madison, WI, USA). The DNA obtained was used for both 16S rRNA gene amplicon sequencing (1.5 K reads on average) and shotgun metagenomic sequencing (an average of 7 M single-end reads of 75 bps per sample) on an Illumina HiSeq 2500 platform as described above (Supplementary Table 1). 16S rRNA gene sequencing data were processed using Qiime as described above, and metagenomic data were assembled using SPAdes and processed as described below.
Metagenomic binning
Scaffolds derived from the assembly of the shotgun metagenomic sequencing data of the flow fractions were filtered to a minimum length of 500 bps. The GC content of each scaffold was calculated using the script calc_gc.pl from Albertsen et al59. The coverage of each scaffold was obtained from the results of the SPAdes assembly. BLASTn (e-value cutoff of 1 × 10−20 and identity cutoff of 95%) was used to identify the scaffolds that belong to the Ca. E. renieramycinifaciens chromosome, p-ren, and the sponge mitochondrion. To assign scaffolds that belong to the sponge genome, BLASTx was used to query the scaffolds using a protein sequence database for coding sequences annotated from the genome of the sponge Amphimedon queenslandica at an e-value cutoff of 1 × 10−100 60. Scaffolds were then plotted in R according to their GC content and log of coverage (Supplementary Fig. 6)61.
Fluorescence in situ hybridization
Small cuts of the paraformaldehyde fixed Ren-PNG-Bali-16–03 sample were used for FISH experiments. The samples were hydrated sequentially in 50% ethanol in PBS (30 min, room temperature), 30% ethanol in PBS (30 min, room temperature) and 100% PBS (30 min, room temperature). The samples were then transferred into 30% sucrose in PBS and left at 4 °C overnight, after which they were transferred into 1:1 of 30% sucrose in PBS:OCT for another 48 hours at 4 °C before they were finally embedded into 100% OCT solution (OCT: Optimal Cutting Temperature embedding medium, Tissue-tek, Sakura). Embedded tissues were stored at −80 °C. To prepare sections, the frozen tissue was cut into 20 μm sections into precut cryofilm (Section-LAB Co Ltd, Japan), and stored at −20 °C until use.
Ca. E. renieramycinifaciens-specific 16S rRNA probes were designed using ARB and further analyzed by BLAST in NCBI62. Corresponding mismatched or negative probes were designed by including a single nucleotide mismatch in the middle of the sequence. Clone-FISH analysis was performed as follows to optimize the hybridization conditions of the probes and confirm that the negative probe has diminished signal63. Briefly, the 16S rDNA of Ca. E. renieramycinifaciens was cloned into a pET28 vector under the control of the T7 promoter and transformed into E. coli BL21 cells. Cells were grown under induced (with IPTG) and uninduced (without IPTG) conditions. The cell pellets were then collected and suspended in RNALater for storage at −20. Aliquots of the suspension were used for the following clone-FISH experiments. Ca. E. renieramycinifaciens-specific probe, CE75 (5’-CCTACGGGCCTGTTACCGTT-3’) and the corresponding mismatch, CE75neg (5’- CCTACGGACCTGTTACCGTT-3’) (final concentration 5 ng/μl) were each mixed with different sets of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl pH 7.4, 0.01% SDS) containing different formamide concentrations (20%, 25%, 30%, 35%). A mixture of universal bacterial probes, EU338 I, EU338 II, and EU338 III (final concentration 5 ng/μl) was added to each reaction34,35. Hybridization was performed at 46 °C for 2–3 hours in pre-humidified tubes with the same hybridization buffers. The samples were then mounted on slides and imaged. The optimal formamide concentration, 35%, as determined from this experiment was used for the hybridization on sponge sections using the same procedure as above except for the additional counter staining with DAPI before mounting. CE75 and CE75neg probes were labeled with 6-FAM and universal eubacterial probes were labeled with Cy3. Imaging was performed on a Leica SP5 laser scanning confocal microscope, using 405, 480, and 581 nm lasers and a 63× magnification oil objective.
Transmission electron microscopy sample preparation and imaging
A small cube (~0.5 cm3) was cut from a paraformaldehyde-fixed sample of Ren-Bali-16–03 and placed into a 2% osmium tetroxide solution (1 mL). Staining was done overnight on a rotator after which the sample was washed five times with ddH2O and then stained with 2% uranyl acetate for another 12 hours. After washing with ddH2O, the sample was then dehydrated sequentially in 30%, 50%, 70%, 85%, and 100% ethanol for 1 hr at each step. The dehydrated sample was embedded in 1:1 LR white resin (Electron Microscopy Sciences, USA): ethanol overnight on a rotator and then transferred to 100% LR white resin for stationary embedding for 48 hrs. Polymerization was performed on a heat block at 65 °C overnight. Ultrathin sections were observed on a Talos F200× Scanning/Transmission Electron Microscope.
Laser capture microdissection of sponge chemobacteriocytes
A small cube (~0.5 cm3) of RNALater preserved sample was washed in sterile water three times to reduce salt crystals. The tissue was then crushed gently on a 70 μm filter which was then washed with sterile water (500 μL). The filtrate was centrifuged at 500 × g for 5 min at 4 °C to pellet larger sponge cells and particles. The resulting pellet was then suspended in water (100 μL) and a 10% dilution (50 μL) was spread on an LCM PEN membrane slide (ThermoFisher Scientific, USA) and allowed to air dry. Chemobacteriocytes were identified as distinct round particles with diameter of 15–25 μm and isolated by laser microdissection on an MMI Cell Cut LCM system (mmi). 100 bacteriocytes were collected and 50 background membrane cuts were further collected as a negative control. DNA was extracted from both the cells and negative control using the Masterpure complete DNA extraction kit following manufacturer’s protocol (Epicenter, Madison, WI, USA) and used for Illumina library preparation and metagenomic sequencing as described above (Supplementary Table 1). BLASTn (e-value cutoff of 1 × 10−20) was used to assign metagenomic reads to the Ca. E. renieramycinifaciens chromosome, p-ren, and sponge mitochondrion.
Chemical extraction and analysis
Frozen sponge samples (~2 cm3) were cut and extracted twice with ethyl acetate (20 mL) and twice with methanol (20 mL). The extracts were combined, dried under vacuum, and analyzed by HPLC-HR-LC/MS/MS on an Agilent QTOF instrument (Agilent Technologies). HPLC elution was done using the following gradient: 0–100% A, 0–25 min; 100% A, 25–30 min; 100–0% A, 30–35 min. The buffer system used was: A - 0.1% Formic acid in acetonitrile, B - 0.1% Formic acid in ddH2O. To purify renieramycin E, frozen Ren-Bali-16–03 sponge (~5 grams) was extracted as described above. The dried extract was subjected to fractionation using a semi-preparative HPLC column (Agilent Poroshell 120 EC-C18, 9.4 × 250 mm, 4 μm) on an Agilent 1260 Infinity HPLC system (Agilent Technologies). The following gradient was used for fractionation: 0.5–100% A, 0–20 min; 100% A, 20–25 min; 100–0.5 % A, 25–30 min. The flow rate for fractionation was 1.5 mL/min. Fractions were analyzed immediately using HPLC-MS and the fraction containing the major peak and the m/z corresponding to renieramycin E (549. 22 m/z (M+H-H2O)+, was dried and submitted for proton NMR. NMR spectra was obtained on an Avance III, 500 MHz (Bruker, USA) in CD3OD. We successfully obtained proton NMR of a semi-pure renieramycin E sample showing distinctive renieramycin signatures (e.g., O-methyl groups (3.91 ppm) and angelic acid methyl groups (1.66 ppm, 1.47 ppm) (Supplementary Fig. 2), but due to the typical instability of renieramycins and THQs in general, further purification attempts yielded degraded products.
Heterologous expression of renB and ren in E. coli
The C-methyltransferase gene, renB, was amplified from the Ren-Bali-16–03 metagenomic DNA using primers (F 5’ GGGTACCGGTAGAAAAAATGTTTGTTGAAGAAGAAAAAGT, R 5’ GCTCAGTTGGAATTCGGATGCTTAATGATTCTTAATCGC), and cloned into pGFP-UV backbone by isothermal assembly using the In-fusion cloning kit (Takara Bio, CA, USA), under the control of lac promoter. For heterologous expression, the pGFP-renB vector was transformed into Stellar E. coli cells (Takara Bio, CA, USA). pGFP-UV empty vector was used as negative control. Colonies were picked from the transformations and grown overnight at 30 °C, 200 rpm. Seed cultures (100 μL) were inoculated into LB broth (100 mL) that was supplemented with carbenicillin (100 μg mL−1) and L-tyrosine (5 mM) (Thermo-Fisher Scientific, MA, USA). Expression cultures were grown shaking at 200 rpm, 30 °C, for five days and then harvested by centrifugation. To obtain the extracts, pre-equilibrated (5 mL) Diaion HP20 adsorbent resin (Millipore-Sigma, MO, USA) was added to the supernatant and allowed to bind by gently shaking at 150 rpm at room temperature for 1 hour. The resin was then filtered and washed with 3 volumes of ddH2O and finally eluted with methanol (20 mL). The methanolic extracts were dried and analyzed using HPLC-HR-MS as described above. Authentic standards of 3-methyl-L-tyrosine and O-methyl-L-tyrosine (Millipore-Sigma, MO, USA) were purchased and compared to the extracts of renB expression. To express the entire ren BGC, 3 overlapping fragments of ~9 Kbps were amplified from Ren-Pal-02 metagenomic DNA using the following primers: natren_514F - 5’ AATGTATGAAGACTGGCCAG and natren_9594R – 5’ GCAAGACTCTCTATTGTCGT, natren_8409F – 5’ CAACTTCCTCGATATCACGT and natren_17633R – 5’ CTGGTATCAAAGTGTTTGGC, and natren_16615F – 5’ TGCTAGATCCAGCAAATCTC and natren_25404R – 5’ CAGCTAAATCTCCATCCCAA. Transformation associated recombination (TAR) in Saccharomyces cerevisiae was performed using established methods to assemble all 3 fragments (ren) into a plasmid designed based on the pGFP-UV backbone and the yeast elements from pLLX1364,65. This construct was expressed in E. coli EPI300 cells using the native ren promoters in LB medium for five days at 30 °C and 200 rpm. To co-express the ren gene cluster with a phosphopantetheinyl transferase and an MbtH-like protein identified in Ca. E. renieramycinifaciens, genes encoding them were amplified from the metagenomic DNA of Ren-Pal-02. For the phosphopantetheinyl transferase, primers F 5’AATTTCACACAGGAAACAGCTATGAAACAACTTCGATGGA and R 5’ ACAGCTTATCATCGATAAGCTTAATTTTCAGAAGCTTGCC were used and the fragment was cloned into pSTV28 backbone (Takara Bio, CA, USA) downstream of the lac promoter. The gene encoding an MbtH-like protein was amplified using primers F 5’ GCGCTCAGTTGGAATTCATCAACGCTTGTTAGCACG and R 5’ CGGGTACCGGTAGAAAAAATGTATAAGTTTGTTGATAGGGATG and cloned into the pGFPUV plasmid after the lac promoter (Takara Bio, CA, USA). The fragment including the lac promoter and the MbtH gene from the pGFUV-mbth construct was subsequently transferred to the pSTV28-pptase construct to yield pSTV28-pptase-mbth plasmid. Plasmids were constructed using Gibson isothermal assembly. The resulting pSTV28-pptase-mbth plasmid was transformed into E. coli EPI300 containing the ren pathway for expression. Expression was performed in LB medium for five days at 30 °C and 200 rpm, and extracts were obtained and analyzed as described above.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Ca. E. renieramycinifaciens genomes have been deposited to the Integrated Microbial Genomes (Joint Genome Institute, Department of Energy) public repository, under IMG submission IDs 151197, 151198, 119799, 119800.
Supplementary Material
Acknowledgments
We would like to thank Eric W. Schmidt, Mary Kay Harper-Ireland, and Chris Ireland at the University of Utah for providing samples Ren-PNG-07060, Ren-PNG-07113, and Ren-Pal-02, and the Republic of Palau, Papua New Guinea, and the Republic of Indonesia as original sources for the sponge samples studied here. We thank Mary Kay Harper-Ireland for the under-water photograph of the Haliclona sponge shown in Fig.1. We are grateful to Christina DeCoste and the Molecular Biology Flow Cytometry Resource Facility (partially supported by the Cancer Institute of New Jersey Cancer Center Support Grant P30CA072720) for assistance with flow cytometry, Paul Shao and the Molecular Biology Electron Microscopy Core Facility for assistance with TEM, Dr. Gary Laevsky, the Molecular Biology Confocal Microscopy Core Facility (a Nikon Center of Excellence) and Jindong Zan for assistance with FISH and microscopy experiments, Gabriela Hrebikova and Alexander Ploss for assistance with LCM, Wei Wang and the Lewis Sigler Institute Sequencing Core Facility for assistance with high-throughput sequencing, Matthew Cahn for assistance with metagenomic data analysis, Seema Chatterjee for general assistance, Yuki Sugimoto and Pranatchareeya Chankhamjon for assistance with NMR and HPLC-HR-MS, and the rest of the Donia lab for useful discussions. We would also like to thank the anonymous Nature Microbiology reviewer who suggested the name Ca. E. renieramycinifaciens for the symbiont discovered in this study. Funding for this project has been provided by Princeton University, and M.S.D. is funded by an NIH Director’s New Innovator Award (ID: 1DP2AI124441).
Footnotes
Competing financial interests
Mohamed S. Donia is a member of the Scientific Advisory Board for Deepbiome Therapeutics, and a consultant for Flagship Pioneering.
Supplementary Information
Supplementary Note, Tables, and Figures are provided.
References
- 1.Thomas T et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun 7, 11870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentschel U, Piel J, Degnan SM & Taylor MW Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol 10, 641 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Nguyen MT, Liu M & Thomas T Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol 23, 1635–1645, doi: 10.1111/mec.12384 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Burgsdorf I et al. Lifestyle evolution in cyanobacterial symbionts of sponges. MBio 6, e00391–00315, doi: 10.1128/mBio.00391-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor MW, Radax R, Steger D & Wagner M Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev 71, 295–347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan L et al. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. U S A 109, E1878–1887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garate L, Sureda J, Agell G & Uriz MJ Endosymbiotic calcifying bacteria across sponge species and oceans. Sci. Rep 7, 43674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F et al. Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges. Proc. Natl. Acad. Sci. U S A 112, 4381–4386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt EW, Obraztsova AY, Davidson S, Faulkner DJ & Haygood M Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel delta-proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol 136, 969–977 (2000). [Google Scholar]
- 10.Wilson MC et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Agarwal V et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol 13, 537–543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blunt JW et al. Marine natural products. Nat. Prod. Rep 35, 8–53, doi: 10.1039/c7np00052a (2018). [DOI] [PubMed] [Google Scholar]
- 13.Amnuoypol S et al. Chemistry of renieramycins. Part 5. Structure elucidation of renieramycin-type derivatives O, Q, R, and S from thai marine sponge Xestospongia species pretreated with potassium cyanide. J. Nat. Prod 67, 1023–1028 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Davidson BS Renieramycin G, a new alkaloid from the sponge Xestospongia caycedoi. Tetrahedron Letters 33, 3721–3724 (1992). [Google Scholar]
- 15.Oku N, Matsunaga S, van Soest RW & Fusetani N Renieramycin J, a highly cytotoxic tetrahydroisoquinoline alkaloid, from a marine sponge Neopetrosia sp. J. Nat. Prod 66, 1136–1139 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Suwanborirux K et al. Chemistry of renieramycins. Part 3.(1) isolation and structure of stabilized renieramycin type derivatives possessing antitumor activity from Thai sponge Xestospongia species, pretreated with potassium cyanide. J. Nat. Prod 66, 1441–1446 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Frincke JM & Faulkner DJ Antimicrobial metabolites of the sponge Reniera sp. J. Am. Chem. Soc 104, 265–269 (1982). [Google Scholar]
- 18.Lopanik NB & Clay K Chemical defensive symbioses in the marine environment. Functional Ecology 28, 328–340 (2014). [Google Scholar]
- 19.Darumas U, Chavanich S & Suwanborirux K Distribution Patterns of the Renieramycin-Producing Sponge, Xestospongia sp., and Its Association with Other Reef Organisms in the Gulf of Thailand. Zoological Studies 46, 695–704 (2007). [Google Scholar]
- 20.Arai T et al. The structures of novel antibiotics, saframycin B and C. Tetrahedron Letters 20, 2355–2358 (1979). [Google Scholar]
- 21.Arai T, Takahashi K, Nakahara S & Kubo A The structure of a novel antitumor antibiotic, saframycin A. Experientia 36, 1025–1027 (1980). [DOI] [PubMed] [Google Scholar]
- 22.Irschik H, Trowitzsch-Kienast W, Gerth K, Hofle G & Reichenbach H Saframycin Mx1, a new natural saframycin isolated from a myxobacterium. J. Antibiot. (Tokyo) 41, 993–998 (1988). [DOI] [PubMed] [Google Scholar]
- 23.Ikeda Y, Matsuki H, Ogawa T & Munakata T Safracins, new antitumor antibiotics. II. Physicochemical properties and chemical structures. J. Antibiot. (Tokyo) 36, 1284–1289 (1983). [DOI] [PubMed] [Google Scholar]
- 24.Ikeda Y, Shimada Y, Honjo K, Okumoto T & Munakata T Safracins, new antitumor antibiotics. III. Biological activity. J. Antibiot. (Tokyo) 36, 1290–1294 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Rinehart KL et al. Ecteinascidins 729, 743, 745, 759A, 759B, and 770: potent antitumor agents from the Caribbean tunicate Ecteinascidia turbinata. J. Org. Chem 55, 4512–4515 (1990). [Google Scholar]
- 26.Rath CM et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol 6, 1244–1256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schofield MM, Jain S, Porat D, Dick GJ & Sherman DH Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol 17, 3964–3975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Recine F et al. Update on the role of trabectedin in the treatment of intractable soft tissue sarcomas. Onco. Targets Ther. 10, 1155–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pospiech A, Cluzel B, Bietenhader J & Schupp T A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology 141 (Pt 8), 1793–1803 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Weber T et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodhouse JN, Fan L, Brown MV, Thomas T & Neilan BA Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J. 7, 1842–1851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu C-Y Biosynthesis of 3-Hydroxy-5-Methyl-O-Methyltyrosine in the Saframycin/Safracin Biosynthetic Pathway. J. Microbiol. Biotechnol 19, 439–446 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Velasco A et al. Molecular characterization of the safracin biosynthetic pathway from Pseudomonas fluorescens A2–2: designing new cytotoxic compounds. Mol. Microbiol 56, 144–154 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Amann RI et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol 56, 1919–1925 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daims H, Bruhl A, Amann R, Schleifer KH & Wagner M The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol 22, 434–444, doi: 10.1016/S0723-2020(99)80053-8 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Webster NS et al. Same, same but different: symbiotic bacterial associations in GBR sponges. Front. Microbiol 3, 444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Checcucci A & Mengoni A The integrated microbial genome resource of analysis. Methods Mol. Biol 1231, 289–295 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lackner G, Peters EE, Helfrich EJ & Piel J Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc Natl Acad Sci U S A 114, E347–E356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori T et al. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc Natl. Acad. Sci. U S A 115, 1718–1723, doi: 10.1073/pnas.1715496115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Florez LV, Biedermann PH, Engl T & Kaltenpoth M Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep 32, 904–936, doi: 10.1039/c5np00010f (2015). [DOI] [PubMed] [Google Scholar]
- 42.Kwan JC et al. Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. U S A 109, 20655–20660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopera J, Miller IJ, McPhail KL & Kwan JC Increased Biosynthetic Gene Dosage in a Genome-Reduced Defensive Bacterial Symbiont. mSystems 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engl T et al. Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc. Natl. Acad. Sci. U S A 115, E2020–E2029, doi: 10.1073/pnas.1719797115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakabachi A et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol 23, 1478–1484, doi: 10.1016/j.cub.2013.06.027 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Maldonado M Intergenerational transmission of symbiotic bacteria in oviparous and viviparous demosponges, with emphasis on intracytoplasmically-compartmented bacterial types. J. Mar. Biol. Ass. U. K 87, 1701–1713, doi: 10.1017/S0025315407058080 (2007). [DOI] [Google Scholar]
- 47.Vacelet J & Donadey C Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol 30, 301–314, doi: 10.1016/0022-0981(77)90038-7 (1977). [DOI] [Google Scholar]
- 48.Uriz MJ, Agell G, Blanquer A, Turon X & Casamayor EO Endosymbiotic calcifying bacteria: a new cue to the origin of calcification in metazoa? Evolution 66, 2993–2999, doi: 10.1111/j.1558-5646.2012.01676.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmieder R & Edwards R Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864, doi: 10.1093/bioinformatics/btr026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bankevich A et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kearse M et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624, doi: 10.1038/ismej.2012.8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magoc T & Salzberg SL FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price MN, Dehal PS & Arkin AP FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albertsen M et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol 31, 533–538 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Srivastava M et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham H ggplot2: Elegant Graphics for Data Analysis. (Springer, New York, NY, 2009). [Google Scholar]
- 62.Ludwig W et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schramm A, Fuchs BM, Nielsen JL, Tonolla M & Stahl DA Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol 4, 713–720 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Wolfgang MC et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A 100, 8484–8489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamanaka K et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl. Acad. Sci. U S A 111, 1957–1962, doi: 10.1073/pnas.1319584111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Ca. E. renieramycinifaciens genomes have been deposited to the Integrated Microbial Genomes (Joint Genome Institute, Department of Energy) public repository, under IMG submission IDs 151197, 151198, 119799, 119800.