Abstract
Bivalent chromatin is characterized by the simultaneous presence of H3K4me3 and H3K27me3, histone modifications generally associated with transcriptionally active and repressed chromatin, respectively. Prevalent in embryonic stem cells (ESCs), bivalency is postulated to poise/prime lineage-controlling developmental genes for rapid activation during embryogenesis while maintaining a transcriptionally repressed state in the absence of activation cues; however, this hypothesis remains to be directly tested. Most gene promoters DNA hypermethylated in adult human cancers are bivalently marked in ESCs, and it was speculated that bivalency predisposes them for aberrant de novo DNA methylation and irreversible silencing in cancer, but evidence supporting this model is largely lacking. Here, we show that bivalent chromatin does not poise genes for rapid activation but protects promoters from de novo DNA methylation. Genome-wide studies in differentiating ESCs reveal that activation of bivalent genes is no more rapid than that of other transcriptionally silent genes, challenging the premise that H3K4me3 is instructive for transcription. H3K4me3 at bivalent promoters—a product of the underlying DNA sequence—persists in nearly all cell types irrespective of gene expression and confers protection from de novo DNA methylation. Bivalent genes in ESCs that are frequent targets of aberrant hypermethylation in cancer are particularly strongly associated with loss of H3K4me3/bivalency in cancer. Altogether, our findings suggest that bivalency protects reversibly repressed genes from irreversible silencing and that loss of H3K4me3 may make them more susceptible to aberrant DNA methylation in diseases such as cancer. Bivalency may thus represent a distinct regulatory mechanism for maintaining epigenetic plasticity.
The DNA in eukaryotic cells is organized into chromatin consisting of repeating units of nucleosome, an octamer of histone proteins wrapped with approximately 147 bp of DNA. Consequently, chromatin plays a central role in regulating accessibility to DNA in many DNA-templated processes including transcription. Histone modifications and DNA methylation are key epigenetic mechanisms that modulate chromatin structure and thus regulate gene expression programs controlling cell fate decisions and cell identity during development (Jaenisch and Bird 2003; Kouzarides 2007; Li et al. 2007; Allis and Jenuwein 2016).
Histones are subject to a vast array of post-translational modifications including acetylation and methylation (Kouzarides 2007; Li et al. 2007). Whereas histone acetylation is generally associated with gene activation, histone methylation, depending on the residue modified, is associated with either activation or repression. Trimethylation of histone H3 on lysine 4 (H3K4me3) and lysine 27 (H3K27me3) are two of the most extensively studied histone modifications associated with transcriptionally active and repressed chromatin, respectively (Barski et al. 2007). H3K4me3 and H3K27me3, respectively, are catalyzed by the Trithorax group (TrxG) and Polycomb group (PcG) of proteins (Simon and Kingston 2009; Margueron and Reinberg 2011; Di Croce and Helin 2013; Piunti and Shilatifard 2016; Schuettengruber et al. 2017). Because TrxG and PcG proteins act antagonistically to regulate, respectively, the activated and repressed states of gene expression, H3K4me3 and H3K27me3 were thought to be mutually exclusive. But this assumption was challenged by the discovery of bivalent domains—genomic regions characterized by the simultaneous presence of H3K4me3 and H3K27me3—found predominantly at developmentally regulated gene promoters in embryonic stem cells (ESCs) (Azuara et al. 2006; Bernstein et al. 2006; Voigt et al. 2012; Harikumar and Meshorer 2015; Shema et al. 2016; Blanco et al. 2020). Although H3K4me3 and H3K27me3 occupy essentially nonoverlapping regions within bivalent domains (Barski et al. 2007), with H3K27me3 domains typically flanking a H3K4me3 domain, it was later established that nucleosomes that bear both “active” H3K4me3 and repressive H3K27me3 do exist in vivo, albeit on opposite H3 tails in nearly all cases (Voigt et al. 2012; Shema et al. 2016), consistent with direct allosteric inhibition of PRC2 activity by H3K4me3 (Schmitges et al. 2011). Despite the presence of H3K4me3, bivalently marked promoters are transcriptionally inactive, if not expressed at very low levels (Mikkelsen et al. 2007). This initial observation led to the elegant and inherently appealing concept that bivalency poises/primes lineage-controlling developmental genes for rapid activation during embryogenesis while maintaining a repressed state in the absence of activation cues (Azuara et al. 2006; Bernstein et al. 2006; Voigt et al. 2013); yet, this hypothesis remains to be directly tested, and the function of bivalent domains in development remains a mystery.
Bivalency was initially hypothesized to be an ESC-specific chromatin state. During ESC differentiation, bivalency is thought to resolve into either H3K4me3-only or H3K27me3-only state depending on whether the gene is activated or silenced, respectively. Later observations, however, confirmed the existence of bivalent domains in terminally differentiated cell types (Barski et al. 2007; Mikkelsen et al. 2007; Mohn et al. 2008), raising the question about its need and functional relevance in cell types with no differentiation potential.
DNA methylation, as a heritable epigenetic mark, adds an additional level of stability by serving as an enduring “lock” to reinforce a previously silenced state by subjecting genes to irreversible transcriptional silencing even in the presence of all of the factors required for their expression (Deaton and Bird 2011; Jones 2012; Bestor et al. 2015; Schübeler 2015). Most gene promoters DNA hypermethylated in adult human cancers are bivalently marked in ESCs, and it was speculated that bivalency predisposes these genes for aberrant de novo DNA methylation and irreversible silencing in cancer (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007), but evidence supporting this model is largely lacking. Here, we set out to decode the function of bivalent chromatin in physiological and pathological settings.
Results
Bivalent chromatin does not poise genes for rapid activation
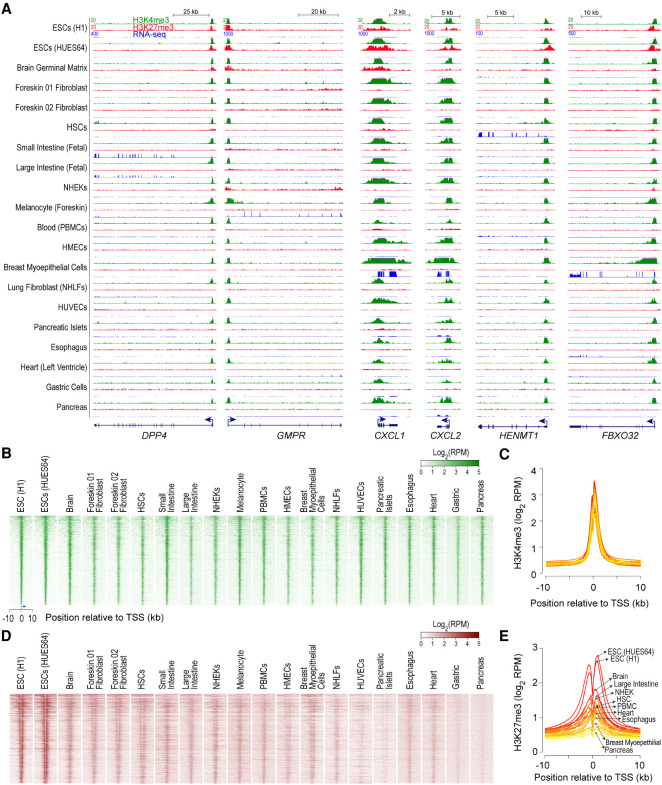
To assess whether bivalent chromatin represents a distinct epigenetic state and/or a regulatory mechanism, we investigated genes with bivalently marked promoters in pluripotent human embryonic stem cells (ESCs) (Supplemental Table S1; Methods); henceforth, we use “bivalent genes” to refer to genes whose promoters are bivalently marked in ESCs, unless stated otherwise. To gain insight into the functional significance of bivalent chromatin, we first sought to determine the chromatin and expression status of bivalent genes in lineage-restricted multipotent and terminally differentiated cells. Using publicly available NIH Roadmap Epigenomics project data from a large and diverse collection of human tissues and progenitor cells (http://www.roadmapepigenomics.org/data/), we noted that H3K4me3 enrichment at bivalently marked gene promoters in ESCs persists in nearly all other cell types irrespective of transcriptional activity (Fig. 1A–C; Supplemental Fig. S1). In contrast, H3K27me3 enrichment is more dynamic across cell types (Fig. 1A,D,E; Supplemental Fig. S1), with its absence not necessarily accompanied by gene activation. Resolution of bivalent state in ESCs to H3K4me3-only state in lineage-restricted cell types coupled with no guarantees of transcription—in the absence of H3K27me3 (Supplemental Fig. S1)—calls into question the premise that the H3K4me3 component of bivalent chromatin poises genes for rapid activation.
Figure 1.
H3K4me3, observed at bivalent promoters in ESCs, persists in nearly all cell types irrespective of gene expression. (A) Genome browser shots of select genes, bivalently marked in human embryonic stem cells (ESCs), showing ChIP-seq read density profiles for H3K4me3 (green) and H3K27me3 (red) in various cell types. Also shown are read density profiles for gene expression (blue; RNA-seq). (HSCs) hematopoietic stem cells; (PBMCs) peripheral blood mononuclear cells; (HMECs) human mammary epithelial cells; (NHEKs) normal human embryonic kidneys; (NHLFs) normal human lung fibroblasts; (HUVECs) human umbilical vein endothelial cells. (B,D) Heatmap representation of H3K4me3 (B) and H3K27me3 (D) ChIP-seq read density, in various cell types, near transcription start sites (TSSs) of genes bivalently marked in human ESCs. Genes were ordered by decreasing order of H3K4me3 signal in ESCs (top to bottom). Read density is represented as reads per million mapped reads (RPKM). (C,E) Average H3K4me3 (C) and H3K27me3 (E) ChIP-seq read density, in various cell types, near TSSs of genes shown in B. Shades of color represent individual cell types. Select cell types, of the 20 plotted, are highlighted.
To determine whether H3K4me3 at bivalent genes confers them rapid or higher activation potential compared with other transcriptionally silent genes that lack the H3K4me3 mark, we sought to investigate the transcriptional fate of bivalent genes during early embryonic development using a previously validated differentiation system (Fig. 2A; Hayashi et al. 2011; Buecker et al. 2014; Shirane et al. 2016; Yang et al. 2019), wherein naive mouse ESCs—representing the preimplantation mouse embryo from approximately embryonic day E3.75 to E4.5—can be induced to epiblast-like cells (EpiLCs), which most closely resemble the early post-implantation epiblast (E5.5–E6.5). Using the data that we previously generated from chromatin immunoprecipitation sequencing (ChIP-seq) analyses of the chromatin from naive mouse ESCs using antibodies against histone modifications H3K4me3 and H3K27me3 (Yang et al. 2019), we identified 2163 genes with bivalently marked promoters (Fig. 2B–D; Supplemental Table S2; Methods). For comparison purposes, we also identified genes whose promoters are enriched for H3K4me3 but not H3K27me3 (H3K4me3-only) and vice versa (H3K27me3-only). Genes with neither H3K4me3 nor H3K27me3 enrichment at their promoters were grouped as “unmarked.” Consistent with H3K27me3's role in maintaining the transcriptionally repressed chromatin state (Riising et al. 2014), nearly all the bivalent (94%) and H3K27me3-only (99%) genes are transcriptionally inactive, if not expressed at very low levels (<1 FPKM), in ESCs (Fig. 2E). Although the expression of bivalent genes is approximately two- to threefold higher compared to transcriptionally silent H327me3-only or unmarked genes, it is still very low (<1 FPKM) to be considered expressed, consistent with mutual exclusivity between PRC2 activity and active transcription (Brookes et al. 2012; Riising et al. 2014).
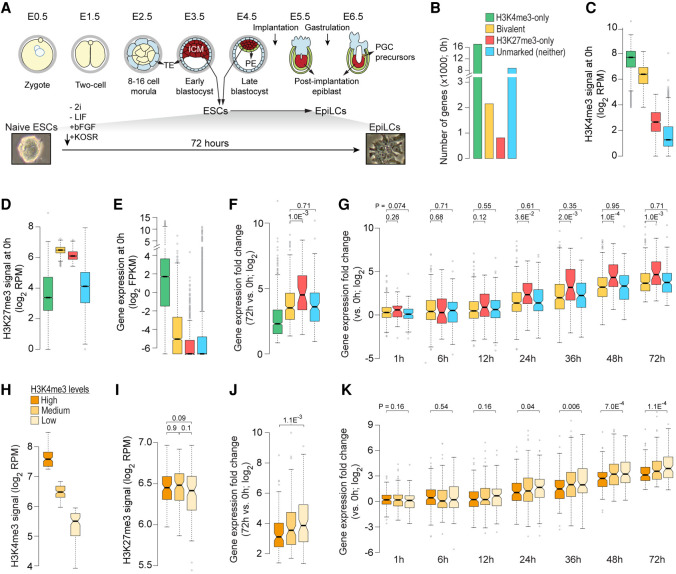
Figure 2.
Bivalent chromatin does not poise genes for rapid activation. (A, top) Developmental events during early embryogenesis in mouse embryo. (Bottom) Schematic showing in vitro differentiation of naive ESCs to EpiLCs. (ICM) inner cell mass; (ESCs) embryonic stem cells; (TE) trophectoderm; (PE) primitive endoderm; (EpiLC) post-implantation epiblast-like cells; (PGCs) primordial stem cells. (B) Number of genes within each of the four classes, defined based on H3K4me3 (±500 bp of TSS) and/or H3K27me3 (±2 kb of TSS) enrichment at gene promoters in naive ESCs (0 h) (Yang et al. 2019). (Bivalent) Positive for H3K4me3 and H3K27me3; (H3K4me3-only) positive for H3K4me3 and negative for H3K27me3; (H3K27me3-only) positive for H3K27me3 and negative for H3K4me3; (Unmarked) negative for both H3K4me3 and H3K27me3. (C–E) Box plots showing the distribution of ChIP-seq read densities for H3K4me3 (C) and H3K27me3 (D) at gene promoters and gene expression (E) in naive ESCs for each of the four gene classes defined in B. (RPM) Reads per million mapped reads; (FPKM) fragments per kilobase per million mapped reads. (F) Box plot showing the distribution of gene expression fold changes for genes up-regulated in EpiLCs (72 h vs. 0 h; n = 1372) (Yang et al. 2019). Genes grouped based on their chromatin states in naive ESCs (0 h). (G) Box plot showing the distribution of gene expression fold changes over time (vs. 0 h) for genes up-regulated in EpiLCs (Yang et al. 2019). Genes grouped based on their chromatin states in naive ESCs (0 h). (H,I) Bivalent genes up-regulated in EpiLCs (72 h vs. 0 h; n = 1372) were binned into three equal-sized sets based on H3K4me3 enrichment at gene promoters in naive ESCs (0 h). Box plots showing the distribution of ChIP-seq read densities for H3K4me3 (H) and H3K27me3 (I) in naive ESCs for each of the three defined sets. (J) Box plot showing the distribution of gene expression fold changes (72 h vs. 0 h) for bivalent genes up-regulated in EpiLCs. Genes grouped based the three sets defined in H. (K) Box plot showing the distribution of gene expression fold changes over time (72 h to 0 h) for bivalent genes up-regulated in EpiLCs (72 h vs. 0 h). Genes grouped based on high/low H3K4me3 signal, as defined in H. All the P-values were calculated using two-sided Wilcoxon rank-sum test. See also Supplemental Figures S2–S4.
To evaluate whether bivalent genes are activated any faster or any more than other transcriptionally silent genes (H3K27me3-only or unmarked), we focused on genes up-regulated (Q-value <0.05) during ESC to EpiLC differentiation (Supplemental Fig. S2A; Supplemental Table S3). Our analysis revealed that up-regulated bivalent genes are no more activated, measured as either fold change or absolute difference in expression, compared to up-regulated H3K27me3-only or unmarked genes (Fig. 2F; Supplemental Fig. S2B). Next, to address whether bivalent chromatin confers rapid activation potential, we examined gene expression changes at various time points (0, 1, 6, 12, 24, 36, 48, and 72 h) during ESC to EpiLC differentiation. We found that activation of up-regulated bivalent genes is no more rapid than that of up-regulated H3K27me3-only or unmarked genes (Fig. 2G; Supplemental Fig. S2C,D), challenging the notion that H3K4me3 at bivalent promoters poises them for rapid activation. This observation holds true irrespective of how stringently up-regulated genes are defined (Supplemental Fig. S3). Nonetheless, if it is true that H3K4me3 at bivalent genes poises them for rapid activation, we reasoned that bivalent genes with higher levels of H3K4me3 must be activated much sooner or much more compared to those with relatively lower levels of H3K4me3. Our analysis of the activation dynamics of up-regulated bivalent genes, divided into three equal-sized groups based on their H3K4me3 levels (Fig. 2H,I; Supplemental Fig. S4A,B) revealed that activation of bivalent genes with higher levels of H3K4me3 is neither greater nor faster compared to those with lower levels of H3K4me3 (Fig. 2J,K; Supplemental Fig. S4C–E). Grouping up-regulated bivalent genes slightly differently based on the distribution of H3K4me3 levels yielded the same result (Supplemental Fig. S4F–J). Together, these results indicate that bivalent chromatin does not poise genes for rapid activation any more than chromatin marked with just H3K27me3 or chromatin marked with neither H3K4me3 nor H3K27me3.
To assess the fate of bivalent genes in later developmental stages, we analyzed gene expression profiles of representative lineage-restricted populations from the three germ layers (ectoderm, mesoderm, and endoderm), derived through directed differentiation of human ESCs (Gifford et al. 2013). Consistent with our findings from ESC to EpiLC differentiation, up-regulated bivalent genes in lineage-restricted cells are no more activated compared to up-regulated H3K27me3-only or unmarked genes (Supplemental Fig. S5).
Transcriptional competence of poised RNA polymerase II at bivalent genes
Although about two-thirds of the bivalent genes lack transcriptionally engaged RNA polymerase II (RNAPII) (Williams et al. 2015), the presence of poised RNAPII, preferentially phosphorylated at serine 5 but not serine 2 and serine 7, at a subset of bivalent gene promoters has lent some credence to the conceptually appealing notion that bivalency poises genes for rapid activation while keeping them repressed (Stock et al. 2007; Brookes et al. 2012; Ferrai et al. 2017). Phosphorylation of serine 5 on RNAPII, largely mediated by the TFIIH complex, promotes transcription initiation, whereas phosphorylation of serine 2 on RNAPII mediates transition of RNAPII from initiation into productive elongation (Phatnani and Greenleaf 2006). At a cohort of PRC2-targeted developmental genes, it is MAPK1 (also known as ERK2) but not TFIIH that phosphorylates serine 5 on RNAPII, and it was hypothesized that, at these PRC2 target genes, MAPK1's phosphorylation of serine 5 on RNAPII establishes a poised/stalled form of RNAPII that is competent for transcription (Tee et al. 2014).
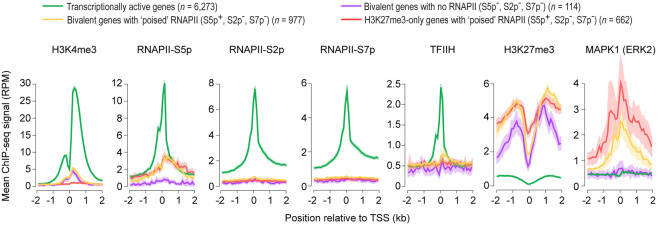
To gain further insight into the transcriptional competence and poising potential of RNAPII observed at bivalent promoters, we focused on bivalent genes that harbor poised RNAPII (Supplemental Table S4), defined as RNAPII phosphorylated at serine 5 (S5p+) but not serine 2 (S2p−) and serine 7 (S7p−) (Brookes et al. 2012). We noted that the expression of bivalent genes with poised RNAPII is no more than that of bivalent genes with no RNAPII (Supplemental Fig. S6A). Unlike transcriptionally active gene promoters, which are enriched for TFIIH but not MAPK1, bivalent gene promoters with poised RNAPII are enriched for MAPK1 but not TFIIH (Fig. 3). Consistent with this observation, RNAPII-S5p levels at active promoters correlate with TFIIH levels (R = 0.61), whereas RNAPII-S5p levels at bivalent promoters correlate with MAPK1 levels (R = 0.58). During early stages of transcription, serine 5 phosphorylation on RNAPII, together with the PAF complex, is known to recruit the histone methyltransferase, Set1/COMPASS, to trimethylate H3K4 (Shilatifard 2012), which is reflected in the correlation between RNAPII-S5p and H3K4me3 levels at active genes (R = 0.6) (Supplemental Fig. S6B,C). However, RNAPII-S5p levels at bivalent promoters with poised RNAPII show no such correlation with H3K4me3 levels (R = 0.13) (Supplemental Fig. S6B,C); instead, RNAPII-S5p levels correlate with repression-associated H3K27me3 levels (R = 0.4), which is in marked contrast to the inverse correlation observed between RNAPII-S5p and H3K27me3 at transcriptionally active genes (R = −0.31), but is similar to the correlation observed between RNAPII-S5p and H3K27me3 at H3K27me3-only genes with poised RNAPII (R = 0.57). Given that (1) MAPK1 binds exclusively to a subset of PRC2 targets and phosphorylates serine 5 on RNAPII (Tee et al. 2014), (2) bivalent promoters with poised RNAPII are devoid of TFIIH and thus any serine 5 phosphorylation on RNAPII at bivalent promoters with poised RNAPII is attributable to MAPK1 (Fig. 3), and (3) PRC2 binding and active transcription are mutually exclusive (Riising et al. 2014), these data suggest that the MAPK1-mediated serine 5 phosphorylation on RNAPII at poised bivalent genes may be incompatible with transcription.
Figure 3.
Bivalent promoters with poised RNA polymerase II are enriched for MAPK1 (also known as ERK2) but not TFIIH. ChIP-seq read density profiles of histone modifications H3K4me3 and H3K27me3 (Marks et al. 2012), phosphorylated forms (S2p, S5p, or S7p) of RNA polymerase II (RNAPII) (Brookes et al. 2012), general transcription factor TFIIH (ERCC3) (Tee et al. 2014), and mitogen-activated protein kinase MAPK1 (Tee et al. 2014) near transcription start site (TSS) of indicated gene classes in mouse ESCs grown in serum-containing medium. Mean ChIP-seq signal is shown. Confidence interval (95%) of the mean is shown as a cloud. (RPM) reads per million mapped reads. See also Supplemental Figure S6.
Next, to assess whether the presence of poised RNAPII at bivalent genes confers them rapid or higher activation potential during differentiation, we examined their expression during ESC to EpiLC differentiation. Our analysis revealed that the activation dynamics of bivalent genes with poised RNAPII is no greater or no more rapid than those of bivalent genes with no RNAPII (Supplemental Fig. S6D,E), raising questions about the poising potential and transcriptional competence of poised RNAPII at bivalent genes.
Bivalent chromatin is a product of PRC2 activity at CpG-rich sequences
Enrichment for developmental genes and regulators among bivalent genes is yet another characteristic that has been used to make the case for the biological relevance of bivalent chromatin in poising lineage-controlling genes for rapid activation during early embryogenesis (Bernstein et al. 2006). Although it is true that bivalent genes are enriched for genes associated with developmental processes, this characteristic is not unique to bivalent genes because it also holds true for H3K27me3-only genes (Supplemental Fig. S7; Supplemental Table S5), making it a general feature of genes targeted by PRC2.
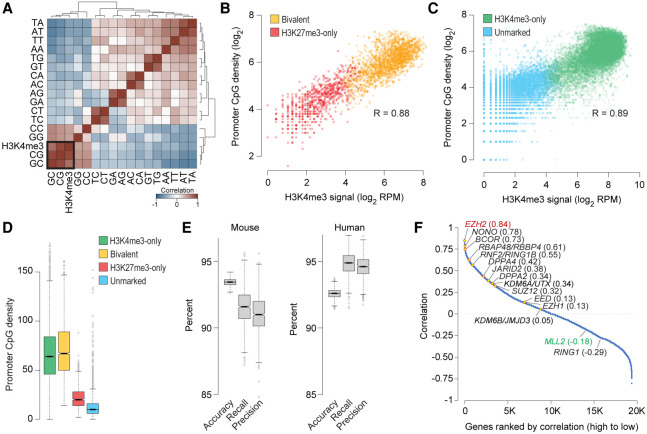
With bivalent chromatin conferring no more poising or activation potential than chromatin decorated with just H3K27me3 (Fig. 2G), we asked why some promoters targeted by PRC2 are also coenriched for H3K4me3, making them bivalent, whereas others are not. Because H3K4me3 enrichment at bivalent promoters in ESCs persists in nearly all cell types irrespective of transcriptional activity (Fig. 1), we reasoned that H3K4me3 at bivalent chromatin is perhaps a product of the underlying DNA sequence features. Indeed, analysis of dinucleotide frequency at H3K27me3-enriched promoters (bivalent and H3K27me3-only) revealed a high correlation (R = 0.88) between CpG density and H3K4me3 levels (Fig. 4A,B; Supplemental Table S2), which is somewhat expected given that CpG-rich sequences, when unmethylated, per se are sufficient to establish H3K4me3 domains (Thomson et al. 2010). This observation indicates that CpG density can discriminate between PRC2 targets that are bivalent versus those that are H3K27me3-only. Moreover, this association between CpG density and H3K4me3 levels holds true even for gene promoters that are not PRC2 targets (Fig. 4C), suggesting that CpG density alone can be an excellent predictor of H3K4me3 levels. A corollary to this conclusion would be that CpG density, together with H3K27me3 levels, can predict bivalent chromatin (Fig. 4B,D). Indeed, a machine learning approach based on multinomial logistic regression of dinucleotide frequencies and H3K27me3 levels predicted, with >90% accuracy, chromatin status of promoters into one of the four classes (bivalent, H3K27me3-only, H3K4me3-only, or unmarked) (Fig. 4E; Supplemental Methods). Promoters with bivalent chromatin are predicted with >90% sensitivity (recall) and precision.
Figure 4.
Bivalent chromatin is a product of PRC2 activity at CpG-rich sequences. (A) Heatmap showing unsupervised hierarchical clustering of pairwise Pearson's correlations between H3K4me3 levels and dinucleotide frequency (5′ to 3′) at the promoters (±500 bp of TSS) of genes enriched for H3K27me3 in naive mouse ESCs. (B,C) Scatter plot showing correlation between CpG dinucleotide frequency and H3K4me3 read density at promoters (±500 bp of TSS) of genes with (B) or without (C) H3K27me3 enrichment in naive mouse ESCs. (D) Box plots showing the distribution of CpG dinucleotide frequency at promoters (±500 bp of TSS) of genes within each the four classes defined in Figure 2B. (E) Statistics summarizing the performance of the multinomial logistic regression-based machine learning method for predicting chromatin state of mouse or human gene promoters (Bivalent, H3K27me3-only, H3K4me3-only, or unmarked) using only H3K27me3 data and dinucleotide frequencies at gene promoters. Box plots show the distribution of indicated performance measures over 1000 models. (Accuracy) fraction of predictions that are correct; (Recall [sensitivity]) fraction of bivalent genes correctly predicted as such; (Precision [positive predictive value]) fraction of predicted bivalent genes that are correct. (F) Plot showing Pearson's correlation between number of bivalent genes and expression of individual genes, calculated based on data from various cell types. Genes, denoted as individual data points, are sorted (left to right, x-axis) based on their correlation values (y-axis). See also Supplemental Figure S7.
With DNA sequence features and CpG density in particular seeming to have such an outsized influence on H3K4me3 levels and thus establishment of bivalent domains, we next sought to understand why bivalent domains are more prevalent in pluripotent ESCs compared to terminally differentiated or lineage-restricted multipotent cells (Supplemental Fig. S8A). Because bivalently marked promoters across various cell types are CpG-rich (Supplemental Fig. S8B) but not vice versa (Fig. 4B–D) and because they almost always harbor H3K4me3 (Fig. 1; Supplemental Fig. S8C), we reasoned that the prevalence of bivalent domains in a given cell type is presumably a function of the extent of PRC2 activity and/or targeting. Consistent with this notion, analysis of gene expression across various cell types revealed that the number of bivalent genes in a given cell type correlates the best with expression levels of EZH2 (R = 0.84), encoding the catalytic subunit of the PRC2 complex (Fig. 4F; Supplemental Table S6). Expression of KMT2B (also known as, and hereafter referred to as MLL2), which is a gene that is on Chromosome 19 in the human genome and Chromosome 7 in the mouse genome, and encodes the catalytic subunit of the Set1/MLL complex chiefly responsible for H3K4me3 at bivalent domains (Hu et al. 2013; Denissov et al. 2014), correlates negatively (R = −0.18), if at all, with the number of bivalent genes (Fig. 4F; Supplemental Fig. S8D). Given that CpG-rich sequences, when unmethylated, per se are sufficient to establish H3K4me3 domains (Thomson et al. 2010) and that EZH2 gain/loss-of-function strongly correlates with number of bivalent genes (Shema et al. 2016), these data suggest that bivalent domains are likely a product of PRC2 activity at genomic regions with high CpG density.
H3K4me3 at bivalent chromatin protects promoters from de novo DNA methylation
Gene promoters targeted by PRC2 in ESCs—mostly bivalent genes—are often found to be DNA hypermethylated in adult human cancers, and it was speculated that bivalent chromatin and/or the presence of Polycomb proteins might predispose these genes for aberrant de novo DNA methylation and irreversible silencing in cancer (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). Acquisition of promoter DNA methylation at these genes is thought to lock in stem cell phenotypes—at the expense of ability to respond to appropriate lineage commitment and differentiation cues—and initiate abnormal clonal expansion and thereby predispose to cancer (Jones and Baylin 2007; Widschwendter et al. 2007; Easwaran et al. 2014).
To determine whether bivalent genes in ESCs are more susceptible to de novo DNA methylation during normal development, we investigated promoter DNA methylation levels in naive mouse ESCs and EpiLCs. ESCs represent the preimplantation epiblast (approximately E3.75–E4.5), and EpiLCs most closely resemble early post-implantation epiblast (E5.5–E6.5) (Fig. 2A; Hayashi et al. 2011; Yang et al. 2019). This time period spanning pre- to post-implantation epiblast differentiation during early embryonic development is noteworthy because naive ESCs are associated with global DNA hypomethylation, with a major wave of global de novo methylation occurring after implantation (∼E5.0) (Okano et al. 1999; Smith et al. 2012; Leitch et al. 2013; Shirane et al. 2016), when de novo methylatransferases Dnmt3a and Dnmt3b—not expressed in naive ESCs—get induced by about 500- to 1000-fold (Supplemental Fig. S9A).
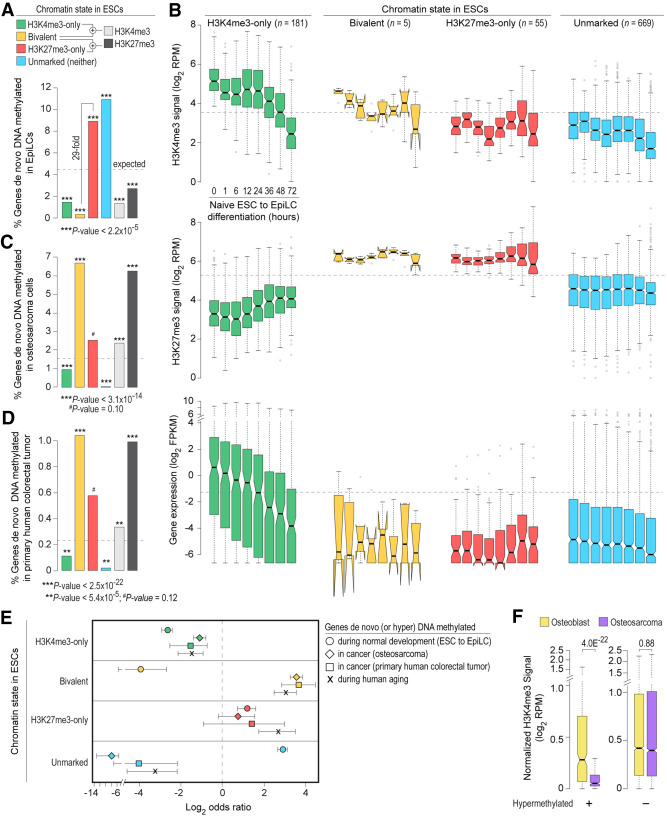
Our analysis of DNA methylation and H3K4me3 levels in naive ESCs revealed that promoters enriched for H3K4me3 are devoid of DNA methylation (Supplemental Fig. S9B). Focusing on the 910 genes whose promoters are hypermethylated in EpiLCs compared to naive ESCs (Supplemental Fig. S9C,D), we noted that bivalent genes in ESCs are significantly underrepresented among genes hypermethylated in EpiLCs (Fig. 5A; Supplemental Fig. S9E; Supplemental Table S7). In contrast, H3K27me3-only genes in ESCs are significantly overrepresented among the hypermethylated genes. Although H3K27me3-only genes in ESCs are 29-fold more likely to be hypermethylated compared with bivalent genes, PRC2 targets in ESCs (bivalent and H3K27me3-only), as a group, are still underrepresented among hypermethylated genes (Fig. 5A), indicating that PRC2 targets are less susceptible to de novo DNA methylation at least during early stages of embryonic development. Moreover, because genes hypermethylated in EpiLCs are relatively CpG poor (Supplemental Fig. S9F), overrepresentation of H3K27me3-only genes among the hypermethylated set is more likely a reflection of their CpG-poor promoters—known to undergo extensive and dynamic methylation and demethylation during normal development (Meissner et al. 2008)—than their being PRC2 targets. Although the generally inverse correlation between CpG density and hypermethylation (Figs. 4D, 5A) suggests that protection from de novo methylation is perhaps a direct function of the local CpG density, studies in mouse have shown that, for most promoters, high CpG density alone cannot account for their unmethylated state in vivo (Lienert et al. 2011), underscoring our limited understanding of the mechanisms that protect most CpG island (CGI) promoters from de novo methylation.
Figure 5.
Bivalent chromatin protects promoters from de novo DNA methylation. (A) Percentage of genes, within each of the four classes of genes defined in naive mouse ESCs, whose promoters are DNA hypermethylated in EpiLCs (Shirane et al. 2016) are shown. See also Supplemental Fig. S9C,D. (B) Box plots showing changes in promoter H3K4me3/H3K27me3 levels and gene expression (Yang et al. 2019), for each of the four classes of genes shown in A, during ESC to EpiLC differentiation. (FPKM) Fragments per kilobase per million mapped reads; (RPM) reads per million mapped reads. (C,D) Percentage of genes, within each of the four classes of genes defined in human ESCs, whose promoters are aberrantly DNA hypermethylated in human osteosarcoma (U2OS) (Easwaran et al. 2012) (C) or primary human colorectal tumor (Widschwendter et al. 2007) (D). (E) Log2 odds ratio, with 95% confidence intervals, of promoter de novo DNA methylation during normal development (mouse ESC to EpiLC differentiation; circle), in cancer (human osteosarcoma and colorectal cancer; diamond and square, respectively), and during aging (Rakyan et al. 2010) (X mark) based on their chromatin state in ESCs. (F) Genes bivalently marked in human ESCs were divided into those that are aberrantly DNA hypermethylated in human osteosarcoma (left) and those that are not (right). Box plots show the distribution of H3K4me3 levels at these gene promoters in human osteoblasts (yellow) and osteosarcoma (purple) (Easwaran et al. 2012). All the P-values were calculated using Fisher's exact test (A,C,D) or two-sided Wilcoxon rank-sum test (F,E). See also Supplemental Figure S9.
To gain insight into mechanisms that underlie protection of bivalent promoters from de novo DNA methylation, we examined chromatin and expression changes that accompany hypermethylation in EpiLCs. High-temporal resolution profiles of H3K4me3, H3K27me3, and gene expression during ESC to EpiLC differentiation (Yang et al. 2019) revealed no major changes in H3K4me3, H3K27me3, or expression levels for hypermethylated H3K27me3-only and unmarked genes (Fig. 5B), suggesting that de novo DNA methylation merely reinforces the previously silenced state at these genes. In contrast, hypermethylated H3K4me3-only genes show a gradual decrease in gene expression and H3K4me3 levels (Fig. 5B), which would be consistent with the notion that transcriptional activity protects promoters from DNA methylation and that hypermethylation of these genes likely reflects consequence rather than cause of transcription inactivation (Jones 2012; Bestor et al. 2015; Schübeler 2015). Although almost all the bivalent genes in ESCs are protected from de novo DNA methylation (Fig. 5A; Supplemental Fig. S9E), the very few that get hypermethylated in EpiLCs also show a gradual loss of H3K4me3 and thus bivalency, but no change in expression (which was negligible to begin with) or H3K27me3 (Fig. 5B). Given that (1) de novo methylases cannot act on H3K4me3 modified nucleosomes in vitro (Ooi et al. 2007), (2) complete erasure of H3K4 methylation elevates DNA methylation levels, and restoration of H3K4 methylation levels reduces DNA methylation levels back to wild-type levels (Hu et al. 2009), and (3) reversible DNA methylation has no impact on H3K4me3 levels at gene promoters (King et al. 2016), these findings suggest that H3K4me3 at bivalent promoters, either directly or indirectly, protects transcriptionally repressed yet permissive CpG-rich promoters from de novo DNA methylation (also see Supplemental Text).
Loss of H3K4me3 at bivalent promoters is associated with aberrant DNA hypermethylation in cancer
To address whether bivalent chromatin predisposes genes for (or protects genes from) aberrant de novo methylation in adult cancer, we analyzed genes hypermethylated in osteosarcoma and colorectal tumor (Widschwendter et al. 2007; Easwaran et al. 2012). About two-thirds of the genes hypermethylated in cancer are bivalent in ESCs (Supplemental Fig. S9G). Unlike in EpiLCs, genes bivalent in ESCs are significantly overrepresented among genes hypermethylated in cancer (Fig. 5A,C,D). Whereas genes that are either H3K4me3-only or H3K27me3-only in ESCs are equally susceptible to hypermethylation during normal development and in cancer, genes bivalent in ESCs are more likely to be hypermethylated in cancer (odds ratio [OR] 11.87, 95% CI: 9.66–14.65 for osteosarcoma, and OR 12.48, 95% CI: 7.21–22.34 for colorectal tumor) than during normal development (OR 0.063, 95% CI: 0.02–0.15) (Fig. 5E).
To determine whether predisposition of bivalent genes in ESCs to acquire aberrant methylation in cancer might be due to loss of their bivalent status, we examined H3K4me3 and H3K27me3 levels for genes hypermethylated in osteosarcoma. We found that bivalent genes in ESCs that are hypermethylated in osteosarcoma, as opposed to those that are not, show significantly reduced levels of H3K4me3 (Fig. 5F). No such specificity was observed for H3K27me3 levels; all bivalent genes in ESCs show reduced levels of H3K27me3 irrespective of their methylation status (Supplemental Fig. S9H). Altogether, these data suggest that bivalent chromatin protects promoters from de novo DNA methylation and irreversible silencing while maintaining a reversibly repressed state, and that loss of H3K4me3 may make these genes more susceptible to aberrant DNA methylation in cancer.
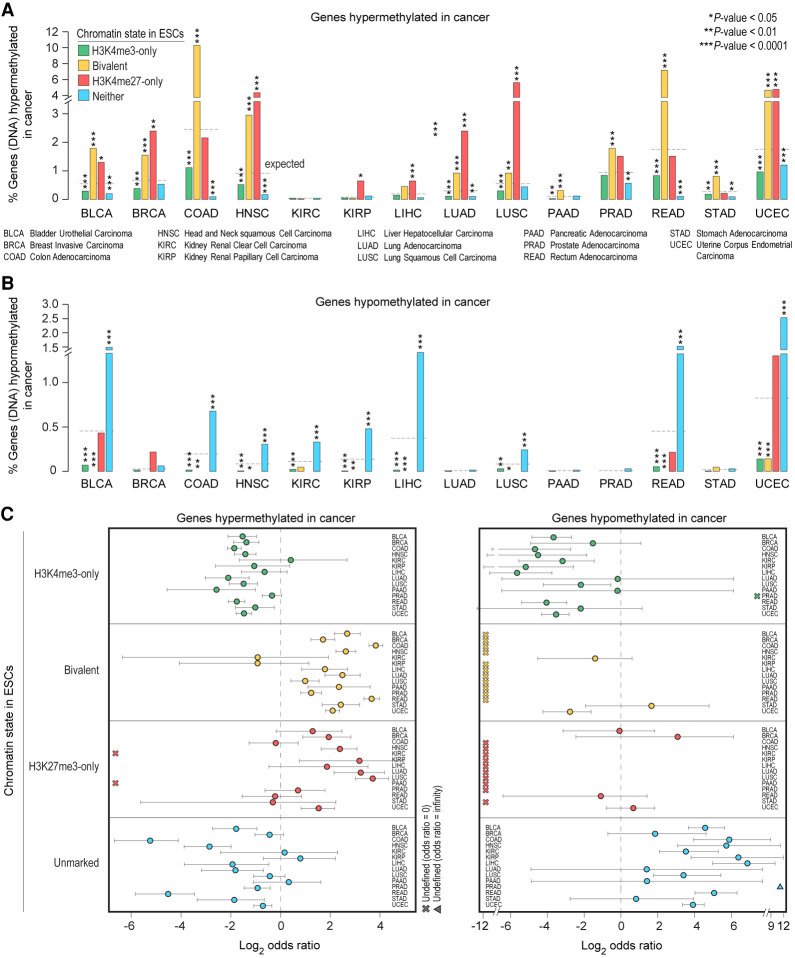
Our analysis of hypermethylated genes in 14 other cancer types further corroborated our observation that genes bivalent in ESCs, which are CpG-rich (Supplemental Fig. S8B) and less likely to be methylated during normal development (Fig. 5A), are significantly overrepresented among genes hypermethylated in cancer (Fig. 6A,C; Supplemental Table S8). In contrast, bivalent genes in ESCs are not among those hypomethylated in cancer (Fig. 6B,C; Supplemental Table S8). Genes unmarked in ESCs, which are CpG poor and more likely to be methylated during normal development (Fig. 5A), are significantly overrepresented among genes hypomethylated in cancer.
Figure 6.
Genes hypermethylated in cancer are more likely to be PRC2 targets in ESCs. (A,B) Percentage of genes, within each of the four classes of genes defined in human ESCs, whose promoters are aberrantly DNA hypermethylated (A) or hypomethylated (B) in various cancer types. All the P-values were calculated using Fisher's exact test. (C) Log2 odds ratio, with 95% confidence intervals, of promoter DNA hypermethylation (left) or hypomethylation (right) in various cancer types based on their chromatin state in human ESCs.
Age is the single biggest risk factor for most diseases including cancer (Niccoli and Partridge 2012). Because genes frequently hypermethylated and silenced in many adult human cancers show aging-associated hypermethylation and because aging-associated hypermethylation occurs predominantly at promoters bivalently marked in ESCs (Rakyan et al. 2010), we surmised that bivalent genes in ESCs are more susceptible to hypermethylation during the aging process. Indeed, our analysis of aging-associated hypermethylated genes (Supplemental Table S7) revealed that genes bivalent in ESCs are more likely to be hypermethylated during aging—to the same extent as in cancer—than during ESC to EpiLC differentiation (Fig. 5E; Supplemental Fig. S9I,J). Moreover, unlike in EpiLCs, promoters of bivalent genes in ESCs that get hypermethylated in cancer and/or during aging are CpG-rich (Supplemental Fig. S9F,K). Given that CpG-rich promoters are mostly unmethylated in all cell types at all stages of development, even when transcriptionally inactive (Deaton and Bird 2011; Jones 2012; Bestor et al. 2015; Schübeler 2015), these findings suggest that aging-associated hypermethylation of genes that are bivalently marked in ESCs can serve as a potential biomarker for carcinogenesis in the elderly.
Establishment and fate of bivalent chromatin
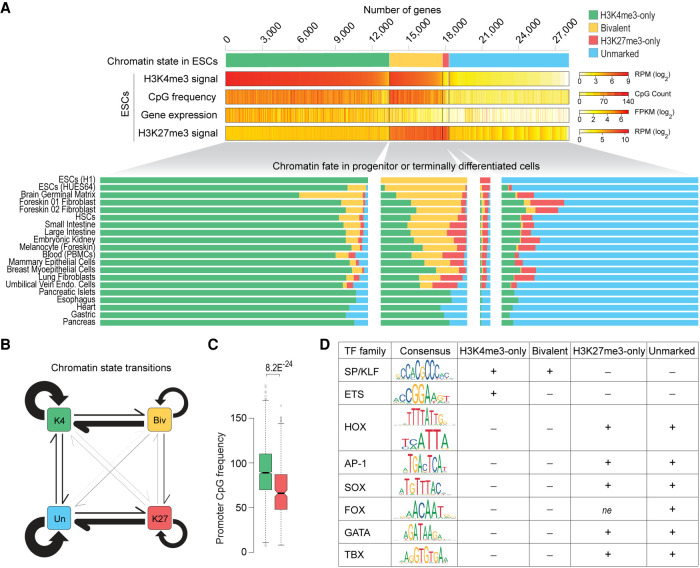
During ESC differentiation and embryonic development, bivalent chromatin is postulated to resolve into either H3K4me3-only or H3K27me3-only state depending on whether the gene is activated or silenced, respectively (Azuara et al. 2006; Bernstein et al. 2006; Voigt et al. 2013). To definitively determine the fates of bivalent genes, we examined chromatin states of gene promoters across various cell types. Our analysis revealed that a vast majority (83%) of genes that are bivalent in ESCs retain H3K4me3 in other cell types, with 52% resolving into H3K4me3-only chromatin state and 31% remaining bivalent (Fig. 7A; Supplemental Table S9). Only a small fraction (10%) of bivalent genes in ESCs resolve into H3K27me3-only state in other cell types. Because bivalent promoters are CpG-rich (Fig. 4D; Supplemental Fig. S8B) and because CpG-rich sequences, when unmethylated, are sufficient to establish H3K4me3 (Thomson et al. 2010), these data suggest that bivalent genes have a predilection to resolve into their presumably default H3K4me3-only state in the absence of PRC2 activity (Fig. 7B). Consistent with this conclusion, we find bivalent promoters that resolve into H3K4me3-only state in most cell types are more CpG-rich compared to those that resolve into H3K27me3-only state in most cell types (Fig. 7C; Supplemental Table S9).
Figure 7.
Chromatin fate and sequence characteristics of bivalent promoters. (A, top) Genes are grouped into four classes based on their chromatin state, defined based on H3K4me3 (±500 bp of TSS) and/or H3K27me3 (±2 kb of TSS) enrichment at gene promoters in human ESCs. H3K4me3 and H3K27me3 levels, CpG density (±500 bp of TSS), and gene expression are shown. (Bottom) Chromatin states of the same four gene classes in various cell types. (B) Schematic summarizing chromatin state transitions of gene promoters from one state to another. Arrows represent state transitions. The thicker the arrow, the more frequently that transition is observed. (K4) H3K4me3-only; (Biv) bivalent; (K27) H3K27me3-only; (Un) unmarked. (C) Box plot showing distribution of CpG density at promoters of bivalent genes (in ESCs) that predominantly resolve into H3K4me3-only (green, left) or H3K27me3-only (red, right) state in other cell types. P-value calculated using two-sided Wilcoxon rank-sum test. (D) Binding motifs for various transcription factor (TF) families and their enrichment within promoters (±500 bp of TSS) of the four genes classes defined in A. (±) Over/under-enrichment; (ne) no enrichment. See also Supplemental Figure S10 and Supplemental Table S10.
Next, to understand the establishment of the bivalent chromatin state, we focused on genes that acquire bivalency in cell types with restricted potency and found that an overwhelming majority (81%) of these genes are H3K4me3-only in ESCs and are CpG-rich (Fig. 7A). Our analysis of chromatin fates during reconstituted pre- to post-implantation epiblast differentiation in mouse revealed similar results (Supplemental Fig. S10A). Specifically, nearly all the genes that acquire bivalency in EpiLCs were H3K4me3-only previously. These data further support our conclusion that bivalent chromatin is the culmination of PRC2 activity at regions with high CpG density. Our findings are consistent with studies linking EZH2 to the acquisition of 910 (of 1026; 88%) new bivalent genes in germinal center B cells, almost all of which were previously H3K4me3-only in naive B cells (Béguelin et al. 2013).
Last, to gain further insight into sequence features (besides CpG density) that underlie bivalent promoters, we explored transcription factor (TF) binding motifs overrepresented in H3K4me3-only, bivalent, H3K27me3-only, and unmarked promoter classes in ESCs (Supplemental Methods). We found motifs for ubiquitously expressed SP/KLF family of TFs generally overrepresented (approximately two- to fivefold) within mostly CpG-rich H3K4me3-only and bivalent promoters compared with mostly CpG-poor H3K27me3-only and unmarked promoters (Fig. 7D; Supplemental Fig. S10B,C; Supplemental Table S10). An exception to this is a small fraction of bivalent promoters (relatively CpG poor) that mostly resolve into H3K27me3-only state in other cell types (Fig. 7C); they show no such enrichment for a subset of SP/KLF TF motifs (Supplemental Fig. S10D). Unlike CpG-rich promoters, the largely CpG-poor H3K27me3-only and unmarked promoters are characterized by overrepresentation of motifs recognized by families of TFs that are tissue-specific (e.g., HOX, AP-1, SOX, FOX, GATA, and TBX) (Fig. 7D; Supplemental Fig. S10B,C; Supplemental Table S10). Our analysis also revealed that H3K4me3-only but not bivalent promoters are characterized by overrepresentation of motifs for ETS family of TFs (Supplemental Fig. S10B), known to activate genes associated with a variety of cellular housekeeping processes including cell cycle control, cell proliferation, and cellular differentiation. These data suggest that CpG-rich promoters that are enriched for motifs for SP/KLF but not ETS factors, when transcriptionally inactive, provide a fertile ground for PRC2 activity and establishment of bivalent chromatin, consistent with a causal role for GC-rich sequences (lacking activating TF motifs) in PRC2 recruitment (Mendenhall et al. 2010).
Discussion
Bivalent genes, by virtue of their exhibiting features of both transcriptionally active and repressed chromatin, are posited as being in a poised state—enabling them to be rapidly activated upon appropriate activation cues during development—while maintaining a transcriptionally repressed state (Azuara et al. 2006; Bernstein et al. 2006; Voigt et al. 2013).
Collectively, our studies reveal that bivalency does not poise genes for rapid activation but protects promoters from de novo DNA methylation. Activation of bivalent genes is neither greater nor more rapid than that of other transcriptionally silent genes that lack H3K4me3 at their promoters (Fig. 2G), challenging the premise that H3K4me3 at bivalent promoters is instructive for rapid activation of transcription. We find that promoter H3K4me3 levels are a product of the underlying CpG-rich DNA sequence, so much so that CpG density alone can predict H3K4me3 levels/enrichment reasonably accurately (Fig. 4B,C). This likely explains why H3K4me3 at bivalent promoters in one cell type persists in nearly all other cell types irrespective of gene expression (Fig. 1) and why unmethylated CGI promoters harbor H3K4me3 even when transcriptionally inactive (Guenther et al. 2007; Mikkelsen et al. 2007). Our findings are consistent with studies showing that high CpG-rich sequences, when unmethylated, per se are sufficient to establish H3K4me3 domains (Thomson et al. 2010)—even in the absence of RNAPII and sequence specific TFs (Vastenhouw et al. 2010)—but insufficient to induce transcriptional activity on chromatin (Hartl et al. 2019). Because bivalently marked promoters across various cell types overlap CpG-rich sequences (Supplemental Fig. S8B), which inherently are devoid of DNA methylation (Deaton and Bird 2011) and almost always positive for H3K4me3 (Supplemental Fig. S8C), establishment (and dissolution) of bivalent domains likely boils down to PRC2 activity (and inactivity, respectively) at genomic regions with high CpG density. Supporting this notion, the number of bivalent genes in a given cell type strongly correlates with EZH2 expression (catalytic subunit of PRC2) (Fig. 4F), with gain- or loss-of-function EZH2 mutation, respectively, associated with increased or decreased bivalency (Shema et al. 2016).
H3K4me3 and H3K27me3 are loosely referred to as “activating” and “repressive” marks, respectively, but neither has been firmly established to play a causative role in the regulation of gene expression. To the contrary, it was shown that PRC2/H3K27me3 is not required for the initiation of transcriptional repression of its targets but is only required for the maintenance of the repressed state (Riising et al. 2014). Despite the general correlation between H3K4me3 and gene expression, it remains unclear as to whether H3K4me3 is instructive for transcription (Howe et al. 2017). Our results showing that the mere presence or the extent of H3K4me3 at bivalent genes does not confer an added advantage when it comes to rapid or higher activation potential (Fig. 2) suggest that H3K4me3 is not instructive for transcription activation. Consistent with this conclusion, deletion of Mll2—chiefly responsible for H3K4me3 at bivalent chromatin—in mouse ESCs resulted in no substantial disruption in the responsiveness of gene activation after retinoic acid treatment despite the almost complete loss of H3K4me3 and concomitant gain of H3K27me3 at bivalent promoters (Hu et al. 2013; Denissov et al. 2014). Our findings are also consistent with studies in yeast demonstrating that loss of H3K4me3 has no effect on the levels of nascent transcription and, conversely, loss of RNAPII has no effect on H3K4me3 levels (Murray et al. 2019). Together, these observations indicate that H3K4me3 is neither instructive for nor informed by transcription.
Besides its ability to predict transcription or chromatin states, the precise role(s) of H3K4me3 still remains elusive (Piunti and Shilatifard 2016). Our findings suggest that H3K4me3 is a better predictor of unmethylated CpGs than transcriptional activity and may be a general mechanism to maintain the hypomethylated state of CGIs, even when transcriptionally inactive. This would be consistent with studies showing that H3K4me3 repulses de novo methyltransferases in vitro (Ooi et al. 2007), and complete erasure of H3K4me3 elevates DNA methylation levels (Hu et al. 2009; Rose and Klose 2014) but reversible DNA methylation has no impact on H3K4me3 levels (King et al. 2016). About 70% of mammalian promoters overlap with CGIs (Deaton and Bird 2011). Although CpG dinucleotides are substrates for DNA methyltransferases, few CGI promoters gain methylation (even when transcriptionally inactive) during normal development (Deaton and Bird 2011; Jones 2012; Bestor et al. 2015; Schübeler 2015). However, most genes that are hypermethylated in cancer have CGI promoters and are bivalently marked in ESCs, which led to the proposition that bivalency predisposes them for aberrant de novo DNA methylation and irreversible silencing in cancer (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). Recent studies have also shown an association between hypermethylation of bivalent promoters in cancer and acquired resistance to chemotherapy (Curry et al. 2018). Our findings suggest that bivalency and H3K4me3 in particular protects promoters from de novo methylation during pre- to post-implantation epiblast differentiation, and aberrant hypermethylation in cancer may be explained by the loss of H3K4me3/bivalency (Fig. 5). In other words, it may be that it is not the bivalency but the loss of bivalency that make bivalent genes more susceptible to aberrant DNA methylation in diseases such as cancer. This would be consistent with studies showing cancer cell lines exhibiting a general loss of bivalency (Bernhart et al. 2016), and bivalent promoters with high H3K27me3:H3K4me3 ratio being targets for DNA hypermethylation in cancer (Dunican et al. 2020).
CGI promoters, the superset containing bivalent promoters, are relatively nucleosome-deficient, intrinsically accessible without the need for ATP-dependent nucleosome displacement, and transcriptionally permissive (Ramirez-Carrozzi et al. 2009). So, what keeps transcriptionally repressed bivalent promoters from getting transcribed? Most bivalent promoters lack transcriptionally engaged RNAPII (Williams et al. 2015) but harbor what is referred to as poised RNAPII (preferentially phosphorylated at serine 5 but not serine 2), and it has been suggested that poised RNAPII primes bivalent genes for rapid activation (Stock et al. 2007; Brookes et al. 2012; Tee et al. 2014; Ferrai et al. 2017). Because promoter-proximal pausing of RNAPII is not a common mechanism used at bivalent genes (Min et al. 2011; Williams et al. 2015), it is less likely that the poised RNAPII at bivalent promoters represents some form of paused/stalled RNAPII competent for rapid transcription reactivation. Our analyses reveal that bivalent promoters are devoid of TFIIH and that any serine 5 phosphorylation on RNAPII at bivalent promoters is attributable to MAPK1 (Fig. 3), known to bind exclusively to a subset of PRC2 targets and phosphorylate serine 5 on RNAPII (Tee et al. 2014). Because MAPK1 and TFIIH are mutually exclusive at their target promoters and because RNAPII-S5p levels at poised bivalent promoters correlate with H3K27me3 levels (and not H3K4me3, as observed at active promoters), it is conceivable that MAPK1-mediated phosphorylation of serine 5 on RNAPII (or MAPK1's mere presence on chromatin) is refractory to transcription. In this scenario, MAPK1 and/or the substrate it modifies on RNAPII (one or more CTD heptad repeats) may antagonize TFIIH, and activation of transcription likely occurs only upon loss of MAPK1 binding and/or MAPK1-mediated phosphorylation of serine 5 on RNAPII, which may be followed by binding of appropriate transcription factors at promoters and/or enhancers. Further studies are required to ascertain any potential antagonism between MAPK1 and TFIIH.
Mll2 is dispensable for maintaining ESC self-renewal, but Mll2 deficiency is embryonic lethal. Mll2 knock-out (KO) mice show growth defects as early as ∼E6.5 and die at ∼E10.5 (Glaser et al. 2006), suggesting that MLL2 is not required until after implantation, right when Dnmt3a and Dnmt3b get induced to carry out global de novo methylation in early post-implantation embryo. Furthermore, in vitro differentiation of Mll2 KO ESCs results in impaired embryoid body formation, with many bivalent genes with key functions in embryonic development and differentiation failing to activate or exhibiting delayed activation kinetics (Lubitz et al. 2007; Mas et al. 2018), indicating an essential role for MLL2 during ESC differentiation. Our finding that H3K4me3 at transcriptionally repressed bivalent promoters, catalyzed primarily by MLL2 (Hu et al. 2013; Denissov et al. 2014), confers protection against de novo DNA methylation during pre- to post-implantation epiblast differentiation (Fig. 5A,B,E) suggests that the requirement for MLL2 after implantation—when it is no longer the major H3K4 trimethyltransferase (Glaser et al. 2006)—at least in part might have to do with its role in implementing H3K4me3 at bivalent genes to maintain epigenetic plasticity by protecting against de novo DNA methylation and thus irreversible silencing. Because ESCs do not express Dnmt3a or Dnmt3b and are DNA hypomethylated, this could perhaps explain why MLL2 is dispensable in mouse ESCs. Moreover, a recent study showed that MLL2—which also is responsible for H3K4me3 at a vast majority of transcriptionally active genes (Denissov et al. 2014)—protects ∼2% of MLL2-dependent active genes from DNMT1-mediated maintenance methylation (Douillet et al. 2020), highlighting MLL2's multifaceted role in regulating gene expression.
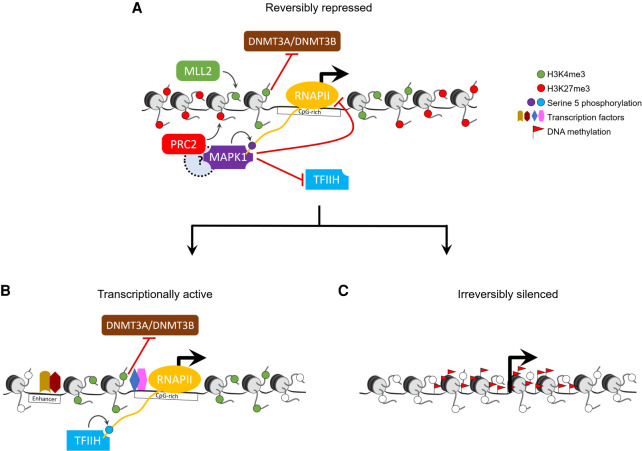
In summary, our findings suggest a unifying model (Fig. 8) wherein bivalency maintains epigenetic plasticity by protecting gene promoters from irreversible silencing while maintaining a reversibly repressed state, and that loss of H3K4me3 may make them more susceptible to aberrant DNA methylation in diseases such as cancer. One limitation of our study is that the ESC to EpiLC differentiation model we used to investigate bivalency only recapitulates events during pre- to post-implantation epiblast development (approximately E3.75–E5.75) and does not provide sufficient time for all of bivalency to resolve. Assessing the fate of all bivalent genes through all stages of development would require further investigation using differentiation models that recapitulate later developmental stages through directed differentiation of ESCs to specific lineages.
Figure 8.
Model for bivalent chromatin maintaining epigenetic plasticity by protecting gene promoters from irreversible silencing while maintaining a reversibly repressed state. (A) H3K4me3, catalyzed primarily by the MLL2/COMPASS complex, protects CpG-rich bivalent promoters from DNA methylation by repelling de novo methyltransferases DNMT3A and DNMT3B. Bivalent promoters, by virtue of their overlapping CGIs, are more averse to assembling into nucleosomes compared to other genomic DNA (Ramirez-Carrozzi et al. 2009); consequently, bivalent promoters are relatively nucleosome-deficient, intrinsically accessible, and transcriptionally permissive (Deaton and Bird 2011; Mas et al. 2018). The assembly of transcription machinery at the promoter triggers PRC2 to recruit—either directly or indirectly through a yet-to-be-determined mechanism—reinforcement in the form of MAPK1 (ERK2), which, in lieu of the usual TFIIH, phosphorylates serine 5 on a particular (or a set of) RNAPII CTD heptad repeat(s) (Tee et al. 2014). The presence of MAPK1 and/or the ensuing serine 5 phosphorylation is refractory to transcription as it antagonizes the recruitment of the TFIIH complex, which, besides its role in phosphorylating serine 5 on RNAPII, is necessary to unwind promoter DNA to form a transcription bubble for RNA synthesis. (B) Activation of transcription at bivalent genes likely occurs only upon loss of PRC2 activity and thus MAPK1 activity, followed by binding of appropriate transcription factors at promoters and/or enhancers. (C) Loss of H3K4me3 at CpG-rich bivalent promoters make them more susceptible to aberrant DNA methylation during aging and in diseases such as cancer.
Poised enhancers, characterized by the co-occurrence of histone modifications generally associated with positive (H3K4me1) and negative (H3K27me3) transcriptional states, are another class of bivalent regions (Blanco et al. 2020). Similar to bivalent promoters, they are considered to be primed for future activation during embryo development, with loss of H3K27me3 and gain of H3K27ac associated with enhancer activation. Although some of our findings on bivalent promoters may be applicable to poised enhancers, further studies are needed to precisely characterize the functional relevance and mechanism of poised enhancers.
Methods
Data sources
All data sets pertaining to this manuscript were obtained from previously published studies (for details, see Supplemental Methods).
Histone modification enrichment at gene promoters
For each gene promoter, read densities (reads per million mapped reads [RPM]) of individual histone modifications (H3K4me3/H3K27me3) and corresponding genomic input were calculated. Promoters were defined as the region spanning TSS ± 500 bp for H3K4me3 and TSS ± 2 kb for H3K27me3. A promoter is deemed to be enriched for a particular histone modification only if its ChIP signal (RPM) is at least (1) threefold greater than its input signal (RPM), and (2) greater than a threshold (1% FDR), estimated as the lowest RPM value at which the number of qualifying promoters (RPM greater than or equal to the threshold) based on the input signal (RPM) is <1% of the number of qualifying promoters based on the ChIP signal (RPM). Promoters enriched for both H3K4me3 and H3K27me3 were defined as bivalent. Promoters enriched for H3K4me3 but not H3K27me3 were defined as H3K4me3-only, and those that are enriched for H3K27me3 but not H3K4me3 were defined as H3K27me3-only. Promoters with neither H3K4me3 nor H3K27me3 enrichment were defined as “unmarked.” Custom scripts used to compute H3K4me3/H3K27me3 enrichment at gene promoters, define promoter chromatin state, and generate data for making tag density plots and heatmaps are available as Supplemental Code and at GitHub (https://github.com/DhirKumar/Biv_ChIP).
RNA-seq data analysis
RNA-seq reads were aligned to the mouse genome (mm9) using STAR aligner (Dobin et al. 2013), allowing up to three mismatches, retaining only reads that align to unique genomic locations. Alignment files were used to quantitate expression (FPKM) of genes and isoforms annotated in the mm9 genome (source: NCBI RefSeq) using the cuffdiff tool (Trapnell et al. 2013) with default parameters and library type defined as fr-firststrand. The RefSeq-GTF file (downloaded from the UCSC database) for the mm9 genome build was supplied to cuffdiff tool as the reference. The resultant “isoforms.fpkm_tracking” file from the cuffdiff run was used to infer differentially expressed genes (Q-value <0.05) and fold changes between two cell types/time points of interest. In the case of multiple promoters giving rise to alternative isoforms, we used one-to-one association of promoter chromatin state and isoform expression. For human, processed RNA-seq signal (FPKM) for various cell types were obtained from the NIH Roadmap Epigenomics project (Roadmap Epigenomics Consortium et al. 2015) (https://egg2.wustl.edu/roadmap/data/byDataType/rna/expression/57epigenomes.RPKM.pc.gz). RefSeq accession (NM/NR ID) was used to integrate gene expression and chromatin-level data.
Mouse DNA methylation analysis
Processed promoter DNA methylation data from naive mouse ESCs and EpiLCs were obtained from a previously published study (Shirane et al. 2016). liftOver tool was used to map gene coordinates between mm9 and mm10 assemblies. A gene promoter is defined as hypermethylated in EpiLCs compared to naive ESCs only if (1) its methylation level in EpiLCs is at least 50%, and (2) its methylation level in EpiLCs is at least twofold greater than that in naive ESCs.
Human DNA methylation analysis
Genes DNA hypermethylated (based on promoter DNA methylation levels) in primary colorectal tumor was obtained from a previously published study (Widschwendter et al. 2007). Genes DNA hypermethylated in human osteosarcoma were inferred from processed probe-level methylation data (Easwaran et al. 2012) using criteria outlined in the original study (for details, see Supplemental Methods). Genes DNA hypermethylated/hypomethylated in 14 TCGA solid epithelial cancer types were inferred from DNA methylation (Illumina 450K array) beta values for 6129 tumors and respective control tissues, generated by the TCGA Research Network (for details, see Supplemental Methods). Genes DNA hypermethylated during aging were inferred by mapping processed probe-level methylation data (Rakyan et al. 2010) to gene promoter coordinates using Infinium manifest file.
Supplementary Material
Acknowledgments
We thank members of the Jothi Laboratory for insightful discussions and comments on the manuscript. We thank J. Rodriguez, P.A. Wade, and S. Yellaboina for critical comments on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (1ZIAES102625, to R.J.).
Author contributions: D.K. and R.J. conceived and designed the study; D.K. performed the research, with contributions from S.C., A.J.O., and P.Y.; D.K. and R.J. analyzed the data and wrote the manuscript; all authors reviewed and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275736.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, et al. 2013. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23: 677–692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Kretzmer H, Holdt LM, Jühling F, Ammerpohl O, Bergmann AK, Northoff BH, Doose G, Siebert R, Stadler PF, et al. 2016. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Sci Rep 6: 37393. 10.1038/srep37393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Edwards JR, Boulard M. 2015. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci 112: 6796–6799. 10.1073/pnas.1415301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, González-Ramírez M, Alcaine-Colet A, Aranda S, Di Croce L. 2020. The bivalent genome: characterization, structure, and regulation. Trends Genet 36: 118–131. 10.1016/j.tig.2019.11.004 [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. 10.1016/j.stem.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. 2014. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14: 838–853. 10.1016/j.stem.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry E, Zeller C, Masrour N, Patten DK, Gallon J, Wilhelm-Benartzi CS, Ghaem-Maghami S, Bowtell DD, Brown R. 2018. Genes predisposed to DNA hypermethylation during acquired resistance to chemotherapy are identified in ovarian tumors by bivalent chromatin domains at initial diagnosis. Cancer Res 78: 1383–1391. 10.1158/0008-5472.CAN-17-1650 [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. 10.1101/gad.2037511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141: 526–537. 10.1242/dev.102681 [DOI] [PubMed] [Google Scholar]
- Di Croce L, Helin K. 2013. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20: 1147–1155. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillet D, Sze CC, Ryan C, Piunti A, Shah AP, Ugarenko M, Marshall SA, Rendleman EJ, Zha D, Helmin KA, et al. 2020. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat Genet 52: 615–625. 10.1038/s41588-020-0618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunican DS, Mjoseng HK, Duthie L, Flyamer IM, Bickmore WA, Meehan RR. 2020. Bivalent promoter hypermethylation in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem cells. BMC Biol 18: 25. 10.1186/s12915-020-0752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, Aryee MJ, Joyce P, Ahuja N, Weisenberger D, et al. 2012. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 22: 837–849. 10.1101/gr.131169.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H, Tsai HC, Baylin SB. 2014. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell 54: 716–727. 10.1016/j.molcel.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Torlai Triglia E, Risner-Janiczek JR, Rito T, Rackham OJ, de Santiago I, Kukalev A, Nicodemi M, Akalin A, Li M, et al. 2017. RNA polymerase II primes Polycomb-repressed developmental genes throughout terminal neuronal differentiation. Mol Syst Biol 13: 946. 10.15252/msb.20177754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153: 1149–1163. 10.1016/j.cell.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. 2006. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423–1432. 10.1242/dev.02302 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88. 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Meshorer E. 2015. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 16: 1609–1619. 10.15252/embr.201541011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D, Krebs AR, Grand RS, Baubec T, Isbel L, Wirbelauer C, Burger L, Schübeler D. 2019. CG dinucleotides enhance promoter activity independent of DNA methylation. Genome Res 29: 554–563. 10.1101/gr.241653.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Howe FS, Fischl H, Murray SC, Mellor J. 2017. Is H3K4me3 instructive for transcription activation? Bioessays 39: 1–12. 10.1002/bies.201600095 [DOI] [PubMed] [Google Scholar]
- Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL. 2009. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci 106: 22187–22192. 10.1073/pnas.0905767106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER, Shilatifard A. 2013. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 20: 1093–1097. 10.1038/nsmb.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254. 10.1038/ng1089 [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AD, Huang K, Rubbi L, Liu S, Wang CY, Wang Y, Pellegrini M, Fan G. 2016. Reversible regulation of promoter and enhancer histone landscape by DNA methylation in mouse embryonic stem cells. Cell Rep 17: 289–302. 10.1016/j.celrep.2016.08.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. 2013. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20: 311–316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128: 707–719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097. 10.1038/ng.946 [DOI] [PubMed] [Google Scholar]
- Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. 2007. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell 18: 2356–2366. 10.1091/mbc.e06-11-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. 2012. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149: 590–604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G, Blanco E, Ballaré C, Sansó M, Spill YG, Hu D, Aoi Y, Le Dily F, Shilatifard A, Marti-Renom MA, et al. 2018. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat Genet 50: 1452–1462. 10.1038/s41588-018-0218-5 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770. 10.1038/nature07107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25: 742–754. 10.1101/gad.2005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. 2008. Lineage-specific Polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Murray SC, Lorenz P, Howe FS, Wouters M, Brown T, Xi S, Fischl H, Khushaim W, Rayappu JR, Angel A, et al. 2019. H3k4me3 is neither instructive for, nor informed by, transcription. bioRxiv 10.1101/709014 [DOI]
- Niccoli T, Partridge L. 2012. Ageing as a risk factor for disease. Curr Biol 22: R741–R752. 10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell–like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242. 10.1038/ng1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717. 10.1038/nature05987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936. 10.1101/gad.1477006 [DOI] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352: aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. 2010. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20: 434–439. 10.1101/gr.103101.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128. 10.1016/j.cell.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. 2014. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55: 347–360. 10.1016/j.molcel.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. 2014. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 1839: 1362–1372. 10.1016/j.bbagrm.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236. 10.1038/ng1950 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517: 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE. 2016. Single-molecule decoding of combinatorially modified nucleosomes. Science 352: 717–721. 10.1126/science.aad7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Kurimoto K, Yabuta Y, Yamaji M, Satoh J, Ito S, Watanabe A, Hayashi K, Saitou M, Sasaki H. 2016. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev Cell 39: 87–103. 10.1016/j.devcel.2016.08.008 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2009. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708. 10.1038/nrm2763 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344. 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435. 10.1038/ncb1663 [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. 2014. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 156: 678–690. 10.1016/j.cell.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086. 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926. 10.1038/nature08866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, LeRoy G, Drury WJ III, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. 2012. Asymmetrically modified nucleosomes. Cell 151: 181–193. 10.1016/j.cell.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27: 1318–1338. 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158. 10.1038/ng1941 [DOI] [PubMed] [Google Scholar]
- Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. 2015. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell 58: 311–322. 10.1016/j.molcel.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Humphrey SJ, Cinghu S, Pathania R, Oldfield AJ, Kumar D, Perera D, Yang JYH, James DE, Mann M, et al. 2019. Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Syst 8: 427–445.e10. 10.1016/j.cels.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.