Abstract
Purpose of review:
Advancements in the next-generation sequencing technologies have identified rare transcripts of long non-coding RNAs (lncRNAs) in the genome of cancers, including in acute myeloid leukemia (AML). The purpose of this review is to highlight the contribution of lncRNAs in AML pathogenesis, prognosis, and chemoresistance.
Recent findings:
Several studies have recently reported that deregulated lncRNAs are novel key players in the development of AML and are associated with AML pathophysiology and may serve as prognostic indicators. A few aberrantly expressed lncRNAs that correlated with the recurrent genetic mutations in AML such as NPM1 and RUNX1 have recently been characterized. Moreover, a few lncRNAs in MLL-rearranged leukemia have been described. Additionally, the involvement of lncRNAs in AML chemoresistance has been postulated.
Summary:
Investigating the functional roles of the non-coding regions including lncRNAs, may provide novel insights into the pathophysiology, refine the prognostic schema, and provide novel therapeutic treatment strategies in AML.
Keywords: Acute myeloid leukemia, lncRNAs, NPM1, RUNX1, FLT3-ITD, MLL, chemoresistance
Introduction
Acute myeloid leukemia (AML) is characterized by an expansion of the abnormal myeloid progenitor cells in the bone marrow and blood resulting from differentiation block (1). Research in AML has traditionally been protein-centric but is now evolving as the involvement of transcripts from the non-coding genome in disease pathogenesis has emerged in recent literature. High throughput analyses demonstrate that human genomes are actively transcribed to produce thousands of lncRNAs that do not encode for functional proteins (2). LncRNAs are arbitrarily defined as RNA transcripts of more than 200 nucleotides in length, transcribed by RNA-polymerase II, which may undergo splicing and polyadenylation (2) (3). LncRNAs exhibit poor sequence conservation across different species but demonstrate higher tissue and development-specific expression than the mRNAs (4). Interestingly, lncRNAs can bind to the DNA, RNAs, and proteins, thereby regulating diverse cellular processes (5). While the number of functional lncRNAs is still debated, it is well established that lncRNAs have important roles in both physiological development and pathological conditions, including cancer initiation, progression, and chemoresistance (5) (6). Figure 1. Illustrates a general overview of lncRNAs’ mechanism(s) that have been reported in AML up to now.
Figure 1. General overview of lncRNA mechanisms involved in AML.
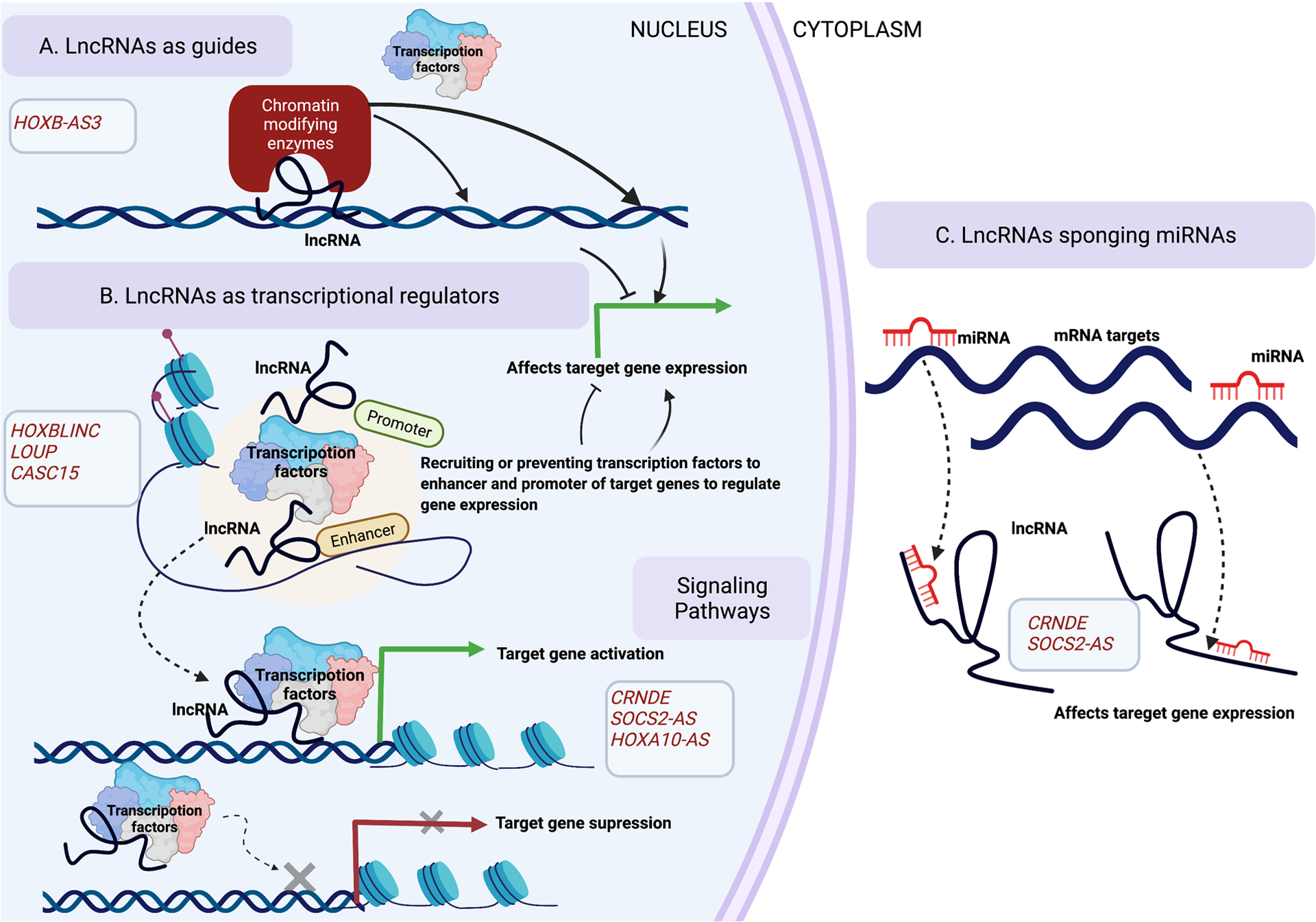
(A) LncRNAs can act as a guide to target chromatin-modifying complexes or transcription factors to specific genomic locations and affect the target gene expression. (B) LncRNAs can serve as transcriptional regulators that can either recruit or prevent transcriptional factors to the enhancers and promoters of target genes, thereby regulating target gene expression. (C) In the cytoplasm, lncRNAs can sequester microRNAs away from their mRNA targets thereby affecting the target gene expression.
In this review, we first discuss lncRNAs in AML pathogenesis and their prognostic significance. Next, we discuss recently characterized lncRNAs that are associated with recurrent mutations in AML including NPM1, RUNX1, FLT3-ITD, and DNMT3A (Table 1). This is followed by a concise discussion on novel lncRNAs involved in MLL-rearranged leukemia (Table 1). Lastly, we discuss lncRNAs involved in AML chemoresistance (Fig. 2, Table 2).
Table 1:
Summary of lncRNAs recently reported in AML
| LncRNAs | Expression in AML | Key findings/mechanism(s) of action | References | |
|---|---|---|---|---|
| LncRNAs associated with NPM1 mutation | HOXB-AS3 | upregulated |
|
(20) |
| HOXBLINC | upregulated |
|
(23) | |
| LONA | upregulated |
|
(26) | |
| CRNDE | upregulated |
|
(28) | |
| LncRNAs associated with RUNX1 | CASC15 | upregulated |
|
(33) |
| LOUP | Downregulated in t(8;21) AML patients |
|
(39) | |
| LncRNAs associated with FLT3-ITD, TET2 mutations | Morrbid | Upregulated in AML patients with FLT3-ITD, TET2 mutations |
|
(44) |
| SOCS2-AS | Upregulated in AML patients with FLT3-ITD mutation |
|
(45) | |
| LncRNAs associated with MLL- rearranged leukemia | LAMP5-AS1 | upregulated |
|
(50) |
| HOXA10-AS | upregulated |
|
(51) |
Figure 2. Mechanisms of lncRNAs involved in AML chemoresistance.
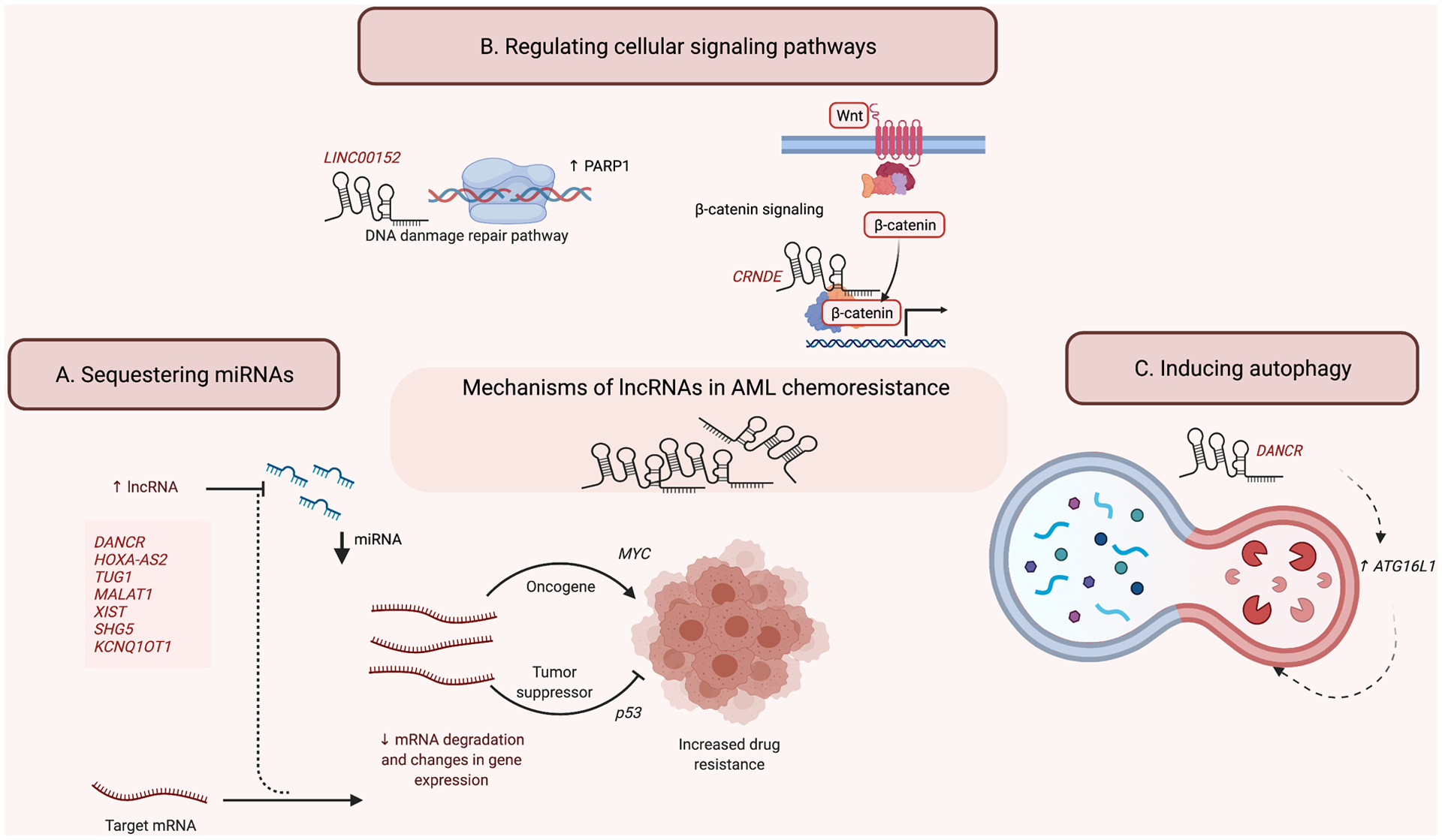
(A) LncRNAs can sequester miRNAs and affect the expression of downstream oncogenes or tumor suppressors to promote drug resistance. (B) LncRNAs can regulate cellular signaling pathways by interacting with transcription factors involved in DNA damage pathways as well as other cell signaling pathways to affect target gene expression. (C) LncRNAs can induce autophagy to maintain chemoresistance.
Table 2:
Recently reported lncRNAs involved in AML chemoresistance
| lncRNA | Chemoresistance status | Mechanism of influence | Target | Changes in gene expression | References |
|---|---|---|---|---|---|
| DANCR | ↑ Chemoresistance to Ara-C | Autophagy, sequestering miRNAs | miR-874–3p | ↑ ATG16L1 | (55) |
| LINC00152 | LSC chemoresistance | DNA damage repair-related signaling | - | ↑ PARP1 | (56) |
| CRNDE | ↑ Chemoresistance to ADR | Wnt/β-catenin pathway | - | - | (57) |
| HOTAIR | ↑ Chemoresistance to ADR | AKT/Notch1 signaling pathways | - | ↑ p21 | (70) |
| HOXA-AS2 | ↑ Chemoresistance to ADR | sequestering miRNAs | miR-520c-3p | ↑ S100A4 | (62) |
| TUG1 | ↑ Chemoresistance to ADR | sequestering miRNAs | miR-34a via recruiting EZH2 | - | (66) |
| MALAT1 | ↑ Chemoresistance to ADR | sequestering miRNAs | miR-96 | - | (67) |
| XIST | ↑ Chemoresistance to ADR | sequestering miRNAs | miR-29a | ↑ MYC | (69) |
| SNHG5 | ↑ Chemoresistance to ADR | Autophagy, sequestering miRNAs | miR-32 | ↑ DNAJB9 | (68) |
| KCNQ1OT1 | ↑ Chemoresistance to ADR | sequestering miRNAs | miR-193a-3p | ↑ TSPAN-3 | (71) |
LncRNAs as potential risk stratification biomarkers in AML
Extensive profiling studies have been a popular strategy to identify distinct lncRNAs associated with AML prognosis and pathogenesis. Several recent studies demonstrated the potential roles of lncRNA signatures in AML risk stratification for patients across a diverse age range. For instance, a combined signature of 48 lncRNAs and 24 lncRNAs can predict the treatment response in older and younger cytogenetically normal AML (CN-AML) patients respectively (7) (8). Mer et al. utilized an RNA-seq based lncRNA profiling of 274 AML patients and the findings were subsequently validated in a TCGA-AML cohort. The authors identified 33 individual lncRNAs associated with overall survival (OS). Based on lncRNAs’ expression, the authors classified all the patients into four distinct molecular subtypes. Remarkably, these lncRNAs-driven subtypes lacked high concordance with any of the conventional genetic or clinical factors or mRNA-based subtypes, suggesting that lncRNAs-subtypes provide independent prognostic information (9). A few studies took a more quantitative approach and formulated a lncRNA scoring system to predict OS and disease-free survival (DFS) in AML, and responses to allogeneic stem cell transplant (10). Relapse after treatment is common in AML. It occurs in 40–50% of younger and even higher in the elderly patients (11). One recent study highlighted the values of lncRNAs’ expression signatures in the prediction of relapse in adult CN-AML patients. Walker et al identified a 10-gene expression signature that is predictive of relapse in CN-AML patients and these are independent of the mutations that are known to impact the outcome in AML, including 3 lncRNA genes (12). The observations from these risk stratification studies await further validation using larger, independent cohorts, but they suggest that perhaps the use of lncRNA signatures could augment the current standard of AML risk stratification based on patient demographics, chromosomal abnormalities, and mutation status. Studies using RNA-seq also demonstrated the intriguing roles of recurrent genetic variants in nucleotide sequences of lncRNAs in AML, although their functional significance awaits further investigation (13).
Association between lncRNAs and recurrent mutations in AML provide mechanistic insights
The interesting observations of the potential prognostic values of lncRNA in AML raise the question of the underlying mechanisms of such associations. While the precise mechanistic roles of many lncRNAs associated with AML remain unknown, we do know a great deal about the mechanisms of master regulators of haematopoiesis and the consequences of their mutations through decades of research. Examples of such regulators include NPM1, RUNX1, FLT3, TET2, DNMT3A, and MLL. Investigating lncRNA associated with mutations in these hematopoietic regulators can provide insights and directions for further investigation of lncRNA’s involvement in AML pathophysiology.
Nucleophosmin (NPM1) associated lncRNAs
NPM1 plays important and multifaceted roles in hematopoiesis and is frequently mutated in AML. Nearly 20%−30% of newly diagnosed AML patients harbor NPM1 mutation (14). Aberrant Homeobox (HOX) expression has been linked to NPM1 mutation in AML (15). The HOXA and HOXB genes are upregulated in NPM1 mutated AML compared to the wild-type NPM1 (16). The mutant NPM1 protein is abnormally localized in the cytoplasm and is recently reported to maintain the leukemic state through HOX expression (17). While a few lncRNAs embedded in the HOX gene loci are reported to participate in cancer pathogenesis by regulating the expression of the protein-coding HOX genes (eg. HOTAIR, HOTTIP) (18,19), others like HOXB-AS3 do not impact the HOX gene expression and function via other mechanisms (20). This section of the review will highlight recently reported, NPM1-associated lncRNAs that are located at the HOX gene loci, HOXB-AS3, and HOXBLINC, as well as lncRNAs that are outside the HOX gene loci.
HOXB-AS3
HOXB-AS3 is upregulated in CN-AML patients with NPM1 mutation. RNA-seq analysis revealed that the expression of HOXB-AS3 in NPM1mut patients (n=223) was approximately three-fold higher than wild-type NPM1 patients (n=120). Knockdown of HOXB-AS3 in OCI-AML3 (NPM1mut AML cell line) led to a decrease in the number of cells in the S-phase as well as a decrease in colony-forming capacity. Reduced expression of HOXB-AS3 also prolonged the survival of xeno-transplanted mice. Mechanistically, HOXB-AS3 interacts with ErbB3-binding protein 1 (EBP1) that can interact with NPM1 in the nucleus to regulate the transcription of rRNA. Thus, HOXB-AS3 guides EBP1 to the rDNA locus and modulates EBP1-NPM1 complex formation which in turn maintains the proliferative phenotype in AML (20). Moreover, higher expression of HOXB-AS3 has also been reported to be an adverse prognostic marker for de novo AML patients as well as primary MDS patients (21).
HOXBLINC
Another lncRNA located at the anterior of HOXB gene locus named HOXBLINC promotes hematopoietic development by upregulating HOXB gene expression via recruitment of SETD1A/MLL histone H3K4 methyltransferases complexes (22). More recently, HOXBLINC was found to be upregulated in AML patients with NPM1 mutation as well as in AML cell lines that carry NPM1 mutation (OCI-AML3 and IMS-M2). Downregulation of HOXBLINC in OCI-AML3 cells significantly decreased cell viability, induced apoptosis, and impaired the transcription of mutant NPM1 (NPM1c+) signature genes like MEIS1 and RUNX1. Moreover, RNA-seq analysis of wild-type LSK cells versus LSK cells derived from NPM1 mutant knock-in (NPM1c/+) mice showed upregulation of HOXBLINC as well as some other signature genes associated with NPM1c+ like MEIS1 and RUNX1. The authors further demonstrated that HOXBLINC mediates long-range chromatin interactions to drive the target genes’ regulatory networks in HSPCs. Consistent with the previous study, HOXBLINC was shown to recruit SETD1A and MLL. Furthermore, recruitment of MLL1 (and not SETD1A) appears to be the key in HOXBLINC overexpression-mediated leukemogenesis (23). This observation was consistent with a previous study suggesting that chromatin binding of MLL1 is crucial for NPM1 mutated leukemias (24). However, how HOXBLINC is upregulated in NPM1c+ AML remains unknown.
Outside of the HOX gene locus, a few other lncRNAs associated with NPM1-mutation in AML have been well characterized in terms of their potential pathogenic mechanisms, including lncRNA overexpressed in NPM1-mutated AML patients (LONA) and lncRNA Colorectal Neoplasia Differentially Expressed (CRNDE)
LncRNA overexpressed in NPM1-mutated AML patients (LONA)
Clara et al. performed RNA-seq on a cohort of 40 AML patients and found 12 distinct lncRNAs that are differentially expressed in wild-type versus mutant NPM1 AML patients (25). In the process of further characterization of these lncRNAs, LONA emerged as a novel lncRNA hypothesized to play a critical role in AML pathogenesis through NPM1. LONA switches its subcellular localization from being cytoplasmic in the wild-type NPM1 AML cells to becoming nuclear in NPM1-mutant AML as the mutant NPM1 is exported in the cytoplasm. Interestingly, knockdown of LONA enhanced Cytarabine (Ara-C)-triggered apoptosis irrespective of the mutational status of NPM1 as evidenced in OCI-AML3 (NPM1-mut) and OCI-AML2 (NPM1-wt) cells. Overexpression of LONA lncRNA reduced the survival of transplanted NOD scid gamma (NSG) mice but did not impact OCI-AML3 cells. Furthermore, manipulation of LONA followed by RNA-seq identified proteins involved in myeloid differentiation such as HSB1, MAFB, and ASB2 as major targets of LONA lncRNA. Taken together, Gourvest et al. reported that the oncogenic functions of LONA lncRNA are through a nucleocytoplasmic cross-transport between mutant NPM1 and LONA lncRNA (26).
lncRNA Colorectal Neoplasia Differentially Expressed (CRNDE)
High expression of CRNDE in AML patients was first reported by Wang et al.(27). The authors demonstrated that knockdown of CRNDE decreased the cell viability and increased apoptosis in U937 cells. More recently, a strong correlation between the expression of CRNDE and NPM1 mutations in AML has been reported. Knockdown of CRNDE in OCI-AML3 cells could promote differentiation as evidenced by the increase of CD11b in CRNDE depleted AML cells. Similar inhibitory effects were also observed when CRNDE was downregulated in FAB M3 AML cells-NB4. Mechanistically, the authors demonstrated that CRNDE promotes leukemogenesis by sponging miR-181 and modulating the NOTCH signaling pathway in acute promyelocytic cells (28). However, the precise functional roles of CRNDE in NPM1 mutated AML remains unclear. Recently, Hola et al. conducted qRT-PCR based expression analysis of CRNDE from the diagnostic blood samples of 200 de novo adult AML patients compared to 50 healthy control samples, which revealed that upregulation of CRNDE was an adverse independent prognostic marker for complete remission in CN-AML patients (29).
RUNX1-associated lncRNAs
Runt-related transcription factor 1 (RUNX1) is a master regulator of hematopoiesis and is crucial for defining the definitive hematopoietic stem cell (30). It is found to be frequently mutated in a variety of hematological malignancies (31). The RUNX1 gene is involved in several chromosomal translocations in leukemia. The most common chromosomal translocation is the t (8;21) which produces a RUNX1-ETO chimeric protein and is found in 30%−40% of FAB-M2 AML patients (32). This section of the review will highlight recently reported RUNX1 associated lncRNAs that have been characterized in AML, including CASC15, and LOUP.
CASC15
Another study by Fernando et al. found upregulation of lncRNA cancer susceptibility candidate 15 (CASC15) in pediatric AML patients with the t (8;21) translocation and in B-cell acute lymphoid leukemia (B-ALL) patients with the t (12;21) translocation. The authors demonstrated that CASC15 affects the expression of its neighboring oncogene-SOX4, by regulating the Yin and Yang-1 (YY1) transcription factor (33). Thus, this is an example demonstrating that lncRNAs can recruit transcription factors and affect gene expression. Notably, although CASC15 is upregulated in RUNX1-translocated leukemia, the mechanism of its action was found to be independent of RUNX1.
LOUP
Trinh et al. have identified a novel lncRNA-Long noncoding RNA originating from the upstream regulatory element (URE) of PU.1 (also known as Spi-1), named as LOUP that mediates active chromatin loop formation at PU.1 locus which is critical for PU.1 expression.
PU.1, a major downstream target of RUNX1, is a hematopoietic lineage-specific ETS-family member and its downregulation is reported to vitiate myeloid cell differentiation leading to AML (34) (35) (36). PU.1 expression is impeded by RUNX1-ETO (AML with t (8;21) translocation) but the precise mechanism of this inhibition is unclear (37). Trinh et al. identified a spliced, and polyadenylated lncRNA-LOUP that was expressed exclusively in the myeloid cells. Depletion of LOUP in U937 cells increased the cell proliferation and decreased the expression of a myeloid marker, CD11b. Interestingly, LOUP expression was correlated with PU.1 expression, and knockdown of LOUP also depleted PU.1 mRNA levels. Previous studies have shown that the formation of chromatin looping facilitated by the URE and proximal promoter (PrPr) interaction is required for PU.1 induction (34) (38). In this study, the authors demonstrated that at the PU.1 locus, LOUP mediates the chromatin loop formation by recruiting RUNX1 to the RUNX1-binding motifs at both the URE and the PrPr. Interestingly, LOUP and PU.1 were downregulated in AML patients with t (8;21) translocation as compared to AML patients with normal karyotype. Last, RUNX1-ETO was demonstrated to inhibit LOUP transcription by deacetylating thereby restricting the promoter accessibility (39).
LncRNAs associated FLT3-ITD, TET2, and DNMT3A mutations
About 30% of AML cases have mutations in the Fms-like tyrosine kinase 3 (FLT3) gene. The more frequent internal tandem duplication (ITD) mutation in the juxtamembrane domain is found in 25% of AML cases while point mutation or deletion in the tyrosine kinase domain (TKD) is found in about 7%−10% of AML cases (40). Also, the genes involved in DNA methylation including ten-eleven translocation 2 (TET2) and DNA methyltransferase 3A (DNMT3A) are frequently mutated in CN-AML patients (41). DNMT3A mutations occur in approximately 20% of AML cases and are independently associated with a poor outcome (41,42). In this section, we discuss some recently reported lncRNAs associated with FLT3-ITD, TET2, and DNMT3A mutations in AML.
MORRBID
MORRBID, a lncRNA specifically expressed in myeloid cells, has been previously shown to repress the pro-apoptotic gene BIM via a DNA loop formation in cis (43). Recently, higher expression of MORRBID was reported in AML patients with TET2 mutation. Also, a three-fold increase in MORRBID expression was observed in patients with FLT3-ITD mutation. To investigate the functional significance of MORRBID in AML, Cai et al. utilized a murine model of AML induced by loss of TET2 and FLT3-ITD mutation (TF mice) and mutants of TF lacking Morrbid (TFM mice). Data from these models suggested that loss of Morrbid increased Bim expression and reduced the leukemic cell percentage in peripheral blood of TFM mice which prevented the infiltration of leukemic cells in the lungs of TFM mice, thus prolonging their survival (44).
SOCS2-AS
Overexpression of lncRNA suppressor of cytokine signaling-2 (SOCS2-AS) was associated with AML patients and cells harboring FLT3-ITD mutation. siRNA-mediated knockdown of SOCS2-AS in FLT3-ITD+ AML cells (Molm-13, MV4–11) decreased the cell viability and induced apoptosis. Also, decreased expression of STAT5/p-STAT5 was observed post knockdown of SOCS2-AS. Moreover, it was reported that SOCS2-AS could regulate genes of the STAT5 pathway by sponging miR221 (45).
Lastly, Yu et al. analyzed TCGA-LAML dataset and reported 619 differentially expressed (DE) lncRNAs as well as 1,428 DEmRNA genes in FLT3-ITD mutant samples versus FLT3-ITD WT samples. Interestingly, high expression of SH3TC2-DT/SH3TC2 gene pairs was associated with FLT3-ITD mutation, high WBC count, intermediate genetic risk, and poor survival in AML. High expression of SH3TC2-DT/SH3TC2 gene pair in FLT3-ITD mutants was also validated in the BeatAML dataset. Additionally, high expression of this gene pair also showed enrichment of transcripts related to stemness, quiescence, and leukemogenesis thereby hinting at a possible role in FLT3-mutant LSCs (40). However, further investigations are essential to confirm this hypothesis.
Analysis of differential gene expression in Dnmt3a R878H mutation conditional knock-in mice compared to WT mice identified 6 murine lncRNAs that were associated with DNMT3A mutation and poor prognosis. However, these lncRNAs were not conserved in humans and hence the expression of these murine lncRNAs could not be evaluated in human AML patients (46). A comprehensive evaluation of specific lncRNAs associated with DNMT3A-mutation is still awaited. Additionally, the expression of HOTAIR and H19 positively correlates with DNMT3A mutated AML (47,48). However, their functional role in the leukemogenesis of DNMT3A-mutated AML is unexplored.
LncRNAs associated with mixed-lineage leukemia (MLL) gene rearrangements
Nearly 70% of infant leukemic patients and about 10% of adult AML cases are reported with rearrangement of mixed-lineage leukemia (MLL) gene (now renamed Lysine [K]-specific MethylTransferase 2A or KMT2A) by 11q23 translocation (49).
Recently, lncRNAs HOXA10-AS and LAMP5-AS1 have been associated with MLL-rearranged leukemia.
LAMP5-AS1
LAMP5-AS1 was significantly upregulated in MLL leukemia patients (n=58) as compared to MLL-WT patients (n=163). Knockdown of LAMP5-AS1 in the primary MLL CD34+ leukemic cells inhibited the self-renewal capacity and increased differentiation in the cells. Also, similar data were obtained from the xenograft models. Mechanistically, LAMP5-AS1 was demonstrated to interact and enhance the enzyme activity of a histone methyltransferase-disruptor of telomeric silencing 1-like (DOT1L) in MLL leukemia (50). In summary, the high expression of LAMP-AS1 in MLL leukemia helps to maintain high methyltransferase activity of DOT1L and global H3K79 methylation thereby upregulating transcription of DOT1L target genes, including genes involved in the self-renewal.
HOXA10-AS
HOXA10-AS is an HSC-specific lncRNA and it is upregulated in various MLL-rearranged AML cell lines. Knockdown of HOXA10-AS reduced the number of cells in the S-phase and induced apoptosis in AML cells. In contrast, the overexpression of HOXA10-AS in CD34+ HSPCs significantly impaired monocytic differentiation. Mechanistically, the authors demonstrated that HOXA10-AS acted in trans via upregulating the target genes of the NF- kB pathway (51).
Potential mechanisms of lncRNAs conferring drug resistance in AML: autophagy, cellular signaling, and microRNA sponging
Numerous studies support the hypothesis that deregulated lncRNAs can contribute to AML chemoresistance. In this section, we discuss some of the recently identified lncRNAs involved in chemoresistance in AML, especially those that act through leukemic stem cells (LSCs), regulate important cellular signaling pathways, and sequestering microRNAs (Fig. 2). Table 2 provides a summary of lncRNAs discussed in this review.
LSCs are an important subset of tumor cells involved in AML pathogenesis (52). The presence of LSCs has been proposed as a major cause of relapse in AML due to their self-renewal property. In this regard, two LSCs-associated lncRNAs- DANCR and LINC00152, have recently been investigated for their contribution to AML chemoresistance. DANCR, previously described as an LSC-associated lncRNA based on RNA-seq profiling studies, was found to be upregulated in AML cells treated with Ara-C. Knockdown of DANCR in LSCs led to a decrease in stem-cell renewal capacity, and in vivo targeting of DANCR increased the survival of mice after serial transplantation (53). Moreover, the overexpression of DANCR confers while siRNA-mediated depletion decreases the Ara-C resistance in human AML cell lines. The induction of autophagy is a mechanism utilized by cancer cells to gain resistance to the anti-cancer drugs (54). DANCR promoted autophagy in Ara-C treated AML cells by sponging miR-874–3P, which in turn upregulated the expression of ATG16L1, a critical component in autophagy and is associated with multiple-drug resistance in solid tumors (55). LINC00152 was highly expressed in CD34+ LSCs in AML patients. The expression of LINC00152 was correlated with 15 genes within a cluster of 17-gene biomarkers that would accurately predict chemoresistance in AML. Interestingly, knockdown of LINC00152 reduced chemoresistance in K562 cells. Moreover, the expression of LINC00152 was correlated with poly (ADP-ribose) polymerase 1 (PARP1) expression and knockdown of LINC00152 resulted in decreased PARP1 expression which consequently resulted in an increased sensitivity of AML cells to the DNA damaging agent-doxorubicin. Thus, LINC00152 potentially contributes to the chemoresistance of LSCs in AML via upregulating PARP1 (56).
LncRNAs can affect various cell signaling pathways and contribute to chemoresistance. For example, lncRNA-CRNDE inhibits the Wnt/β-catenin pathway while HOTAIR promotes drug resistance partially via the AKT pathway. LncRNA-CRNDE was significantly upregulated in patients with Adriamycin (ADR) based chemotherapy. Notably, knockdown of CRNDE increased apoptosis, suppressed proliferation, and increased chemosensitivity of ADR-resistant AML cells. Moreover, the authors demonstrated that in ADR-resistant AML cells, CRNDE functions via inhibiting the Wnt/β-catenin pathway (57). Another lncRNA, HOTAIR, is overexpressed in AML and its expression is decreased post-treatment in AML patients (58). It is consistently reported to be correlated with shorter OS and DFS (47,58,59). It is known to mediate ADR resistance by regulating the expression of P21 and AKT/Notch1 signaling pathways in K562/A02 cells (60).
One emerging mechanism of how lncRNAs mediate chemoresistance is through sequestration of different miRNAs and regulating their target genes. LncRNAs HOXA-AS2, TUG1, MALAT1, XIST, SNHG5, and KCNQ1OT1 are some of such examples, in addition to DANCR as described above. HOXA-AS2 is overexpressed in AML tissues and cell lines (61), and it is upregulated in AML patients after ADR chemotherapy (62). The investigations on ADR-resistant AML cells demonstrated that HOXA-AS2 sponges miR-520c-3p. Additionally, 100 calcium-binding protein A4 (S100A4) was found to be the downstream target of miR-520c-3p (62). S100A4 is overexpressed in the nuclear proteome of AML as compared to normal CD34+ cells and is important for AML survival (63). However, the role of S100A4 in AML chemoresistance remains unknown. Higher expression of lncRNA Taurine upregulated gene 1 (TUG1) has been correlated with poor OS and lower complete response (CR) rates in AML patients (64,65). Recently, Li et al. showed that TUG1 was upregulated in ADR-resistant AML patients as compared to ADR-sensitive AML patients. Knockdown of TUG1 could resensitize HL60/ADR cells to ADR by promoting ADR-induced apoptosis and overcome ADR resistance by epigenetically silencing miR-34a via recruiting EZH2 in AML (66). A negative correlation between MALAT1 expression and miR-96 expression was observed in AML patients as compared to healthy controls. Hu et al. demonstrated that MALAT1 sponges miR-96 and knockdown of MALAT1 could enhance the sensitivity of ADR-resistant AML cells by upregulating miR-96 (67). Similarly, lncRNA XIST, SNHG5, and KCNQ1OT1 mediate chemoresistance by modulating the miR-29a/myc, miR-32/DNAJB9, and miR-326/myc+miR-193a-3p/TSPAN-3 axis, respectively (68,69). Taken together, the current literature has accumulated evidence that lncRNAs could regulate drug resistance in AML via diverse mechanisms (Fig. 2).
Conclusion
Dysregulation of lncRNAs has been postulated as a key player in promoting leukemogenesis and drug resistance in AML. In this review, we summarized recent studies on lncRNAs that are associated with prognosis, mutational signatures, and drug resistance in AML. However, in-depth functional characterization of the majority of these lncRNAs is still lacking. Thus, dissecting the diverse mechanisms specifically shaped by lncRNAs, such as how they promote self-renewal or differentiation block, could be highly beneficial in elucidating novel therapeutic targets in the treatment of AML.
Key Points:
Aberrantly expressed lncRNAs have prognostic potential and may serve as novel biomarkers for risk stratification in AML.
Several lncRNAs’ expression has been positively correlated with AML chemoresistance, with diverse mechanisms ranging from autophagy regulation to microRNA sponging.
LncRNAs associated with recurrent mutations in master regulators of hematopoiesis have provided mechanistic insights into their functional roles in AML pathogenesis.
Financial support and sponsorship:
This work was supported by the Singapore Ministry of Health’s National Medical Research Council (Singapore Translational Research (STaR) Investigator Award STaR18nov-0002 (D.G.T.); the Singapore Ministry of Education under its Research Centres of Excellence initiative; NIH/NCI Grant R35CA197697 and NIH/NHLBI P01HL131477–01A1 (D.G.T); as well as NIH/NHLBI Grant P01HL095489 and Xiu research fund (L.C.).
Footnotes
Conflicts of interest: None
REFERENCES
Papers of particular interest, published within the annual period of review have been highlighted as
∗ of special interest
∗ ∗ of outstanding interest
- 1.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003. February;3(2):89–101. [DOI] [PubMed] [Google Scholar]
- 2.Yao R-W, Wang Y, Chen L-L. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019. May;21(5):542–51. [DOI] [PubMed] [Google Scholar]
- 3.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015. January 19;47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009. February 20;136(4):629–41. [DOI] [PubMed] [Google Scholar]
- 5.Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021. February;22(2):96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izadirad M, Jafari L, James AR, Unfried JP, Wu Z-X, Chen Z-S. Long noncoding RNAs have pivotal roles in chemoresistance of acute myeloid leukemia. Drug Discov Today [Internet]. 2021. March 27; Available from: 10.1016/j.drudis.2021.03.017 [DOI] [PubMed] [Google Scholar]
- 7.Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014. December 30;111(52):18679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaioannou D, Nicolet D, Volinia S, Mrózek K, Yan P, Bundschuh R, et al. Prognostic and biologic significance of long non-coding RNA profiling in younger adults with cytogenetically normal acute myeloid leukemia [Internet]. Vol. 102, Haematologica. 2017. p. 1391–400. Available from: 10.3324/haematol.2017.166215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mer AS, Lindberg J, Nilsson C, Klevebring D, Wang M, Grönberg H, et al. Expression levels of long non-coding RNAs are prognostic for AML outcome. J Hematol Oncol. 2018. April 7;11(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai C-H, Yao C-Y, Tien F-M, Tang J-L, Kuo Y-Y, Chiu Y-C, et al. Incorporation of long non-coding RNA expression profile in the 2017 ELN risk classification can improve prognostic prediction of acute myeloid leukemia patients. EBioMedicine. 2019. February;40:240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thol F, Ganser A. Treatment of Relapsed Acute Myeloid Leukemia. Curr Treat Options Oncol. 2020. June 29;21(8):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker CJ, Mrózek K, Ozer HG, Nicolet D, Kohlschmidt J, Papaioannou D, et al. Gene expression signature predicts relapse in adult patients with cytogenetically normal acute myeloid leukemia. Blood Adv. 2021. March 9;5(5):1474–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papaioannou D, Ozer HG, Nicolet D, Urs AP, Herold T, Mrózek K, et al. Clinical and molecular relevance of genetic variants in the non-coding transcriptome of patients with cytogenetically normal acute myeloid leukemia. Haematologica [Internet]. 2021. July 15; Available from: 10.3324/haematol.2021.266643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011. January 27;117(4):1109–20. [DOI] [PubMed] [Google Scholar]
- 15.Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013. April;27(5):1000–8. [DOI] [PubMed] [Google Scholar]
- 16.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015. June;29(6):1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang Y-H, Ramabadran R, et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell. 2018. September 10;34(3):499–512.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007. June 29;129(7):1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011. April 7;472(7341):120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papaioannou D, Petri A, Dovey OM, Terreri S, Wang E, Collins FA, et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun. 2019. November 25;10(1):5351. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ ∗ This study demonstrates how HOXB-AS3 lncRNA acts as a guide and regulates the rRNA transcription which helps to maintain the proliferative phenotype in NPM1-mutated leukemia.
- 21.Huang H-H, Chen F-Y, Chou W-C, Hou H-A, Ko B-S, Lin C-T, et al. Long non-coding RNA HOXB-AS3 promotes myeloid cell proliferation and its higher expression is an adverse prognostic marker in patients with acute myeloid leukemia and myelodysplastic syndrome. BMC Cancer. 2019. June 24;19(1):617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng C, Li Y, Zhou L, Cho J, Patel B, Terada N, et al. HoxBlinc RNA Recruits Set1/MLL Complexes to Activate Hox Gene Expression Patterns and Mesoderm Lineage Development. Cell Rep. 2016. January 5;14(1):103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu G, Luo H, Feng Y, Guryanova OA, Xu J, Chen S, et al. HOXBLINC long non-coding RNA activation promotes leukemogenesis in NPM1-mutant acute myeloid leukemia. Nat Commun. 2021. March 29;12(1):1956. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ ∗ This is a detailed study reporting the oncogenic roles of HOXBLINC lncRNA in NPM1c+ leukemia and how HOXBLINC is involved in epigenetic gene regulation in HSPSc.
- 24.Kühn MWM, Song E, Feng Z, Sinha A, Chen C-W, Deshpande AJ, et al. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov. 2016. October;6(10):1166–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Clara E, Gourvest M, Ma H, Vergez F, Tosolini M, Dejean S, et al. Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica. 2017. October;102(10):1718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gourvest M, De Clara E, Wu H-C, Touriol C, Meggetto F, De Thé H, et al. A novel leukemic route of mutant NPM1 through nuclear import of the overexpressed long noncoding RNA LONA. Leukemia [Internet]. 2021. June 15; Available from: 10.1038/s41375-021-01307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study has uncovered a novel leukemic path that is NPM1 mutation dependent, involving a nucleocytoplasmic cross-transport between LONA and mutant NPM1.
- 27.Wang Y, Zhou Q, Ma J-J. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. Eur Rev Med Pharmacol Sci. 2018. February;22(3):763–70. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, Zhang W, Zhao M, Li S, Jin W, Wang K. Oncogenic role of lncRNA CRNDE in acute promyelocytic leukemia and NPM1-mutant acute myeloid leukemia. Cell Death Discov. 2020. November 11;6(1):121. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study has highlighted the oncogenic roles of CRNDE in APL and NPM1-mutated AML.
- 29.Hola MAM, Ali MAM, ElNahass Y, Salem TAE, Mohamed MR. Expression and prognostic relevance of long noncoding RNAs CRNDE and AOX2P in adult acute myeloid leukemia. Int J Lab Hematol [Internet]. 2021. June 15; Available from: 10.1111/ijlh.13586 [DOI] [PubMed] [Google Scholar]
- 30.Kishtagari A, Levine RL, Viny AD. Driver mutations in acute myeloid leukemia. Curr Opin Hematol. 2020. March;27(2):49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rejeski K, Duque-Afonso J, Lübbert M. AML1/ETO and its function as a regulator of gene transcription via epigenetic mechanisms. Oncogene [Internet]. 2021. July 30; Available from: 10.1038/s41388-021-01952-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossetti S, Sacchi N. RUNX1: A microRNA hub in normal and malignant hematopoiesis. Int J Mol Sci. 2013. January 14;14(1):1566–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017. July 19;16(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebralidze AK, Guibal FC, Steidl U, Zhang P, Lee S, Bartholdy B, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev. 2008. August 1;22(15):2085–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004. June;36(6):624–30. [DOI] [PubMed] [Google Scholar]
- 36.van der Kouwe E, Heller G, Czibere A, Pulikkan JA, Agreiter C, Castilla LH, et al. Core binding factor leukemia hijacks T-cell prone PU.1 antisense promoter. Blood [Internet]. 2021. May 19; Available from: 10.1182/blood.2020008971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staber PB, Zhang P, Ye M, Welner RS, Levantini E, Di Ruscio A, et al. The Runx-PU.1 pathway preserves normal and AML/ETO9a leukemic stem cells. Blood. 2014. October 9;124(15):2391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staber PB, Zhang P, Ye M, Welner RS, Nombela-Arrieta C, Bach C, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013. March 7;49(5):934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinh BQ, Ummarino S, Zhang Y, Ebralidze AK, Bassal MA, Nguyen TM, et al. Myeloid lncRNA LOUP Mediates Opposing Regulatory Effects of RUNX1 and RUNX1-ETO in t(8;21) AML. Blood [Internet]. 2021. May 10; Available from: 10.1182/blood.2020007920 [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study has identified a novel myeloid-specific lncRNA-LOUP that has provided mechanistic insights into the cross-regulation and molecular interactions of lncRNAs with transcription factors and their oncogenic derivatives.
- 40.Yu P, Lan H, Song X, Pan Z. High Expression of the SH3TC2-DT/SH3TC2 Gene Pair Associated With FLT3 Mutation and Poor Survival in Acute Myeloid Leukemia: An Integrated TCGA Analysis. Front Oncol. 2020. June 19;10:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park DJ, Kwon A, Cho B-S, Kim H-J, Hwang K-A, Kim M, et al. Characteristics of DNMT3A mutations in acute myeloid leukemia. Blood Res. 2020. March;55(1):17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010. December 16;363(25):2424–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016. August 15;537(7619):239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai Z, Aguilera F, Ramdas B, Daulatabad SV, Srivastava R, Kotzin JJ, et al. Targeting Bim via a lncRNA Morrbid Regulates the Survival of Preleukemic and Leukemic Cells. Cell Rep. 2020. June 23;31(12):107816. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study has identified a novel myeloid regulator- Morrbid and with the help of various mouse models it has depicted that Morrbid is a myeloid-specific cell survival regulator both in physiologic as well as in leukemic conditions.
- 45.Zhang R, Huo C-H. Long Noncoding RNA SOCS2-AS Promotes Leukemogenesis in FLT3-ITD+ Acute Myeloid Leukemia Through miRNA-221. Onco Targets Ther. 2020. April 5;13:2925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai Y-J, Hu F, He S-Y, Wang Y-Y. Epigenetic landscape analysis of lncRNAs in acute myeloid leukemia with DNMT3A mutations. Ann Transl Med. 2020. March;8(6):318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y-Y, Huang S-H, Zhou H-R, Chen C-J, Tian L-H, Shen J-Z. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol Rep. 2016. December;36(6):3113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang T-J, Zhou J-D, Zhang W, Lin J, Ma J-C, Wen X-M, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018. April 10;10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winters AC, Bernt KM. MLL-Rearranged Leukemias—An Update on Science and Clinical Approaches. Frontiers in Pediatrics. 2017;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W-T, Chen T-Q, Zeng Z-C, Pan Q, Huang W, Han C, et al. The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia. J Hematol Oncol. 2020. June 17;13(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ ∗ This is the first study that has mechanistically characterized the epigenetic roles of LAMP5-AS in the self-renewal program and differentiation block in MLL leukemia cells by facilitating the methyltransferase activity of DOT1L and global H3K79 methylation.
- 51.Al-Kershi S, Bhayadia R, Ng M, Verboon L, Emmrich S, Gack L, et al. The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia. Blood Adv. 2019. December 23;3(24):4252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study demonstrates that HOXA10-AS acts in trans and induces NF-kB target genes in KMT2A-r AML.
- 52.Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. 2017. March 23;129(12):1627–35. [DOI] [PubMed] [Google Scholar]
- 53.Bill M, Papaioannou D, Karunasiri M, Kohlschmidt J, Pepe F, Walker CJ, et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia. 2019. September;33(9):2169–82. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Zhou Y, Li Y, Yang L, Ma Y, Peng X, et al. Autophagy: A novel mechanism of chemoresistance in cancers. Biomed Pharmacother. 2019. November;119:109415. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Liu L, Chen L, Liu H, Ren S, Tao Y. Long noncoding RNA DANCR confers cytarabine resistance in acute myeloid leukemia by activating autophagy via the miR-874–3P/ATG16L1 axis. Mol Oncol. 2021. April;15(4):1203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study identified lncRNA DANCR as a novel positive regulator of Ara-C resistance in AML cells and shows that miR-874–3P/ATG16L1 axis is the key player in conferring chemoresistance.
- 56.Cui C, Wang Y, Gong W, He H, Zhang H, Shi W, et al. Long non-coding RNA LINC00152 regulates self-renewal of leukemia stem cells and induces chemoresistance in acute myeloid leukemia. Front Oncol. 2021. July 6;11:694021. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study highlights the contribution of LINC00152 in regulating self-renewal of LSCs and induction of drug resistance via PARP1.
- 57.Kang Y, Zhang S, Cao W, Wan D, Sun L. Knockdown of LncRNA CRNDE suppresses proliferation and P-glycoprotein-mediated multidrug resistance in acute myelocytic leukemia through the Wnt/β-catenin pathway. Biosci Rep [Internet]. 2020. June 26;40(6). Available from: 10.1042/BSR20193450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B, et al. Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett. 2015. October 1;10(4):2410–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol. 2015. June 1;8(6):7223–8. [PMC free article] [PubMed] [Google Scholar]
- 60.Li M-L, Wang Y, Xu Y-N, Lu Q-Y. Overexpression of LncRNA-HOTAIR promotes chemoresistance in acute leukemia cells. Int J Clin Exp Pathol. 2020. December 1;13(12):3044–51. [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Y, Hu S, Li L, Peng X, Chen F. Long noncoding RNA HOXA-AS2 functions as an oncogene by binding to EZH2 and suppressing LATS2 in acute myeloid leukemia (AML). Cell Death Dis. 2020. December 2;11(12):1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X, et al. Knockdown of Long Noncoding RNA HOXA-AS2 Suppresses Chemoresistance of Acute Myeloid Leukemia via the miR-520c-3p/S100A4 Axis. Cell Physiol Biochem. 2018. November 22;51(2):886–96. [DOI] [PubMed] [Google Scholar]
- 63.Alanazi B, Munje CR, Rastogi N, Williamson AJK, Taylor S, Hole PS, et al. Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia. Leukemia. 2020. February;34(2):427–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi J, Shi X, Dai R-Q. The prognostic impact of abnormally expressed, long noncoding RNAs in acute myeloid leukemia: a meta-analysis. Hematology. 2020. December;25(1):219–28. [DOI] [PubMed] [Google Scholar]
- 65.Qin J, Bao H, Li H. Correlation of long non-coding RNA taurine-upregulated gene 1 with disease conditions and prognosis, as well as its effect on cell activities in acute myeloid leukemia. Cancer Biomark. 2018;23(4):569–77. [DOI] [PubMed] [Google Scholar]
- 66.Li Q, Song W, Wang J. TUG1 confers Adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2. Biomed Pharmacother. 2019. January;109:1793–801. [DOI] [PubMed] [Google Scholar]
- 67.Hu N, Chen L, Wang C, Zhao H. MALAT1 knockdown inhibits proliferation and enhances cytarabine chemosensitivity by upregulating miR-96 in acute myeloid leukemia cells. Biomed Pharmacother. 2019. April;112:108720. [DOI] [PubMed] [Google Scholar]
- 68.Wang D, Zeng T, Lin Z, Yan L, Wang F, Tang L, et al. Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed Pharmacother. 2020. March;123:109802. [DOI] [PubMed] [Google Scholar]
- 69.Wang C, Li L, Li M, Wang W, Liu Y, Wang S. Silencing long non-coding RNA XIST suppresses drug resistance in acute myeloid leukemia through downregulation of MYC by elevating microRNA-29a expression. Mol Med. 2020. November 24;26(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∗ This study highlights the contribution of XIST in AML chemoresistance.
- 70.Hu L, Liu J, Meng Y, Zheng H, Ding C, Wang H, et al. Long non-coding RNA HOTAIR regulates myeloid differentiation through the upregulation of p21 via miR-17–5p in acute myeloid leukaemia. RNA Biol. 2020. December 9;1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun H, Sun Y, Chen Q, Xu Z. LncRNA KCNQ1OT1 contributes to the progression and chemoresistance in acute myeloid leukemia by modulating Tspan3 through suppressing miR-193a-3p. Life Sci. 2020. January 15;241:117161. [DOI] [PubMed] [Google Scholar]


