Abstract
Alu are high copy number interspersed repeats that have accumulated near genes during primate and human evolution. They are a pervasive source of structural variation in modern humans. Impacts that Alu insertions may have on gene expression are not well understood, although some have been associated with expression quantitative trait loci (eQTLs). Here, we directly test regulatory effects of polymorphic Alu insertions in isolation of other variants on the same haplotype. To screen insertion variants for those with such effects, we used ectopic luciferase reporter assays and evaluated 110 Alu insertion variants, including more than 40 with a potential role in disease risk. We observed a continuum of effects with significant outliers that up- or down-regulate luciferase activity. Using a series of reporter constructs, which included genomic context surrounding the Alu, we can distinguish between instances in which the Alu disrupts another regulator and those in which the Alu introduces new regulatory sequence. We next focused on three polymorphic Alu loci associated with breast cancer that display significant effects in the reporter assay. We used CRISPR to modify the endogenous sequences, establishing cell lines varying in the Alu genotype. Our findings indicate that Alu genotype can alter expression of genes implicated in cancer risk, including PTHLH, RANBP9, and MYC. These data show that commonly occurring polymorphic Alu elements can alter transcript levels and potentially contribute to disease risk.
Complex disease risk loci have been identified throughout the genome, and many of the haplotypes associated with disease occur in noncoding, presumably regulatory loci (Zhang et al. 2014; Lowe and Reddy 2015). Now investigators seek to define the causative variants, which genes they impact, and how they function. We previously showed that commonly occurring structural variants caused by insertions of Alu short interspersed elements (SINEs) frequently occur at disease risk loci (Payer et al. 2017), raising the possibility that they may alter risk by impacting gene expression.
Like other interspersed repeats, Alu sequences are retrotransposons, genetic elements that proliferate through a “copy-and-paste” mechanism with an RNA intermediate (for review, see Batzer and Deininger 2002; Burns and Boeke 2012; Hancks and Kazazian 2016; Payer and Burns 2019). There are about 1.1 million copies of Alu in the human genome (Smit 1999; International Human Genome Sequencing Consortium 2001), and a minor subset of these are polymorphic in human populations (Stewart et al. 2011; Witherspoon et al. 2013; Sudmant et al. 2015; Gardner et al. 2017), meaning that at a specific loci, some individuals have the Alu insertion but others have the preinsertion “empty” allele. Polymorphic Alu elements are prevalent with more than 3200 reported with an allele frequency >5% (Sudmant et al. 2015). Ongoing research aims to evaluate functional effects of these prevalent polymorphic Alu sequences. Recently, we showed that a subset of those mapping to introns can alter mRNA splicing of nearby exons (Payer et al. 2019), and that is likely just one of many functional consequences of polymorphic Alu elements.
Retrotransposons have intrinsic sequences that regulate expression of their own RNAs. These sequences can also affect nearby genes, with the best-known cases being endogenous retroviruses and their flanking long terminal repeats (e.g., Chuong et al. 2013, 2016; Dunn-Fletcher et al. 2018; Fuentes et al. 2018; Jang et al. 2019). Much less is known about Alu regulatory potential. Some Alu elements that have accumulated point mutations over time have become enhancers (e.g., Norris et al. 1995; Gombart et al. 2009; Jacobsen et al. 2009; Zhang et al. 2019). In general, these evolutionarily older Alu elements that are “fixed” in the genome, homozygous present in all individuals, can be epigenetically marked like enhancers including positioned phased nucleosomes, active histone marks including H3K4me1 and H3K27ac, enrichment upstream proximal to genes, and preferential long-range contacts with promoters (Su et al. 2014). A subset of Alu elements functions as enhancers for cell-cycle genes through RNA polymerase III transcription factor C (TFIIIC) recruitment with subsequent altered chromatin looping and histone acetylation resulting in gene expression changes in cis (Ferrari et al. 2020). Alu RNA may also play a role in reducing transcription of RNA polymerase II (Pol II) transcripts in trans (Mariner et al. 2008). Many of these effects are restricted to Alu sequences that have existed for long periods of time in the human genome. Therefore, the question remains as to whether evolutionarily young, polymorphic Alu insertions have this intrinsic regulatory potential. More than 250 Alu insertion variants at expression quantitative trait loci (eQTLs) have been identified (Wang et al. 2017b); similarly, large numbers of Alu insertions map to regions rich with transcription regulators (Wang et al. 2017b; Goubert et al. 2020). These eQTLs have not been delimited to the polymorphic Alu, and no systematic assessments of regulatory effects of these Alu have been reported.
To this end, here, we evaluate the regulatory impact of large numbers of polymorphic Alu elements isolated from other nearby variants. We also focus on mechanisms of this activity and the potential for Alu-regulated transcript levels to alter disease risk.
Results
Polymorphic Alu elements have regulatory potential
To assess if polymorphic Alu elements alter gene regulation, we used standard ectopic enhancer reporter assays in 293T cells. This allowed us to compare Alu insertion and preinsertion alleles for relatively large numbers of loci in a model that was then tractable for follow-up studies to dissect sequence requirements. In all, we tested 110 polymorphic Alu loci (Supplemental Table S1) selected because these are common variants, many of which map to Genome-Wide Association Studies (GWAS) signals and therefore may have a role in disease risk (Payer et al. 2017).
For each locus, we cloned the sequence with (∼600 bp) or without (∼300 bp) the polymorphic Alu element upstream of firefly luciferase driven by a minimal promoter (Fig. 1A). Because ectopic assays are designed to detect short range cis effects, we cloned each locus so that the orientation relative to the nearest transcription start site in the genome was retained relative to luciferase in the vector. Each construct was transfected into 293T cells along with a Renilla luciferase expression plasmid used to normalize for transfection efficiency. We measured the effect of the polymorphic Alu on luciferase expression.
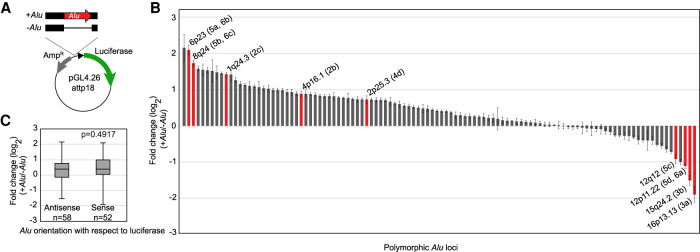
Figure 1.
Polymorphic Alu elements have a continuum of effects on luciferase expression in an ectopic enhancer assay. (A) A genomic locus (black box) with or without the polymorphic Alu element (red) present was cloned upstream of a minimal promoter (triangle) in firefly luciferase (green) expression vector that had been modified to use Gateway cloning. (B) Luciferase measurements shown as the fold change for each locus when the Alu is present compared to when it is absent. Polymorphic Alu loci highlighted in subsequent figures (figure number in parentheses) are shown in red with chromosomal location indicated. (C) Effect of Alu orientation was evaluated by sorting results based on the orientation of the Alu with respect to luciferase in each evaluated construct. Alu orientation does not drive luciferase expression (t-test).
These Alu insertions showed a continuum of effects on transcription (Fig. 1B). The Alu resulting in the greatest up-regulation (Alu-363) caused a 4.46-fold change (log2 = 2.1) (Fig. 1B) when the Alu is present relative to when it is absent. The polymorphic Alu with the greatest negative effect on transcript levels (Alu-534) resulted in a 3.7-fold down-regulation (log2 = −1.9) (Fig. 1B) when the Alu is present relative to when it is absent. More loci showed some degree of up-regulation (fold change < 0; n = 81) than down-regulation (fold change < 0; n = 29) in the presence of the Alu (P < 0.001, χ2 test). This continuum of effects on transcript levels was not dependent on the orientation of the Alu relative to luciferase (P = 0.4917, unpaired t-test) (Fig. 1C); this lack of directionality suggests that interference by the Alu intrinsic RNA Pol III promoter is not driving the ectopic assay results. Similarly, we wanted to consider if the orientation of the entire genomic locus made a significant difference in the assay results. In particular, at several loci the Alu variant maps near a bidirectional promoter (e.g., Alu-793) or to a large intergenic region with distal genes on opposite strands (e.g., Alu-351). For these loci and others (n = 62), we evaluated the genomic locus in the opposite orientation with respect to luciferase. Overall, we found good correlation between luciferase expression levels irrespective of the direction of the cloned locus, although there was some variance (Supplemental Fig. S1). This ectopic assay allows us to evaluate a large number of loci and identify the outliers where polymorphic Alu insertions have the greatest effect on transcript levels in 293T cells. We focus on these outliers throughout the remainder of this study.
Alu variants are associated with changes in gene transcription
To gain additional evidence of polymorphic Alu roles in regulating gene expression, we compared results of the ectopic reporter assay to previously published expression quantitative trait loci (eQTL). Because most eQTL studies are based on SNPs and do not consider polymorphic Alu elements, we found the best proxy SNP for the Alu at each locus. To determine if there are eQTL already reported for genes near the Alu variants assayed, we looked for a precalculated cis-eQTL in the Genotype-Tissue Expression (GTEx) database associated with that proxy SNP. For 90 of the 110 Alu variants evaluated, we identified a strong proxy SNP with r2 > 0.8 (r2 range 0.81–1, average = 0.97). Of these 90 loci, 57 (63%) have at least one eQTL reported in at least one tissue (average P-value of the eQTL = 9.18 × 10−6, range 1.8 × 10−4 to 1.6 × 10−49). The number of genes differentially expressed at each locus varies from one to 11 genes (average = 3, median = 2, mode = 1). For each eQTL, the number of tissues with differential expression ranges from a single tissue to 44 tissues (average = 4, median = 2, mode = 1). The entire list of Alu eQTLs are in Supplemental Table S2. Comparing our ectopic enhancer reporter data with these reported eQTLs, 71.9% (41/57) of Alu eQTLs show a concordant directional effect. That is, addition of Alu to the luciferase reporter has the same effect, either up- or down-regulation, as is associated with the Alu-containing haplotype for at least one gene in the single tissue eQTLs. When focusing on the outliers in our luciferase assay, those with >1.5-fold change in expression, 75.8% of those with associated eQTLs agree in direction of effect with the GTEx eQTL. Further, the strength of effect seen in the luciferase assay corresponds well to whether there is agreement between GTEx eQTL and luciferase results (P < 0.01, unpaired t-tailed t-test). Overall, these findings suggest that some outliers in our ectopic assay may affect transcript levels at the endogenous loci.
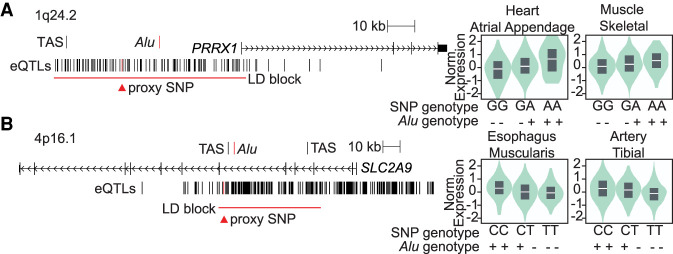
Of particular interest are those polymorphic Alu elements that map to disease risk loci identified by GWAS. One such Alu, Alu-355, maps to 1q24.2 an atrial fibrillation risk locus (P = 8 × 10−14) (Fig. 2A; Ellinor et al. 2012). There is moderate linkage disequilibrium (LD) between the Alu and the GWAS signal (rs3903239, r2 = 0.4, D′ = 0.93) (Payer et al. 2017), indicating that any functional effects of the Alu could have been detected in the GWAS but not necessarily that the Alu is the causative variant for atrial fibrillation risk at this locus. The Alu maps 29 kb upstream of the paired related homeobox 1 (PRRX1) gene. Decreased expression of PRRX1 is associated with increased risk of atrial fibrillation (Tucker et al. 2017). A SNP, rs1048923, which is a strong proxy for the Alu variant (r2 = 1), is an eQTL of PRRX1 in two tissues, including the heart (Supplemental Table S2), where the higher expression of PRRX1 occurs from Alu-containing haplotype than the haplotype with no Alu present (Fig. 2A). This is consistent with our ectopic luciferase assay, where the Alu increases luciferase expression (fold change = 2.69, log2 = 1.43) (Fig. 1B). At the endogenous locus, the Alu may be responsible for affecting PRRX1 expression levels. Thus, more PRRX1 expression directed by the Alu, or other variant on the same haplotype, may decrease the chance of atrial fibrillation.
Figure 2.
Polymorphic Alu outliers in the enhancer assay are associated with known eQTLs. Two example loci are shown, drawn to scale, with GWAS trait-associated SNPs (TAS) and Alu element (red) location marked. GTEx identified eQTLs, including the SNP used as a proxy for the polymorphic Alu element (red triangle), are annotated as well as the extent of the linkage disequilibrium (LD) surrounding the proxy SNP (red bar). The presence (+) or absence (−) of the Alu was phased with the proxy SNP genotype, and GTEx genotype-dependent expression is shown for two example tissues. (A) Polymorphic Alu at PRRX1 is candidate causative variant in atrial fibrillation risk GWAS (TAS = rs39033239, r2 = 0.4, D′ = 0.93). A GTEx eQTL, rs10489231, is a perfect proxy for the Alu (r2 = 1). (B) Polymorphic Alu at SLC2A9 maps to uric acid and gout GWAS signals (TAS = rs3775948 and rs4475146). GTEx eQTL rs4235346 is a perfect proxy for the Alu (r2 = 1).
Another similar example is Alu-330, which maps to 4p16.1, a region linked to uric acid levels and gout risk (P = 2 × 10−65 and P = 4 × 10−26) (Fig. 2B; Okada et al. 2012; Köttgen et al. 2013). The Alu maps to an intron of SLC2A9, a gene that encodes solute carrier family 2 member 9 protein that is involved in transmembrane transport of urate and fructose. A SNP, rs4235346, which is a strong proxy for the Alu variant (r2 = 1), is an eQTL of SLC2A9 in 11 tissues, and the Alu-haplotype-associated SNP allele is associated with up-regulation in eight of those tissues (Supplemental Table S2). In our luciferase assay, the Alu up-regulates luciferase expression (fold change = 1.84, log2 = 0.88) (Fig. 1B). At the endogenous locus, the Alu may be responsible for increased SLC2A9 expression levels and ultimately the risk of developing gout.
Polymorphic Alu elements can disrupt other regulators
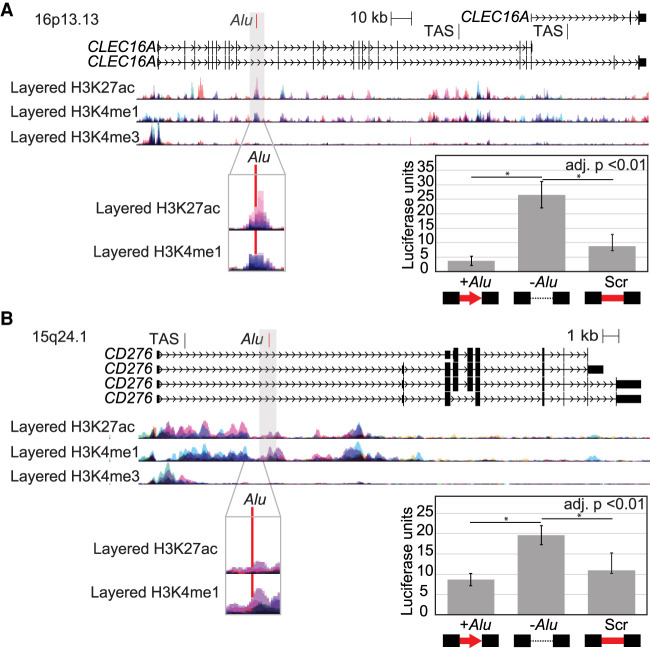
We hypothesized that Alu variants can alter transcript levels by either disrupting other regulators or by introducing new regulatory sequences. To identify cases in which the Alu insertion may disrupt an enhancer, we sought instances in which the preinsertion allele showed high activation of luciferase in the ectopic enhancer assay, and the presence of the Alu decreased luciferase expression. Two loci with the most reduced luciferase expression in the presence of the Alu, Alu-530 and Alu-534 (3- and 3.7-fold decrease, respectively), both fit these criteria. Alu-534 maps to the tenth intron of CLEC16A (Fig. 3A) and is in weak LD (r2 = 0.37, D′ = 0.77) with a GWAS SNP (rs8038465) associated with type 1 diabetes (P = 1 × 10−9) (Hoffman et al. 2012), indicating it is a candidate, albeit unlikely, causative variant leading to disease risk. Alu-350 maps to the first intron of CD276 (Fig. 3B) and is in strong LD (r2 = 0.82, D′ = 0.98) with a SNP (rs12708716) associated with liver enzyme levels (P = 3 × 10−18) (Todd et al. 2007).
Figure 3.
Polymorphic Alu elements mapping to epigenetic marks disrupt other genomic regulators. Genomic loci drawn to scale with layered chromatin marks from ENCODE tracks on UCSC Genome Browser and trait-associated SNPs (TAS) annotated. A magnified view (gray box) highlights the location of the polymorphic Alu (red) relative to chromatin marks. Ectopic luciferase reporter assay results are shown as relative luciferase units for each construct relative to a control vector. Comparisons were made between the locus with the Alu, without any insert, or with a scrambled Alu sequence (Scr) (t-test, adjusted for three comparisons). Error bars are the standard deviation of two clones tested in triplicate in two experiments (n = 12). (A) Polymorphic Alu element mapping to intron of CLEC16A, a region associated with type 1 diabetes. (B) Polymorphic Alu element mapping to an intron of CD276, a region linked to liver enzyme levels.
To build support for the presence of an enhancer element at the Alu insertion site in each case, we used data published by the Encyclopedia of DNA Elements (ENCODE) project (Ernst and Kellis 2010; Hoffman et al. 2012; The ENCODE Project Consortium 2012) and ChromHMM annotation (Supplemental Table S3; Ernst and Kellis 2010). ChromHMM annotates both loci as transcriptionally active in most cell lines. Further, the Alu insertion sites are marked by H3K27ac and H3K4me3, which are associated with regulatory regions, in at least some cell lines (Fig. 3). In all cases, these annotated epigenetic marks most likely come from the preinsertion empty allele for two reasons. First, for these polymorphic Alu loci, the presence of the Alu is the minor allele, that is, the Alu-containing allele is less common. Second, the Alu is not included or annotated in the reference genome, meaning that most standard read mapping pipelines would discard Alu-containing reads from this locus. Therefore, these results are consistent with these Alu elements potentially disrupting active regulatory regions. Although this supported a model wherein the Alu disrupts regulatory elements, we wanted to directly test this hypothesis.
Such a disruption would not rely on sequence-specific features of an Alu insertion. To test this prediction, we scrambled the Alu sequence within the cloned genomic sequences (Supplemental Table S4); the length and GC content of the Alu were retained, but any regulatory sequences intrinsic to the Alu sequence would be disrupted (Fig. 3). For both loci tested, the preinsertion allele results in higher luciferase expression than when the Alu is present (P < 0.0001, t-test). At either, replacing the Alu with scrambled sequence yields similar results to the Alu being present (adjusted P < 0.01, t-test) (Fig. 3). Thus, the change associated with the Alu genotype is consistent with the Alu insertion disrupting an enhancer-like regulator. It is unclear what specific regulator is disrupted by the presence of the Alu at these loci because no ENCODE ChIP-seq transcription factor binding is annotated precisely at the Alu insertion site nor are any putative transcription factor binding sites disrupted by the presence of the Alu. We might speculate that the regulatory change is caused by changes in the relative spacing of known or putative regulatory features that flank the Alu insertion site.
Polymorphic Alu elements have intrinsic ability to alter transcript levels
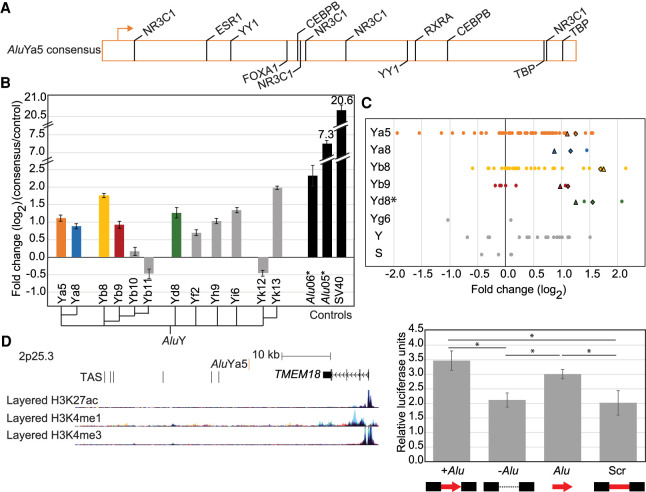
An alternative mechanism by which Alu variants can alter transcript levels would be through regulatory functions encoded by the retrotransposon. Older Alu elements, now fixed in human populations, have acquired nucleic acid substitutions that have made them tissue-specific enhancers (e.g., Norris et al. 1995; Gombart et al. 2009; Jacobsen et al. 2009). It has been suggested that evolutionarily younger Alu elements may also introduce enhancer functions (Wang et al. 2017a). To evaluate one of the youngest and most commonly polymorphic Alu subfamilies, AluYa5, we used PROMO and TRANSFAC (Messeguer et al. 2002; Farre et al. 2003) to identify 14–109 putative transcription factor binding sites (TFBS) for 9–40 transcription factors (Fig. 4A). Using this method, we identify highly stringent (0% dissimilarity) YY1 binding sites, as previously reported in Alu elements (Humphrey et al. 1996; Oei et al. 2004; Polak and Domany 2006). In all, the putative TFBS are well conserved across AluY consensus sequences (Supplemental Table S5); of the 14 sites identified in AluYa5 using the most stringent criteria, all but one are highly conserved across other AluY subfamily sequences (Supplemental Table S5). This could suggest that all AluY subfamily elements would behave similarly in our assay.
Figure 4.
Alu elements have intrinsic regulatory potential. (A) AluYa5 has 14 high confidence putative transcription factor binding sites. (B) Ectopic luciferase assay results with isolated Alu consensus sequences. Cladogram shows the approximate evolutionary relationship between the 12 commonly polymorphic AluY subfamily consensus sequences, including five (colored) represented in our evaluated polymorphic Alu loci. Ectopic assay results for non-polymorphic, evolutionarily older Alu elements (*) previously evaluated in Su et al. (2014), and the strong SV40 enhancer are also shown (black). (C) Results from Figure 1B separated based on the Alu subfamily present at each locus. In cases where the consensus sequence was evaluated in B (matching colors), results for the consensus are shown as triangles (Alu tested sense with respect to luciferase) and diamonds (Alu consensus tested antisense with respect to luciferase). Alu subfamily does not drive the locus-specific results, as only the AluYd8 subfamily gave consistently distinct results (*) P = 0.02, ANOVA. (D, left) Genomic locus for Alu-609 drawn to scale with annotated epigenetic marks as in Figure 3. The AluYa5 (red) does not map to any notable epigenetic marks. This region was identified in GWAS as associated with obesity and body mass index (GWAS trait-associated SNPs [TAS]). Right, luciferase assay results for the locus with (+) and without (−) the Alu present compared to the isolated Alu-609 sequence (Alu). (*) Adjusted P < 0.05, t-test. Error bars are the standard deviation of two clones tested in triplicate in two experiments (n = 12).
To experimentally evaluate the ability of commonly polymorphic AluY subfamilies to alter transcript levels, we tested isolated Alu sequences (i.e., without surrounding genomic context) in the ectopic luciferase reporter assay. We evaluated 12 AluY subfamily consensus sequences including the four most highly represented in the tested polymorphic loci. Alu elements were tested both in sense (Fig. 4B) and antisense (Supplemental Fig. S2) orientation with respect to luciferase with similar results in 293T cells (Supplemental Fig. S2). Again, Alu sequences showed a continuum of effects in this ectopic assay (Fig. 4B). The strongest up-regulation in luciferase expression by polymorphic AluY consensus sequences approaches that of the evolutionarily older AluY (Alu06) that is epigenetically marked consistent with enhancer function (Fig. 4B; Su et al. 2014). Overall, these AluY consensus sequences have effect sizes in the luciferase assay that are much smaller than the strongest Alu sequence with reported enhancer function, an AluSc (Alu05) (Su et al. 2014) or the well-characterized SV40 enhancer (Fig. 4B). AluYk13 results in a fourfold up-regulation (log2 = 1.98) of luciferase expression relative to a control sequence, whereas on the other extreme, AluYb11 results in a 0.75 reduction (log2 = −0.47) in luciferase expression. AluYk13 and AluYb11 share 288 of 309 bp (93%); there are also 8 bp specific to AluYb. Although these two sequences share 12 high confidence (0% dissimilarity) putative transcription factor binding sites, there are four sites that are specific to AluYb11 and one high confidence site in AluYk13 that is less conserved in AluYb11 (Supplemental Table S5). The 8 bp insertion common to AluYb subfamily members cannot be the only determinant because although AluYb11 shows the most down-regulation of luciferase, another AluYb subfamily member, AluYb8, resulted in the second most up-regulation in the ectopic assay (after AluYk13). Despite different results in the ectopic assay, these two AluYb family members, AluYb11 and AluYb8, are nearly identical (306/309 bp, 99%) and share 16 high confidence putative transcription factor sites (Supplemental Table S5). In all, these findings indicate that, despite similar sequence content, different Alu subfamilies can have highly distinct effects on gene regulation.
To evaluate Alu sequences that may have intrinsic enhancer-like effects, we focused on the five subfamily consensus sequences that up-regulate luciferase expression where we had tested representative polymorphic Alu loci (Fig. 4B, colored bars). We compared the putative TFBS between consensus Alu sequences looking for distinguishing features (Supplemental Table S5). For example, of these consensus sequences, AluYb8 and AluYb9 are most distinct from AluYa5 and AluYa8. AluYa5/8 consensus sequences have 11–14 TFBS absent in AluYb8/9, and AluYb8/9 have 20 unique TFBS including a highly stringent (0% dissimilarity) CEBPB (also known as C/EBP-beta) site. Although these differences may account for the degree of luciferase up-regulation, AluYa5, AluYa8, AluYb8, and AluYb9 consensus sequence all increase luciferase expression. Thus, sequences that distinguish AluYb from AluYa subfamily members are unlikely to be the key enhancer-like regulatory sequences.
We next evaluated the presence or absence of each predicted TFBS in the specific polymorphic Alu sequences evaluated in this study. Most Alu elements are ∼300 bp in length, but occasionally on insertion, 5′ truncations of the Alu occur. In particular, two polymorphic AluYb8 elements, Alu-103 and Alu-253, are missing ∼175 bp and ∼190 bp of the 5′ end of the consensus sequence, respectively, and one AluYa8 sequence, Alu-411, is missing ∼180 bp of 5′ Alu sequence (Supplemental Table S5). For all three of these 5′ truncated loci, the presence of the Alu in its genomic context results in up-regulation of luciferase relative to when the Alu is absent (Fig. 1B; Supplemental Table S1), consistent with the effect seen with the consensus sequence. Because of their truncated size, Alu-103, Alu-253, and Alu-411 contain only 38, 30, and 26 putative TFBS, respectively, compared to ∼100 in a full-length Alu sequence. All three truncated elements contain just four highly stringent shared TFBS, including one of the YY1 sites previously reported in Alu sequences (e.g., Humphrey et al. 1996; Oei et al. 2004; Polak and Domany 2006). Because these sequences are present in even the shortest elements with enhancer-like effects, they make attractive candidates for being key regulatory sequences. However, additional studies will be necessary to fully dissect the sufficient and necessary sequences.
To test whether the subfamily of the Alu would be an important determinant of regulatory effects, we subsetted the luciferase results for the evaluated polymorphic loci (Fig. 1) based on the subfamily of the Alu at the locus (Fig. 4C). We compared the results for all loci with a specific Alu subfamily present against the subfamily consensus sequence evaluated in isolation. In all cases, we observed a range of effects for loci within each Alu subfamily not tightly correlated to the subfamily sequence tested in isolation (Fig. 4C). Further, comparing across subfamilies, the Alu effects on luciferase expression for different subfamilies overlapped significantly. The AluYd8 subfamily is notable in that all loci tested show very similar up-regulation; however, a small sample size was considered here. Excluding this AluYd8 subfamily, there is no statistical difference between the subfamilies evaluated (P = 0.946, ANOVA) (Fig. 4C). Thus, some Alu sequences have intrinsic regulatory function (Fig. 4B), but this alone does not account for all of the regulatory effects captured in luciferase assays that include a locus-specific Alu sequence and surrounding sequences. It is likely that a complex combination of molecular mechanisms occurs at each of these loci.
When a naturally occurring Alu variant affects transcript levels by an intrinsic mechanism, we might predict that the genomic locus without the Alu does not alter transcript levels relative to a control empty vector, and when the Alu is present, a significant change in luciferase expression occurs relative to the control. Alu-609 is one of these examples. Alu-609 is an AluYa5 that maps to 2p25.3 and is in strong LD (r2 = 1) with eight GWAS signals for phenotypes such as body mass index (BMI) and obesity (e.g., best P = 3 × 10−49) (Fig. 4D; Speliotes et al. 2010). The Alu maps downstream from TMEM18, a gene long associated with energy levels and BMI although the molecular mechanism is not well understood (e.g., Almén et al. 2010; Larder et al. 2017). The downstream region containing the GWAS signals and Alu variant is void of epigenetic marks across diverse cell types (Fig. 4D). We believe this annotation most likely reflects the preinsertion allele because the Alu variant is not included in the reference genome, making mapping of Alu-containing reads difficult for most standard pipelines. Consistent with this, in our luciferase assay, the Alu-609 locus with no Alu present had very little activation of luciferase compared to a large increase in luciferase expression when the Alu was present (fold change = 1.65, log2 = 0.724) (Fig. 1B). We hypothesized that the increase in luciferase expression was intrinsic to the Alu sequence. To test this, we cloned the specific Alu from this locus into the luciferase reporter construct independent from the context of its genomic locus. The Alu from this locus has intrinsic ability that almost completely recapitulates that of the Alu in the context of its genomic locus; both are significantly different from the genomic locus with no Alu (adjusted P < 0.05, t-test) (Fig. 4D). The Alu-609 is 99.65% identical to the AluYa5 consensus sequence with only 1 bp mismatch. Both yield similar up-regulation, approximately twofold, when tested in isolation (P = 0.611, t-test). Therefore, the polymorphic AluYa5 mapping to TMEM18 has intrinsic enhancer function in the ectopic assay consistent with that of the AluYa5 consensus sequence.
Breast cancer risk loci have Alu variants with regulatory potential
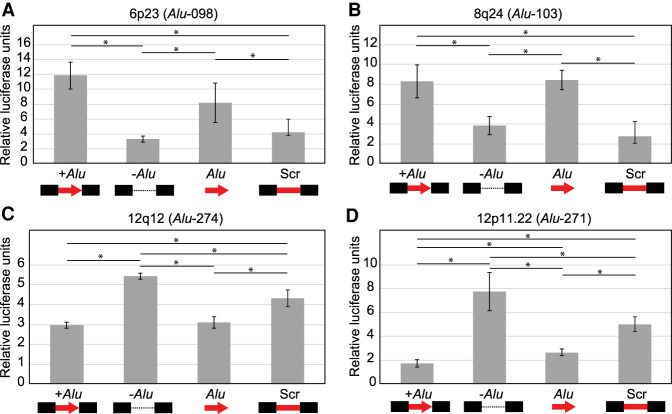
We next wanted to evaluate the function of the Alu insertions that were both associated with disease and “outliers” in their regulatory impact using relevant cellular models. Informed by our initial luciferase assays, we chose to focus on four polymorphic Alu elements all associated with breast cancer risk. At two of these loci, Alu-098 located at 6p23 and Alu-103 located at 8q24.21, the presence of the Alu results in up-regulation of luciferase expression compared to when the Alu is absent, a 4.28 (log2 = 2.10) and 3.32 (log2 = 1.73) fold change, respectively (Fig. 1B). At the other two loci, Alu-271 mapping to 12p11.22 and Alu-274 mapping to 2q35, the presence of the Alu results in a decrease in luciferase levels compared to when the Alu is not present, 0.46 (log2 = −1.11) and 0.53 (log2 = −0.92) fold change, respectively (Fig. 1B).
Because enhancers can be tissue specific, we repeated the ectopic luciferase assays in two cell lines derived from mammary gland, T-47D and MCF10A. The presence of the Alu resulted in similar changes in luciferase expression in all cell lines tested for each of the four loci evaluated (Supplemental Fig. S3A).
We next determined the mechanism by which the Alu alters luciferase expression using a series of ectopic reporter constructs like previous experiments (Fig. 5). For two loci, Alu-098 and Alu-103, the effect of the Alu in genomic context (increasing luciferase expression) is recapitulated when the Alu is evaluated independently (adjusted P < 0.05, t-test) (Fig. 5A,B). Further, scrambling the Alu sequence within the genomic context did not increase luciferase expression. Together, this indicates that the effects of Alu-098 and Alu-103 on expression are intrinsic to the Alu. Alu-098 is an AluYd8 (98.9% identity) and Alu-103 is an AluYb8 (100% identity); both Alu subfamily consensus sequences have intrinsic ability to up-regulate luciferase (Fig. 4B). Alu-103 is truncated yet shares a similar level of up-regulation when tested in isolation to the full-length AluYb8 consensus sequence (Figs. 5A, 4B). This suggests that the intrinsic sequences directing this altered luciferase expression occur in the 114 bp common to the two sequences (see the previous section for putative TFBS in this region).
Figure 5.
Mechanisms for how Alu insertion polymorphisms associated with breast cancer affect gene expression levels. For each of four loci (A–D), a series of ectopic reporter constructs were tested: the Alu in the genomic locus (+Alu); the locus with no Alu present (−Alu); the isolated locus-specific Alu sequence (Alu); and the genomic locus with a scrambled Alu sequence (Scr). (*adjusted P < 0.05, t-test). Error bars are the standard deviation of two clones tested in triplicate in two experiments (n = 12).
For the other two loci, Alu-274 and Alu-271, where the presence of the Alu results in a decrease in luciferase expression, our findings suggest a more complex molecular mechanism. For Alu-274, low luciferase expression is recapitulated when the Alu is tested in isolation (adjusted P < 0.05, t-test), indicating that regulatory features may be intrinsic. However, a scrambled Alu also results in a significant decrease in luciferase expression (adjusted P < 0.05, t-test), albeit to a lesser extent than the Alu (Fig. 5C). Based on these findings, we postulate that Alu-274 contains regulatory potential, but also interacts with regulators at the integration site. Similarly, Alu-271 effects in the luciferase assay are complex and depend both on sequences intrinsic to the Alu and the surrounding locus as each construct evaluated produces a different level of luciferase expression (adjusted P < 0.05, t-test) (Fig. 5D). Altogether, these data shed light on the molecular mechanism by which Alu might regulate gene expression at these loci.
Polymorphic Alu elements at breast cancer risk loci are eQTLs
Because enhancer effects can be sensitive to broader genomic context, we wanted to evaluate Alu regulatory effects at these breast cancer–associated loci in their endogenous context. We used CRISPR to edit 293T cells to generate isogenic cell lines that were identical at the locus of interest except for the Alu genotype. We generated lines that were homozygous for the presence of the Alu and homozygous for the “empty” preinsertion allele with no Alu. These cell lines were sequence verified to be perfect edits, and we maintained the original 293T haplotypes with the only exception being the Alu presence or absence (Supplemental Table S6). In this way, we isolated the effects of the Alu from other variants that naturally occur on the same haplotype. For two of these loci, Alu-098 and Alu-103, we also used CRISPR to edit the endogenous locus in the mammary derived cell line, T-47D-Cas9. At both these loci, the parental T-47D cell line is heterozygous for the presence of the Alu. We deleted either Alu-098 or Alu-103 resulting in cell lines that were homozygous for no Alu present and compared these to the heterozygous cell line.
Alu-274 is in strong LD with a breast cancer GWAS signal (Michailidou et al. 2013), rs16857609, at 2q35 (r2 = 0.953). Fine mapping studies at this locus have narrowed the GWAS signal to a 20-kb region that contacts the IGFBP5 promoter (Wyszynski et al. 2016) and have identified a likely causative variant, rs4442975, that maps to an enhancer and is an eQTL for IGFBP5 (Ghoussaini et al. 2014; Fachal et al. 2020). Because Alu-274 does not map to this region and IGFBP5 is not well expressed in 293T cells, we focused on the other three breast cancer risk loci for these studies.
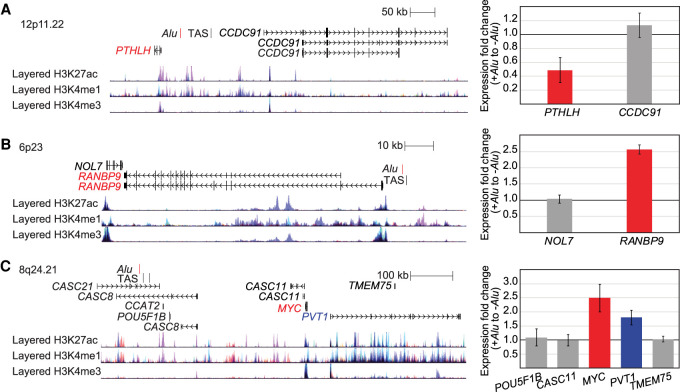
We performed genome editing at one locus where presence of the polymorphic Alu, Alu-271, reduced luciferase expression levels in the ectopic reporter assay (Fig. 1B). Alu-271 is a full-length (292 bp) AluYb8 that maps to intergenic region between, and upstream of both, CCDC91 and PTHLH at 12p11.22 (Fig. 6A). This region has been associated with breast cancer risk (P = 8 × 10−31, OR = 1.16) (Michailidou et al. 2013), and we previously determined that Alu-271 is a good causative variant because it is in strong linkage disequilibrium with the GWAS identified SNP (r2 = 0.918 with rs10771399) (Payer et al. 2017). Fine mapping of this region identified three independent disease risk signals (Zeng et al. 2016). Alu-271 maps to a ∼65 kb implicated region (Signal 2) (Zeng et al. 2016) with no candidate causative variant previously reported (Zeng et al. 2016; Fachal et al. 2020). Further, each of the three signals contributes to disease risk and the effects are cumulative, but the largest effect is seen for the region to which the Alu variant maps. This region interacts with the nearby PTHLH gene (Zeng et al. 2016). PTHLH encodes the parathyroid hormone like hormone protein, which is required for embryonic development of the breast and secreted during lactation (Wysolmerski 2012). It is less clear the role PTHLH transcript and protein play in breast tumors; it may be involved in cell turnover, tumor growth, or clinical outcome, although conflicting results have been reported (e.g., Yin et al. 1999; Fleming et al. 2009; Li et al. 2011; Luparello 2011). Regardless, multiple breast cancer GWAS have highlighted this locus. Based on the ectopic luciferase assay, we hypothesized that the Alu reduces expression of a nearby gene, potentially PTHLH, which in turn leads to cancer risk. We measured expression of the genes near the Alu insertion site in the CRISPR-edited 293T cell lines by qRT-PCR. The presence of the Alu results in a 0.49-fold change in PTHLH expression relative to when the Alu is absent; CCDC91 expression is essentially unchanged (1.13-fold change) by the Alu genotype (Fig. 6A). These results indicate that the polymorphic Alu alters transcript levels of PTHLH in the endogenous context.
Figure 6.
Polymorphic Alu are eQTLs at breast cancer risk loci. Loci, drawn to scale with the trait-associated SNPs (TAS) annotated, were edited by CRISPR to create cell lines that differed in genotype at the indicated Alu (red). qRT-PCR was performed on cell lines homozygous for the Alu insertion and lines homozygous for the absence of the Alu. Error bars are the standard deviation from 12 qRT-PCR measured ratios (two lines homozygous for the Alu relative to two lines homozygous for no Alu, each tested from two different cultures, in technical triplicate). (A) The presence of Alu-271 reduces PTHLH (red) expression. (B) The presence of Alu-098 increases RANBP9 (red) expression. (C) The presence of Alu-103 results in MYC (red) and PVT1 (blue) up-regulation compared to when Alu-103 is absent.
We also edited a locus where presence of the polymorphic Alu, Alu-098, increased luciferase expression in the ectopic reporter assay (Fig. 1B). Alu-098 is a polymorphic full-length (270 bp) AluYd8 that maps upstream of the RAN binding protein 9 gene, RANBP9 (Fig. 6B). A GWAS associated this region with breast cancer risk (P = 8 × 10−9, OR = 1.05) (Michailidou et al. 2013). The role of RANBP9 in breast cancer development and/or progression is not well understood. RANBP9 is expressed in normal breast tissue and up-regulated in cancer (Wang et al. 2002). The RANBP9-encoded protein interacts with androgen receptor (Cochrane et al. 2014) and tumor associated genes (Yan et al. 2015) and has been implicated in activating cell signaling pathways (Wang et al. 2002), including proapoptotic pathways in response to DNA damage (Atabakhsh et al. 2009). We previously determined that the polymorphic Alu-098 was a good causative variant for the GWAS because it is in strong LD with the GWAS SNP (r2 = 1 with rs204247) (Payer et al. 2017). We therefore hypothesized that Alu-098 increases expression of RANBP9, which ultimately increases the risk of breast cancer. We measured expression of RANBP9 and another gene near the Alu insertion site, NOL7, in the 293T and T-47D CRISPR-edited lines using qRT-PCR. In 293T, the presence of the Alu on both alleles resulted in a 2.6-fold change in RANBP9 expression relative to when the Alu is absent; NOL7 expression is unchanged (fold change = 1) between the cell lines (Fig. 6B). In T-47D, a 1.5-fold change in RANBP9 expression was detected between the parental line with one Alu-containing allele relative to the edited line with no Alu present; NOL7 expression was unchanged (Supplemental Fig. S3B). These results indicate that the polymorphic Alu enhances RANBP9 expression.
Polymorphic Alu at 8q24 alters MYC expression and is associated with breast cancer risk
Polymorphic Alu-103 increased luciferase expression in the ectopic reporter assay (Fig. 1B). Alu-103 is a 114-bp 5′ truncated AluYb8 that maps to 8q24, a region associated with different types of cancer risk (Ghoussaini et al. 2008). In particular, Alu-103 maps to a region of breast cancer susceptibility (P = 1 × 10−27 OR = 1.09) (Michailidou et al. 2013). Fine mapping studies have identified five independent signals for breast cancer risk within a ∼1 Mb region at 8q24 (Shi et al. 2016). Candidate causative variants have been identified for two of these signals as SNPs rs7815245 and rs11780156 that map at enhancers, alter TFBS motifs, and are eQTLs (Shi et al. 2016). However, the strongest signal in the region is tagged by rs13281615, and a strong causative variant at this location has yet to be identified. We previously showed that Alu-103 is a good genetic candidate for the GWAS signal (r2 = 0.77 with rs13281615) (Payer et al. 2017).
8q24 has many noncoding transcripts that show tissue specificity and have only been identified in cancers. PVT1 is a long noncoding RNA (lncRNA) that is overexpressed in many cancers, including breast cancer, although its function is not well understood (Colombo et al. 2015); it maps >500 kb away from the polymorphic Alu. At 391 kb away from the Alu variant maps MYC, a highly studied and important oncogene that is up-regulated in many cancers (for review, see Lancho and Herranz 2018).
The Alu could alter expression of any number of genes at this locus. Therefore, in our qRT-PCR analysis of the 293T CRISPR-edited lines, we included all genes mapping to this locus that are expressed in 293T cells. We used qRT-PCR to measure expression levels in cell lines homozygous for either the presence or absence of Alu-103. The Alu genotype did not affect expression levels of most genes (Fig. 6C). However, in the presence of the Alu, MYC was ∼2.5-fold up-regulated and PVT1 was ∼1.8-fold up-regulated. Given the distance between this Alu and MYC, we confirmed the results by evaluating gene expression in additional 293T CRISPR-edited lines (Supplemental Fig. S4). We next evaluated gene expression changes in T-47D CRISPR-edited cell lines. MYC was ∼1.9-fold up-regulated and PVT1 was ∼1.8-fold up-regulated in the parental T-47D line, heterozygous for the presence of Alu-103 relative to the edited line with no Alu-103 present (Supplemental Fig. S3B).
The MYC locus is a classic example of a gene controlled by several long-distance acting enhancers, some located >1 Mb away (Herranz et al. 2014; Bahr et al. 2018). The three-dimensional structure of the MYC locus has been dissected using chromosome conformation capture techniques such as Hi-C. Most of the long-range contacts that the MYC promoter engages in are with enhancers downstream from the MYC gene (for review, see Lancho and Herranz 2018). Alu-103 maps >390 kb upstream of the MYC promoter in a distinct topologically associating domain (TAD) from MYC, suggesting that contacts to this region occur less frequently. However, contacts between MYC and upstream loci decorated with enhancer marks do occur (e.g., Petrovic et al. 2019), and contacts between this region and MYC were observed in MCF-7 cells (Ahmadiyeh et al. 2010). Furthermore, enhancers located in the same TAD as this Alu have been previously identified (Haiman et al. 2007; Yashiro-Ohtani et al. 2014; Gekas et al. 2016), suggesting that even if the MYC promoter is engaging here less often than with its downstream vicinity, contacts established with these upstream enhancers can affect MYC expression.
Discussion
We evaluated the possibility that polymorphic Alu insertions alter gene transcript levels. In particular, we were interested in functional effects of polymorphic Alu elements we previously identified as candidates to contribute to human disease risk through a common disease–common variant paradigm (Payer et al. 2017). We identified a subset of these polymorphic Alu elements associated with disease risk that alter luciferase expression in an ectopic assay and confirmed that many of these are eQTLs. These results are consistent with the disproportionate number of disease haplotypes associated with enhancer sequences (Ernst et al. 2011; Cowper-Sal lari et al. 2012; Maurano et al. 2012; Schaub et al. 2012; Corradin and Scacheri 2014; Wu and Pan 2018) and eQTLs (Nica et al. 2010; Nicolae et al. 2010; Hernandez et al. 2012). To further understand how polymorphic Alu elements alter transcript levels, we identified the molecular mechanism at some of the “outlier” loci where this effect was largest. Testing effects of Alu elements isolated from surrounding sequence, and replacing Alu with random sequence, we find that an Alu may alter transcription either by disrupting other regulators or by introducing intrinsic regulatory sequence. We next focused on three polymorphic Alu insertions that were outliers in our ectopic assay and associated with breast cancer risk. We used CRISPR editing to generate cell lines that differed only in the Alu genotype at the locus of interest, and this showed that presence or absence of the Alu impacts the expression of genes associated with breast cancer and accounts for the reported eQTL. Collectively these data show that Alu insertion variants alter gene transcript levels.
Using ectopic assays that incorporate small intervals of surrounding genomic sequence allowed us to easily manipulate Alu variants, and thus differentiate between Alu insertions that disrupt preexisting regulators and those Alu with intrinsic regulatory properties. Another aspect of our approach is that we did not only focus on sites with known epigenetic features. Epigenetic states determined by aligning ChIP-seq reads to the reference genome often represent the state of the preinsertion allele (i.e., with no Alu present), because many Alu polymorphisms are not incorporated into the reference genome and there is inherent difficulty in mapping short reads that contain Alu sequence. An early focus on these would lead to an underappreciation of Alu that introduce regulatory functions. Our results directly show the ability of some young Alu elements to alter transcript levels in this manner, both by consensus sequences for AluY subfamilies (Fig. 4B) and at specific insertion loci (Figs. 4D, 5). This was previously hypothesized for young AluY elements (Wang et al. 2017a) and documented for fixed, older Alu elements on a locus-specific level (e.g., Norris et al. 1995; Gombart et al. 2009; Jacobsen et al. 2009) or through genome-wide surveys (Su et al. 2014).
This intrinsic potential of Alu sequences could have broad consequences for gene regulation. Retrotransposons distribute regulatory sequences throughout the genome. This so-called plug-and-play regulation has been well documented for human endogenous retroviruses (HERVs), which have strong RNApol II promoters and enhancer functions (Chuong et al. 2016), but any retrotransposon might similarly disperse regulatory sequence. Given the rate of Alu expansion that has taken place in primate lineages, the overall effects of Alu on gene regulation is potentially very significant. Some effects will be indirect. A previously reported polymorphic Alu is a cis-eQTL of the transcription factor gene, PAX5, and a trans-eQTL for several PAX5-target genes (Wang et al. 2017b).
Intrinsic functions delivered to a locus by an Alu insertion are likely mediated through transcription factor binding to Alu sequences. We identify putative binding sites within young Alu sequences that have intrinsic function in our luciferase reporter assay (Fig. 4A). Despite the high similarity of Alu sequences tested, we observed varying effects in this assay. A single base pair difference at a key site or combinatorial effects of a few base pair substitutions may be underlying sometimes significant differences in regulatory potential between Alu sequences. Previous analysis of binding motifs in older Alu sequences shows that these sites tend to occur in clusters (Polak and Domany 2006). It is likely that locus-specific context and this precise transcription factor binding compliment dictate the regulatory potential of a particular Alu sequence. For older, fixed insertions—where there has been time for a single ancestral allele to diversify into an allelic series—additional functional variants may exist in human populations. This is an important, but technically challenging, question that long-read sequencing may help to address. Long-read technologies will also enable characterizations of the epigenetic status of polymorphic Aluelements, which may be complex and variable. Detailed studies of AluDNA methylation patterns and associated histone marks may reveal implications for gene regulatory functions and are highly interesting future directions to pursue.
The ectopic reporter assay we used allows for semi-high-throughput analyses of many loci in a selected cell type, and similar systems have been used to evaluate other transposable element sequences for enhancer function (Su et al. 2014; Nguyen et al. 2018; Cao et al. 2019). Another type of SINE, the mammalian-wide interspersed repeat (MIR), and a long interspersed element (LINE) L2 can function as enhancers, and in a tissue-dependent manner reduce luciferase expression when present (Cao et al. 2019). Overall, these elements show a continuum of effects (Cao et al. 2019) similar to the polymorphic Alu elements evaluated in this study and consistent with the idea that repeats are often proto-enhancers (Su et al. 2014). When evolutionarily older, non-polymorphic AluJ and AluS elements that are epigenetically marked similar to an enhancer were tested in a luciferase assay, luciferase was up-regulated 1.2- to 207-fold (Su et al. 2014). Although the polymorphic Alu elements evaluated in our study fall on that lower end of that range, a 1.5-fold change is often considered significant (e.g., Cao et al. 2019). Fifty-four of the polymorphic Alu elements we evaluated (49%) reach this threshold with 47 up-regulating and seven down-regulating luciferase expression. Of the polymorphic Alu elements we assessed, the one with the strongest effect induces a 4.5-fold up-regulation of luciferase. Although it is not possible to equate the absolute magnitude of effect of a variant in an ectopic expression assay with differential gene expression at the endogenous locus, we view the assay as a means to identify “outliers” with greater likelihood to have biologic effect. However, the assay also has limitations. Some of the Alu elements with little or no effect on this reporter may alter transcription in the context of the endogenous locus, or show large effects that depend on cell type or developmental stage (Heintzman et al. 2009; Creyghton et al. 2010; Whyte et al. 2013; Huang et al. 2016) not captured in our experimental system.
A strength of our study is that we corroborate some of the ectopic assay results at endogenous loci. This is a significant step forward, building on our own results and previously published ectopic (e.g., Su et al. 2014; Nguyen et al. 2018; Cao et al. 2019) and computational analyses (Wang et al. 2017b; Goubert et al. 2020) that stopped short of assigning regulatory effects to the transposable element at the endogenous locus. We see excellent correspondence between the effect of the Alu in the ectopic reporter assay and at the endogenous locus in our three genome editing experiments.
In some cases, gene expression appears to reflect both Alu sequence properties and insertion site properties (i.e., Alu-271) (Fig. 5D). Regulation at any locus is likely complex and will encompass more than one enhancer or silencer over a larger distance than is included in our ectopic assays. Similarly, the combined effect of several variants on the same haplotype, that is, the Alu and surrounding SNPs in LD, may act synergistically to alter gene expression level. Further, the repetitive nature of Alu elements in the genome may also lead to interactive effects. For example, at CD8A, an Alu harboring transcription factor binding sites, especially GATA3, acts as an enhancer (Hambor et al. 1993), while a nearby inverted, truncated Alu causes a cruciform structure to form encompassing the enhancer Alu and impairs transcription factor binding (Hanke et al. 1995), so overall regulation of CD8A expression is a balance between these two Alu-derived regulators.
We have previously shown that Alu elements near exons can interfere with mRNA splicing (Payer et al. 2019), and our current work highlights the potential for Alu insertions to impact gene function through regulatory mechanisms that may be more far-reaching. Because intrinsic regulatory potential resides in young, polymorphic Alu elements and because Alu elements have accumulated near genes during primate evolution, their functional impact may be significant. Collectively, these Alu may be important determinants of species-specific traits and, within our species, of phenotypic variation and differences in heritable disease risk.
Methods
Genome editing
We edited three loci in 293T cells (Alu-103, Alu-098, Alu-271) and two loci in T-47D (Alu-098 and Alu-103). Guide RNAs (gRNAs) were cloned into a vector with Cas9 and a GFP marker (pSpCas9(BB)-2A-GFP). gRNA-Cas9 vectors were cotransfected with repair templates (Supplemental Methods; Supplemental Table S6). Single cell outgrowths from genome editing were screened for perfect edits with no extra or missing sequence at the edited site. When possible, at least two perfectly edited lines were derived (Supplemental Table S6). Gene expression was measured by qRT-PCR, with primers listed in Supplemental Table S7, calculated by the 2−ΔΔCt method and normalization to the housekeeping gene actin beta (ACTB) (Supplemental Methods). Results are shown as expression in the 293T cell lines with the Alu present to when it is absent (Fig. 6) or in T-47D cell lines as when the Alu is heterozygous to the homozygous no Alu present cell lines (Fig. 3B).
Enhancer assays
Each genomic region of interest was cloned into a modified pGL4.26 (Promega) vector upstream of the minimal promoter and luciferase. Primers flanking each polymorphic Alu insertion site amplified ∼300 bp of genomic DNA (Supplemental Table S1). Additional cloning details can be found in Supplemental Methods. Although the Alu-containing allele (∼600 bp) and the preinsertion allele (∼300 bp) are different sizes, we saw no indication that this difference in size consistently affected reporter expression. Further, for some outlier loci, we include a scrambled Alu placeholder in place of the Alu so that constructs for each allele are the same size. As other controls, at some loci, we replaced the Alu sequence with two scrambled Alu sequence (scr1, scr3) (Supplemental Table S4; Supplemental Fig. S5A,B). In other cases, we tested the locus-specific Alu or consensus Alu sequence, in isolation in our ectopic luciferase assay; the specific sequences tested are in Supplemental Table S4.
All clones were sequence verified and, in most cases, two independent clones were evaluated for each construct. Luciferase levels were measured using Dual-Glo Luciferase Assay System (Promega) and the GloMax-Multi Detection System (Promega) per manufacturer's protocol. Additional normalization details can be found in Supplemental Methods and in Supplemental Figure S5C. T-tests were performed to compare different constructs.
Epigenetic analysis
The ENCODE genome segmentations using ChromHMM (Ernst and Kellis 2010) for each of the six analyzed cell lines (GM12878, H1-hESC, K562, HeLa-S3, HepG2, and HUVEC) were downloaded from UCSC Genome Browser (https://genome.ucsc.edu, hg19). The Alu polymorphism coordinates were intersected with this data using BEDTools intersect (Quinlan and Hall 2010). Results are in Supplemental Table S3. To examine epigenetic state at some loci more carefully, we viewed the ChromHMM tracks and Integrated Regulation from ENCODE tracks on UCSC Genome Browser. We viewed the layered H3K4me1, H3K4me3, and H3K27ac tracks as well as DNase Clusters and Transcription Factor ChIP E3 (The ENCODE Project Consortium 2011, 2012; Gerstein et al. 2012; Wang et al. 2012,2013). Because both the ENCODE data (e.g., The ENCODE Project Consortium 2011, 2012) and The 1000 Genomes Project annotation of Alu polymorphisms (The 1000 Genomes Project Consortium 2015; Sudmant et al. 2015) were performed on the GRCh37/hg19 human reference genome build, we used this build throughout our analysis and manuscript. Prediction of transcription factor binding sites in Alu sequences were determined with PROMO utilizing TRANSFAC version 8.3 (Messeguer et al. 2002; Farre et al. 2003). We considered high quality calls, to be those with a 0% dissimilarity rate. Supplemental Table S5 contains all putative binding sites with <15% dissimilarity rate.
Supplementary Material
Acknowledgments
We thank Kelsie L. Thu and David W. Cescon (Princess Margaret Cancer Centre/University of Toronto) for providing the T-47D-Cas9 cell line. This work was supported by the National Institutes of Health (R01GM124531 and R01GM130680) (to K.H.B.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.261305.120.
Competing interest statement
The authors declare no competing interests.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci 107: 9742–9746. 10.1073/pnas.0910668107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén MS, Jacobsson JA, Shaik JH, Olszewski PK, Cedernaes J, Alsiö J, Sreedharan S, Levine AS, Fredriksson R, Marcus C, et al. 2010. The obesity gene, TMEM18, is of ancient origin, found in majority of neuronal cells in all major brain regions and associated with obesity in severely obese children. BMC Med Genet 11: 58. 10.1186/1471-2350-11-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabakhsh E, Bryce DM, Lefebvre KJ, Schild-Poulter C. 2009. RanBPM has proapoptotic activities that regulate cell death pathways in response to DNA damage. Mol Cancer Res 7: 1962–1972. 10.1158/1541-7786.MCR-09-0098 [DOI] [PubMed] [Google Scholar]
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. 2018. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553: 515–520. 10.1038/nature25193 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379. 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- Burns KH, Boeke JD. 2012. Human transposon tectonics. Cell 149: 740–752. 10.1016/j.cell.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chen G, Wu G, Zhang X, McDermott J, Chen X, Xu C, Jiang Q, Chen Z, Zeng Y, et al. 2019. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res 29: 40–52. 10.1101/gr.235747.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45: 325–329. 10.1038/ng.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, D'Amato NC, Spoelstra NS, Edgerton SM, Jean A, Guerrero J, et al. 2014. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res 16: R7. 10.1186/bcr3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo T, Farina L, Macino G, Paci P. 2015. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int 2015: 304208. 10.1155/2015/304208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Scacheri PC. 2014. Enhancer variants: evaluating functions in common disease. Genome Med 6: 85. 10.1186/s13073-014-0085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. 2012. Breast cancer risk–associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 44: 1191–1198. 10.1038/ng.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn-Fletcher CE, Muglia LM, Pavlicev M, Wolf G, Sun MA, Hu YC, Huffman E, Tumukuntala S, Thiele K, Mukherjee A, et al. 2018. Anthropoid primate–specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol 16: e2006337. 10.1371/journal.pbio.2006337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, et al. 2012. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 44: 670–675. 10.1038/ng.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. 10.1038/nbt.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachal L, Aschard H, Beesley J, Barnes DR, Allen J, Kar S, Pooley KA, Dennis J, Michailidou K, Turman C, et al. 2020. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat Genet 52: 56–73. 10.1038/s41588-019-0537-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31: 3651–3653. 10.1093/nar/gkg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, de Llobet Cucalon LI, Di Vona C, Le Dilly F, Vidal E, Lioutas A, Oliete JQ, Jochem L, Cutts E, Dieci G, et al. 2020. TFIIIC binding to Alu elements controls gene expression via chromatin looping and histone acetylation. Mol Cell 77: 475–487.e11. 10.1016/j.molcel.2019.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming NI, Trivett MK, George J, Slavin JL, Murray WK, Moseley JM, Anderson RL, Thomas DM. 2009. Parathyroid hormone–related protein protects against mammary tumor emergence and is associated with monocyte infiltration in ductal carcinoma in situ. Cancer Res 69: 7473–7479. 10.1158/0008-5472.CAN-09-0194 [DOI] [PubMed] [Google Scholar]
- Fuentes DR, Swigut T, Wysocka J. 2018. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife 7: e35989. 10.7554/eLife.35989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Lam VK, Harris DN, Chuang NT, Scott EC, Pittard WS, Mills RE, 1000 Genomes Project Consortium, Devine SE. 2017. The mobile element locator tool (MELT): population-scale mobile element discovery and biology. Genome Res 27: 1916–1929. 10.1101/gr.218032.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C, D'Altri T, Aligué R, González J, Espinosa L, Bigas A. 2016. β-Catenin is required for T-cell leukemia initiation and MYC transcription downstream of Notch1. Leukemia 30: 2002–2010. 10.1038/leu.2016.106 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966. 10.1093/jnci/djn190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Edwards SL, Michailidou K, Nord S, Lari RCS, Desai K, Kar S, Hillman KM, Kaufmann S, Glubb DM, et al. 2014. Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat Commun 5: 4999. 10.1038/ncomms5999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombart AF, Saito T, Koeffler HP. 2009. Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics 10: 321. 10.1186/1471-2164-10-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubert C, Zevallos NA, Feschotte C. 2020. Contribution of unfixed transposable element insertions to human regulatory variation. Philos Trans R Soc Lond B Biol Sci 375: 20190331. 10.1098/rstb.2019.0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, et al. 2007. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet 39: 638–644. 10.1038/ng2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambor JE, Mennone J, Coon ME, Hanke JH, Kavathas P. 1993. Identification and characterization of an Alu-containing, T-cell-specific enhancer located in the last intron of the human CD8 α gene. Mol Cell Biol 13: 7056–7070. 10.1128/mcb.13.11.7056-7070.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke JH, Hambor JE, Kavathas P. 1995. Repetitive Alu elements form a cruciform structure that regulates the function of the human CD8α T cell-specific enhancer. J Mol Biol 246: 63–73. 10.1006/jmbi.1994.0066 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, et al. 2012. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis 47: 20–28. 10.1016/j.nbd.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, Xu L, Castillo-Martin M, Llobet-Navás D, Cordon-Cardo C, et al. 2014. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med 20: 1130–1137. 10.1038/nm.3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. 2012. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 9: 473–476. 10.1038/nmeth.1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, et al. 2016. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell 36: 9–23. 10.1016/j.devcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey GW, Englander EW, Howard BH. 1996. Specific binding sites for a pol III transcriptional repressor and pol II transcription factor YY1 within the internucleosomal spacer region in primate Alu repetitive elements. Gene Expr 6: 151–168. [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jacobsen BM, Jambal P, Schittone SA, Horwitz KB. 2009. ALU repeats in promoters are position-dependent co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors. Mol Endocrinol 23: 989–1000. 10.1210/me.2009-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 51: 611–617. 10.1038/s41588-019-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O'Seaghdha CM, Haller T, et al. 2013. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45: 145–154. 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancho O, Herranz D. 2018. The MYC enhancer-ome: long-range transcriptional regulation of MYC in cancer. Trends Cancer 4: 810–822. 10.1016/j.trecan.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Sim MFM, Gulati P, Antrobus R, Tung YCL, Rimmington D, Ayuso E, Polex-Wolf J, Lam BYH, Dias C, et al. 2017. Obesity-associated gene TMEM18 has a role in the central control of appetite and body weight regulation. Proc Natl Acad Sci 114: 9421–9426. 10.1073/pnas.1707310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Karaplis AC, Huang DC, Siegel PM, Camirand A, Yang XF, Muller WJ, Kremer R. 2011. PTHrp drives breast tumor initiation, progression, and metastasis in mice and is a potential therapy target. J Clin Invest 121: 4655–4669. 10.1172/JCI46134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe WL Jr, Reddy TE. 2015. Genomic approaches for understanding the genetics of complex disease. Genome Res 25: 1432–1441. 10.1101/gr.190603.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luparello C. 2011. Parathyroid hormone-related protein (PTHrP): a key regulator of life/death decisions by tumor cells with potential clinical applications. Cancers (Basel) 3: 396–407. 10.3390/cancers3010396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509. 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334. 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, et al. 2013. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45: 353–361, 361e1–2. 10.1038/ng.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THM, Carreira PE, Sanchez-Luque FJ, Schauer SN, Fagg AC, Richardson SR, Davies CM, Jesuadian JS, Kempen MHC, Troskie RL, et al. 2018. L1 retrotransposon heterogeneity in ovarian tumor cell evolution. Cell Rep 23: 3730–3740. 10.1016/j.celrep.2018.05.090 [DOI] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET. 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6: e1000895. 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888. 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP. 1995. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem 270: 22777–22782. 10.1074/jbc.270.39.22777 [DOI] [PubMed] [Google Scholar]
- Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV. 2004. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics 83: 873–882. 10.1016/j.ygeno.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, Takahashi A, Maeda S, Tsunoda T, Chen P, et al. 2012. Meta-analysis identifies multiple loci associated with kidney function–related traits in east Asian populations. Nat Genet 44: 904–909. 10.1038/ng.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, Burns KH. 2019. Transposable elements in human genetic disease. Nature Reviews Genetics 20: 760–772. 10.1038/s41576-019-0165-8 [DOI] [PubMed] [Google Scholar]
- Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH. 2017. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc Natl Acad Sci 114: E3984–E3992. 10.1073/pnas.1704117114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer LM, Steranka JP, Ardeljan D, Walker J, Fitzgerald KC, Calabresi PA, Cooper TA, Burns KH. 2019. Alu insertion variants alter mRNA splicing. Nucleic Acids Res 47: 421–431. 10.1093/nar/gky1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic J, Zhou Y, Fasolino M, Goldman N, Schwartz GW, Mumbach MR, Nguyen SC, Rome KS, Sela Y, Zapataro Z, et al. 2019. Oncogenic notch promotes long-range regulatory interactions within hyperconnected 3D cliques. Mol Cell 73: 1174–1190.e12. 10.1016/j.molcel.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E. 2006. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7: 133. 10.1186/1471-2164-7-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. 2012. Linking disease associations with regulatory information in the human genome. Genome Res 22: 1748–1759. 10.1101/gr.136127.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhang Y, Zheng W, Michailidou K, Ghoussaini M, Bolla MK, Wang Q, Dennis J, Lush M, Milne RL, et al. 2016. Fine-scale mapping of 8q24 locus identifies multiple independent risk variants for breast cancer. Int J Cancer 139: 1303–1317. 10.1002/ijc.30150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9: 657–663. 10.1016/S0959-437X(99)00031-3 [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, et al. 2010. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42: 937–948. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Kural D, Strömberg MP, Walker JA, Konkel MK, Stütz AM, Urban AE, Grubert F, Lam HY, Lee WP, et al. 2011. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet 7: e1002236. 10.1371/journal.pgen.1002236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Han D, Boyd-Kirkup J, Yu X, Han JJ. 2014. Evolution of Alu elements toward enhancers. Cell Rep 7: 376–385. 10.1016/j.celrep.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39: 857–864. 10.1038/ng2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker NR, Dolmatova EV, Lin H, Cooper RR, Ye J, Hucker WJ, Jameson HS, Parsons VA, Weng LC, Mills RW, et al. 2017. Diminished PRRX1 expression is associated with increased risk of atrial fibrillation and shortening of the cardiac action potential. Circ Cardiovasc Genet 10: e001902. 10.1161/CIRCGENETICS.117.001902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing EM, Wu G. 2002. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J Biol Chem 277: 36216–36222. 10.1074/jbc.M205111200 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812. 10.1101/gr.139105.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, Moore J, Pierce BG, Dong X, Virgil D, et al. 2013. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res 41: D171–D176. 10.1093/nar/gks1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Norris ET, Jordan IK. 2017a. Human retrotransposon insertion polymorphisms are associated with health and disease via gene regulatory phenotypes. Front Microbiol 8: 1418. 10.3389/fmicb.2017.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Rishishwar L, Marino-Ramirez L, Jordan IK. 2017b. Human population-specific gene expression and transcriptional network modification with polymorphic transposable elements. Nucleic Acids Res 45: 2318–2328. 10.1093/nar/gkw1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Zhang Y, Xing J, Watkins WS, Ha H, Batzer MA, Jorde LB. 2013. Mobile element scanning (ME-scan) identifies thousands of novel Alu insertions in diverse human populations. Genome Res 23: 1170–1181. 10.1101/gr.148973.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Pan W. 2018. Integration of enhancer-promoter interactions with GWAS summary results identifies novel schizophrenia-associated genes and pathways. Genetics 209: 699–709. 10.1534/genetics.118.300805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysolmerski JJ. 2012. Parathyroid hormone-related protein: an update. J Clin Endocrinol Metab 97: 2947–2956. 10.1210/jc.2012-2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski A, Hong CC, Lam K, Michailidou K, Lytle C, Yao S, Zhang Y, Bolla MK, Wang Q, Dennis J, et al. 2016. An intergenic risk locus containing an enhancer deletion in 2q35 modulates breast cancer risk by deregulating IGFBP5 expression. Hum Mol Genet 25: 3863–3876. 10.1093/hmg/ddw223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Jiao X, Zou H, Li K. 2015. Prognostic significance of c-Met in breast cancer: a meta-analysis of 6010 cases. Diagn Pathol 10: 62. 10.1186/s13000-015-0296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. 2014. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci 111: E4946–E4953. 10.1073/pnas.1407079111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massagué J, Mundy GR, Guise TA. 1999. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest 103: 197–206. 10.1172/JCI3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Guo X, Long J, Kuchenbaecker KB, Droit A, Michailidou K, Ghoussaini M, Kar S, Freeman A, Hopper JL, et al. 2016. Identification of independent association signals and putative functional variants for breast cancer risk through fine-scale mapping of the 12p11 locus. Breast Cancer Res 18: 64. 10.1186/s13058-016-0718-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bailey SD, Lupien M. 2014. Laying a solid foundation for Manhattan—‘setting the functional basis for the post-GWAS era’. Trends Genet 30: 140–149. 10.1016/j.tig.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Gingeras TR, Weng Z. 2019. Genome-wide analysis of polymerase III–transcribed Alu elements suggests cell-type–specific enhancer function. Genome Res 29: 1402–1414. 10.1101/gr.249789.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.