Abstract
NGLY1 deficiency is a rare recessive genetic disease caused by mutations in the NGLY1 gene which codes for N-glycanase 1 (NGLY1). Here, we report the generation of two gene corrected iPSC lines using a patient-derived iPSC line (NCATS-CL6103) that carried a homozygous p.R401X mutation in the NGLY1 gene. These lines contain either one (NCATS-CL6104) or two (NCATS-CL6105) CRISPR/Cas9 corrected alleles of NGLY1. This pair of NGLY1 mutation corrected iPSC lines can be used as a control for the NCATS-CL6103 which serves as a cell-based NGLY1 disease model for the study of the disease pathophysiology and evaluation of therapeutics under development.
Resource utility
These two patient-derived iPSC lines contain either one or two CRISPR/Cas9 corrected alleles that restore endogenous NGLY1 levels. These lines will accelerate therapeutic development by serving as isogenic controls to determine how partial or full restoration of NGLY1 expression can address the observed pathobiology of NGLY1 deficiency.
Resource Details
NGLY1 deficiency is a rare autosomal recessive disorder caused by mutations in the NGLY1 gene. This gene encodes the enzyme N-glycanase 1, which hydrolyzes the N-linked glycans from glycosylated protein, particularly for misfolded protein in the ER-associated degradation pathway.1 This enzyme has also been shown to transcriptionally regulate some protein like AQP1.2 The variety of functions carried out by this enzyme may explain the diversity and varying severity of symptoms caused by mutations in this gene. Severe clinical symptoms include intellectual disability, movement disorders, seizures, liver disease, and alacrima.1
Human dermal fibroblasts of a 16-year-old female patient (GM26612, Coriell Institute) with a homozygous nonsense mutation of p.R401X (c.1201A > T) in exon 8 of the NGLY1 gene (3p24.2) were reprogrammed into an iPSC line (NCATS-CL6103/TRNDi010-D). We previously reported a NGLY1 iPSC line that was from a different iPSC colony (TRNDi010-C).3 This mutation represents the most common mutation in patients with the most severe phenotype associated with NGLY1 deficiency.1 The fibroblasts were reprogrammed using the non-integrating CytoTune™-iPS 2.0 Sendai Reprogramming Kit containing four pluripotency transcription factors, OCT3/4, KLF4, SOX2, and c-MYC.4 The NCATS-CL6103 line was then used to generate two gene corrected lines that have either a heterozygous (NCATS-CL6104/TRNDi010-D-1) or homozygous (NCATS-CL6105/TRNDi010-D-2) correction of this mutant allele using Cas9-mediated homology directed repair (Fig. 1A). In brief, passage 20 iPSCs were electroporated with Cas9 complexed with a single guide RNA (sgRNA) that specifically recognized the mutant sequence along with the donor template (Table 2). Individual colonies were isolated and screened for either heterozygous or homozygous gene correction by PCR amplification and Sanger sequencing (Fig. 1B). The original mutant line was shown to be clear of Sendai virus and exogenous reprogramming factors at passage 20 (Fig. 1C), and all lines were then further expanded maintenance of pluripotency (Table 1). All lines displayed the standard pluripotent stem cell morphology under phase contrast microscopy and maintained expression of pluripotency markers OCT4, NANOG and SOX2 in their nuclei and SSEA4 on their plasma membranes (Fig. 1D). Quantitative analysis of expression markers by flow cytometry showed that NCATS-CL6103 had a 99% and a 90% expression rate of TRA-1-60 and NANOG, respectively, while NCATS-CL6104 had a rate of 93% and 85%, and NCATS-6105 had a rate of 90% and 75% (Fig. 1E). The ability of the three iPSC lines to generate all three germ layers (ectoderm, mesoderm, endoderm) in vivo was demonstrated using teratoma formation (Fig. 1F). G-banded karyotyping analysis of each line confirmed the karyotype, which showed the normal diploid 46, XX, without any detectable abnormalities (Supplementary Fig. S1A). The isogenic identity of all lines was determined by STR DNA profile of NCATS-CL6103, NCATS-CL6104 and NCATS-CL6105, which matched their parental GM26612 fibroblast line (data not shown). Additionally, no mycoplasma contamination was found in any cell line (Supplementary Fig. S1B). Lastly, we confirmed that our gene corrected lines had restored levels of NGLY1 expression by Western blot and RT-qPCR in comparison to the mutant line and two previously published iPSC wildtype lines (TRNDi023-D, TRNDi024-D) (Supplementary Fig. S1C,D).5
Figure 1.
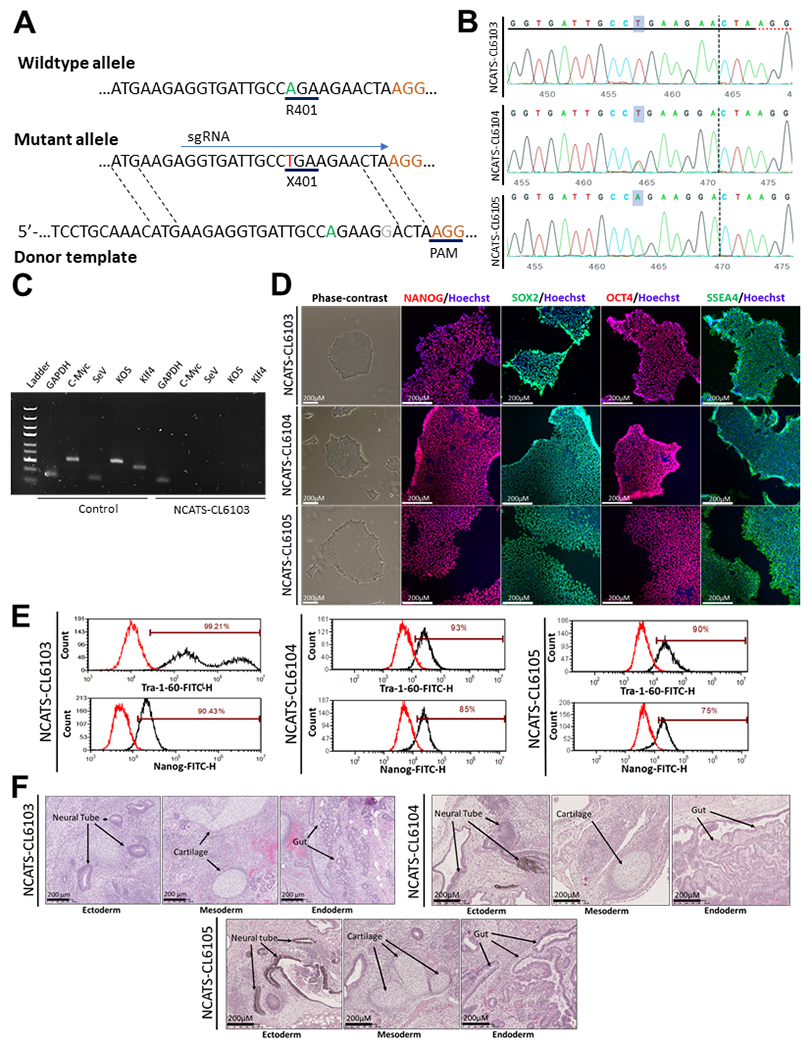
Generation and characterization of patient and gene corrected NGLY1 (c.1201 T > A) lines (NCATS-CL6103, NCATS-CL6104, NCATS-CL6105). (A) Schematic of editing approach, the mutant residue is highlighted in red with the desired edit in green. The sgRNA recognizes the mutant target sequence inducing a cut before the C residue before the PAM. The donor template then guides the repair of the sequence with the desired edit (T > A) in addition to a silent mutation (A > G) highlighted in gray. (B) Sequencing of PCR amplicons confirming presence either the mutant allele or heterozygous and homozygous correction and a silent homozygous mutation. (C) PCR result showing the clearance of Sendai virus and exogenous reprogramming factors in 592D iPSCs. (D) Phase contrast and immunofluorescence images showing pluripotent phenotype and the expression of pluripotency markers (NANOG, SOX2, OCT4 and SSEA4). (E) Flow cytometry data showing the percentage of cells expressing pluripotency markers (Tra-1–60 and NANOG). (F) Teratoma formation demonstrating the normal ectodermal, mesodermal and endodermal differentiation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2:
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:400 | Cell signaling, Cat# 4900S, RRID: AB_10560516 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# A13998, RRID: AB_2534182 |
| Pluripotency Markers | Mouse anti-TRA-1-60 | 1:500 | Cell signaling, Cat# 4746, RRID: AB_2119059 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:400 | Cell signaling, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Markers | Mouse anti-SSEA4 | 1:500 | Cell signaling, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fisher, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fisher, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies | Anti-Tra-1-60-DyLight 488 | 1:50 | Thermo Fisher, Cat# MA1-023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies | Anti-NANOG-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| NGLY1 Expression primary antibody | Rabbit Anti-NGLY1 | 1:1000 | Sigma-Aldrich, Cat# HPA036825, RRID: AB_10672231 |
| NGLY1 Expression primary antibody | Mouse Anti-GAPDH | 1:1000 | Santa Cruz, Cat# sc-47724, RRID: AB_627678 |
| NGLY1 Expression secondary antibody | Goat anti-Rabbit IgG HRP | 1:5000 | Thermo Fisher, Cat# G-21234, RRID: AB_2536530 |
| NGLY1 Expression secondary antibody | Anti-Mouse IgGκ HRP | 1:5000 | Santa Cruz, Cat# sc-516102, RRID: AB_2687626 |
| Site-specific nuclease | |||
| Nuclease information | CRISPR/Cas9 | ||
| Delivery method | Electroporation | ||
| Selection/enrichment strategy | N/A | ||
| Primers and Oligonucleotides used in this study | |||
| Target | Forward/Reverse primer (5′-3′) | ||
| Targeted mutation analysis/sequencing | NGLY1 (c.1201T > A) | Forward: ACCAGGCCCCATGACTTTTA Reverse: AGGCTAATAAACGGCTATTCTCG |
|
| sgRNA | NGLY1, cut site at Chr3: 25,733,925 | GGTGATTGCCTGAAGAACTA | |
| Donor template | NGLY1 (c.1201T > A) | CACTTGGCGATATTCCTGCAAACATGAAGAGGTGATTGCCAGAAGGACTAAGGTTAAAGAAGCATTACTTCGAGACACTAT | |
| FAM-MGB NGLY1 TaqMan Probe (RT-qPCR) | NGLY1 | Thermo Fisher, Cat#4448484, Assay ID: Hs01046153_m1 | |
| VIC-MGB GAPDH TaqMan Probe (RT-qPCR) | GAPDH | Thermo Fisher, Cat#4448489, Assay ID: Hs02786624_g1 | |
Table 1:
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 Panel D |
| Pluripotency status evidence for the described cell line | Qualitative analysis: Immunocytochemistry | SOX2, OCT4, NANOG, SSEA-4 | Fig. 1 Panel D |
| Quantitative analysis: Flow cytometry | NCATS-CL6103 (TRA-1-60: 99%, NANOG: 90%), NCATS-CL6104 (TRA-1-60: 93%, NANOG: 85%), NCATS-CL6105 (Tra-1-60: 90% NANOG: 75%) | Fig. 1 Panel E | |
| Karyotype | Karyotype (G-banding) and resolution | 46XX Resolution: 400-500 bhps |
Supplementary Fig. S1 Panel A |
| Verification of the absence of random plasmid integration events | PCR/Southern | N/A | N/A |
| Parental and modified cell line genetic identity evidence | STR analysis | 15 loci tested, all sites matched. | Archived with journal |
| Mutagenesis / genetic modification outcome analysis | Sanger sequencing | Homozygous and heterozygous correction of mutant allele p.R401X (c.1201A > T) in NGLY1 (3p24.2) | Fig. 1 Panel B |
| Specific pathogen-free status | Mycoplasma | Mycoplasma testing by luminescence. Negative. | Supplementary Fig. S1B |
| Multilineage differentiation potential | Teratoma formation | Teratoma with three germ layer formation: ectoderm, mesoderm and endoderm | Fig. 1 Panel F |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype - additional histocompatibility info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Materials and Methods
Cell culture and reprogramming
The NGLY1 patient fibroblast line (GM26612) was acquired from Coriell Cell Repositories. The cells were cultured in DMEM medium (Thermo Fisher) supplemented with 10% fetal bovine serum, 100 ug/ml streptomycin, 100 units/ml penicillin, and incubated at 37°C with 5% CO2 and 5% O2. Patient fibroblasts were reprogrammed into iPSCs using the non-integrating Sendai virus technology.4 All iPSCs lines were cultured on Matrigel (Corning)-coated six-well plates in StemFlex medium (Gibco) at 5% CO2 and 5% O2. The iPSCs were passaged at 70% confluency using EZ-Lift (Millipore) at a 1:5 to 1:10 ratio. The iPSCs were cryopreserved using StemFlex medium supplemented with 10% DMSO (Amresco) and 10 μM Rock Inhibitor (Sigma-Aldrich).
Gene editing and clone selection
Genome editing of NCATS-CL6103 to generate NGLY gene corrected (c.1201T > A) heterozygous (NCATS-CL6104) and homozygous lines (NCATS-CL6105) was contracted through Synthego (Synthego, Redwood City, CA). The sgRNA was selected based on strict specificity requirements of at least 2 mismatches and the donor template design included the necessary homology arms of sufficient length to ensure knock-in efficiency (Table 2). Additionally, a silent A > G mutation was included in the donor template to reduce cutting of edited sequences and maximize the efficiency of the knock-in. The sgRNA was complexed with SpCas9 and delivered into iPSCs via electroporation along with the donor template. Positive control sgRNA was transfected at the same time. To assess knock-in efficiency the edited site was PCR-amplified, and the amplicons were Sanger sequenced (Table 2). The sequence data were then analyzed using Synthego’s Inference of CRISPR Edits (ICE) software tool (Synthego Performance Analysis, ICE Analysis. 2019. v2.0. Synthego). ICE identified the indel frequency and the specific indels present in the pool. Additionally, ICE calculated the frequency of the desired knock-in (knock-in scores). Single cell colonies were selected based on knock-in scores and further expanded to yield the heterozygous and homozygous lines, which were again confirmed by Sanger sequencing. Both homozygous and heterozygous lines contained a homozygous silent mutation due to its closer proximity to the cutting site and thus higher efficiency of knock-in.
Immunocytochemistry
The iPSCs were grown and fixed in 96-well plates with 4% paraformaldehyde in DPBS for 15 mins at room temperature (RT), permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) in Dulbecco’s phosphate-buffered saline (DPBS) for 15 mins and washed with DPBS. Next, the cells were blocked using Cell Staining Buffer (Biolegend) for 30 mins at RT. After the removal of the Cell Staining Buffer, cells were incubated overnight at 4°C with primary antibodies (Table 2) diluted in the blocking buffer. The next day cells were washed with DPBS and incubated with corresponding secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Table 2) diluted in Cell Staining Buffer for 1 hour at RT. Cells were washed in DPBS, the nuclei were stained with Hoechst 33342 for 15 mins and imaged using an INCell Analyzer 2500 imaging system (Cytiva, Marlborough, MA) with 20x objective lens and Texas Red, FITC and DAPI filter sets.
Sendai virus detection
NCATS-CL6103 iPSCs were cultured until passage 20 and then total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). The cDNA was reverse-transcribed from 0.4 μg RNA by SuperScript™ III First-Strand Synthesis SuperMix (Thermo Fisher Scientific). The Platinum II Hot-Start PCR Master Mix (Thermo Fisher Scientific) was used to amplify the target sequence with a PCR program: 94 °C, 2 min; 30 cycles of 94 °C, 15 s, 60 °C, 15 s, and 68 °C, 15 s on Mastercycler pro S (Eppendorf) with the specific primers (Table 2). The human fibroblasts (GM05659, Coriell Institute) transfected with Sendai virus for 4 days was used as the positive control.
G-banding karyotyping and short tandem repeat (STR) analysis
The G-banding karyotype analysis was performed at WiCell Research Institute (Madison, WI, USA) using standard cytogenetic protocols. A total of 20 cells at metaphase were examined and analyzed to check for potential clonal abnormalities.
The STR analysis of the fibroblast and mutant iPSC lines was conducted by the Johns Hopkins University Genetic Resources Core Facility using the Promega PowerPlex 18D Kit. The ABI Prism® 3730xl Genetic Analyzer was used to electrophorese the PCR products and GeneMapper® v 4.0 software (Applied Biosystems) was used to analyze the data. The STR analysis for the fibroblasts and corrected iPSCs were conducted at WiCell Research Institute (Madison, WI, USA) using the PowerPlex® 16 HS System.
Flow cytometry analysis
The iPSCs were detached using TrypLE Express (Thermo Fisher Scientific) and fixed with 4% paraformaldehyde for 10 min at RT. The cells were then washed with DPBS and permeabilized with 0.2% Tween-20 in DPBS for 10 min at RT. They were then stained with fluorophore-conjugated antibodies (Table 2) for 1 hour at 4°C on a shaker. After incubation, cells were analyzed on a BD Accuri™ C6 Flow Cytometry system (BD Biosciences).
Teratoma formation assay
iPSCs cultured in 6-well plates were dissociated using EZ-Lift (Sigma-Aldrich). For each line, Approximately 1 to 5 × 106 dissociated cells were resuspended in 400 μl culture medium supplemented with 25mM HEPES (pH 7.4) and chilled on ice. Next, 200 μl of cold Matrigel (Corning, 354277) was added to the cells. This mixture was injected subcutaneously into NSG mice (JAX No. 005557) at 150 μl per injection site. Visible tumors were removed 6–8 weeks post injection, immediately fixed in 10% neutral buffered formalin and embedded in paraffin. Finally, the tissue was stained with hematoxylin and eosin for visualization of teratoma formation.
Mycoplasma detection
NCATS-CL6103 iPS cells was grown to passage 17 and tested for mycoplasma. The NCATS-CL6104 and NCATS-CL6105 iPS cells were tested after gene editing at passage 25. Mycoplasma levels were detected and analyzed using the Lonza MycoAlert Kit following manufacturer’s instruction (Ratio B/A > 1.2 mycoplasma positive; 0.9 - 1.2 ambiguous result; < 0.9 mycoplasma negative).
NGLY1 expression analysis
For Western blot, iPSC lines were lysed in RIPA Buffer (Enzo) supplemented with Protease Inhibitor Cocktail (Roche). Debris was removed by centrifugation and protein concentration was quantified by BCA (Thermo Fisher). Lysate was separated by 4-20% TGX gel (Bio-Rad) and transferred onto PVDF membrane which was blocked for 30 minutes at RT using StartingBlock buffer (Thermo Fisher) and then incubated overnight at 4°C with primary antibody (Table 2). Membrane was then washed using TBS-T buffer and then incubated with secondary antibody for 1 hour at RT (Table 2). Finally, membrane was washed using TBS-T, incubated with Immobilon HRP substrate (Millipore) for 2 minutes and imaged using G:Box (Syngene).
For RT-qPCR, total RNA was extracted using Cells-to-CT 1-Step TaqMan Kit (Thermo Fisher) and cDNA synthesis and quantitative PCR was performed in a single step by multiplexing VIC-MGB GAPDH and FAM-MGB NGLY1 TaqMan probes (Table 2) using a ViiA 7 Real-Time PCR System (Applied Biosystems). The relative expression of genes was normalized to GAPDH and calculated using the 2−ΔΔCt method.
Supplementary Material
Resource Table:
| Unique stem cell lines identifier | TRNDi010-D, TRNDi010-D-1, TRNDi010-D-2 |
|---|---|
| Alternative name(s) of stem cell lines | NCATS-CL6103 (TRNDi010-D), NCATS-CL6104 (TRNDi010-D-1), NCATS-CL6105 (TRNDi010-D-2) |
| Institution | National Institutes of Health National Center for Advancing Translational Sciences Bethesda, Maryland, USA |
| Contact information of the reported cell line distributor | Dr. Wei Zheng Wei.Zheng@nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info (applicable for human ESC or iPSC) | Age: 16-year-old Sex: Female Ethnicity: Caucasian |
| Cell Source | Skin fibroblasts |
| Method of reprogramming | Integration-free Sendai viral vectors |
| Clonality | Clonal |
| Type of Genetic Modification | Gene correction (isogenic control) |
| Associated disease | NGLY1 Deficiency |
| Gene/locus | NGLY1R401X |
| Method of modification | CRISPR/Cas9 |
| Site-specific nuclease (SSN) delivery method | Electroporation |
| All genetic material introduced into the cells | Donor template, sgRNA |
| Analysis of the nuclease-targeted allele status | Sequencing of the targeted allele |
| Name of transgene | N/A |
| Eukaryotic selective agent resistance (including inducible/gene expressing cell-specific) | N/A |
| Inducible/constitutive system details | N/A |
| Date archived/stock date | May 2021 |
| Cell line repository/bank | https://hpscreg.eu/cell-line/TRNDi010-D, https://hpscreg.eu/cell-line/TRNDi010-D-1, https://hpscreg.eu/cell-line/TRNDi010-D-2 |
| Ethical/GMO work approvals | NIGMS Informed Consent Form was obtained from patient at time of sample submission. Confidentiality Certificate: CC-GM-15-004 |
Acknowledgment
We want to thank Dr. Zu-xi Yu of the Pathology Core of National Heart, Lung and Blood Institute, National Institutes of Health for sectioning and staining the teratoma. We also would like to thank Research Services Section at National Center for Advancing Translational Sciences for coordinating the STR DNA analysis and mycoplasma testing service. This work was supported by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health, and was a CRADA collaboration between NCATS, CDG CARE (cdgcare.org), and Travere Therapeutics.
References
- 1.Enns GM et al. Mutations in NGLY1 Cause an Inherited Disorder of the Endoplasmic Reticulum-Associated Degradation (ERAD) Pathway. Genet Med 16, 751–758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tambe MA, Ng BG & Freeze HH N-Glycanase 1 Transcriptionally Regulates Aquaporins Independent of Its Enzymatic Activity. Cell Reports 29, 4620–4631.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Yang S et al. An induced pluripotent stem cell line (TRNDi010-C) from a patient carrying a homozygous p.R401X mutation in the NGLY1 gene. Stem Cell Res 39, 101496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beers J et al. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci Rep 5, 11319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X et al. Four induced pluripotent stem cell lines (TRNDi021-C, TRNDi023-D, TRNDi024-D and TRNDi025-A) generated from fibroblasts of four healthy individuals. Stem Cell Research 49, 102011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


