Abstract
The change in the formal charge of 34 SARS-CoV-2 lineages from September 2020 to June 2021 was analyzed according to the monthly evidence of the European agency. The reported point mutations and small insertions are electrically neutral (17), positive (12), or negative (3). They had been found in the spike glycoprotein S, in the RBD and S1/S2 regions, crucial for initiation of viral infection. The most often observed were positive mutations, especially D614G and E484K, located in the region of S1/S2 junction, and in the receptor-binding domain (RBD), respectively. They are related to G and A switching. Positive mutations are stretching equally in both areas, but in the RBD region, they are more dispersed. In the set of analyzed virus variants, the increasing tendency in the number of positively charged residues in spike protein was observed. Furthermore, the well-documented WHO classes show an increase in the COVID-19 percentage case fatality with the positive increase in the spike crucial region’s total charge. The data mining, applying classifier algorithm based on the artificial neuronal network, confirms that the value and the distribution of additional positive charge in S may be important factors enabling virus impact to immunity. This may be promoted by the stronger long-range electrostatic attraction of the virus particle to the host cell, preceding the infection. The estimation of the potential energy for the RBD approaching the angiotensin-converting enzyme (ACE2) was presented.
Keywords: coronavirus, COVID-19, SARS-CoV-2, spike protein S, amino acids, charge, potential energy, electrostatic interactions
Introduction
It was recently discussed the role of the net positive electric charge in the influenza A virus binding to, or released from the infected, host cell,1,2 and the possible anionic prevention.3 Similarly, electrical features of the structure of COVID-19 coronavirus have aroused interest since the beginning of the pandemic.4 An adequate concept was presented that special distributions of charged amino acids (Figure 1) can, in an electrostatic manner, facilitate both the SARS-CoV-2 infection of the cells and the interaction with potential drugs.5 In the area of biochemical research, the electrostatic inhibition of interplaying furin6 and the electrostatic interactions of therapeutic antibodies with RBD was considered.7 In the field of practical applications, the physical basis of electrostatic filter prevention was discussed.8
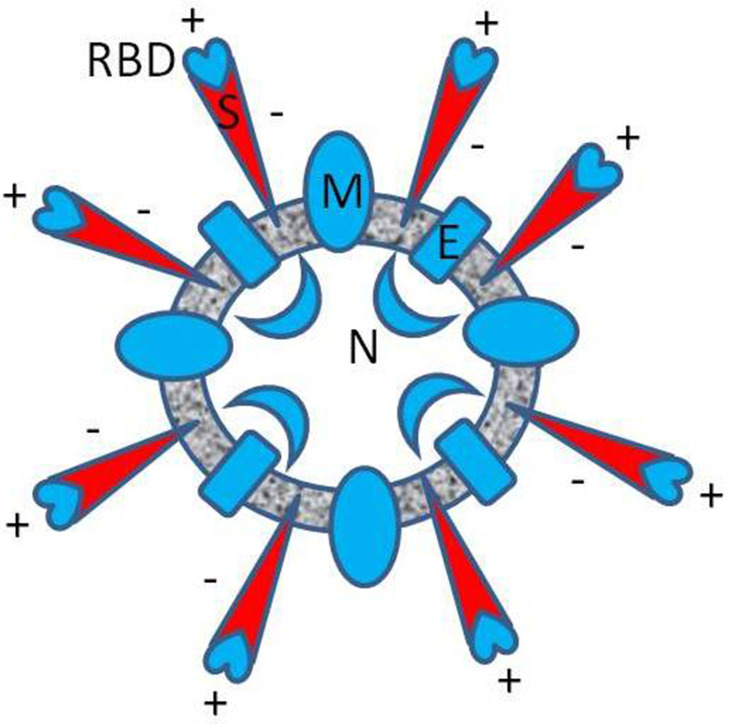
Figure 1.
Approximate diagram of the spatial distribution of electric charge in SARS-CoV-2 (B.1 variant). The resultant formal charge, estimated from the charged amino acid content of the envelope (E), the membrane (M), and the nucleocapsid protein (N) is positive (blue), respectively 2, 8, 24 elementary charge units [e]. The entire charge of spike protein (S) is negative (red), −12 [e], but locally, in the RBD, the resultant charge is positive, +7 [e], as was reported in Pawłowski’s study.5
This manuscript is a continuation of the previous paper.5 Current work examines the number of positive charges in the spike proteins of SARS-CoV-2 and concludes that systematically new coronavirus variants have more positive charges in them. The importance of the consequences of this phenomenon is emphasized.
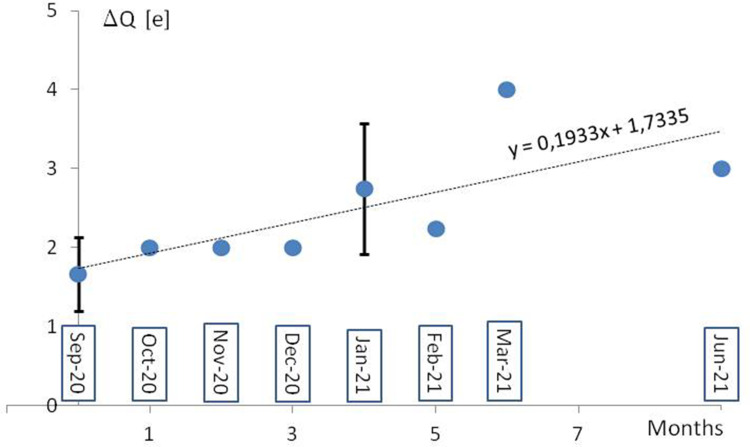
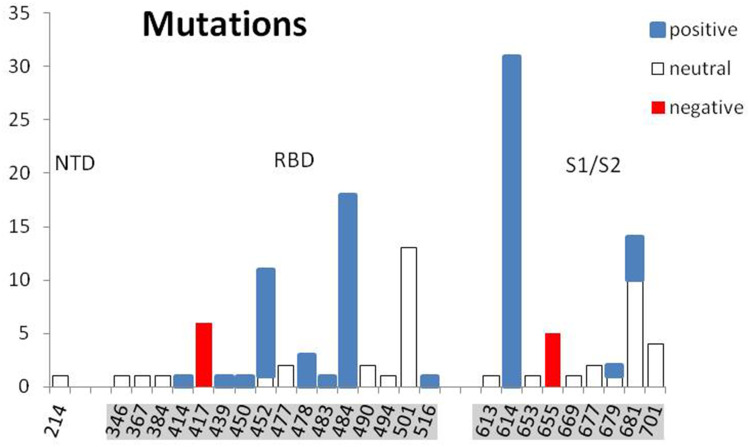
How it was recently postulated, the long-range electrostatic interactions of positive and negative charges of RBD and ACE2, respectively, and the short-range interactions of the dipoles tangent to the border of contacting molecular surfaces can make it much easier for a virus to reach and bind to the cell.9 The evidence of the European Centre for Disease Prevention and Control (ECDC) on SARS-CoV-2 variants detected as of 8th July 202110 shows a systematic increase of positive charge in RBD and S1/S2 cleavage areas of S protein in successive monthly reported new lineages (Figure 2). The 32 mutations repeated in 34 new lineages are indicated in Table 1, with the protein S region, the number of cases, the change of the formal charge, and the time of first detection. The positive formal charge in mutations (72) dominates over the neutral (43) and the negative (11) ones (Figure 3). The number of positive cases in the RBD region is the same as for the S1/S2 cleavage area.
Figure 2.
A mean increase of formal electric charge in RBD and S1/S2 cleavage regions in the lineages monthly reported by ECDC from September 2020 to June 2021. The formal charge means the charge of the amino acids at neutral pH, in elementary charge units. The reference “wild-type’” virus B.1 was assumed (with no mutation D614G and other spike protein changes). The standard deviation of the population (vertical bar) was shown.
Table 1.
Mutations in SARS-CoV-2 Variants of Concern as of 8 July 2021 (ECDC Report) the Data in Columns Were Collected for All Reported Lineages and Their Mutations. Many Mutations Occur in Several Lineages
| Region | Mutation | Number of Cases (Modified Lineages) | Change of Formal Charge ΔQ [e] | Time of First Detection |
|---|---|---|---|---|
| N-terminal domain | ins214TDR | 1 | 0 | Dec-20 |
| RBD | R346K | 1 | 0 | Jan-21 |
| V367F | 1 | 0 | Dec-20 | |
| P384L | 1 | 0 | Dec-20 | |
| Q414K | 1 | 1 | Dec-20 | |
| K417N | 4 | –1 | Sep-20 | |
| K417T | 2 | -1 | Dec-20 | |
| N439K | 1 | 1 | Mar-21 | |
| N450K | 1 | 1 | Dec-20 | |
| L452Q | 1 | 0 | Dec-20 | |
| L452R | 10 | 1 | Sep-20 | |
| S477N | 2 | 0 | Dec-20 | |
| T478K | 3 | 1 | Nov-20 | |
| V483A | 1 | 1 | Feb-21 | |
| E484K | 16 | 2 | Sep-20 | |
| E484Q | 2 | 1 | Dec-20 | |
| F490S | 2 | 0 | Dec-20 | |
| S494P | 1 | 0 | Jan-21 | |
| N501T | 1 | 0 | Dec-20 | |
| N501Y | 12 | 0 | Sep-20 | |
| E516Q | 1 | 1 | Jan-21 | |
| S1/S2 junction |
Q613H | 1 | 0 | Dec-20 |
| D614G | 31 | 1 | Sep-20 | |
| A653V | 1 | 0 | Dec-20 | |
| H655Y | 5 | –1 | Dec-20 | |
| G669S | 1 | 0 | Feb-21 | |
| Q677H | 2 | 0 | Dec-20 | |
| ins679GIAL | 1 | 0 | Jan-21 | |
| N679K | 1 | 1 | Jan-21 | |
| P681H | 10 | 0 | Sep-20 | |
| P681R | 4 | 1 | Dec-20 | |
| A701V | 4 | 0 | Sep-20 |
Figure 3.
The distribution of reported by ECDC 32 mutations (126 cases in 34 lineages). Mutations and the formal charge change are listed in Table 1.
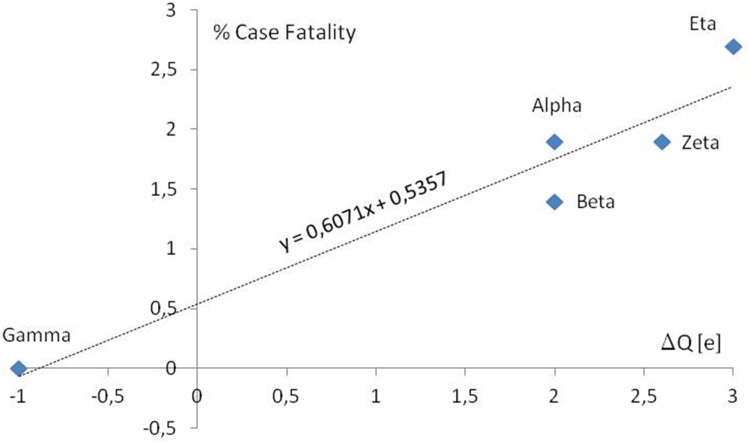
According to the data published by Public Health England (PHE),11 the COVID-19 percentage case fatality increases with the formal charge increase in discussing areas of S protein of new investigated variants (Figure 4).
Figure 4.
The COVID-19 percentage case fatality vs formal charge changes in RBD and S1/S2 cleavage regions according to PHE. In the case of WHO labeled variants, Alpha and Zeta the mean change of the charge were calculated for 2 and 5 lineages. Only well-documented variants first detected in the UK, South Africa, Japan ex Brazil, and Brazil were presented. According to the Public Health England (PHE).11
Materials and Methods
Neuronal Analysis of S Charge Impact on Immunity
To determine how much the additional charge of spike protein determines the impact of SARS-CoV-2 variants on immunity, the classification process with the use of three machine learning classification algorithms in Weka 3.8.5 was carried out. There were deep learning Dl4jMlpClassifier, Logistic regression model, and the neuronal network algorithm, MultilayerPerceptron. The 28 well-documented lineages (instances) with the clear evidence for some impact on immunity, described by 37 attributes (32 possible mutations, characterized by the occurrence nominal labels{1, 0}, and 4 numeric parameters: the time (month) of the first detection, the total change of charge in S, the charge change in RBD, and the charge change in the S1/S2 cleavage area, followed by the class decision attribute with the nominal labels {no, neutralization, escape}) were used to develop the classification model, relating classes of the evidence for impact on immunity to the other attributes of virus lineages.10
The final model offering the best classification of instances (conceptual visualization in Figure 5) was the neuronal network obtained with the MultilayerPerceptron classifier (weka.classifiers.functions.MultilayerPerceptron -L 0.3 -M 0.2 -N 5000 -V 0 -S 0 -E 20 -H a -R).12 In this case, the weights for the inputs of each attribute to a given neuronal node and the threshold for the sigmoid transfer function were fitted.
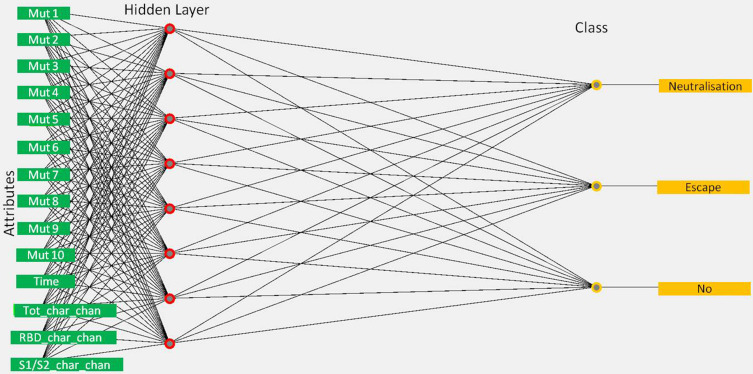
Figure 5.
Conceptual visualization of the final model built using Weka 3.8.5 MultilayerPerceptron classifier for decision making, relating attributes of virus lineages to the evidence for impact on immunity {no, neutralization, escape}. For simplicity only selected 10 mutations are shown in the input layer of neuronal networks. Circles represent neurons and lines represent synapses. The model contains the input layer of considered attributes (32 mutations, time of the first detection, the total change of charge in S, the charge change in RBD, and the charge change in the S1/S2 cleavage area), one hidden layer to process the classification decision, and the output layer of classes for impact on immunity. Synapses take the input and multiply it by a fitted weight. Neurons add the outputs from all synapses and apply a sigmoid transfer function with a fitted threshold.
Calculation of the Potential Electrostatic Screened Energy
The potential electrostatic screened energy U was estimated for charged RBD and ACE2 as a function of a distance d. Point model and screened Coulomb energy formula U=Q1Q2Exp(-d/LD)/(4Πε0εd) was assumed, with Q1= +7 [e] or Q1= +10 [e], Q2= −28 [e], ε= 80, and Debye screening length LD= 0.7 nm. The ε0 is the dielectric permittivity of a vacuum.
Results
Neuronal Analysis
The final MultilayerPerceptron model correctly relates 89% instances (cross-validation 10) with the considered classes of evidence for impact on immunity when they are characterized by the variants of mutation, the time of first detection, total charge change, and charge change in chosen S1/S2 area. Charge change in RBD may be omitted. Fitted parameters fall in the range, for the weights, −1.99 to 2.19, and the threshold, from −2.95 to 1.41.
Changing area of interest S1/S2 for RBD, or both areas, decrease correctness of decisions to 86%. Leaving a single attribute, the charge change in S1/S2 cleavage area offers 82%, which is more than 79%, for the mutations or the time of the first detection, and 75%, for the total charge change or the charge change in RBD. Other tested algorithms offer less correctly classified instances, maximum 79% - Logistic, and 54% - Dl4jMlpClassifier.
Potential Energy
The results of the calculation are presented in Figure 6.
Figure 6.
The potential electrostatic screened energy U is estimated for charged RBD and ACE2 as a function of a distance d. Point model and screened Coulomb energy U=Q1Q2Exp(-d/LD)/(4Πε0εd) was assumed, with Q1= +7 [e] or Q1= +10 [e], Q2= −28 [e], ε= 80, and Debye screening length LD= 0.7 nm. The ε0 is the dielectric permittivity of a vacuum. Two curves represent different RBD charges, referential (+7 [e]) and after positive charge mutation (+10 [e]), as in virus variant Zeta A.V1. The arrow indicates the direction of attraction of the positive electric charge.
Discussion
There is a wealth of publications showing structural data on how mutations in the SARS-CoV-2 spike protein affect the spike structure, the interaction with the receptor ACE2, and infectivity in pseudovirus or authentic virus systems. Structural changes are crucial for PDB crystallographic predictions. However, they seem to be less important than long-range electrostatic interactions in vivo, at the distance of a few nanometers, when the virus approaches the host cell in the mucus environment.
A data report (Figures 2 and 3) indicating accumulation of amino acids carrying formally positive electrical charge in crucial areas of spike S protein, in new variants of SARS-CoV-2, raises the question about its impact on immunity. The shortage of data (Figure 4) may raise some doubts about the broadcasting of the importance of this phenomenon. However, neuronal analysis (Figure 5) of the S charge impact on immunity shows that this question is worth an answer. Variant Delta of the virus has raised the charge +2 both in the RBD and S1/S2 cleavage areas. According to the MultilayerPerceptron classifier model, the change in the S1/S2 cleavage area seems to be more important. It is probably due to the strengthening of the interaction of spike residuals with negatively charged furin.13 The change of the total charge or the charge in RBD is also important for discussing classification, but less than supported with mutations and the time of the first detection. This reminds us that the force of electrostatic attraction between the positive RBD and the negative ACE2 (Figure 6), is an important but not the only factor. The distributions of charged amino acids in RBD and the time of epidemic development may be significant, too. Furthermore, each positive increase of virus surface charge, especially near the epitopic sites, can decrease the effectiveness of positively charged antibodies.14 This requires further investigations of the virus proteins15 and the soft matter physicochemical interactions16 preceding drugs implementation. The author realizes that this analysis and finding are too presumptive and need data support from the wet lab. Some experimental support for the idea of the essentiality in virus infectivity, the mutations increasing the number of positive residues comes from the work on negatively charged polysulfates binding to the spike protein via electrostatic interactions.17 Although the isoelectric point of the spike glycoprotein, pI = 6.24,18 is less than physiological, ph = 7, so the stalk part of the spike is negatively charged,19 the top part of the spike molecule, especially the RBD, remains positively charged for a broad range of pH. Such a charge distribution promotes the spike corona stability and enhances the virion attachment to receptors and surfaces, mostly negatively charged,20 eg, gangliosides-rich lipid rafts.21
Summing up, it seems reasonable to conclude that additional charges may implicate three important consequences:
The increase of the long-range electrostatic interactions of positive parts of SARS-CoV-2 with the negatively charged receptor ACE2 on the surface of the host cells.
The increase of the interaction of the region S1/S2 with negatively charged furin enzyme catalyzing protein S cleavage. Inhibition of the action of positively charged antibodies.
Inhibition of the action of positively charged antibodies
The above can seriously enhance the toxicity of the virus.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Saad-Roy CM, Arinaminpathy N, Wingreen NS, Levin SA, Akey JM, Grenfell BT. Implications of localized charge for human influenza A H1N1 hemagglutinin evolution: insights from deep mutational scans. PLoS Comput Biol. 2020;16(6):e1007892. doi: 10.1371/journal.pcbi.1007892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi Y, Suzuki Y. Compensatory evolution of net-charge in influenza a virus hemagglutinin. PLoS One. 2012;7(7):e40422. doi: 10.1371/journal.pone.0040422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagbom M, Nordgren J, Nybom R, et al. Ionizing air affects influenza virus infectivity and prevents airborne-transmission. Sci Rep. 2015;5:11431. doi: 10.1038/srep11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W. Structurally Observed Electrostatic Features of the COVID-19 Coronavirus-Related Experimental Structures Inside Protein Data Bank: A Brief Update. Preprints.org; 2020. doi: 10.20944/preprints202003.0081.v1. [DOI] [Google Scholar]
- 5.Pawłowski PH. Charged amino acids may promote coronavirus SARS-CoV-2 fusion with the host cell. AIMS Biophys. 2021;8(1):111–120. doi: 10.3934/biophy.2021008 [DOI] [Google Scholar]
- 6.López-Vallejo F, Martínez-Mayorga K. Furin inhibitors: importance of the positive formal charge and beyond. Bioorg Med Chem. 2012;20(14):4462–4471. PMID: 22682919. doi: 10.1016/j.bmc.2012.05.029 [DOI] [PubMed] [Google Scholar]
- 7.Corrêa Giron C, Laaksonen A, Barroso da Silva FL. On the interactions of the receptor-binding domain of SARS-CoV-1 and SARS-CoV-2 spike proteins with monoclonal antibodies and the receptor ACE2. Virus Res. 2020;285:198021. doi: 10.1016/j.virusres.2020.198021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Javidpour L, Božič A, Naji A, Podgornik R. Electrostatic interactions between the SARS-CoV-2 virus and a charged electret fibre. Soft Matter. 2021;17:4296–4303. [DOI] [PubMed] [Google Scholar]
- 9.Luisetto M, Tarro G, Edbey K, et al. Coronavirus COVID-19 surface properties: electrical charges status. Int J Clin Microbiol Biochem Technol. 2021;4:016–027. [Google Scholar]
- 10.European Centre for Disease Prevention and Control. Available from: https://www.ecdc.europa.eu/en/covid-19/variants-concern. Accessed November 17, 2021.
- 11.Public Health England. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/993879/Variants_of_Concern_VOC_Technical_Briefing_15.pdf. Accessed November 17, 2021.
- 12.Frank E, Hall MA, Witten IH. The WEKA Workbench. Online Appendix for Data Mining: Practical Machine Learning Tools and Techniques. 4th ed. Morgan Kaufmann; 2016. [Google Scholar]
- 13.Hasan A, Paray BA, Hussain A, et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J Biomol Struct Dyn. 2021;39(8):3025–3033. doi: 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M, Zhu D, Zhu J, Nussinov R, Ma B. Local and global anatomy of antibody-protein antigen recognition. J Mol Recognit. 2018;31(5):e2693. doi: 10.1002/jmr.2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Yang C, Xu X, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherstvy AG, Petrov EP. Modeling DNA condensation on freestanding cationic lipid membranes. February 2014. Phys Chem Chem Phys. 2020;16. doi: 10.1039/C3CP53433B. [DOI] [PubMed] [Google Scholar]
- 17.Nie C, Pouyan P, Lauster D, et al. Polysulfates Block SARS-CoV-2 Uptake through Electrostatic Interactions. Angew Chem Int Ed. 2021;60:15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheller C, Krebs F, Minkner R, Astner I, Gil-Moles M, Wätzig H. Physicochemical properties of SARS-CoV-2 for drug targeting, virus inactivation and attenuation, vaccine formulation and quality control. Electrophoresis. 2020;41:1137–1151. doi: 10.1002/elps.202000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javidpour L, Božič A, Naji A, Podgornik R. Electrostatic interactions between the SARS-CoV-2 virus and a charged electret fibre. Soft Matter. 2021;17(16):4296–4303. PMID: 33908595 doi: 10.1039/d1sm00232e. [DOI] [PubMed] [Google Scholar]
- 20.Adamczyk Z, Batys P, Barbasz J. SARS-CoV-2 virion physicochemical characteristics pertinent to abiotic substrate attachment. Curr Opin Colloid Interface Sci. 2021;55:101466. doi: 10.1016/j.cocis.2021.101466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fantini J, Yahi N, Azzaz F, Chahinian H. Structural dynamics of SARS-CoV-2 variants: a health monitoring strategy for anticipating Covid-19 outbreaks. J Infect. 2021;83(2):197–206. doi: 10.1016/j.jinf.2021.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]