Abstract
Hepatocyte nuclear factor 4α (HNF4α) is an orphan receptor of the nuclear receptor superfamily and expressed in vertebrates as a tissue-specific transcription factor in liver, kidney, intestine, stomach, and pancreas. It also plays a crucial role in early embryonic development and has been identified as a maternal component in the Xenopus egg. We now report on an activity present in Xenopus embryos that inhibits the DNA binding of HNF4. This HNF4 inhibitor copurifies with a 25-kDa protein under nondenaturing conditions but can be separated from this protein by sodium dodecyl sulfate treatment. Protease treatment of the inhibitor results in a core fragment of about 5 kDa that retains full inhibitory activity. The activity of the HNF4 inhibitor can also be monitored in the absence of DNA, as it alters the mobility of the HNF4 protein in native polyacrylamide gels and the accessibility of antibodies. Comparing the activity of the HNF4 inhibitor with acyl coenzyme A's, recently proposed to be ligands of HNF4, we observe a more stringent specificity for the HNF4 inhibitor activity. Using deletion constructs of the HNF4 protein, we could show that the potential ligand-binding domain of HNF4 is not required, and thus the HNF4 inhibitor does not represent a classical ligand as defined for the nuclear receptor superfamily. Based on our previous finding that maternal HNF4 is abundantly present in Xenopus embryos but the target gene HNF1α is only marginally expressed, we propose that the HNF4 inhibitor functions in the embryo to restrict the activity of the maternal HNF4 proteins.
Hepatocyte nuclear factor 4 (HNF4) constitutes transcription factor subfamily 2A (28), whose first member, HNF4α (NR2A1), has been identified as a factor interacting with promoter elements mediating liver-specific transcription (35). Based on the zinc finger motif of the DNA-binding domain and on a potential ligand-binding domain, HNF4 is classified as a member of the nuclear orphan receptor superfamily (33). Recently, it has been reported that acyl coenzyme A's (acyl-CoAs) are potential ligands of HNF4α: acyl-CoAs containing fatty acids with 16 C residues or shorter act as agonists by increasing the DNA-binding potential of HNF4α, whereas acyl-CoAs with 18 C residues or longer have antagonistic properties and inhibit DNA binding of HNF4α (11). HNF4α turned out to be present as well in nonhepatic cells such as kidney, intestine, stomach, and pancreas (23, 38, 45). The importance of HNF4α in gene control in tissues distinct from the liver has been documented by the fact that an inherited human disease is based on the expression of a mutated HNF4α gene in the β cells of the endocrine pancreas, leading to maturity-onset diabetes of the young (MODY1 [42]). Most interestingly, another MODY gene identified in humans represents the tissue-specific transcription factor HNF1α (43), known to be tightly regulated by HNF4 (17, 39, 44).
In addition to its role as a tissue-specific transcription factor, HNF4α is also a maternal component in the egg of Drosophila melanogaster (46) as well as of the amphibian species Xenopus laevis (12). In the mouse, an early embryonic function is implied by the fact that HNF4α transcripts are present in the primary endoderm at day 4.5 (6) and can be detected in totipotent embryonic stem cells (25). Disruption of the gene encoding HNF4α in the mouse established that HNF4α plays an essential role for the function of the visceral endoderm prior to gastrulation, leading to early embryonic death (2, 7). This embryonic lethality can be rescued by complementing the defective embryo with wild-type-derived visceral endoderm, leading to specification and early differentiation of the liver but to a dramatic failure in full hepatic differentiation (21).
In the Xenopus egg, we identified the HNF4α and the related HNF4β proteins as maternal transcription factors (13). Both proteins (12) are believed to contribute to the zygotic activation of the gene encoding HNF1α, a distinct tissue-specific transcription factor of the homeodomain family. Consistent with this assumption, we have identified a functional HNF4 binding site in the HNF1α promoter (12, 13, 30). Furthermore, overexpression of HNF4α or HNF4β in Xenopus embryos leads to a dramatic increase in expression of the endogenous HNF1α gene as early as the late blastula stage (26). This demonstrates that the HNF1α promoter is accessible for artificial activation at these early embryonic stages. Surprisingly, in early Xenopus embryogenesis, HNF1α transcription is very low (1, 26), although the transcription factors HNF4α and HNF4β are abundantly present (12, 13). In addition, the HNF4 proteins are distributed in a gradient from the animal to the vegetal pole, with the highest concentration in the animal region that does not differentiate into tissues expressing the target gene HNF1α (12, 29). The discrepancy between the presence of the transcription factor HNF4 and the inactivity of its target gene HNF1α might now be explained by our finding that a novel component in the Xenopus embryo inhibits the DNA-binding activity of HNF4.
MATERIALS AND METHODS
Purification of HNF4 inhibitor from egg extracts.
Xenopus eggs were dejellied in 2.5% cysteine hydrochloride (pH 7.8) and washed several times in H2O. Eggs were homogenized in buffer A (20 mM Tris-HCl [pH 8], 10% glycerol, 0.1% EDTA) using 2.5 μl per egg, and the soluble components were recovered by centrifugation at 13,000 rpm. The supernatant was incubated for 10 min at 95°C, and the precipitated proteins were removed by high-speed centrifugation (50,000 rpm). The supernatant was bound to DEAE-Sepharose, step eluted with 50% buffer B (20 mM Tris-HCl [pH 8], 1 M NaCl, 10% glycerol, 0.1% EDTA) in buffer A and fractionated by anion-exchange chromatography on a MonoQ column (Pharmacia) using a gradient from 20% buffer B in buffer A to 100% buffer B. Fractions containing HNF4 inhibitor were identified in a gel retardation assay. Protein-free HNF4 inhibitor was prepared by digesting a MonoQ preparation with pronase (1 mg/ml) (Roche) according to the manufacturer's instructions, extracting the digested inhibitor with phenol-chloroform, and fractionating on a MonoQ column. This protein-free inhibitor preparation eluted at the same salt concentration as the undigested sample.
Elution of HNF4 inhibitor from SDS-polyacrylamide gels.
Two microliters of a MonoQ inhibitor preparation was separated in a standard sodium dodecyl sulfate (SDS)–15% polyacrylamide gel, the lane was cut into slices, and the gel slices were shaken in 0.4 ml of H2O for 2 h at 37°C. Eluted substances were precipitated with 1.6 ml of acetone (−80°C), recovered by centrifugation, dried in a vacuum concentrator, and redissolved in 20 μl of H2O. HNF4 inhibitor was localized by adding 1 μl of the fractions to gel retardation reactions.
Plasmid constructions.
Expression vectors encoding HNF4α (44), Mt-HNF4α (19), and Mt-HNF4ΔEF (R154X in reference 19) have been described. To construct HNF4ΔAB, the region coding for amino acids 55 to 464 of rat HNF4α was amplified using the primer pair 5′-CACCAAGCTTCCACCATGGGTGTCAGTGCCC-3′ (forward) and 5′-GGGGTACCTAGATGGCTTCCTGC-3′ (reverse), introducing an HindIII site (italics). The PCR product was cloned into the SmaI site of pUC18, excised with HindIII, and cloned into the HindIII site of Rc/CMV.
Cell culture, transfection, and preparation of nuclear extracts.
HEK 293 cells were cultured at 37°C in Dulbecco's modified Eagle's medium containing penicillin (100 U/ml), streptomycin (100 U/ml), l-glutamine (2 mM), and 10% heat-inactivated fetal calf serum (Biochrome). Cells were transfected with 30 μg of DNA per 10-cm cell culture dish using the DNA calcium phosphate coprecipitation method (9). High-salt nuclear extracts were obtained as described (5).
Gel retardation assays.
Gel retardation assays were performed as described (16) using the following oligonucleotide probes: the HNF4 binding site of the Xenopus HNF1α promoter (44), the HNF1 binding site of the Xenopus albumin promoter (31), an AP1 binding site (37), an ATF/CREB binding site (37), an Sp1 site (22), and the HNF3 binding site of the transthyretin promoter (−111/−85 [18]).
Gel retardation reactions contained 1.3 μg of liver nuclear protein (kindly provided by L. Klein-Hitpass and F. Esser), an optimized amount of salmon sperm DNA, and 20,000 cpm of labeled oligonucleotide in GRBB (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 4% Ficoll 400) for detection of HNF1, HNF3, HNF4, and CREB. For detection of AP1 and Sp1, 4.5 μg of liver nuclear protein was used and the reaction was run in buffer P (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 4% glycerol). Inhibitor activity under all these experimental conditions was confirmed. For gel retardation involving proteins of transfected HEK 293 cells, reactions contained 1 μl of nuclear extract of transfected cells, 2 μl of high-salt nuclear extraction buffer (20 mM HEPES [pH 7.6], 25% [vol/vol] glycerol, 420 mM KCl, 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM DTT), and 100 ng of salmon sperm DNA in GRBB without KCl. HNF4 inhibitor was diluted in H2O and added to the reactions in a 1-μl volume.
Trypsin (sequencing grade; Roche) digestion of MonoQ inhibitor preparations was performed according to the manufacturer's instructions. Trypsin was inactivated by adding 0.5 μl of aprotinin (Roche; 10 mg/ml) and 0.1 mM PMSF to the gel retardation reactions.
Digestion of HNF4 inhibitor was performed with pronase (0.2 mg/ml) (Roche) according to the manufacturer's instructions. Pronase was inactivated by mixing 5 μl of the reaction with an equal volume of complete protease inhibitor (Roche) and adding 0.3 μl of PMSF (10 mM), leupeptin (1 mg/ml), and pepstatin (1 mg/ml). One microliter of the final mixture was added to gel retardation reactions.
Native gel electrophoresis and Western blotting.
For native gel electrophoresis, 3 μl of nuclear extract of transfected HEK 293 cells and 1 μl of a MonoQ preparation of HNF4 inhibitor were incubated in GRBB (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 1 mM DTT, 4% Ficoll 400) for 15 min at room temperature. Probes were separated for 1.5 h at 100 V in a 4% polyacrylamide gel containing 0.25× TBE (90 mM Tris base, 90 mM boric acid, 1 mM EDTA). After electrophoresis, gel denaturation of proteins (see Fig. 6, lanes 5 to 8) was achieved by incubating the gel in a buffer containing 62.5 mM Tris (pH 6.7), 100 mM β-mercaptoethanol, and 2% SDS for 20 min at 50°C.
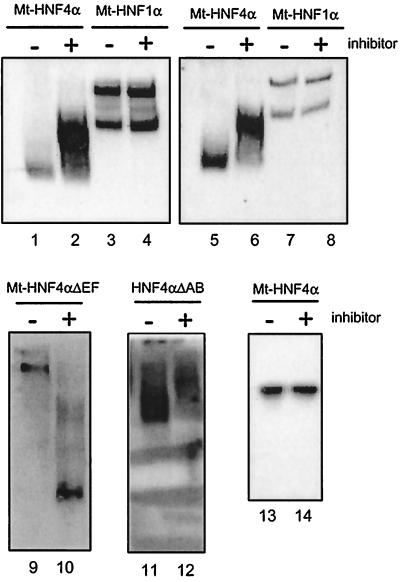
FIG. 6.
Effect of the HNF4 inhibitor on the electrophoretic mobility of various HNF4 deletion constructs. Nuclear extracts (3 μl) from HEK 293 cells transfected with an expression vector encoding either Myc-tagged HNF4α (lanes 1, 2, 5, and 6), Myc-tagged HNF1α (lanes 3, 4, 7, and 8), Myc-tagged C-terminal deletion construct Mt-HNF4αΔEF (lanes 9 and 10), or the N-terminal deletion construct HNF4αΔAB (lanes 11 and 12) were subjected to electrophoresis on a native 4% polyacrylamide gel. Prior to electrophoresis, the extracts were mixed with 1 μl of MonoQ-purified HNF4 inhibitor as indicated. The gels were blotted and probed with the Myc-specific antibody 9E10 (lanes 1 to 10) or the HNF4α-specific monoclonal antibody H4/39f (32) (lanes 11 and 12). The gel with lanes 5 to 8 was denatured before blotting. Lanes 13 and 14 are blotted from an SDS–15% polyacrylamide gel containing Myc-tagged transfected HNF4α and probed with the Myc tag-specific antibody 9E10.
Western blotting of native or SDS gels was performed using as primary antibodies the anti-Myc tag antibody (9E10) for detection of Myc-tagged constructs and monoclonal antibody H4/39f directed against rat HNF4α (32) for detection of HNF4α and HNF4ΔAB. Peroxidase-coupled rabbit anti-mouse immunoglobulin antibody (Dianova) was used as the secondary antibody and detected using the ECL system (Amersham).
RESULTS
Identification of the HNF4 inhibitor.
In our previous experiments, we have shown by Western blotting that the transcription factor HNF4α is present in the fertilized egg as well as throughout the entire embryogenesis of Xenopus (12, 13). Surprisingly, using gel retardation assays, no HNF4α DNA-binding activity could be detected in embryonic extracts (data not shown).
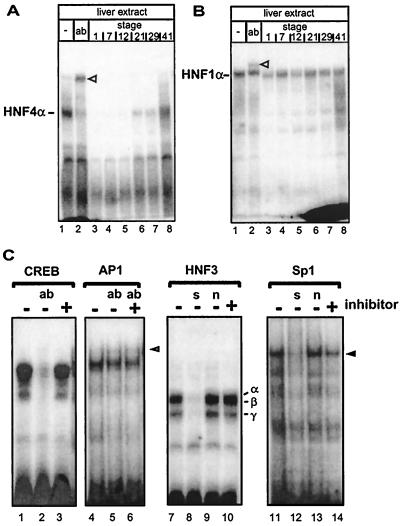
By adding egg extracts to gel retardation reactions containing hepatic HNF4α, we observed that the extract inhibits the binding of HNF4α to its DNA recognition element (compare lane 3 with lane 1 in Fig. 1A). This inhibitory activity persists throughout embryogenesis (lanes 4 to 7), but disappears gradually in extracts of later stages and is essentially absent in swimming larvae at stage 41 (lane 8). In contrast, the DNA-binding activity of the homeodomain transcription factor HNF1α present in the hepatic extract was not inhibited by adding embryonic extracts (Fig. 1B). From these experiments, we conclude that Xenopus embryonic extracts contain a component (HNF4 inhibitor) that inhibits HNF4α DNA binding in a specific way.
FIG. 1.
Identification of an HNF4 inhibitor in Xenopus eggs that gradually disappears during development. (A) Xenopus liver extract (3 μl) was incubated with the labeled oligonucleotide containing the HNF4 binding site of the human apolipoprotein B promoter and analyzed in a gel retardation assay. Monoclonal antibody H4/55, specific for HNF4α (32), is in lane 2 (ab). Lanes 3 to 8 contain in addition 1 μl of embryonic extracts from the developmental stages given (27). The HNF4α-DNA complex is indicated, and the supershifted complex containing the antibody is marked with an open triangle. (B) Gel retardation assay essentially as in panel A, but using the HNF1 binding site of the albumin promoter in combination with monoclonal antibody XAD5, specific for HNF1α (1). (C) Gel retardation assay using rat liver nuclear extract and a CREB binding site (lanes 1 to 3), an AP1 site (lanes 4 to 6), an HNF3 site (lanes 7 to 10), and an Sp1 site (lanes 11 to 14). Protein-free HNF4 inhibitor preparation (1 μl) was added to the reactions as indicated. We used this protein-free inhibitor preparation to avoid a change in protein concentration that may alter the DNA binding in the gel retardation assays. To identify protein-DNA complexes, antibodies (ab) were added as indicated: lane 2, 2 μg of antibody CREB-1 (Santa Cruz; C-21) directed against the DNA-binding domain of CREB; lanes 5 to 6, 2 μg of antibody against c-Fos (Santa Cruz; K-25). The complex supershifted by antibody to c-Fos (K-25) is marked by an open triangle in lane 6. In lanes 8 and 12, a 100-fold excess of unlabeled specific competitor (s) was added (HNF3 site and Sp1 site, respectively). In lanes 9 and 13, a 100-fold excess of unlabeled nonspecific competitor (n) was included in the reaction (Sp1 and HNF3 site, respectively). HNF3α, -β, and -γ protein-DNA complexes are indicated in lane 10 (18), and the Sp1 protein-DNA complex is marked by an arrowhead in lane 14 (22).
To verify the specificity of the HNF4 inhibitor, its action on DNA binding of some other transcription factors was monitored (Fig. 1C). To avoid nonspecific effects of protein or lipid components of the egg extracts, a protein-free inhibitor preparation was used for these experiments. DNA binding of CREB (lanes 1 to 3), AP1 (lanes 4 to 6), and HNF3 (lanes 7 to 10) was not affected by adding HNF4 inhibitor. On the other hand, Sp1 DNA binding was slightly decreased by HNF4 inhibitor (lanes 11 to 14).
Purification of the HNF4 inhibitor from egg extracts.
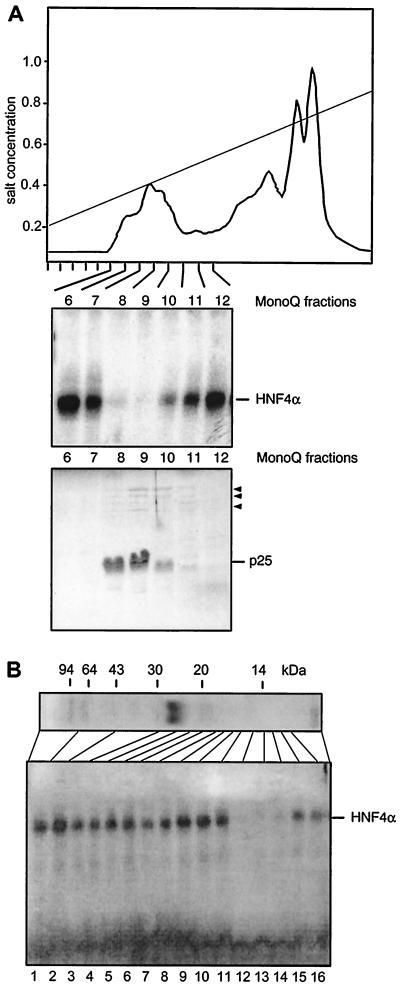
Xenopus eggs were homogenized in a low-salt buffer, and the soluble components were recovered by centrifugation. As the activity of the HNF4 inhibitor is heat resistant (data not shown), we incubated the supernatant at 95°C for 10 min and removed the precipitated proteins by high-speed centrifugation. The supernatant was bound to DEAE-cellulose, step eluted with 0.5 M salt, and fractionated by anion-exchange chromatography on a MonoQ column (Fig. 2A). Individual fractions were tested for the presence of HNF4 inhibitor activity in gel retardation assays containing hepatic HNF4α. As summarized in Fig. 2A, the inhibitory activity coelutes in MonoQ fractions 8 to 10 with a major protein with an apparent molecular mass of 25 kDa. Further purifications on Superose, Mono-S, phenyl-Sepharose, and octyl-Sepharose revealed copurification of the 25-kDa protein with the HNF4 inhibitor (data not shown).
FIG. 2.
Purification and characterization of the HNF4 inhibitor. (A) Elution profile of a heat-treated Xenopus egg extract on a MonoQ column using a salt gradient from 0.2 to 1.0 M NaCl. An aliquot of 0.1 μl of each column fraction was added to a gel retardation assay using rat liver extract and the HNF4 binding site of the Xenopus HNF1α promoter (44). The results are given for fractions 6 to 12 only, as the other fractions did not contain any inhibitory activity. In the lower panel, a Coomassie blue-stained SDS–15% polyacrylamide gel of the same frac- tions is shown using 5 μl of each fraction. The major protein of 25 kDa is indicated, and the minor bands of between 60 and 120 kDa are marked by arrowheads. (B) MonoQ-purified inhibitor preparation (2 μl) was separated on an SDS–15% polyacrylamide gel and stained with Coomassie blue. The migration of molecular size markers is shown. An adjacent lane containing the same amount of inhibitor was cut into individual fractions as indicated. An aliquot representing 5% of each fraction was added to a gel retardation assay using the HNF4 binding site, as given in Fig. 1.
To test whether the 25-kDa protein represents the HNF4 inhibitor, we eluted and renatured various fractions of an SDS-polyacrylamide gel containing the MonoQ-purified 25-kDa protein. As shown in Fig. 2B, the inhibitory activity has an apparent molecular mass of 10 to 14 kDa and clearly does not comigrate with the 25-kDa protein detectable in the SDS gel.
Inhibitory function is protease resistant.
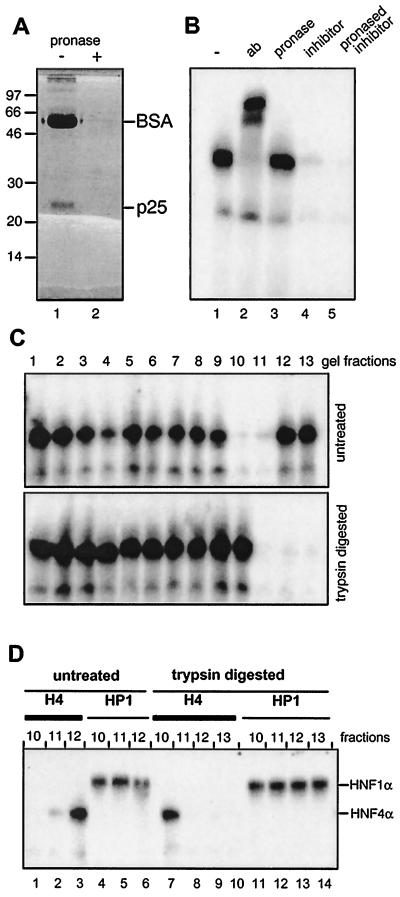
The HNF4 inhibitor purified by MonoQ-Sepharose chromatography as given in Fig. 2A was digested with pronase. To monitor protease activity, bovine serum albumin (BSA) was added as an internal control to the HNF4 inhibitor. SDS-polyacrylamide gel electrophoresis revealed that BSA and the 25-kDa protein were completely digested (Fig. 3A, compare lanes 1 and 2). After digestion, the protease was inactivated by adding a protease inhibitor cocktail, and the mixture was analyzed for HNF4 inhibitor activity in a gel retardation assay using hepatic extract. Figure 3B illustrates that the protease-digested HNF4 inhibitor (lane 5) retained its inhibitory function compared to untreated inhibitor sample (lane 4). In a control experiment involving pronase-digested BSA, no inhibition was observed (lane 3), indicating that no pronase activity remained in the gel retardation assay. Similar results were obtained using trypsin and proteinase K digestion (data not shown).
FIG. 3.
Protease digestion of the HNF4 inhibitor. (A) MonoQ-purified inhibitor preparation (2 μl) was combined with 20 μg of BSA and either separated directly on an SDS–15% polyacrylamide gel (lane 1) or digested with pronase prior to electrophoresis (lane 2). (B) Gel retardation assay using rat liver nuclear extract and the labeled HNF4 binding site as oligonucleotide is given in lane 1 and supplemented in lane 2 with the polyclonal antibody (ab) H4/66 specific for HNF4α (32). In lanes 3, 4, and 5, pronase-digested BSA, untreated HNF4 inhibitor, and pronase-digested inhibitor were added, respectively. (C) Untreated and trypsin-digested HNF4 inhibitor was separated in adjacent lanes of an SDS–15% polyacrylamide gel. The gel was cut into slices, and the HNF4 inhibitor was eluted and assayed in a gel retardation assay as given in Fig. 2B. (D) Aliquots of the indicated gel fractions of the untreated and trypsin-digested lanes of panel C were assayed in a gel retardation assay using the labeled oligonucleotide representing either the HNF4 (H4) or the HNF1 (HP1) binding site. The DNA-protein complexes containing HNF1α and HNF4α are indicated.
To analyze whether protease digestion alters the mobility of the HNF4 inhibitor on an SDS-polyacrylamide gel, we compared the distribution of the inhibitory activity between an untreated and a trypsin-digested sample separated on adjacent lanes of an SDS-polyacrylamide gel. As shown in Fig. 3C, the inhibitory activity was present in fractions 10 and 11 of the untreated sample, whereas the activity was shifted to fractions 11 to 13 in the lane containing the digested inhibitor. In gel retardation assays specific for HNF1α, the addition of the same fractions did not affect the binding activity of HNF1α (Fig. 3D, lanes 4 to 6 and 11 to 14), showing the specificity of the inhibitory effect. This finding clearly establishes that the mobility of the HNF4 inhibitor is sensitive to protease digestion but that the function of the inhibitor itself is not affected. Obviously, the HNF4 inhibitor contains a protease-resistant core that prevents the DNA-binding activity of HNF4. A reduction in size of the HNF4 inhibitor upon protease digestion was also deduced from ultrafiltration experiments, because the inhibitory activity passed through a 5-kDa membrane exclusively after pronase digestion (data not shown).
HNF4 inhibitor is distinct from acyl-CoAs, the potential ligands of HNF4.
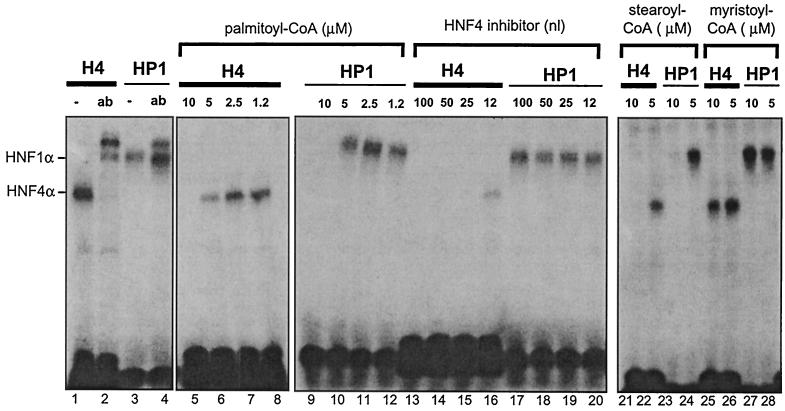
Recently, Hertz et al. (11) reported that acyl-CoAs are potential ligands of HNF4α and that acyl-CoAs containing long-chain fatty acids inhibit DNA binding of HNF4α. To investigate whether the HNF4 inhibitor has similar properties, we compared the effect of acyl-CoAs on the DNA binding of HNF4α in the gel retardation assays used to identify the HNF4 inhibitor. Figure 4 illustrates that with increasing amounts, palmitoyl (C16:0)-CoA inhibits HNF4α binding to DNA (lanes 5 to 8). However, this effect is not specific, as a very similar inhibition is seen on the DNA binding of HNF1α (lanes 9 to 12). In contrast, a serial dilution of the HNF4 inhibitor specifically inhibits HNF4α DNA binding (lanes 13 to 16), but has no effect on the DNA-binding activity of HNF1α (lanes 17 to 20). Stearoyl (C18:0)-CoA also shows a nonspecific effect, because HNF4α and HNF1α are equally affected by this compound (lanes 21 to 24). In contrast, myristoyl (C14:0)-CoA has no effect on the DNA binding of either of these two transcription factors (lanes 25 to 28). From these experiments, we conclude that the HNF4 inhibitor is more specific than the acyl-CoAs tested and thus represents a distinct effector molecule.
FIG. 4.
Comparison of the action of the HNF4 inhibitor with the effect of acyl-CoAs. Gel retardation assays using the labeled oligonucleotide representing either the HNF4 (H4) or the HNF1 (HP1) binding site were performed in the presence of various concentrations of palmitoyl-CoA (lanes 5 to 12), stearoyl-CoA (lanes 21 to 24), and myristoyl-CoA (lanes 25 to 28). The DNA-protein complexes containing HNF1α and HNF4α are indicated, and the reaction with the corresponding antibodies (ab) is given in lanes 2 and 4. For comparison, the effect of the addition of HNF4 inhibitor is shown in lanes 14 to 20.
Ligand-binding domain of HNF4 is not required for mediating the inhibitory effect.
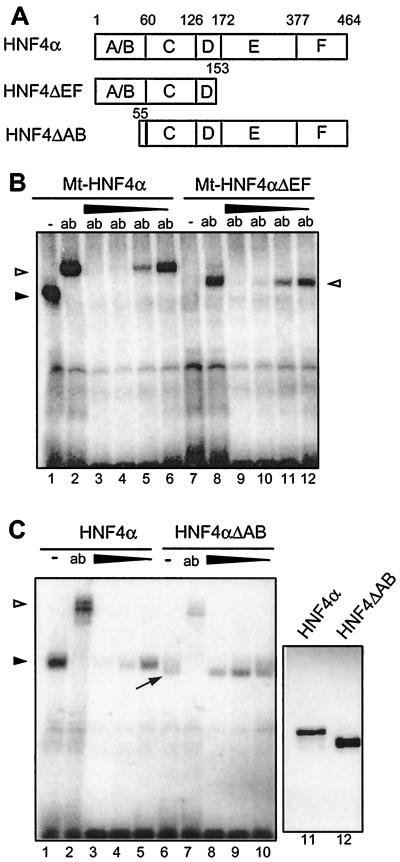
To test whether the HNF4 inhibitor acts via ligand-binding domain E of HNF4α, extending from amino acids 172 to 377, we used the deletion construct Mt-HNF4αΔEF containing the N-terminal part of HNF4α (amino acids 1 to 153), as given in Fig. 5A. This construct and the wild-type protein used as a control (Mt-HNF4α) contain a Myc tag at the N terminus that allows efficient identification of the protein in transfected HEK 293 cells. Since binding of the Mt-HNF4αΔEF protein to DNA is weak and can only be detected in the presence of the Myc tag-specific antibody (lane 8 in Fig. 5B) (19), HNF4 inhibitor was added in the presence of the Myc tag-specific antibody. As Fig. 5B illustrates, the truncated protein Mt-HNF4αΔEF and the full-length protein Mt-HNF4α are inhibited in complex formation by the same HNF4 inhibitor concentrations (compare lanes 9 to 12 with lanes 3 to 6). Therefore, the ligand-binding domain is not necessary to convey inhibitor sensitivity to HNF4.
FIG. 5.
Effect of the HNF4 inhibitor on various HNF4 deletion constructs in gel retardation assays. (A) Schematic drawing of the deletion constructs used. Domains C and E represent the DNA-binding and the potential ligand-binding domains of HNF4α, respectively. (B) Nuclear extracts from HEK 293 cells transfected with the Myc-tagged full-length HNF4α (lanes 1 to 6) or with the C-terminal deletion construct Mt-HNF4αΔEF (lanes 7 to 12) were analyzed in gel retardation assays using the labeled HNF4 binding site. The addition of the monoclonal Myc tag-specific antibody 9E10 is shown (ab). A protein-free inhibitor preparation was added (1, 0.3, 0.1, or 0.03 μl) in lanes 3 to 6 and 9 to 12, respectively. The arrowhead marks the HNF4α-DNA complex, and the open triangles indicate the complex supershifted by the antibody. (C) Nuclear extracts from HEK 293 cells transfected with the full-length HNF4α (lanes 1 to 5) or with the N-terminal deletion construct HNF4αΔAB were analyzed in gel retardation assays using the labeled HNF4 binding site. The polyclonal antibody XH4α specific for HNF4α (13) was added in lanes 2 and 7 (ab). The protein-free inhibitor preparation was used at 1, 0.3, or 0.1 μl in lanes 3 to 5 and 8 to 10, respectively. The arrowhead marks the HNF4α-DNA complex, and the open triangle marks the complex supershifted by the antibody. The unusually broad protein-DNA complex containing HNF4αΔAB is marked by an arrow. Lanes 11 and 12 show the abundance of the transfected HNF4α proteins in 3 μl of HEK 293 cell extract in a Western blot of an SDS–12% polyacrylamide gel using monoclonal antibody H4/39f (32).
To evaluate the role of the N-terminal region, a deletion construct of HNF4α lacking the A/B domain (HNF4αΔAB in Fig. 5A) was analyzed. Because the binding of the HNF4αΔAB construct was lower (Fig. 5C, lane 6) compared to the full-length protein (lane 1), we measured the amount of HNF4α proteins in the transfected cells. The Western blot in Fig. 5C (lanes 11 and 12) showed even a slightly increased level of the HNF4αΔAB protein, and therefore deletion of the A/B domain reduces the DNA binding of HNF4α. Furthermore, the DNA-HNF4 complex containing the HNF4αΔAB protein formed an unusual broad band (lane 6). Whereas the strong binding of HNF4α is greatly reduced by adding inhibitor at different concentrations (Fig. 5C, lanes 1 to 5), the much weaker binding of HNF4αΔAB is not further reduced by equal concentrations of HNF4 inhibitor (Fig. 5C, lanes 6 to 10). However, the broad band sharpened upon the addition of the highest concentration of inhibitor (lane 8). From this observation, we conclude that the inhibitor retains its ability to act on the HNF4 protein lacking the A/B domain but that in this case the reduced DNA binding of HNF4αΔAB is not further decreased. This implies that the strength of DNA binding and the response to inhibitor action are not strictly correlated.
HNF4 inhibitor alters the electrophoretic mobility of HNF4.
To elucidate whether the activity of the HNF4 inhibitor requires DNA binding, we separated nuclear extracts from HEK 293 cells transfected with Myc-tagged HNF4α constructs on native polyacrylamide gels and located the HNF4α protein by Western blotting using the Myc tag-specific antibody. Figure 6 illustrates that HNF4α migrates faster than the same protein that has been incubated with the HNF4 inhibitor (compare lanes 1 and 2). In contrast, the mobility of the Myc-tagged HNF1α protein that migrates as a doublet is not affected by addition of the HNF4 inhibitor (lanes 3 and 4). Notably, a weak signal was consistently seen in the Western blot for the untreated HNF4α protein (lane 1) compared to the inhibitor-treated sample (lane 2). In contrast, when the gel was denatured in SDS prior to transfer to the membrane, similar intensities for the HNF4α protein were obtained (Fig. 6, lanes 5 and 6). From this observation, we conclude that properties of HNF4α are altered by addition of the HNF4 inhibitor. This alteration leads to decreased mobility as well as to an increase in accessibility to the Myc tag-specific antibody. This effect was only seen on native polyacrylamide gels, whereas an equal mobility of HNF4α with or without HNF4 inhibitor incubation was observed on SDS gels (Fig. 6, lanes 13 and 14).
To define the region involved in the inhibitor-induced mobility shift, we analyzed extracts derived from HEK 293 cells transfected with various HNF4α deletion constructs. Using the Mt-HNF4αΔEF construct, which lacks the ligand-binding domain (see Fig. 5A), we observed a dramatic increase in electrophoretic mobility upon HNF4 inhibitor addition (Fig. 6, compare lanes 9 and 10). Similarly, the mobility of the HNF4α construct lacking the N-terminal A/B domain was altered (Fig. 6, lanes 11 and 12), but as for the full-length protein, addition of the inhibitor decreased the mobility of HNF4ΔAB. We conclude that the HNF4 inhibitor acts on the HNF4 proteins in the absence of DNA and that this interaction does not require either the A/B or E and F domains.
DISCUSSION
We have characterized a novel component that specifically inhibits the DNA-binding activity of HNF4. DNA binding of unrelated transcription factors, namely the POU/homeobox protein HNF1α (Fig. 1B), the leucine zipper proteins CREB and AP1, and the forkhead/winged helix protein HNF3 (Fig. 1C), is clearly not affected by HNF4 inhibitor. In contrast, DNA binding of Sp1 seems to be weakened by HNF4 inhibitor to some extent (Fig. 1C). Although this effect is much less obvious than the inhibition of HNF4 DNA binding, it could point to a partial action of HNF4 inhibitor on other zinc finger proteins as well. The inhibitor also acts on the progesterone receptor as well as on the retinoic acid RAR/RXR, but does not affect the estrogen receptor (data not shown). Therefore, not all members of the nuclear receptor superfamily are affected.
The HNF4 inhibitor is hydrophilic and resistant to protease (Fig. 3B), RNase, DNase, and N-glycosidase (data not shown). As the activity of the HNF4 inhibitor is protease resistant, the activity resides in a nonprotein component. On the other hand, the inhibitor contains a peptide bond, as the electrophoretic mobility is increased upon protease digestion (Fig. 3C). In fact, we assume that there are two distinct peptide bonds, because the inhibitory activity is sensitive to trypsin as well as pronase digestion, as assayed on a denaturing polyacrylamide gel, but passes through a 5-kDa membrane only after pronase treatment (data not shown). In conclusion, our data support a model in which the inhibitor consists of a protease-resistant core carrying the inhibitory activity and a distinct protease-sensitive region.
Although the chemical identity of the HNF4 inhibitor is not yet known, it is most unlikely that it represents a member of the acyl-CoAs, which have recently been proposed to be ligands for HNF4 (11). In fact, the HNF4 inhibitor is more specific, as it does not inhibit the transcription factor HNF1α, a member of the homeodomain proteins, whereas various acyl-CoAs were found to inhibit DNA binding of HNF1α as well (Fig. 4). More importantly, we could show that the HNF4 inhibitor does not require the potential ligand-binding domain of HNF4 to exert its action (Fig. 5B and Fig. 6, lanes 9 and 10).
The inhibitor is a relatively small molecule whose protease-resistant core of about 5 kDa retains full inhibitory function and thus resembles the relatively small size of the well-known ligands of the nuclear receptor superfamily. However, the inhibitor would be a novel type of ligand, as it acts on an HNF4 derivative lacking the ligand-binding domain. Based on our finding that the inhibitor interferes with the DNA-binding property of HNF4, it acts as an antagonist and may thus be similar to androstane, which inhibits the constitutive androstane receptor (8). However, such an interpretation is quite speculative, because at present it remains unclear whether the inhibitor binds to HNF4 or just induces the observed changes without binding. In fact, several different attempts made to isolate the inhibitor using the HNF4 protein as a bait failed, and thus we favor a model in which the inhibitor binds only transiently to the HNF4 protein.
It is well established that HNF4 is a phosphoprotein and that the extent of phosphorylation is modulated (14, 40). However, we exclude an inhibitor-induced change in phosphorylation because the mobility of HNF4α in SDS-polyacrylamide gels is known to be influenced by phosphorylation (14), but we could not see any change in the mobility of HNF4 on SDS-polyacrylamide gels upon incubation with the inhibitor (Fig. 6, lanes 13 and 14).
The action of the HNF4 inhibitor can be traced in three distinct ways. In the first, the DNA-binding activity of HNF4α is inhibited in the gel retardation assay and is therefore measured in the presence of the HNF4 DNA-binding site (Fig. 1 to 5). However, the inhibitor also affects HNF4 in the absence of the DNA-binding site by altering the mobility in a native polyacrylamide gel (Fig. 6). In these native polyacrylamide gels, HNF4 inhibitory action is evident in a third way, because inhibitor treatment leads to an altered accessibility of HNF4α to the antibody in the Western blot. This is most evident using the Myc tag-specific antibody that reacts to the artificial N-terminal tag of HNF4α, but also occurs using a monoclonal antibody raised against the ABCD domains of HNF4α (36).
Using various deletion constructs, it is clear that domain E, the potential ligand-binding domain of HNF4α, is not required to get the DNA-binding inhibition in the gel retardation assay (Fig. 5B). In contrast, the HNF4α deletion construct HNF4αΔAB, lacking the A/B domains, is apparently not affected in its binding affinity by the inhibitor. However, this construct has a low binding activity compared to the full-length protein, implying some effect of the A/B domain on the DNA-binding domain. Surprisingly, this N-terminal truncation of HNF4α leads to an unusual broad DNA-protein band in the gel retardation assay (Fig. 5C, lane 6). As this diffuse band is sharpened by the addition of HNF4 inhibitor (Fig. 5, lane 8), we conclude that the inhibitor is still able to act upon the truncated HNF4α derivative lacking the A/B domain. This interpretation agrees with our finding that the same construct is affected in its mobility in native gel electrophoresis in the absence of the DNA-binding site (Fig. 6, lanes 11 and 12).
The addition of the inhibitor in the absence of DNA leads to decreased mobility of HNF4 in the native polyacrylamide gel, implying either some conformational change, multimerization, or a decrease in negative charge (Fig. 6, lanes 1 and 2). Whereas decreased mobility was also obtained for the deletion construct HNF4αΔAB lacking the A/B domain (Fig. 6, lanes 11 and 12), the mobility of construct Mt-HNF4αΔEF lacking the ligand-binding domain is increased (Fig. 6, lanes 9 and 10). This opposite effect of the HNF4 inhibitor on the electrophoretic mobility of HNF4 derivatives is quite unexpected. In this context, it is noteworthy that Mt-HNF4αΔEF migrates much more slowly than the full-length Mt-HNF4α in a native polyacrylamide gel, although the theoretic isoelectric point of Mt-HNF4αΔEF is lower, 4.5 compared to 4.9 for full-length Mt-HNF4α, and thus should migrate faster at the pH used for electrophoresis. Possibly during electrophoresis under native conditions, HNF4 interacts with other components of the extract that influence the electrophoretic migration. Therefore, not only a change in the charge or in the conformation of HNF4 upon inhibitor addition has to be considered, but an influence on the interaction with other components as well. Indeed, it is well established that HNF4 interacts with a series of other factors involved in transcriptional regulation (10, 15, 20, 34, 41).
The presence of an HNF4 inhibitor in the Xenopus embryo gives a most attractive explanation for our finding that the HNF4 target gene HNF1α is only marginally expressed in the early embryo, although maternal HNF4 proteins α and β are abundantly present (12, 13). From injections of RNAs encoding HNF4 into fertilized Xenopus eggs, we know that the endogenous HNF1α gene is competent for activation and can be activated as early as the blastula stage by overexpression of HNF4 (26). Indeed, under these conditions, overexpressed HNF4 can be found in embryonic extracts in gel retardation assays (data not shown), implying that the HNF4 inhibitor is titrated out by the introduced HNF4 protein. Therefore, we assume that the HNF4 inhibitor inactivates the maternal HNF4 transcription factor and that this inhibition is gradually relieved during development. This release of suppression during development is supported by our finding that the inhibitory activity is lost in the process of development (Fig. 1). During this time period of development, the amount of HNF4 α and β proteins does not change significantly (12, 13), but the expression of the HNF1α gene increases dramatically (1, 26). Thus, we propose that the decrease in the HNF4 inhibitor is responsible for the activation of HNF4-dependent target genes during Xenopus development. The balance of positive and negative regulators crucial for the concerted activation of the developmental program is a well-established phenomenon during embryogenesis (3, 4, 24). We further speculate that the distribution of the inhibitor within the embryo is a crucial determinant in the patterning of the HNF4 response. Clearly, firm conclusions concerning the regulatory role of the HNF4 inhibitor awaits its chemical identification, which will allow us to determine its distribution within the embryo and to test its function in injected embryos.
The identification of an HNF4 inhibitor in Xenopus embryos opens up a potential way to manipulate the activity of HNF4α, a gene product mutated in MODY1 patients. Recent analysis of the mutated HNF4α factors found in MODY patients have shown that these naturally occurring mutations impair the function of the transcription factor to greatly varying degrees (19 and references therein), but that most of these mutants retain the C and D domain that is responsive to the HNF4 inhibitor. Therefore, compounds with properties of the HNF4 inhibitor may constitute a novel approach to interfering with HNF4α function.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Ry5/3–4).
REFERENCES
- 1.Bartkowski S, Zapp D, Weber H, Eberle G, Zoidl C, Senkel S, Klein-Hitpass L, Ryffel G U. Developmental regulation and tissue distribution of the liver transcription factor LFB1 (HNF1) in Xenopus laevis. Mol Cell Biol. 1993;13:421–431. doi: 10.1128/mcb.13.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W S, Manova K, Weinstein D C, Duncan S A, Plump A S, Prezioso V R, Bachvarova R F, Darnell J E., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 3.Cho K W, Blitz I L. BMPs, Smads and metalloproteases: extracellular and intracellular modes of negative regulation. Curr Opin Genet Dev. 1998;8:443–449. doi: 10.1016/s0959-437x(98)80116-0. [DOI] [PubMed] [Google Scholar]
- 4.Dale L, Jones C M. BMP signalling in early Xenopus development. Bioessays. 1999;21:751–760. doi: 10.1002/(SICI)1521-1878(199909)21:9<751::AID-BIES6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 5.Dignam J D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 6.Duncan S A, Manova K, Chen W S, Hoodless P, Weinstein D C, Bachvarova R F, Darnell J E., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc Natl Acad Sci USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan S A, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of hnf-4(−/−) embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- 8.Forman B M, Tzameli I, Choi H S, Chen J, Simha D, Seol W, Evans R M, Moore D D. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- 9.Gorman C, Padmanabhan R, Howard B H. High efficiency DNA-mediated transformation of primate cells. Science. 1983;221:551–553. doi: 10.1126/science.6306768. [DOI] [PubMed] [Google Scholar]
- 10.Green V J, Kokkotou E, Ladias J A. Critical structural elements and multitarget protein interactions of the transcriptional activator AF-1 of hepatocyte nuclear factor 4. J Biol Chem. 1998;273:29950–29957. doi: 10.1074/jbc.273.45.29950. [DOI] [PubMed] [Google Scholar]
- 11.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 12.Holewa B, Pogge E, Strandmann V, Zapp D, Lorenz P, Ryffel G U. Transcriptional hierarchy in Xenopus embryogenesis: HNF4 a maternal factor involved in the developmental activation of the gene encoding the tissue specific transcription factor HNF1 alpha (LFB1) Mech Dev. 1996;54:45–57. doi: 10.1016/0925-4773(95)00460-2. [DOI] [PubMed] [Google Scholar]
- 13.Holewa B, Zapp D, Drewes T, Senkel S, Ryffel G U. HNF4beta, a new gene of the HNF4 family with distinct activation and expression profiles in oogenesis and embryogenesis of Xenopus laevis. Mol Cell Biol. 1997;17:687–694. doi: 10.1128/mcb.17.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang G Q, Nepomuceno L, Yang Q, Sladek F M. Serine/threonine phosphorylation of orphan receptor hepatocyte nuclear factor 4. Arch Biochem Biophys. 1997;340:1–9. doi: 10.1006/abbi.1997.9914. [DOI] [PubMed] [Google Scholar]
- 15.Ktistaki E, Talianidis I. Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science. 1997;277:109–112. doi: 10.1126/science.277.5322.109. [DOI] [PubMed] [Google Scholar]
- 16.Kugler W, Kaling M, Ross K, Wagner U, Ryffel G U. BAP, a rat liver protein that activates transcription through a promoter element with similarity to the USF/MLTF binding site. Nucleic Acids Res. 1990;18:6943–6951. doi: 10.1093/nar/18.23.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo C J, Conley P B, Chen L, Sladek F M, Darnell J E, Jr, Crabtree G R. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 18.Lai E, Prezioso V R, Tao W F, Chen W S, Darnell J E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- 19.Lausen J, Thomas H, Lemm I, Bulman M, Borgschulze M, Lingott A, Hattersley A T, Ryffel G U. Naturally occurring mutations in the human HNF4alpha gene impair the function of the transcription factor to a varying degree. Nucleic Acids Res. 2000;28:430–437. doi: 10.1093/nar/28.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y K, Dell H, Dowhan D H, Hadzopoulou-Cladaras M, Moore D D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Ning G, Duncan S A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Shapiro R A, Nie S, Zhu D, Vodovotz Y, Billiar T R. Characterization of rat CD14 promoter and its regulation by transcription factors AP1 and Sp family proteins in hepatocytes. Gene. 2000;250:137–147. doi: 10.1016/s0378-1119(00)00179-7. [DOI] [PubMed] [Google Scholar]
- 23.Miquerol L, Lopez S, Cartier N, Tulliez M, Raymondjean M, Kahn A. Expression of the L-type pyruvate kinase gene and the hepatocyte nuclear factor 4 transcription factor in exocrine and endocrine pancreas. J Biol Chem. 1994;269:8944–8951. [PubMed] [Google Scholar]
- 24.Miyazono K. Positive and negative regulation of TGF-beta signaling. J Cell Sci. 2000;113:1101–1109. doi: 10.1242/jcs.113.7.1101. [DOI] [PubMed] [Google Scholar]
- 25.Nakhei H, Lingott A, Lemm I, Ryffel G U. An alternative splice variant of the tissue specific transcription factor HNF4alpha predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998;26:497–504. doi: 10.1093/nar/26.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nastos A, Pogge van Strandmann E, Weber H, Ryffel G U. The embryonic expression of the tissue-specific transcription factor HNF1alpha in Xenopus: rapid activation by HNF4 and delayed induction by mesoderm inducers. Nucleic Acids Res. 1998;26:5602–5608. doi: 10.1093/nar/26.24.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieuwkoop P, Faber J. Normal table of Xenopus laevis (Daudin). Amsterdam, The Netherlands: Elsevier/North-Holland Publishing Co.; 1975. [Google Scholar]
- 28.Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 29.Pogge V. Strandmann E, Nastos A, Holewa B, Senkel S, Weber H, Ryffel G U. Patterning the expression of a tissue-specific transcription factor in embryogenesis: HNF1 alpha gene activation during Xenopus development. Mech Dev. 1997;64:7–17. doi: 10.1016/s0925-4773(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 30.Ryffel G U, Lingott A. Distinct promoter elements mediate endodermal and mesodermal expression of the HNF1alpha promoter in transgenic Xenopus. Mech Dev. 2000;90:65–75. doi: 10.1016/s0925-4773(99)00230-0. [DOI] [PubMed] [Google Scholar]
- 31.Schorpp M, Kugler W, Wagner U, Ryffel G U. Hepatocyte-specific promoter element HP1 of the Xenopus albumin gene interacts with transcriptional factors of mammalian hepatocytes. J Mol Biol. 1988;202:307–320. doi: 10.1016/0022-2836(88)90460-3. [DOI] [PubMed] [Google Scholar]
- 32.Sel S, Ebert T, Ryffel G U, Drewes T. Human renal cell carcinogenesis is accompanied by a coordinate loss of the tissue specific transcription factors HNF4 alpha and HNF1 alpha. Cancer Lett. 1996;101:205–210. doi: 10.1016/0304-3835(96)04136-5. [DOI] [PubMed] [Google Scholar]
- 33.Sladek F M. Hepatocyte nuclear factor 4 (HNF-4) In: Tronche F, Yaniv M, editors. Liver-specific gene expression. R. G. Austin, Tex: Landes Co.; 1994. pp. 207–230. [Google Scholar]
- 34.Sladek F M, Ruse M D J, Nepomuceno L, Huang S M, Stallcup M R. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sladek F M, Zhong W M, Lai E, Darnell J E., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 36.Stumpf H, Senkel S, Rabes H M, Ryffel G U. The DNA binding activity of the liver transcription factors LFB1 (HNF1) and HNF4 varies coordinately in rat hepatocellular carcinoma. Carcinogenesis. 1995;16:143–145. doi: 10.1093/carcin/16.1.143. [DOI] [PubMed] [Google Scholar]
- 37.Tacchini L, Radice L, Bernelli-Zazzera A. Differential activation of some transcription factors during rat liver ischemia, reperfusion, and heat shock. J Cell Physiol. 1999;180:255–262. doi: 10.1002/(SICI)1097-4652(199908)180:2<255::AID-JCP13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 38.Taraviras S, Monaghan A P, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 39.Tian J M, Schibler U. Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- 40.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J C, Stafford J M, Granner D K. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–30850. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagata K, Furuta H, Oda N, Kaisaki P J, Menzel S, Cox N J, Fajans S S, Signorini S, Stoffel M, Bell G I. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 43.Yamagata K, Oda N, Kaisaki P J, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox R D, Lathrop G M, Boriraj V V, Chen X, Cox N J, Oda Y, Yano H, Le B M, Yamada S, Nishigori H, Takeda J, Fajans S S, Hattersley A T, Iwasaki N, Hansen T, Pedersen O, Polonsky K S, Bell G I. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 44.Zapp D, Bartkowski S, Holewa B, Zoidl C, Klein-Hitpass L, Ryffel G U. Elements and factors involved in tissue-specific and embryonic expression of the liver transcription factor LFB1 in Xenopus laevis. Mol Cell Biol. 1993;13:6416–6426. doi: 10.1128/mcb.13.10.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong W, Mirkovitch J, Darnell J E., Jr Tissue-specific regulation of mouse hepatocyte nuclear factor 4 expression. Mol Cell Biol. 1994;14:7276–7284. doi: 10.1128/mcb.14.11.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong W, Sladek F M, Darnell J E., Jr The expression pattern of a Drosophila homolog to the mouse transcription factor HNF-4 suggests a determinative role in gut formation. EMBO J. 1993;12:537–544. doi: 10.1002/j.1460-2075.1993.tb05685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]