Abstract
Base editors are dedicated engineered deaminases that enable directed conversion of specific bases in the genome or transcriptome in a precise and efficient manner, and hold promise for correcting pathogenic mutations. A major concern limiting application of this powerful approach is the issue of off-target edits. Several recent studies have shown substantial off-target RNA activity induced by base editors and demonstrated that off-target mutations may be suppressed by improved deaminases versions or optimized guide RNAs. Here, we describe a new class of off-target events that are invisible to the established methods for detection of genomic variations and were thus far overlooked. We show that nonspecific, seemingly stochastic, off-target events affect a large number of sites throughout the genome or the transcriptome, and account for the majority of off-target activity. We develop and employ a different, complementary approach that is sensitive to the stochastic off-target activity and use it to quantify the abundant off-target RNA mutations due to current, optimized deaminase editors. We provide a computational tool to quantify global off-target activity, which can be used to optimize future base editors. Engineered base editors enable directed manipulation of the genome or transcriptome at single-base resolution. We believe that implementation of this computational approach would facilitate design of more specific base editors.
Base editors are dedicated engineered deaminases that enable directed conversion of specific bases in the genome or transcriptome in a precise and efficient manner and hold promise for correcting pathogenic mutations (Komor et al. 2016; Rees and Liu 2018). A major concern limiting application of this powerful approach is the issue of off-target edits (Gehrke et al. 2018; Zuo et al. 2019). Several recent studies (Rees et al. 2019; Zhou et al. 2019; Grünewald et al. 2019a) have shown substantial off-target RNA activity induced by base editors and demonstrated that off-target mutations may be suppressed by improved deaminases versions or optimized guide RNAs. These studies have employed one of the established methods to find genomic variations reoccurring in multiple copies of the edited transcript (McKenna et al. 2010; Garrison and Marth 2012), which are well-suited for detection of genomic polymorphisms and reoccurring mutations but are insensitive to weakly edited sites. Accordingly, these studies have mainly focused on specific off-target sites, where the guided deaminase binds efficiently and edits a substantial fraction of DNA/RNA molecules.
Endogenous deaminases (on which the engineered versions are based) are known to induce abundant low-level RNA modifications (Rosenberg et al. 2011; Bazak et al. 2014a). In fact, most ADAR (also known as ADAR1) A-to-I mRNA editing activity occurs at sites edited up to a few percent level or even lower (Bazak et al. 2014a) and is therefore undetectable in a single sample. It is only natural to ask whether engineered deaminases, too, exhibit nonspecific, seemingly stochastic, off-target activity in addition to the previously studied specific off-target sites. As is the case with endogenous base editors, the nonspecific activity may affect a large number of sites throughout the genome or the transcriptome, so that, although these sites are edited at a low probability per site per molecule, nonspecific deamination events may outnumber the specific ones (Fig. 1A).
Figure 1.
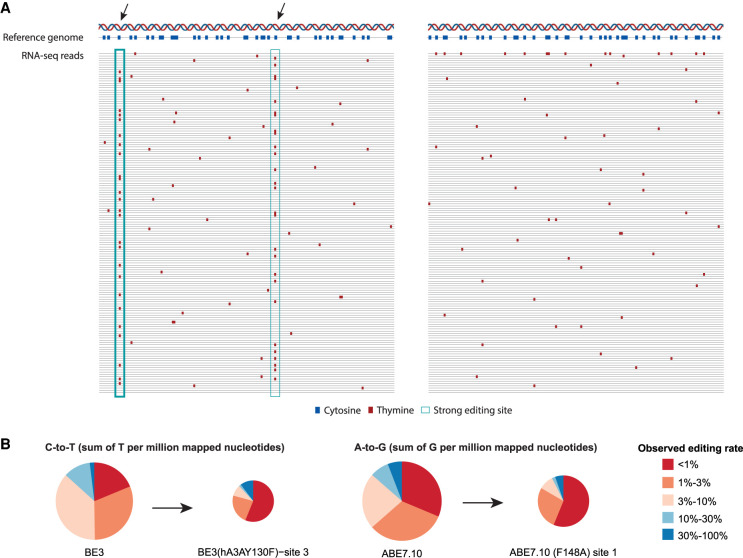
Most RNA mutations due to off-target base editing are nonspecific. (A) Engineered guided deaminases may target efficiently some off-target locations (marked with arrows). These strongly edited sites result in a DNA-RNA mismatch seen in a large fraction of RNA molecules, resembling the mismatch profile observed at genomic polymorphism and clonally selected somatic mutation sites. In parallel, nonspecific off-target base editing activity affects multiple additional sites but a small fraction of RNA molecules per site. However, due to their large number, the total number of nonspecific events could surpass that of specific editing sites, as illustrated in the figure. Hyperedited reads, where multiple mismatches are seen in the same read (right panel, top read), occasionally appear. They provide a strong indication of off-target activity but account for a small minority of mismatches. (B) Relative contribution of editing sites to the editing index, by their observed editing rates, for two engineered base editors. Most off-target activity occurs at weakly edited sites in agreement with the scenario depicted in panel A. Pie area is proportional to the number of detected off-target events. Four base editors from Study H are shown (two cytidine deaminases: BE3, BE3 [hA3AY130F]−site 3; two adenine base editors: ABE7.10 and ABE7.10 [F148A]−site 1). See Supplemental Figure S1 for similar data for all enzymes.
Global quantification methods have been devised to study endogenous deaminases activity, including nonspecific ones (Bazak et al. 2014b; Roth et al. 2019). Inspired by this approach, we develop here a method to quantify the global variation rate induced by engineered base editors, including low-level variations that cannot be individually resolved. We apply it to various state-of-the-art base editors, either DNA or RNA editors, engineered ADAR and APOBEC, or other deaminases, including ones that are fused to CAS proteins or to other guiding systems (Supplemental Table S1; Vallecillo-Viejo et al. 2018; Vogel et al. 2018; Abudayyeh et al. 2019; Katrekar et al. 2019; Rees et al. 2019; Zhou et al. 2019; Zuo et al. 2019; Grünewald et al. 2019a,b; Doman et al. 2020; Lee et al. 2020; Yu et al. 2020).
Results
Most RNA mutations due to off-target base editing are nonspecific
Genomic variations of interest typically occur in a sizable fraction of chromosome copies. For example, heterozygous polymorphisms are present in half of the molecules, and cancer-related somatic mutations of interest are those that have been selected and appear in multiple clonal copies. Methods for calling variations and mutations based on DNA sequencing data (McKenna et al. 2010; Garrison and Marth 2012) use this fact to filter out sequencing and alignment technical noise and analyze multiple alignment of many DNA reads to the reference genome, looking for reoccurring mismatches. At typical read coverage, these approaches are insensitive to low-level variations that affect only a small fraction of the transcript copies and are thus almost indistinguishable from the technical noise level (Martincorena et al. 2018).
Most of the endogenous editing activity occurs in such low-level editing sites that are “invisible” to standard variant calling methods. Looking at several examples of engineered base editors, we observe a similar phenomenon. In addition to specific off-target sites where the guided deaminase binds efficiently and edits a sizable fraction of DNA/RNA molecules, most off-target activity is nonspecific and seemingly stochastic, affecting a large number of sites at a low editing level. Whereas these sites are edited at a low probability per site per molecule, nonspecific deamination events outnumber the specific ones. Recent optimization efforts have considerably lowered the volume of nonspecific off-target activity, but many state-of-the-art base editors exhibit sizable amounts of RNA off-targets in weakly edited sites (Fig. 1B; Supplemental Fig. S1).
Technical errors in the sequencing process are still a major source for deviations between the original biological information and the output sequencing reads (Zaranek et al. 2010; Alioto et al. 2015). Therefore, a single read supporting an isolated mismatch cannot be reliably attributed to deaminase activity and may result from sequencing errors. However, endogenous deaminases often edit multiple sites in the same molecule (hyperediting), resulting in clusters of mismatches of the same type that may be identifiable even at the single-read level (Carmi et al. 2011). Scanning RNA-seq data representing dozens of recently developed base editors (Supplemental Table S1; Vallecillo-Viejo et al. 2018; Vogel et al. 2018; Abudayyeh et al. 2019; Katrekar et al. 2019; Rees et al. 2019; Zhou et al. 2019; Grünewald et al. 2019a,b; Yu et al. 2020), we found in some of the samples up to a million off-target hyperediting sites (Supplemental Fig. S2A) in excess of the endogenous A-to-I editing signal observed in control samples (Porath et al. 2014). Only a small fraction of these were previously identified for the same samples (Supplemental Fig. S2B), indicating that the classical SNV detection schemes may overlook a large fraction of sites (see also Supplemental Table S6). Note that, for some editors, no excess of hyperediting sites is observed. However, the hyperediting analysis is not sensitive enough and the clusters of sites it finds may be just the tip of the iceberg, attesting for a much wider nonclustered editing activity that is overlooked by standard variant-calling methods. Therefore, a different approach is required to explore nonspecific off-target activity.
In the following, we apply global quantification methods for the editing activity, which are complementary to the above-mentioned variant-calling approaches, to reveal the full scope of off-target editing activity.
Editing activity is globally enhanced following introduction of base editors
To quantify the total off-target activity, we follow an approach developed previously for studying global endogenous RNA editing that takes into account the loads of editing activity occurring at low levels (Roth et al. 2019). Briefly, we apply a strict alignment approach and look at all mismatches to the reference genome, not trying to determine whether each one of them can reliably be attributed to deaminase activity. We then compare the editing index, the average mismatch level weighted by expression level, between samples (Methods). Clearly, this approach includes contributions from sequencing and other errors. However, the excess signal seen in samples expressing the deaminases over the baseline (control) signal attests to global editing activity.
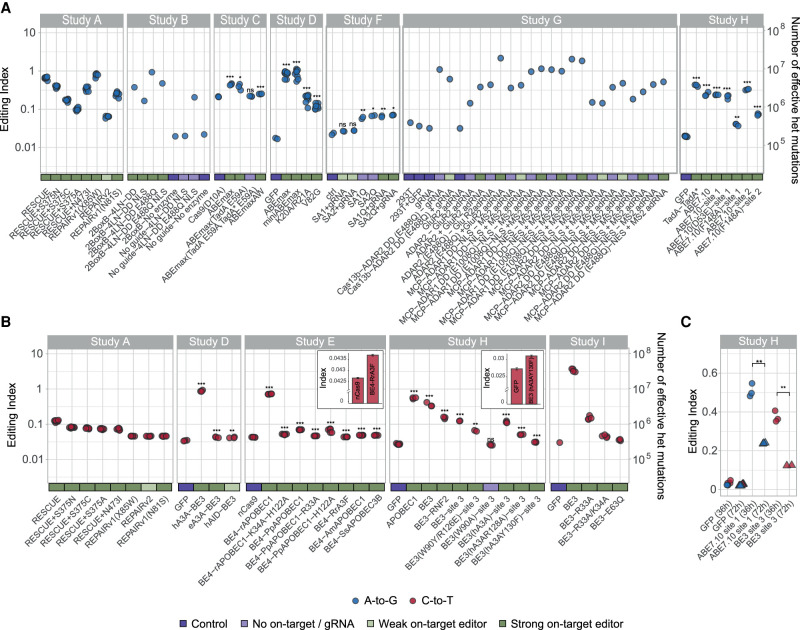
Applying this approach, we find a statistically significant excess for 35 out of the 37 active enzymes analyzed, indicating widespread off-target RNA editing for current best optimized A-to-I (Fig. 2A) and C-to-U (Fig. 2B) DNA and RNA base editors. Even for adenosine base editors, where endogenous A-to-I activity is known to be widespread (Roth et al. 2019), the effect of the engineered deaminase is clearly noticeable. This excess is seen genome-wide (Supplemental Fig. S3) as well as in coding sequence regions. The sites harboring the mismatches exhibit a clear sequence motif, supporting their being targeted by the base editors (Supplemental Fig. S4). Of the enzymes screened in this work, adenosine base editors seem to be noisier, showing up to 0.1% off-target editing in some of the optimized deaminases (i.e., one in a thousand adenosines is deaminated into an inosine). As expected, there is only a little overlap between the sites contributing to the global A-to-I signal and those detected by previous methods which are designed to locate specific editing sites. The excess signal comes mostly from weakly edited sites which are usually invisible to standard SNV detection tools (Supplemental Fig. S1). Studies B and D provide matched data for RNA base editors with and without a nuclear localization signal (NLS). Enzymes containing NLS exhibit reduced nonspecific off-target levels compared with matched enzymes containing a nuclear export signal (NES) (Supplemental Fig. S5), as they have lower chances to meet other mRNAs.
Figure 2.
Editing activity is globally enhanced following introduction of base editors. The editing index is a global measure of editing activity, quantifying the fraction (percent) of the RNA nucleotides exhibiting a DNA-RNA mismatch (i.e., A-to-G index of 1 means that 1% of the RNA nucleotides mapped to a genomic adenine are guanosines). (A) For adenine base editors (Studies A, B, C, D, F, G, H), the A-to-G index (blue circles) per sample over the coding sequence is presented (see Supplemental Fig. S3 for whole-genome calculation). In almost all cases tested, the index is significantly elevated for base editors compared to the control samples. Two-sided t-test for log(index); (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. Significance was not assessed for Study A (did not include untreated controls) and Studies B and G (only one replicate per condition). The two cases in Study F that do not show a significant difference exhibit weak on-target activity as well. In order to appreciate the significance of the high index values obtained, the index values are translated into an equivalent number of heterozygous mutations (Methods), right axis. Note that the index cannot be directly compared between samples of different reads’ lengths (Methods). (B) Same as A; C-to-U index (red) for cytidine deaminase samples (Studies A, D, E, H, I). (C) Samples sequenced 36 h posttreatment show a two- to threefold higher level of induced mutations compared to ones sequenced 72 h posttreatment (as are all samples in panels A,B). The data per sample are available in Supplemental Table S2. Exact P-values are presented in Supplemental Table S3.
Adenosine base editors target preferentially the endogenous targets of the ADAR enzymes. The vast majority of endogenous A-to-I editing occurs within the million Alu copies in the genome (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004). Consistently, the index calculated over Alu sequences is considerably elevated for all adenosine base editors (Supplemental Fig. S3; cf. Supplemental Tables S7, S8) and, in some cases, is as high as 10%, namely 1/10 of all Alu adenosines are deaminated. In contrast, editing of well-covered endogenous recoding sites, mostly edited by ADARB1 (also known as ADAR2) (Tan et al. 2017), is not generally elevated (Supplemental Fig. S6; cf. Supplemental Table S9).
The deamination rates observed in the coding regions of the human genome (excess of index over the control baseline level) vary considerably across the different active base editors and range between 0.004% (Study G, BE3 [hA3AY130F]–site 3) and 3% (Study G, BE3) (Fig. 2). Note that these rates are based on RNA collected 72 h posttreatment. The deamination rate is actually two- to threefold higher 36 h posttreatment (Fig. 2C). Study E provides the expression level of the enzymes. The index values observed are not correlated to the expression level. In addition, we did not find a significant correlation between on-target and off-target editing levels for any of the studies for which on-target data are available (A, C, D, E, F, G).
In terms of the total transcriptome mutation load, the observed range of rates (0.004%–3%) is equivalent to a range of 675 up to 658,000 heterozygous genomic mutations in the coding sequence alone (Fig. 2). The nonsynonymous to synonymous ratio, the prevalence of nonconservative amino acid substitutions, the distribution over genes with different expression levels, the level in essential genes and oncogenes, and the occurrence of known harmful mutations (Supplemental Fig. S7; cf. Supplemental Table S10) are all consistent with these nonspecific off-target editing events being spread randomly and uniformly over the coding sequence of all genes.
Endogenous genetic information flow is not perfect and introduces errors at all levels. The sources of information are more tightly controlled than the end-point products. Thus, the genomic information itself is most tightly regulated, with replication error rates as low as ∼10−10 mutations per base pair per cell division (Drake et al. 1998), whereas protein production, the end point of this process, can tolerate error rates as high as 10−4 (Mordret et al. 2019). Due to the transient nature of RNA, transcriptional fidelity is much lower than replication but still much higher than translation, with error rates estimated to be 4 × 10−6 (Gout et al. 2013, 2017) in eukaryotic cells. The evolutionary pressure to optimize eukaryotic transcription fidelity, in comparison to the bacterial one which is an order of magnitude higher (Traverse and Ochman 2016), suggests that higher rates of RNA substitutions are detrimental. Accordingly, the off-target activity identified here, which even for the best active base-editor analyzed here is an order of magnitude higher than the endogenous eukaryotic error rate, could be potentially harmful.
Low-level nonspecific base editing affects only a small fraction of the copies of each protein. For the low end of the deamination rates (0.004%), a typical 1000-bp-long coding mRNA sequence harboring ∼250 cytosines will show an off-target deamination in only 1% of its copies. For highly expressed genes, where multiple copies of the transcript exist in each cell, the effect would probably be minimal. However, thousands of genes are expressed at a level of 1–2 transcripts per cell or less (Melé et al. 2015). Thus, each cell would have dozens of genes for which the only transcript in the cell is mutated. Furthermore, a low level of RNA mutations may have a harmful impact even if most copies of the transcript are not affected. Furthermore, 3.7% of A-to-G and 4.7% of C-to-T edits in coding regions are expected to create neoantigens (Methods) that can provoke the immune system to attack self-tissue. In addition, accumulation of misfolded proteins may lead to aggregation, a key contributor to neurodegenerative diseases.
Further support for the potential damaging effect of low-level off-target RNA editing comes from the observation that the index over A-to-G harmful mutation sites is somewhat lower than the overall index (Supplemental Fig. S7D, blue). This depletion may indicate that weak off-target editing of these critical adenosines by the endogenous A-to-I editor is deleterious and therefore selected against. A similar effect was recently reported for mammalian housekeeping genes and viruses that adapted to avoid editing by endogenous APOBEC enzymes (Chen and MacCarthy 2017).
Finally, we have looked for signs of a deleterious effect at the cellular level and used the GSEA tool and the hallmark gene-set (Liberzon et al. 2015) to look for overexpression of the apoptosis related gene-group (Methods). Significant overexpression is observed in 28 of the 33 enzymes analyzed (Supplemental Table S11). For example, CDKN1A (also known as p21) is a key DNA damage-inducible protein whose transcriptional induction can occur dependent on TP53 and is considered an archetype of the cell response to genotoxic damage. We find significantly elevated expression of CDKN1A for most enzymes analyzed (Supplemental Fig. S8; Supplemental Table S11). These results further support the need to control the level of off-target activity and to understand its cellular effects.
Off-target DNA editing activity following introduction of base editors
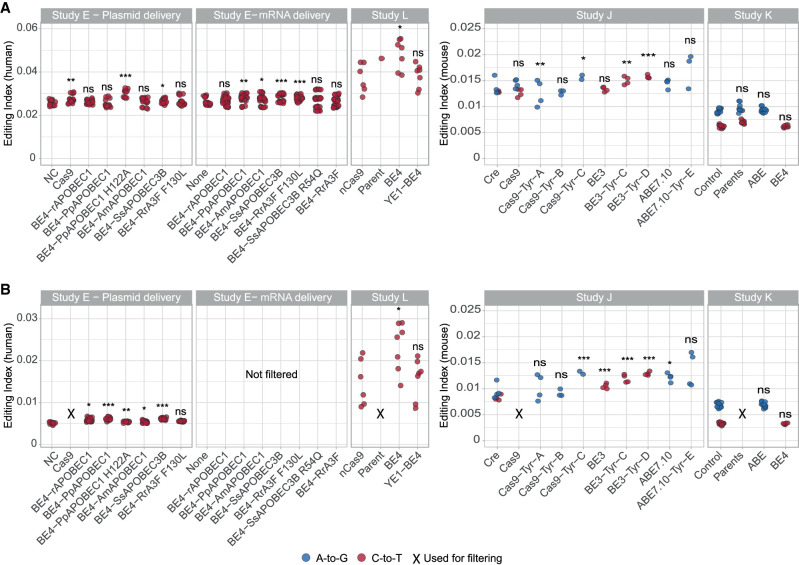
The same approach may be used to quantify off-target activity not only in the transcriptome but also in the genome. We have analyzed four recent studies for which DNA-seq data were available (Zuo et al. 2019; Doman et al. 2020; Lee et al. 2020; Yu et al. 2020) and found detectable off-target DNA editing activity for nine of the 22 enzymes studied (Studies D, K, I, J) (Fig. 3A). Removing putative polymorphisms in the untreated samples (see Methods) improves the sensitivity and reveals a significant signal for DNA off-target activity in 10 of the 15 enzymes (Fig. 3B).
Figure 3.
Off-target DNA editing activity following introduction of base editors. (A) The editing index is a global measure of editing activity, quantifying the fraction (percent) of the DNA nucleotides exhibiting a mismatch with respect to the reference genome. Common polymorphic sites are excluded. Four studies are analyzed (E, L, J, K), examining adenine base editors and cytidine deaminases. For each enzyme, the relevant indices (A-to-G, blue; and C-to-T, red) are calculated over the coding region. The relevant index (A-to-G and C-to-T for adenine and cytidine deaminases, respectively) is compared with control samples (Study E: NC/None; Study L: nCas9; Study J: Cre; Study K: Control). A significant increase is observed for nine enzymes. (B) In order to suppress baseline contributions to the index due to genomic polymorphisms unrelated to the base editor, we repeated the calculation excluding mismatch sites appearing in at least half of the untreated samples not used for the statistical tests (Study E: Cas9; Study L: parent; Study J: Cas9; Study K: parents). Ten of the 15 examined editors exhibit a significant increase in the index, indicating off-target DNA editing. In Study E (mRNA delivery), there were no samples suitable for excluding shared SNVs, and therefore filtering was not applied. Two-sided t-test for log(index); (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. The data per sample are available in Supplemental Table S4, and exact P-values are presented in Supplemental Table S5.
The absolute level of the excess index, representing off-target DNA mutations, is 2.2–4.6 × 10−5 (Fig. 3), lower than the one observed in RNA. However, it is still orders of magnitude higher than the natural DNA polymerase error rate of ∼10−10, and the impact of these heritable mutations is much more severe. To put it into a physiological context, one may compare the base editors’ off-target activity to ionizing radiation, a different heavily studied source of mutations. The mutation load due to (low rate) radiation is estimated to be 7.3 ± 0.8 × 10−6 mutations/bp/cell/Sievert (Russell and Kelly 1982a,b). Note that our detection limit for mutation rate is roughly 10−5, equivalent to ∼1 Sievert, and is still much higher than the accepted ionizing radiation safety limit. Thus, the mutation load detected in some of the editors calls for a more accurate sequencing and quantification methods to assess the risk due to off-target DNA editing, even if it is too weak a signal to be detected using current standard sequencing protocols and our approach.
Discussion
We present here a new method to quantify nonspecific off-target activity. Off-target RNA mutations are found to be abundant even for current optimized deaminase editors, and some of these editors result in abundant DNA mutations as well. We provide a computational tool (https://github.com/a2iEditing/BEIndexer) to quantify global off-target activity, which can be used to optimize future base editors.
Note that this approach does not replace currently used methods, which are designed to identify specific off-target sites, but is presented as a complementary approach, focusing on a different manifestation of off-target activity. This method reveals varying levels of appreciable off-target activity induced by state-of-the-art base editors. It is not straightforward to compare the base editors using these data, gathered under varying experimental conditions. However, these results emphasize the need for further optimization of base editors with respect to their nonspecific off-target activity, genomic and transcriptomic. The approach presented here may be utilized for these optimization efforts. In addition, cutoff values for tolerable RNA and DNA mutation rate in the context of genetic therapy should be established.
Methods
Sequencing data and alignment
To analyze off-target editing of RNA by recently developed base editors, we downloaded RNA-seq data from seven different studies (Supplemental Table S1; Vallecillo-Viejo et al. 2018; Vogel et al. 2018; Abudayyeh et al. 2019; Katrekar et al. 2019; Zhou et al. 2019; Grünewald et al. 2019a,b; Yu et al. 2020). In total, we analyzed 306 RNA-seq samples, representing 48 adenosine base editors, 29 cytidine base editors, and controls. Details are provided in Supplemental Table S1. RNA-seq reads were aligned to the human (hg38) genome using STAR v2.6.0 (default parameters) (Dobin et al. 2013), keeping only uniquely aligned reads.
To analyze genomic off-target editing by recently developed base editors, we downloaded DNA-seq data from four recently published studies (Supplemental Table S1; Zuo et al. 2019; Doman et al. 2020; Lee et al. 2020; Yu et al. 2020). In total, we analyzed 493 DNA-seq samples, representing three base editors (BE3, BE4, and ABE7.10), and controls. DNA-seq reads were uniquely aligned to the mouse (mm10) or human (hg38) genomes using STAR v2.6.0 (default parameters, except of alignIntronMin = 2, scoreDelOpen = −10,000, scoreInsOpen = −10,000 in order to avoid spliced alignments) (Dobin et al. 2013).
It is worth noting that Studies E and L have applied single-cell sequencing to the DNA samples and show low alignment levels (<40%).
Genomes (hg38 and mm10) and gene annotations (RefSeq data) were downloaded from the UCSC Genome Browser (Table ncbiRefSeq) (Haeussler et al. 2019).
Hyperediting
We used the hyperediting algorithm as previously described (Porath et al. 2014) to identify heavily edited RNA-seq reads which the aligner fails to align to the genome. Many of the off-targets identified by this tool occur within coding regions. Our computation tool for detecting hyperedited reads is available at GitHub (https://github.com/hagitpt/Hyper-editing).
Off-target index calculation
To measure the total off-target activity, we followed the previously developed approach for calculating the Alu editing index (Roth et al. 2019). This measure is robust and takes into account low-level variations that cannot be individually determined. The details of the present calculation are identical to those specified for the Alu editing index, except that (1) we did not look at Alu elements alone but also at the coding regions or the whole genome, and (2) we calculated the A-to-G index or the C-to-U index, depending on the base editor analyzed. Most of our results deal with the coding region, for which we assumed all reads were expressed from the annotated coding strand (see Roth et al. 2019 for analysis of the accuracy of this approach). Briefly, we calculated the weighted average over millions of genomic cytosines (9,700,565 C locations in coding regions for C-to-T) and adenosines (8,754,152 A locations in coding regions for A-to-G), and the weights are the total number of reads in these sites. To estimate the noise level, we measured the abundance of A-to-T substitution, which is the substitution type (other than A-to-G and C-to-T) with the highest noise level in most studies. Genomic sites overlapping common single nucleotide polymorphisms (SNPs) (human:dbSNP150; mouse:dbSNP142) were excluded.
We have made the computation tool for calculating the base editing index available at GitHub (https://github.com/a2iEditing/BEIndexer). Importantly, the editing index is sensitive to reads’ length (Roth et al. 2019). Longer reads may be mapped even if they include multiple editing events and thus lead to higher values of the index. The different studies examined here have used reads of varying lengths, and therefore the index values should not be compared across studies. We decided not to trim the reads of all studies to a uniform length, because some of them are very short (35 bp).
In addition, we calculated the index for sites or regions of special interest: (1) housekeeping exons (Eisenberg and Levanon 2013), which are essential for the existence of a cell; (2) oncogenes (Tate et al. 2019) with known mutations that have been causally implicated in cancer; and (3) reported pathogenic clinical variations with known human phenotype (ClinVar [Landrum et al. 2014]).
We annotated the potential substitutions in the coding regions using ANNOVAR (Wang et al. 2010). Amino acid substitutions were classified as nonconservative if the substitution results in an amino acid of a different group, using the following classification: electrically charged side chain (R, H, K, D, and E), polar uncharged (S, T, N, and Q), hydrophobic (A, I, L, M, F, W, Y, and V), and three amino acids with special side chain cases—cysteine (C), glycine (G), and proline (P). These mismatches can lead to changes in the protein structure and function. Amino acid substitutions within the same group are termed conservative.
For DNA off-target editing, we took an additional step to improve our sensitivity and discarded mismatch sites that were detected in at least half of a set of untreated samples (Cas9 for Study H and parental samples for Study I) from all other samples. These sites are likely to be enriched in genomic polymorphisms that are common to all samples and are not due to the base editors’ activity.
Conserved recoding site editing
Editing levels at the endogenous mammalian conserved RNA editing sites (Pinto et al. 2014) were calculated using the REDIToolsKnown.py script that is part of the REDItools package (Picardi and Pesole 2013). We calculated the editing level only if site coverage exceeds 10 reads. The weighted average of all these sites was used to calculate the conserved recoding index for each case.
Gene expression analysis
Gene expression levels of all genes were calculated by using the Salmon tool (Patro et al. 2017).
Neoantigen simulation
Neoantigen creation by off-target editing was simulated as follows. All 57,204 coding mRNA sequences were downloaded from the UCSC Table Browser (table “UCSC RefSeq [refGene]”, filtered by “name does match NM_*” and “chrom doesn't match alt* fix* random* chrUn*”). These sequences represent the transcriptome (including splice-variants) and include 58,472,242 adenosines and 50,119,924 cytosines. A Python script was used to perform 100,000 A-to-G or C-to-T substitutions at randomly chosen locations. The edited mRNA sequences were translated to proteins, and the resulting peptides were analyzed using netMHCpan (Lundegaard et al. 2008) to identify potentially immunogenic peptides. Binding was evaluated for 9-mer sequences with the human HLA-A02:01 MHC-I allele. For amino acid substitutions, peptide sequences ranging eight amino acids upstream of and downstream from the edited amino acid were used for binding prediction. For stop-loss edits, the sequence up to the next stop codon (or the end of the UTR if no stop codon occurred) was examined. “Strong binders” were determined using default netMHCpan parameters and cutoffs, with the “Binding Affinity” option selected. We then excluded strong binding 9-mers that exist in the naturally occurring human proteome (downloaded from the UniProtKB database [Bateman 2019] using the following query: “organism:”Homo sapiens [Human] [9606]” AND proteome:up000005640”). The resulting strong-binders 9-mers were designated as neoantigens.
Gene-set enrichment analysis
Gene expression levels were computed (using Salmon) for all genes and analyzed for enrichment of the apoptosis-related gene-set using GSEA software and version 4.0.2 (10,000 permutations) and the hallmark gene-set collection (Liberzon et al. 2015).
Data analysis
The statistical analysis was calculated using R v.3.5.1 (R Core Team 2018). All tests conducted were two-sided, and the significant difference was considered as P-value < 0.05.
Software availability
Editing index software is available at GitHub (https://github.com/a2iEditing/BEIndexer) and as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Ayal Hendel, Shay Ben-Aroya, and the Levanon lab members for fruitful discussions. This work was supported by the Israel Science Foundation (grant numbers 2673/17, 1945/18 to E.E. and 2039/20, 231/21 to E.Y.L.), as well as an International Collaboration Grant from the Jacki and Bruce Barron Cancer Research Scholars’ Program, a partnership of the Israel Cancer Research Fund and City of Hope, as supported by The Harvey L. Miller Family Foundation (grant number 205467 to E.Y.L.).
Author contributions: I.B. performed most of the bioinformatics data analyses. S.H.R. designed and wrote the software. E.Ka. initiated the bioinformatics work. A.F. contributed to specific data analyses. E.Y.L. and E.E. conceived the study, designed the analyses, and wrote the paper. All authors read and approved the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275770.121.
Competing interest statement
E.E. is a consultant to Korro Bio, a company that develops RNA editors. E.Y.L. is a consultant to ADARx.
References
- Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, Joung J, Kirchgatterer P, Cox DBT, Zhang F. 2019. A cytosine deaminase for programmable single-base RNA editing. Science 365: 382–386. 10.1126/science.aax7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto TS, Buchhalter I, Derdak S, Hutter B, Eldridge MD, Hovig E, Heisler LE, Beck TA, Simpson JT, Tonon L, et al. 2015. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun 6: 10001. 10.1038/ncomms10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47: D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014a. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Levanon EY, Eisenberg E. 2014b. Genome-wide analysis of Alu editability. Nucleic Acids Res 42: 6876–6884. 10.1093/nar/gku414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal AP, Wooster R, Stratton MR. 2004. A survey of RNA editing in human brain. Genome Res 14: 2379–2387. 10.1101/gr.2951204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi S, Borukhov I, Levanon EY. 2011. Identification of widespread ultra-edited human RNAs. PLoS Genet 7: e1002317. 10.1371/journal.pgen.1002317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, MacCarthy T. 2017. The preferred nucleotide contexts of the AID/APOBEC cytidine deaminases have differential effects when mutating retrotransposon and virus sequences compared to host genes. PLoS Comput Biol 13: e1005471. 10.1371/journal.pcbi.1005471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doman JL, Raguram A, Newby GA, Liu DR. 2020. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol 38: 620–628. 10.1038/s41587-020-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. 10.1093/genetics/148.4.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2013. Human housekeeping genes, revisited. Trends Genet 29: 569–574. 10.1016/j.tig.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907v2 [q-bio.GN].
- Gehrke JM, Cervantes O, Clement MK, Wu Y, Zeng J, Bauer DE, Pinello L, Joung JK. 2018. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 36: 977–982. 10.1038/nbt.4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J-F, Thomas WK, Smith Z, Okamoto K, Lynch M. 2013. Large-scale detection of in vivo transcription errors. Proc Natl Acad Sci 110: 18584–18589. 10.1073/pnas.1309843110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J-F, Li W, Fritsch C, Li A, Haroon S, Singh L, Hua D, Fazelinia H, Smith Z, Seeholzer S, et al. 2017. The landscape of transcription errors in eukaryotic cells. Sci Adv 3: e1701484. 10.1126/sciadv.1701484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, Joung JK. 2019a. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569: 433–437. 10.1038/s41586-019-1161-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J, Zhou R, Iyer S, Lareau CA, Garcia SP, Aryee MJ, Joung JK. 2019b. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat Biotechnol 37: 1041–1048. 10.1038/s41587-019-0236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, et al. 2019. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 47: D853–D858. 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katrekar D, Chen G, Meluzzi D, Ganesh A, Worlikar A, Shih Y-R, Varghese S, Mali P. 2019. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat Methods 16: 239–242. 10.1038/s41592-019-0323-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DDY, Kim TTY, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725. 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. 2014. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42: D980–D985. 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Smith HE, Liu C, Willi M, Hennighausen L. 2020. Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun Biol 3: 19. 10.1038/s42003-019-0745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. 10.1038/nbt996 [DOI] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1: 417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36: W509–W512. 10.1093/nar/gkn202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, et al. 2018. Somatic mutant clones colonize the human esophagus with age. Science 362: 911–917. 10.1126/science.aau3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348: 660–665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordret E, Dahan O, Asraf O, Rak R, Yehonadav A, Barnabas GD, Cox J, Geiger T, Lindner AB, Pilpel Y. 2019. Systematic detection of amino acid substitutions in proteomes reveals mechanistic basis of ribosome errors and selection for translation fidelity. Mol Cell 75: 427–441.e5. 10.1016/j.molcel.2019.06.041 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Pesole G. 2013. REDItools: high-throughput RNA editing detection made easy. Bioinformatics 29: 1813–1814. 10.1093/bioinformatics/btt287 [DOI] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol 15: R5. 10.1186/gb-2014-15-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY. 2014. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun 5: 4726. 10.1038/ncomms5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rees HA, Liu DR. 2018. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19: 770–788. 10.1038/s41576-018-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, Wilson C, Doman JL, Liu DR. 2019. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv 5: eaax5717. 10.1126/sciadv.aax5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol 18: 230–236. 10.1038/nsmb.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SH, Levanon EY, Eisenberg E. 2019. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat Methods 16: 1131–1138. 10.1038/s41592-019-0610-9 [DOI] [PubMed] [Google Scholar]
- Russell WL, Kelly EM. 1982a. Mutation frequencies in male mice and the estimation of genetic hazards of radiation in men. Proc Natl Acad Sci 79: 542–544. 10.1073/pnas.79.2.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL, Kelly EM. 1982b. Specific-locus mutation frequencies in mouse stem-cell spermatogonia at very low radiation dose rates. Proc Natl Acad Sci 79: 539–541. 10.1073/pnas.79.2.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550: 249–254. 10.1038/nature24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse CC, Ochman H. 2016. Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles. Proc Natl Acad Sci 113: 3311–3316. 10.1073/pnas.1525329113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallecillo-Viejo IC, Liscovitch-Brauer N, Montiel-Gonzalez MF, Eisenberg E, Rosenthal JJC. 2018. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol 15: 104–114. 10.1080/15476286.2017.1387711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Moschref M, Li Q, Merkle T, Selvasaravanan KD, Li JB, Stafforst T. 2018. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat Methods 15: 535–538. 10.1038/s41592-018-0017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM. 2020. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun 11: 2052. 10.1038/s41467-020-15887-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaranek AW, Levanon EY, Zecharia T, Clegg T, Church GM. 2010. A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing. PLoS Genet 6: e1000954. 10.1371/journal.pgen.1000954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, Wei Y, Hu X, Zeng R, et al. 2019. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571: 275–278. 10.1038/s41586-019-1314-0 [DOI] [PubMed] [Google Scholar]
- Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, Yang H. 2019. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364: 289–292. 10.1126/science.aav9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.