Abstract
The major genetic risk factors for Hirschsprung disease (HSCR) are three common polymorphisms within cis-regulatory elements (CREs) of the receptor tyrosine kinase gene RET, which reduce its expression during enteric nervous system (ENS) development. These risk variants attenuate binding of the transcription factors RARB, GATA2, and SOX10 to their cognate CREs, reduce RET gene expression, and dysregulate other ENS and HSCR genes in the RET–EDNRB gene regulatory network (GRN). Here, we use siRNA, ChIP, and CRISPR-Cas9 deletion analyses in the SK-N-SH cell line to ask how many additional HSCR-associated risk variants reside in RET CREs that affect its gene expression. We identify 22 HSCR-associated variants in candidate RET CREs, of which seven have differential allele-specific in vitro enhancer activity, and four of these seven affect RET gene expression; of these, two enhancers are bound by the transcription factor PAX3. We also show that deleting multiple variant-containing enhancers leads to synergistic effects on RET gene expression. These, coupled with our prior results, show that common sequence variants in at least 10 RET enhancers affect HSCR risk, seven with experimental evidence of affecting RET gene expression, extending the known RET–EDNRB GRN to reveal an extensive regulatory code modulating disease risk at a single gene.
It is now well established that most human complex traits and diseases arise from the additive genetic effects of hundreds to thousands of variants distributed across the genome (Visscher et al. 2017). At each locus, multiple statistically significant variants are detected, but it is unknown how many of them make functionally independent contributions to the phenotype. The widespread existence of genetic association (linkage disequilibrium [LD]) between local sequence variants makes this a difficult question to answer by statistical methods alone and requires experimental perturbation and assessment of each candidate variant (Chatterjee et al. 2016). This is because genetic associations between variants depend on their recombination frequency, not their functional effects, thereby confusing causal with innocent variants.
The majority of causal variants that contribute to trait variation reside within cis-regulatory elements (CREs) and enhancers of a target gene, thereby modulating its gene expression, usually in a cell type–specific manner (Maurano et al. 2012; Chakravarti and Turner 2016). Such gene expression control is assumed to occur within a topologically associating domain (TAD) (Dixon et al. 2012; Rao et al. 2014), defining the physical locus within which CREs function. However, three major questions remain unanswered. First, because a TAD usually harbors multiple CREs and genes, which CREs affect which gene's expression? Second, do different CREs of a specific gene have unique functions in space, time, and cellular states, or are they redundant (shadow enhancers)? Do they act independently, or are they synergistic and require clustering (super-enhancers) for function (Chakravarti and Turner 2016; Chatterjee and Ahituv 2017; Kvon et al. 2021)? Third, because most CRE effects are small, how do such small gene expression effects modulate phenotypes? The existence of many experimental methods to identify CREs comprehensively now allows us to address these questions (Inoue et al. 2019; Kapoor et al. 2019).
In this study, we use Hirschsprung disease (HSCR; congenital colonic aganglionosis) as an exemplar to ask how many ENS enhancers with disease-associated variants at its major gene, the receptor tyrosine kinase RET, are involved. HSCR is a complex neurodevelopmental disorder in which failure of differentiation of enteric neural crest cell (ENCC) precursors during ENS development leads to aganglionosis; more than 33 genes/loci explaining 62% of its population attributable risk have been identified (Tilghman et al. 2019). Significantly, most of this risk arises from coding and enhancer variants at RET with smaller contributions from other genes, all of whose functions in ENS development are united through a gene regulatory network (GRN) coregulating RET and EDNRB (Kapoor et al. 2015; Chatterjee et al. 2016; Chatterjee and Chakravarti 2019; Tilghman et al. 2019). Thus, we asked how many HSCR-associated noncoding sequence variants identified in genetic screens are individually sufficient to perturb RET gene expression as well as GRN activity. We also examined if perturbing multiple CREs containing causal variants leads to additive or synergistic effects on RET gene expression and other genes of the GRN.
Results
Enhancers at the RET locus
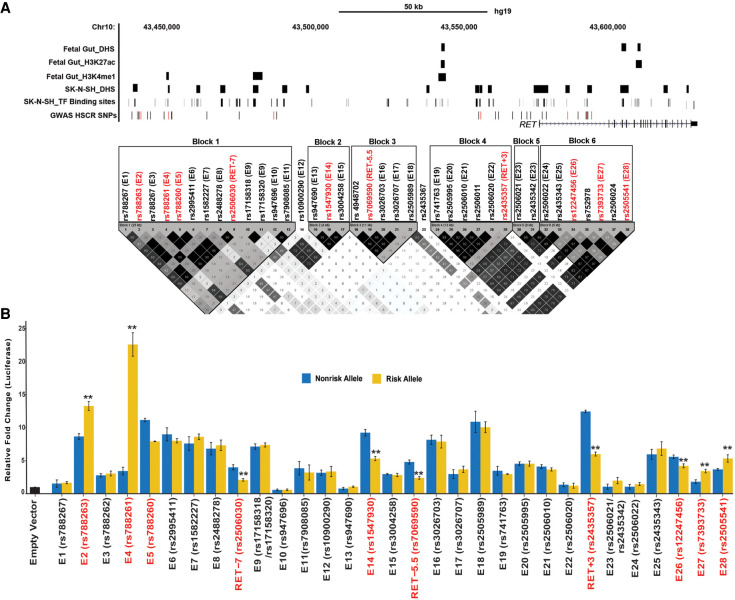
To create a complete catalog of common (MAF ≥ 10%) RET regulatory variants associated with HSCR, we began with the analysis of all 38 genome-wide significant noncoding single-nucleotide polymorphisms (SNPs) discovered in a genome-wide association study (GWAS) of 220 HSCR trios comprising a proband and both of her/his parents (Jiang et al. 2015). All these SNPs were genotyped in our previous studies (Jiang et al. 2015; Chatterjee et al. 2016) and were distributed across six LD blocks in a 155-kb TAD (Chr 10: 43,434,933–43,590,368; hg19) containing RET as the sole gene (Fig. 1A). We had previously analyzed eight of these SNP-containing genomic elements because they each disrupted a predicted TF binding site (TFBS) determined from ENCODE chromatin immunoprecipitation (ChIP)–seq data (Chatterjee et al. 2016).
Figure 1.
The RET regulatory landscape in the enteric nervous system. (A) The 155-kb RET locus (Chr 10: 43,434,933–43,590,368; hg19) contains 38 HSCR-associated polymorphisms in six linkage disequilibrium (LD) blocks. LD between all 38 SNPs was estimated as by Gabriel et al. (2002). Multiple enhancer-associated epigenetic marks (DNase I hypersensitivity [DHS], H3K27ac, H3K4me1) in 108-day human fetal large intestine and the SK-N-SH neuroblastoma cell line and transcription factor (TF) binding sites (TFBSs) from public sources are noted. All common (minor allele frequency ≥10%) variants associated with HSCR are shown, with those showing allelic difference in in vitro transcription assays marked in red. (B) Allele-specific in vitro luciferase assays of 28 CREs containing 30 polymorphisms plus three previously tested controls in SK-N-SH cells are shown: 22 CREs act as enhancers, compared with a promoter-only control, of which seven also show allelic difference in luciferase activity (boxed) for its cognate HSCR-associated polymorphism in addition to RET-7, RET-5.5, and RET+3 positive controls. Error bars are SEM of three independent biological replicates: (**) P < 0.001.
We first asked if the remaining 30 SNPs reside within CREs. We conducted in vitro functional tests of enhancer activity by cloning ∼500-bp elements centered on each risk variant (Table 1) into a pGL4.23 luciferase vector, with a minimal TATA-box of the hemoglobin subunit beta (HBB) gene, and transfecting them into the human neuroblastoma SK-N-SH cell line. SK-N-SH expresses all known members of the RET–EDNRB GRN and is an appropriate cell model system for studies of ENS transcriptional regulation (Chatterjee et al. 2016; Chatterjee and Chakravarti 2019). Two pairs of SNPs (rs17158318/rs17158320 and rs2506021/rs2435342) were only 64 bp and 108 bp apart and were cloned into the same elements (E9 and E23, respectively) (Table 1). As positive controls, we reanalyzed three HSCR-associated SNPs (rs2506030, rs7069590, rs2435357) previously shown to be RET enhancer variants (Table 1; Chatterjee et al. 2016). Our reporter assays showed that 22 new elements had significant enhancer activity (P < 0.001 and >2× reporter activity over the promoter only control vector), of which seven (E2, E4, E5, E14, E26, E27, and E28) also displayed differential reporter activity between the risk and nonrisk alleles (Table 1; Fig. 1B). Among the latter, 71% (E2, E4, E26, E27, and E28) overlapped an open chromatin region or an enhancer-associated epigenetic mark in the human fetal gut and SK-N-SH cells (Bernstein et al. 2010), whereas 20% (E9, E16, E21) of the remaining 15 elements with reporter activity that had no allelic difference overlapped a potential enhancer mark (Fig. 1A). Thus, at least 10 functionally distinct CREs within the RET TAD can potentially affect HSCR risk.
Table 1.
Genomic coordinates (hg19) of 31 elements containing 33 single-nucleotide polymorphisms (SNPs) within six linkage disequilibrium (LD) blocks at the RET locus that are associated with Hirschsprung disease (HSCR)
Haplotype-specific effect of causal RET polymorphisms
The identification of 10 CRE-associated risk variants (rs788263, rs788261, rs788260, rs2506030, rs1547930, rs7069590, rs2435357, rs12247456, rs7393733, and rs2505541) (Table 1) prompted us to ask which allelic combinations were associated with disease risk. We first estimated haplotypes and their frequencies for all 10 SNPs in 220 unrelated HSCR cases (Jiang et al. 2015) and 503 unrelated controls from The 1000 Genomes Project Consortium (2015), all of non-Finnish European ancestry. Second, we estimated the odds ratio (OR) for all haplotypes with a frequency ≥1% in controls. We observed 10 distinct haplotypes for which CTGAACCACT (risk allele in bold) was used as the reference because it had the smallest number (one) of risk alleles (Table 2) and we have previously shown that HSCR risk scales with increasing number of CRE variants (Kapoor et al. 2015; Chatterjee et al. 2016; Chatterjee and Chakravarti 2019; Tilghman et al. 2019): Significant risk was observed for two haplotypes, GCAGGTTGGT (OR 12.2, 95% CI: 5.97 – 24.93, P = 7.02 × 10−12) and CTGAGTTGGT (OR 7.2, 95% CI: 3.26 – 15.91, P = 1.02 × 10−6) (Table 2). These two haplotypes contain our previously identified risk-increasing ATT and GTT haplotypes (for rs2506030, rs7069590, rs2435357) (Chatterjee et al. 2016). The 10-SNP risk haplotypes differ only for the first four SNPs (rs788263, rs788261, rs788260, rs2506030), which occur within the most 5′ LD block. SNPs within this LD block do contribute to HSCR, but we do not have the statistical power to test the hypothesis that GCAGGTTGGT (OR 12.2) has significantly higher risk than CTGAGTTGGT (OR 7.2), although their estimates suggest this, a feature expected from the larger numbers of risk alleles in the former than in the latter haplotype (Kapoor et al. 2015). Whether this increased risk is from additive or from synergistic effects is unknown from these haplotype data.
Table 2.
Haplotypes for 10 RET enhancer polymorphisms (rs788263, rs788261, rs788260, rs2506030, rs1547930, rs7069590, rs2435357, rs12247456, rs7393733, and rs2505541) with risk alleles denoted in bold
It is evident that HSCR risk is clearly spread over at least three LD blocks, suggesting multiple independent enhancer variants contributing to risk (Fig. 1). To replicate these findings, we reassessed allele frequencies of these 10 SNPs (or proxy SNPs in near perfect linkage disequilibrium) in 235 independent HSCR cases of European ancestry (Kapoor et al. 2021) and their control frequencies in 9400 European ancestry individuals in the Genome Aggregation Database (gnomAD) (Supplemental Table S1; Karczewski et al. 2020). All our SNPs have near-identical allele frequencies for the risk allele in both new and previous cases (Supplemental Fig. S1A) and controls (Supplemental Fig. S1B), and all have higher frequency in cases than in controls (Supplemental Fig. S1C), providing additional evidence of role of these variants in HSCR.
The association of specific haplotypes containing multiple independent noncoding polymorphisms suggests that risk of or protection from HSCR depends on the simultaneous binding or the lack of binding of multiple independent TFs at RET CREs, implying a RET regulatory code that gets disrupted during HSCR.
Transcription factors regulating RET
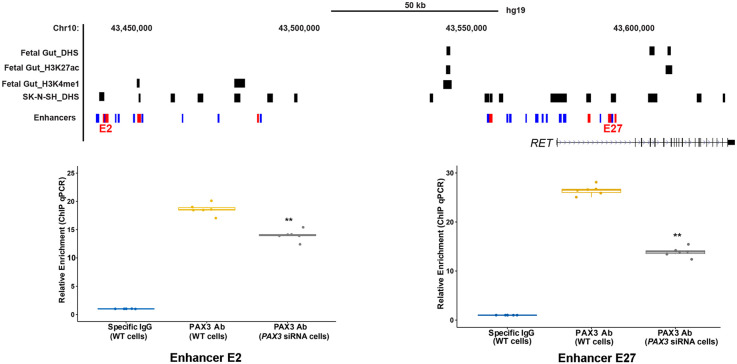
To identify the TFs that underlie this code, we searched for transcription factor (TF) binding sites (TFBSs) using FIMO (Bailey et al. 2009; Grant et al. 2011) and using 890 validated TF motifs in TRANSFAC (Wingender et al. 1996), within the seven-novel risk-associated CREs centered on the polymorphisms, and identified three candidate TFs: (1) PAX3 binding to E2 (AATAAACCC; P = 4.67 × 10−5) and E27 (TCGTCACTCTTAC; P = 9.99 × 10−5), (2) ZBTB6 to E5 (TGGCTCCATCATG; P = 2.387 × 10−6), and (3) ZNF263 binding to E14 (GCCTCACTGCTCCAG; P = 8.09 × 10−5). To determine their relevance for HSCR, we performed qPCR in SK-N-SH cells and observed no expression for ZNF263 and ZBTB6. Further, their expression was absent in the developing mouse gut (Chatterjee et al. 2019), where Ret expression is critical for ENS development (Natarajan et al. 2002), making it unlikely that these TFs control RET expression via specific enhancers. Moreover, ZNF263 and ZBTB6 are both zinc finger domain-containing proteins that are GC-rich, a feature that overestimates their statistical significance owing to their rarity. In contrast, we detected expression of PAX3 in both SK-N-SH and the developing mouse gut (Chatterjee et al. 2019). We performed ChIP-qPCR for PAX3 in SK-N-SH cells and detected significant binding at both E2 (18-fold enrichment; P = 10−3) and at E27 (26-fold enrichment, P = 5 × 10−4) compared with the appropriate nonspecific IgG control (Fig. 2). We further showed the specificity of this binding by performing ChIP-qPCR after siRNA-mediated knockdown of PAX3 in SK-N-SH cells to show a 1.3-fold (P = 8 × 10−3) reduced binding at E2 and twofold (P = 4 × 10−4) reduced binding at E27 (Fig. 2).
Figure 2.
Identification of cognate TFs bound to RET enhancers. Genome map of the RET locus with locations of the E2 and E27 CREs together with ChIP-qPCR results using a PAX3 antibody in SK-N-SH cells shows enrichment of binding compared with the background. The specificity of binding is shown by siRNA knockdown of PAX3 with concomitant reduction in ChIP-qPCR signals at both CREs. (**) P < 0.001 for two technical replicates for three independent biological replicates (n = 6).
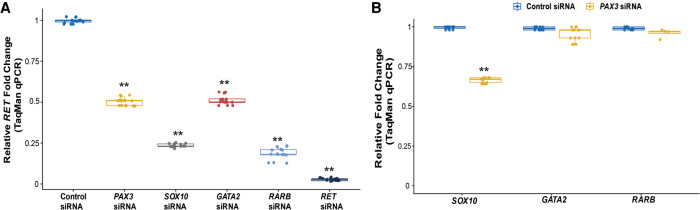
To further prove that PAX3 does indeed control RET, we also measured RET gene expression after siRNA-mediated knockdown of PAX3. As positive controls, we measured RET levels after siRNA-mediated knockdown of the established RET TFs SOX10, GATA2, and RARB. These experiments showed that decreasing PAX3 led to a 49% (P = 4 × 10−4) reduction in RET in comparison to 76% (P = 2.3 × 10−6), 50% (P = 3.1 × 10−3), and 81% (P = 4.1 × 10−5) decreases consequent to SOX10, GATA2, and RARB knockdown, respectively; as a control, knockdown of RET by its specific siRNA reduced its expression by 96% (P = 4.4 × 10−6) (Fig. 3A). We have previously shown that there is considerable cross talk between the established RET TFs (Chatterjee et al. 2016); hence, we measured gene expression of SOX10, GATA2, and RARB after siRNA-mediated knockdown of PAX3: We observed only a significant drop in SOX10 gene expression (32% decrease, P = 3 × 10−3); GATA2 and RARB levels were decreased but not significantly so (Fig. 3B).
Figure 3.
TF-mediated in vitro and in vivo effects on gene expression. (A) siRNA-mediated knockdown of PAX3, SOX10, GATA2, RARB, and RET in SK-N-SH cells decreases RET gene expression significantly. (B) siRNA-mediated knockdown of PAX3 has significant transcriptional effects on SOX10 but small yet statistically insignificant decreases on GATA2 and RARB. (**) P < 0.001 in three technical replicates for five independent biological replicates in all experiments.
In vivo evidence for RET enhancers
The human genetic evidence for HSCR-associated polymorphisms within CREs identified from in vitro (reporter activity) and ex vivo (siRNA in SK-N-SH) experiments can be buttressed by deletion analysis of each enhancer rather than knockdown of its cognate TF. To do so, we designed a single guide RNA close to each HSCR-associated SNP for all 10 target enhancers to introduce nonhomologous end joining–induced deletions in SK-N-SH cells. We screened five independently transfected pools of cells (wells) for each guide and detected, by Sanger sequencing, successful small deletions within all the CREs except E5. We used the Inference of CRISPR Edits (ICE) tool (Hsiau et al. 2019) to estimate that individual guides introduced deletions >3 bp in 10%–50% of the cells in all successfully targeted CREs in the pools of cells and none of the guides introduced deletions >10 bp (Supplemental Table S2).
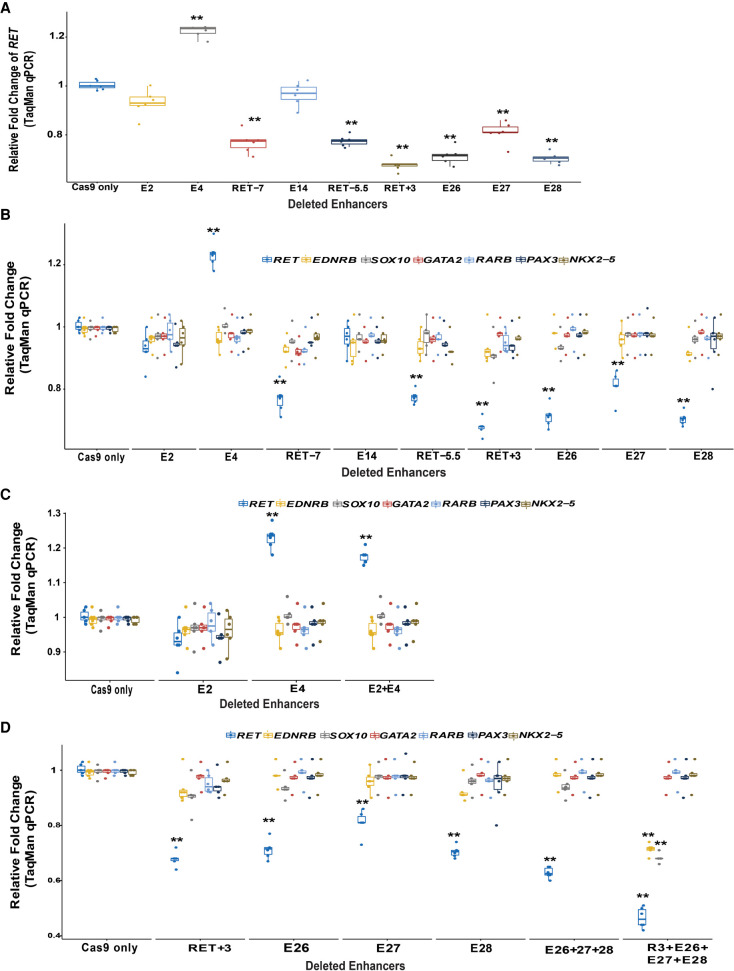
We subsequently measured RET expression in these enhancer-deleted cells. Our results show that except for enhancers E2 and E14, deletion of DNA sequences surrounding the HSCR-associated SNPs in all other CREs led to changes in RET expression. Thus, deletion of E4 (24%; P = 3.2 × 10−4) leads to higher expression, whereas deletion of E26 (28%; P = 3.7 × 10−4), E27 (19%; P = 1.2 × 10−3), and E28 (29%; P = 3.2 × 10−4) all lead to lower RET expression (Fig. 4A). The positive controls, RET-7 (22%; P = 1.3 × 10−3), RET-5.5 (22%; P = 2 × 10−3), and RET+3 (32%; P = 4.1 × 10−4), reduced RET gene expression as expected. All four intronic enhancers (RET+3, E26, E27, and E28) reside within a 8.5-kb region (Chr 10: 43,581,812–43,590,347) in the first intron of RET to control RET gene expression. Thus, these elements might comprise an enhanceosome critical for spatiotemporal expression of RET, which is also supported by the fact that the risk-associated variants at these sites are on a single haplotype (Fig. 1A).
Figure 4.
CRISPR-Cas9-induced deletions of RET enhancers with HSCR-associated variants reduce RET gene expression in vitro. (A) There is significant loss of RET gene expression from seven of nine CREs with small (≤10-bp) deletions centered on the variant site; only the E4 enhancer shows increased gene expression. (B) The expression of other RET GRN genes is unaffected by these CRE deletions likely owing to RET gene expression not decreasing below 50%. (C) Simultaneous deletion of enhancers E2 and E4 does not lead to any additional effect on RET gene expression compared with individual deletions. (D) Simultaneous deletions of E26/E27/E28 with and without deletion of RET+3 lead to a significantly greater decrease in RET gene expression compared with individual deletions. The deletion of all four enhancers also leads to decreased expression of EDNRB and SOX10. (**) P < 0.001 in two technical replicates each for three independent biological replicates in all experiments.
We next asked whether these changes in RET expression had concomitant changes in the expression of the remaining members of the RET GRN by quantifying expression of EDNRB, SOX10, GATA2, RARB, NKX2-5, and PAX3, the members of the GRN that express in this cell line, in the individual enhancer-deleted cells. Our results show no significant changes in gene expression (Fig. 4B). This result is not altogether unexpected given that GRN transcriptional dysregulation occurs only when RET gene expression decreases below 50% of its wild-type levels (Chatterjee et al. 2016; Chatterjee and Chakravarti 2019).
HSCR is associated with loss of RET function (Angrist et al. 1995; Pasini et al. 1995), and diminished Ret expression leads to loss of ENS during gut development in mice, the hallmark of HSCR (Uesaka et al. 2008; Chatterjee et al. 2019). Thus, we predicted that the cumulative effect of the disruption of all HSCR-associated CRE at E4, RET-7, RET-5.5, RET+3, E26, E27, and E28 should lead to reduced RET expression. To address this, we deleted multiple enhancers in close physical proximity to each other: (1) E2 and E4, which are within 200 bp of each other, and (2) E26, E27, and E28 with or without RET+3 deletion within the first intron of RET.
The pool of cells containing both the E2 and E4 deletion leads to an 18% (P = 2.8 × 10−3) increase in RET expression, which is not significantly different (P = 0.8) compared with the 24% increase with E4 deletion alone (Fig. 4C). Note that the individual deletion of E2 alone had no measurable effect on RET transcription (Fig. 4A,C), and it does not seem to affect the activity of its nearest variant-containing RET enhancer, E4. Second, the pool of cells with simultaneous deletion of E26, E27, and E28 led to a drop in RET expression by 40% (P = 3.8 × 10−4), which is not significantly lower than individual deletions of E26 or E28 but is 20% (P = 0.01) lower than the effect on RET owing to deletion of E27 alone (Fig. 4D). Additional deletion of RET+3 along with E26, E27, and E28 leads to a 56% (P = 3.6 × 10−4) drop in RET gene expression compared with Cas9-only cells. This 24%–37% greater loss of RET expression than from individual deletion of each enhancer (Fig. 4D) is a synergistic effect. Further, these joint enhancer deletions lead to RET expression decrease below 50% of wild-type expression, with a consequent loss of expression of EDNRB (28%, P = 2.1 × 10−3) and SOX10 (30%, P = 1.8 × 10−3) but no change in the expression of other TFs of the GRN.
Discussion
It is evident that a multiplicity of enhancers controls a gene's expression: This feature has many implications for complex disease genetics and its mechanisms. The data reported here, based on human genetics, siRNA, ChIP, and CRISPR-Cas9 deletion analyses in the SK-N-SH cell line, together with our prior studies (Kapoor et al. 2015; Chatterjee et al. 2016; Chatterjee and Chakravarti 2019; Tilghman et al. 2019), have identified 30 distinct CREs around RET with common sequence variants that are associated with HSCR. But only 10 CREs show allelic difference in enhancer activity. Thus, many risk allele–containing CREs are not causal for HSCR but may be for other phenotypes outside the ENS. CRISPR-Cas9 deletion analyses of these CREs show that at least seven of these have demonstrable effects on RET transcription through its control via the TFs SOX10, RARB, GATA2, and PAX3 and as-yet-unknown TFs (Table 3). Given that none of these experimental approaches are 100% efficient to capture all CREs, there are yet other enhancers that will regulate RET expression in the ENS, as evidenced by the presence of sequences with epigenetic signatures of enhancers in the human fetal gut, but that fail to act as enhancers in our assay (Fig. 1; Table 3). Conversely, the many enhancers at the RET locus, identified through in vitro analyses, do not imply that all of them are involved in transcriptional control of RET in the ENS as opposed to other RET-expressing tissues (mid- and fore-brain, kidney, and dorsal root ganglia). We are also unaware whether all these CREs have primary control on RET during ENS development (Chatterjee et al. 2016) or are merely shadow enhancers (Kvon et al. 2021).
Table 3.
Thirty-eight Hirschsprung disease (HSCR)–associated polymorphisms in six LD blocks, contained within 36 DNA elements at the RET locus annotated with respect to epigenetic marks (DNase I hypersensitivity [+a] in the SK-N-SH cell line, H3K4me1 [+b] marks in human fetal gut), luciferase reporter assays of alleles in the SK-N-SH cell line, allelic differences in luciferase assays in the SK-N-SH cell line, the TF binding the indicated regulatory element, and whether deletion of the element affected RET gene expression
Our multiple deletion experiments provide evidence that disruption of multiple variant-containing enhancers has synergistic effects on RET transcription and, hence, disease severity. Because many of these human enhancers do not have sequence conservation in mice, definitive in vivo proof of our hypothesis that disruption in multiple RET enhancers causes HSCR will require the creation of humanized mouse models with multiple regulatory variants at the same, which is far more feasible now using new methods of synthetic biology (Richardson et al. 2017) in addition to deletion screens using CRISPR-Cas9 genome editing. Nevertheless, this hypothesis is supported by the observation that the TFs that implicated RARB, GATA2, SOX10, and PAX3 binding to five of these enhancers have known roles in ENS development and HSCR (Bondurand et al. 2000; Lang et al. 2000; Lang and Epstein 2003). In other words, if the trans factors lead to HSCR and the cis factors that bind them are HSCR-associated, then these cis factors are direct risk factors of HSCR. Their distinct nature and LD relationships also suggest that they contribute independently to RET gene expression and, therefore, to HSCR.
The multiplicity of noncoding variants in CREs all controlling the same gene should give us pause in interpreting the effect size and functional effects of individual GWAS variants. As we have shown, the cumulative effect of all variant-bearing enhancers is to significantly lower RET gene expression to levels expected from RET coding mutations (Emison et al. 2005, 2010). As we have also shown, the largest risks are associated with haplotypes with multiple risk alleles, even across LD blocks (Chatterjee et al. 2016). This accumulation of multiple CRE variants is expected to lead to a larger effect through reduced binding of multiple TFs to multiple enhancers; given the role of multiple TFs on the promoter, this is likely synergistic. Note that RET also controls the gene expression of its own TFs PAX3, GATA2, and SOX10. This feedback may be a secondary but important cause of reduced RET gene expression, further exacerbating the enhancers’ effect. We do not yet know whether this diversity of genetic control with feedback is typical or not. RET is a highly dosage-sensitive gene with higher and lower than wild-type levels being associated with neuroendocrine tumors and aganglionosis, respectively. Extensive regulatory control is common for developmental genes (Bolt and Duboule 2020); hence, these genetic lessons are likely to be universal.
The human genetic implications of these data, beyond understanding HSCR, are that genetically independent SNPs at a specific GWAS locus are not the only candidate variants for understanding a phenotype. Additional SNPs may be involved, even those perfectly associated with one another, provided they affect functionally independent enhancers, as has been shown by the recent discovery of additional RET variants and CREs that control its expression (Fu et al. 2020; Kapoor et al. 2021). Additionally, multivariant disruption of gene expression has also been shown in other traits like the electrocardiographic QT interval (Kapoor et al. 2019) and expression of adiponectin, which is critical for glucose regulation (Spracklen et al. 2020), highlighting that this is a more widespread phenomenon.
Finally, the nature of gene regulation dictates that such regulatory control by the individual variant allele will also be quite varied (like increase in enhancer activity owing to risk alleles at enhancers E4, E27, and E28 compared with others) depending on activator versus repressor TFs and their coregulators, thereby decreasing or increasing target gene expression. This suggests that understanding the regulatory contributions to GWAS will require experimental data on enhancer effects beyond what statistical analysis can provide. Furthermore, we need broader enhancer screens to define the full enhancer architecture of RET and do so in vivo at different developmental stages and by sex. We also need to elucidate the full repertoire of TFs that regulate RET. These pieces of information are crucial to understand the full extent and composition of the RET–EDNRB GRN, which in turn will identify new genes that then become mutational targets of HSCR.
Methods
Cell lines
The human neuroblastoma cell SK-N-SH, purchased from ATCC (HTB-11), was grown under standard conditions (DMEM + 10% FBS and 1% penicillin–streptomycin). It was maintained in 10-cm culture dishes and passaged every 48 h when it reached ∼80% confluency.
ChIP-seq peak calling
Three epigenomic data sets generated from a 108-day human fetal large intestine, histone H3K27ac ChIP-seq (GSM1058765), histone H3K4me1 ChIP-seq (GSM1058775), and DNase-seq (GSM817188), were downloaded from the NIH Roadmap Epigenomics Project (Bernstein et al. 2010; The ENCODE Project Consortium et al. 2020). For the SK-N-SH cell line, DNase-seq data (GSM736559) were obtained from The ENCODE Project Consortium (2020). For each data set, MACS software v1.4 (Zhang et al. 2008) with default settings was used to call peaks at genomic sites where sequence reads were significantly enriched over background. With the default peak-calling threshold (P < 10−5), 51,771, 61,689, 66,930, and 52,534 genomic regions were identified in the GSM1058765, GSM1058775, GSM817188, and GSM736559 data sets, respectively.
Reporter assays
Four hundred nanograms of firefly luciferase vector (Promega Corporation pGL4.23) containing the DNA sequence of interest and 2 ng of Renilla luciferase vector (transfection control) were transiently transfected into the SK-N-SH cell line (5 × 104–6 × 104 cells/well), using 6 µL of FuGENE HD transfection reagent (Roche Diagnostic) in 100 µL of Opti-MEM medium (Invitrogen). Cells were grown for 48 h and luminescence measured using a dual luciferase reporter assay system on a Tecan multidetection system luminometer per the manufacturer's instructions.
ChIP-qPCR assays
A mammalian expression vector containing the full-length PAX3 cDNA (Origene SC309286) was transfected at 500 ng into SK-N-SH cells, and ChIP was performed 48 h post transfection, thrice independently, using 1 × 106 SK-N-SH cells using the EZ-Magna ChIP kit (MilliporeSigma) per the manufacturer's instructions, with the following modifications: The chromatin was sonicated for 30 sec on and 30 sec off for 10 cycles; sheared chromatin was preblocked with unconjugated beads for 4 h; and specific antibodies separately conjugated to the beads for 4 h before immunoprecipitation were performed with the preblocked chromatin. A polyclonal antibody was used against PAX3 (Invitrogen 16HCLC) at 15 µg concentration. ChIP assays were also performed on cells 48 h after transfection with the PAX3 siRNAs (Dharmacon/Horizon Discovery L-012399-00-0005) at 25 nM to assess specificity of TF binding. qPCR assays were performed using SYBR Green (Thermo Fisher Scientific) and specific primers against enhancer E2 (E2_FWD 5′-GCTGCAGATATGCAACTTCCAA-3′ and E2_REV 5′-AGATATGCTGGTGAGGGGCT-3′) and enhancer E27 (E27_FWD 5′-AGGAAGGTAGGCACCCTGTA-3′ and E27_REV 5′-AGCCCTGTGTTAACTGTCCG-3′). The data were normalized to input DNA, and enrichment was calculated by fold excess over ChIP performed with specific IgG as background signal. All assays were performed in triplicate for each independent ChIP assay (n = 6).
siRNA assays
PAX3 (L-012399-00-0005), RET (L-003170-00-0005), SOX10 (L-017192-00), GATA2 (L-009024-02), and RARB (L-003438-02) SMARTpool siRNAs (combination of four distinct siRNAs targeting each gene; Dharmacon/Horizon Discovery) were transfected at 20 nM in SK-N-SH cells at a density of 104–105 cells using the FuGENE HD transfection reagent (Promega Corporation) per the manufacturer's instructions. The ON-TARGET plus nontargeting siRNAs (D-001810-10, negative control) was always transfected at 25 nM concentration.
Gene expression assays
Total RNA was extracted from SK-N-SH cells using TRIzol (Thermo Fisher Scientific) and cleaned on RNeasy columns (Qiagen). Five hundred nanograms of total RNA was converted to cDNA using SuperScript III reverse transcriptase (Thermo Fisher Scientific) using oligo(dT) primers. The diluted (1/5) total cDNA was subjected to TaqMan gene expression (Thermo Fisher Scientific) using the following transcript-specific probes and primers: RET (Hs01120032_m1), EDNRB (Hs00240747_m1), PAX3(Hs00992437_m1), SOX10 (Hs00366918_m1), GATA2 (Hs00231119_m1), and RARB (Hs00977140_m1). Human ACTB was used as an internal loading control for normalization.
For siRNA knockdown experiments, five independent wells of SK-N-SH cells were used for RNA extraction, and each assay was performed in triplicate (n = 15). Relative fold change was calculated based on the 2ΔΔCt (threshold cycle) method. For siRNA experiments, 2ΔΔCt for negative control nontargeting control siRNA was set to unity. P-values were calculated from pairwise two-tailed t-tests.
CRISPR-Cas9-induced deletions
Each enhancer region centered on a polymorphic site was targeted using a single guide RNA (Supplemental Table S3) by transfecting a ribonucleoprotein complex containing 100 pmol of specific gRNA, coupled with 5 µg/µL of TrueCut Cas9 nuclease (Thermo Fisher Scientific) in Lipofectamine CRISPRMAX solution (Thermo Fisher Scientific). For each enhancer, three wells containing approximately 30,000 SK-N-SH cells were independently transfected. To increase the efficiency of deletion, we retransfected the cells with the same ribonucleoprotein mix a second time after 72 h. The cells were further grown for 48 h and then equally split into two tubes for DNA and RNA extraction. To confirm disruption of the enhancer regions, specific primers (Supplemental Table S4) were used to verify deletions by PCR followed by Sanger sequencing (Supplemental Figs. S2, S3). We used the ICE tool (Hsiau et al. 2019) to estimate the percentage of cells carrying various insertion/deletions (indels).
For multiple deletion experiments, the guides targeting E2 and E4 and guides targeting E26, E27, and E27 along with RET+3 were transfected simultaneously in the cells and experiments repeated as stated above. The data represent the percentage of cells in the pool of cells in which indels have been detected in the enhancer(s).
Estimating haplotype-specific HSCR risk
Genotypes at 10 RET CRE variants in 220 S-HSCR cases and 503 European ancestry controls were obtained from our published HSCR GWAS (Jiang et al. 2015) and The 1000 Genomes Project Consortium (2015), respectively. Haplotypes were generated from unphased genotypes using Beagle (Browning and Browning 2007) and were filtered to retain only those that had a frequency >1% in controls. Standard methods using χ2 statistics were used to calculate haplotype-count-based OR, their upper and lower confidence limits, and significance of their deviation from the null hypothesis of no association (OR = 1) (Kapoor et al. 2015).
For replication study, we looked at an independent patient cohort of 235 S-HSCR cases on which we have performed genotyping (Kapoor et al. 2021) and used reported allele frequencies from 9400 European ancestry controls in the gnomAD (Karczewski et al. 2020). In our new cases, rs2506030 was not genotyped so we used allele frequencies of rs788260, which is in near perfect LD (r2 = 0.988). Similarly, rs2505541 was not genotyped, and hence, we used rs2506024, which is in perfect LD with it (r2 = 1).
LD analyses
LD between all 38 SNPs was estimated using a previously described method (Gabriel et al. 2002) and plotted using Haploview (Barrett et al. 2005) with its default settings. In brief, 95% confidence bounds on D prime are generated, and each comparison is called “strong LD,” “inconclusive,” or “strong recombination.” A block is created if 95% of informative (i.e., noninconclusive) comparisons are “strong LD.” This method by default ignores markers with MAF < 0.05.
Identifying TFs for candidate enhancers
We searched for TFBSs within all putative CREs using FIMO (Bailey et al. 2009; Grant et al. 2011) and 890 validated TF motifs in TRANSFAC (Wingender et al. 1996). We used the setting of “minimize false positives” and a stringent cut off of P < 10−4 to identify candidate cognate TFs.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health (Eunice Kennedy Shriver National Institute of Child Health and Human Development) R01 award HD028088 to A.C.
Author contributions: S.C. and A.C. conceived and designed the study. K.M.K. conducted all in vitro luciferase assays, A.K. conducted all genotyping assays, and S.C. and L.E.F. conducted all in vivo CRISPR assays. S.C. and A.C. wrote and edited the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275667.121.
Competing interest statement
The authors declare no competing interests.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrist M, Bolk S, Thiel B, Puffenberger EG, Hofstra RM, Buys CH, Cass DT, Chakravarti A. 1995. Mutation analysis of the RET receptor tyrosine kinase in Hirschsprung disease. Hum Mol Genet 4: 821–830. 10.1093/hmg/4.5.821 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol 28: 1045–1048. 10.1038/nbt1010-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt CC, Duboule D. 2020. The regulatory landscapes of developmental genes. Development 147: dev171736. 10.1242/dev.171736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Le Caignec C, Wegner M, Goossens M. 2000. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 9: 1907–1917. 10.1093/hmg/9.13.1907 [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81: 1084–1097. 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Turner TN. 2016. Revealing rate-limiting steps in complex disease biology: the crucial importance of studying rare, extreme-phenotype families. Bioessays 38: 578–586. 10.1002/bies.201500203 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Ahituv N. 2017. Gene regulatory elements, major drivers of human disease. Annu Rev Genomics Hum Genet 18: 45–63. 10.1146/annurev-genom-091416-035537 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Chakravarti A. 2019. A gene regulatory network explains RET–EDNRB epistasis in Hirschsprung disease. Hum Mol Genet 28: 3137–3147. 10.1093/hmg/ddz149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Kapoor A, Akiyama JA, Auer DR, Lee D, Gabriel S, Berrios C, Pennacchio LA, Chakravarti A. 2016. Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell 167: 355–368.e10. 10.1016/j.cell.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Nandakumar P, Auer DR, Gabriel SB, Chakravarti A. 2019. Gene- and tissue-level interactions in normal gastrointestinal development and Hirschsprung disease. Proc Natl Acad Sci 116: 26697–26708. 10.1073/pnas.1908756116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. 2005. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434: 857–863. 10.1038/nature03467 [DOI] [PubMed] [Google Scholar]
- Emison ES, Garcia-Barcelo M, Grice EA, Lantieri F, Amiel J, Burzynski G, Fernandez RM, Hao L, Kashuk C, West K, et al. 2010. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet 87: 60–74. 10.1016/j.ajhg.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu AX, Lui KN, Tang CS, Ng RK, Lai FP, Lau ST, Li Z, Garcia-Barcelo MM, Sham PC, Tam PK, et al. 2020. Whole-genome analysis of noncoding genetic variations identifies multiscale regulatory element perturbations associated with Hirschsprung disease. Genome Res 30: 1618–1632. 10.1101/gr.264473.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Conant D, Rossi N, Maures T, Waite K, Yang J, Joshi S, Kelso R, Holden K, Enzmann BL, et al. 2019. Inference of CRISPR Edits from Sanger trace data. bioRxiv 10.1101/251082 [DOI] [PubMed]
- Inoue F, Kreimer A, Ashuach T, Ahituv N, Yosef N. 2019. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell 25: 713–727.e10. 10.1016/j.stem.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Arnold S, Heanue T, Kilambi KP, Doan B, Kapoor A, Ling AY, Sosa MX, Guy M, Jiang Q, et al. 2015. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with Ret are critical to Hirschsprung disease liability. Am J Hum Genet 96: 581–596. 10.1016/j.ajhg.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Jiang Q, Chatterjee S, Chakraborty P, Sosa MX, Berrios C, Chakravarti A. 2015. Population variation in total genetic risk of Hirschsprung disease from common RET, SEMA3 and NRG1 susceptibility polymorphisms. Hum Mol Genet 24: 2997–3003. 10.1093/hmg/ddv051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Lee D, Zhu L, Soliman EZ, Grove ML, Boerwinkle E, Arking DE, Chakravarti A. 2019. Multiple SCN5A variant enhancers modulate its cardiac gene expression and the QT interval. Proc Natl Acad Sci 116: 10636–10645. 10.1073/pnas.1808734116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Nandakumar P, Auer DR, Sosa MX, Ross H, Bollinger J, Yan J, Berrios C, Hirschsprung Disease Research Collaborative (HDRC), Chakravarti A. 2021. Multiple, independent, common variants at RET, SEMA3 and NRG1 gut enhancers specify Hirschsprung disease risk in European ancestry subjects. J Pediatr Surg S0022-3468(21)00312-2. 10.1016/j.jpedsurg.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Waymack R, Gad M, Wunderlich Z. 2021. Enhancer redundancy in development and disease. Nat Rev Genet 22: 324–336. 10.1038/s41576-020-00311-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Epstein JA. 2003. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet 12: 937–945. 10.1093/hmg/ddg107 [DOI] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. 2000. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest 106: 963–971. 10.1172/JCI10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan D, Marcos-Gutierrez C, Pachnis V, de Graaff E. 2002. Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development 129: 5151–5160. 10.1242/dev.129.22.5151 [DOI] [PubMed] [Google Scholar]
- Pasini B, Borrello MG, Greco A, Bongarzone I, Luo Y, Mondellini P, Alberti L, Miranda C, Arighi E, Bocciardi R, et al. 1995. Loss of function effect of RET mutations causing Hirschsprung disease. Nat Genet 10: 35–40. 10.1038/ng0595-35 [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CL, Chandrasegaran S, Cai Y, et al. 2017. Design of a synthetic yeast genome. Science 355: 1040–1044. 10.1126/science.aaf4557 [DOI] [PubMed] [Google Scholar]
- Spracklen CN, Iyengar AK, Vadlamudi S, Raulerson CK, Jackson AU, Brotman SM, Wu Y, Cannon ME, Davis JP, Crain AT, et al. 2020. Adiponectin GWAS loci harboring extensive allelic heterogeneity exhibit distinct molecular consequences. PLoS Genet 16: e1009019. 10.1371/journal.pgen.1009019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman JM, Ling AY, Turner TN, Sosa MX, Krumm N, Chatterjee S, Kapoor A, Coe BP, Nguyen KH, Gupta N, et al. 2019. Molecular genetic anatomy and risk profile of Hirschsprung's disease. N Engl J Med 380: 1421–1432. 10.1056/NEJMoa1706594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka T, Nagashimada M, Yonemura S, Enomoto H. 2008. Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J Clin Invest 118: 1890–1898. 10.1172/JCI34425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 2017. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. 1996. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24: 238–241. 10.1093/nar/24.1.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.