ABSTRACT
Multidrug-resistant bacterial infections are becoming increasingly common, with only few last-resort antibiotics such as colistin available for clinical therapy. An alternative therapeutic strategy gaining momentum is phage therapy, which has the advantage of not being affected by bacterial resistance to antibiotics. However, a major challenge in phage therapy is the rapid emergence of phage-resistant bacteria. In this work, our main aim was to understand the mechanisms of phage-resistance used by the top priority pathogen Acinetobacter baumannii. We isolated the novel phage Phab24, capable of infecting colistin-sensitive and -resistant strains of A. baumannii. After co-incubating Phab24 with its hosts, we obtained phage-resistant mutants which were characterized on both genotypic and phenotypic levels. Using whole genome sequencing, we identified phage-resistant strains that displayed mutations in genes that alter the architecture of the bacterial envelope at two levels: the capsule and the outer membrane. Using an adsorption assay, we confirmed that phage Phab24 uses the bacterial capsule as its primary receptor, with the outer membrane possibly serving as the secondary receptor. Interestingly, the phage-resistant isolates were less virulent compared to the parental strains in a Galleria mellonella infection model. Most importantly, we observed that phage-resistant bacteria that evolved in the absence of antibiotics exhibited an increased sensitivity to colistin, even though the antibiotic resistance mechanism per se remained unaltered. This increase in antibiotic sensitivity is a direct consequence of the phage-resistance mechanism, and could potentially be exploited in the clinical setting.
KEYWORDS: Phage, phage therapy, phage-resistance, phage adsorption, colistin resistance, antibiotic resensitation, virulence
Introduction
Antimicrobial resistance (AMR) is a global concern. The overuse of antibiotics in human medicine and the misuse of even last resort antimicrobial compounds such as colistin in agriculture, is contributing to the increasing number of antibiotic resistant bacterial pathogens [1]. Molecular mechanisms of resistance have evolved for most antibiotics, which are then quickly distributed throughout bacterial populations by horizontal gene transfer. This is primarily mediated by plasmids and integrative and conjugative elements [2–4]. At the same time, the slow and expensive discovery process and clinical development of antimicrobial compounds together with the lack of monetary incentives have resulted in continuously decreasing numbers of effective drugs to treat bacterial infections [5,6]. Even the last resort antibiotic colistin, often associated with strong renal and neurological side effects, is now being deployed for infections by multidrug resistant pathogens, regaining clinical importance. However, colistin resistance in pathogens is increasing as well.
Phage therapy has emerged as a promising strategy to treat drug resistant bacterial infections, as viruses are not affected by resistance to antimicrobial compounds [7–9]. Phage therapy is the use of lytic phages that have the ability to inactivate pathogens. However, phage-resistance, i.e. the emergence of bacterial mutants that are resistant to a therapeutic phage, is commonly observed [10]. Several solutions have been explored in the past, such as combinational phage-antibiotic therapeutic courses, where synergistic effects are often observed, or the deployment of phage mixtures (“phage cocktails”). Yet, in the majority of clinical trials phage-resistance occurs [11]. Therefore, it is important to understand the mechanisms that enable bacteria to gain resistance to phages and the consequences of selection. In addition, identifying target molecules that facilitate phage infection and deploying phages that do not bind the same receptors has been proposed to decrease the likelihood of phage-resistance [12].
In order to understand molecular mechanisms of phage-resistance, we investigated a phage-pathogen system consisting of the type strain of A. baumannii ATCC17978, and a colistin-resistant mutant derived from the isolate. We employed whole genome sequencing of phage-escape mutants that emerged after co-incubating the novel phage Phab24 that infects both strains. We found that genes abolishing infection are primarily involved in the biogenesis of the envelope of the bacterial host, namely the lipooligosaccharide (LOS) of the outer membrane and capsular polysaccharides. Specifically, phage resistance is mediated by mutations in gene gtr9, which is putatively involved in capsule formation, and gtrOC3, which plays a role in LOS biosynthesis. While a gtr9 deletion leads to decreased adsorption, the mutation in the gtrOC3 gene, by itself, does not alter binding of the phage to the bacterial surface. Gene engineered strains introducing the observed mutations one at a time in the parental strain, and their complementation with the wildtype gene in trans, confirmed the role of the genes in phage-resistance. In vitro evolution experiments resulted in the selection of escape mutants with decreased antibiotic resistance, with mutations in gtr9, and possibly gtrOC3, contributing to this effect.
Results
Isolation of phage Phab24 that infects colistin-resistant A. baumannii strain XH198
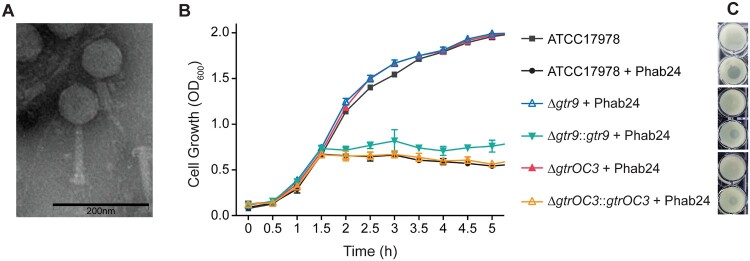
Colistin resistance is mediated by a fundamental change in the bacterial outer membrane composition. A mutation in gene pmrB (G315D) results in surface modification of the bacterial envelope, preventing efficient colistin binding and thus mediating resistance to the antibiotic [13–17]. To study phage-resistance of bacteria in vitro, we used the colistin resistant Acinetobacter baumannii strain XH198 and its non-resistant parental strain ATCC17978. Strain XH198 was obtained by an in vitro evolution experiment and exhibits altered LOS molecules, which have also been observed in clinical isolates, rendering colistin ineffective [14]. We isolated several phages that are able to infect both XH198 and ATCC17978. To focus on one particular virus-host system, we selected a phage that we named Phab24, for Phage Acinetobacter baumannii number 24 [18] (Figure 1(A)). Whole genome analysis using the programme PhageAI (https://phage.ai/) revealed that bacteriophage Phab24 is virulent (lytic) with a 93.76% prediction confidence [19].
Figure 1.
Phage Phab24 and effect on A. baumannii strain growth dynamics. (A) Transmission electron micrograph of Phab24 (negative staining). Phage Phab24 belongs to Myoviridae which have contractile tails (as seen on the rightside virion). Bar: 200 nm. (B) Growth curves of the A. baumannii reference strain ATCC17978 and phage-resistant reconstructed strains with introduced genomic mutations in the putative capsular biosynthesis gene gtr9 or in the putative LOS biogenesis gene gtrOC3, and their plasmid-complementations (::gtr9 and::gtrOC3) in the presence or absence of phage Phab24. (C) Spot testing of Phab24 on agar containing either phage-resistant or susceptible strains.
Isolation and characterization of phage-resistant bacterial mutants
We first used the phage-host system of Phab24 and ATCC17978, to study the emergence of phage-resistant bacterial mutants, by co-incubating both in liquid media. Subsequently, bacteria were plated on solid media from which we randomly picked 80 bacterial colonies (R1-R80, genome accession data: Supplemental Table 1). Surprisingly, two isolates displayed susceptibility to Phab24, possibly due to persister cells [20], while the remaining 78 were resistant to Phab24. Similarly, from co-incubating Phab24 with XH198, the colistin-resistant derivative of the ATCC reference strain, we isolated 400 colonies at random. Next, we sequenced the whole genome of six of the phage-resistant isolates of ATCC17978 (R5, R2, R13, R10, R22, R23, R39, R70) and nine of XH198 (R81, R83, R86, R115, R125, R130, R132, R134, R137). Using Breseq [21], we found several point mutations as well as deletions or insertions in genes that might mediate phage-resistance, which were confirmed by PCR and subsequent sequencing (Table 1 and Supplemental Table 2). The most common gene to have mutations was gtrOC3 [22,23], which codes for a putative glycosyltransferase (gtr) present in the type-2 OC locus, responsible for the biosynthesis of the outer core of A. baumannii’s lipooligosaccharide. Therein, we observed a single nucleotide deletion or an IS insertion (by the IS701-like element from the ISAba11 family), indicating parallel emergence of phage resistance. We also found mutations in genes predicted to code for other glycosyltransferases (gtr1, AUO97_06920), phosphohydrolase phoH (AUO97_03925) and the putative ABC transporter abcT (AUO97_07355). While we found many mutations in genes that were later identified to be irrelevant for phage-resistance, we also identified two types of mutations (a frameshift or an insertion) in the glycosyltranferase gene gtr9 (AUO97_06900), which is found in the type-3 capsule biosynthesis (K) locus.
Table 1.
Resistant strains and mutations conveying phage resistance and outcome of wildtype gene complementations in trans.
| Resistant Isolate | Mutation | Putative gene function | Complementation | |
|---|---|---|---|---|
| Yes | No | |||
| R1 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R1 | actP (A1144G, p.Thr382Ala) | acetate permease | actP | |
| R1 | gtrOC3 (IS4 family insertion) | LPS/ LOS biosynthesis | gtrOC3 | |
| R1 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R1 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R1 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R1 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R2 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R2 | actP (A1144G, p.Thr382Ala) | acetate permease | actP | |
| R2 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R2 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R2 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R2 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R2 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R5 | gtr9 (IS4 insertion) | Capsule biosynthesis | gtr9 | |
| R5 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R5 | actP (A1144G, p.Thr382Ala) | acetate permease | actP | |
| R5 | gtr0C3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtr0C3 | |
| R5 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R5 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R5 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R5 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R6 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R6 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R6 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R6 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R6 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R6 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R7 | gtr9 (IS5 insertion) | Capsule biosynthesis | gtr9 | |
| R7 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R7 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R7 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R7 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R7 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R7 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine | ||
| R8 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R8 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R8 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R8 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R8 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R8 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R8 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R10 | gtr9 (559th T loss, p. Leu187Tyr fs Ter3) | Capsule biosynthesis | gtr9 | |
| R10 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R10 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R10 | phoH (1011nt, 103th A loss) | phosphohydrolase | ||
| R10 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R10 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R10 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R10 | isoS (1698nt, 655th T loss, p.Ser 219 Pro fs Ter23) | 2-isopropylmalate synthase | ||
| R10 | LptD (2439nt, 2395th G to T, p.Val 799 Phe) | LPS- assembly protein | ||
| R10 | long-chain fatty acid--CoA ligase (1680nt,1240th T to A, p.Phe 414 Ile) | long-chain fatty acid--CoA ligase | ||
| R12 | abcT (1A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R12 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R12 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R12 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R12 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R12 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R12 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R13 | No PCR product of gtr9 | Capsule biosynthesis | gtr9 | |
| R13 | abcT (A1844T, p.Glu615Val) | ABC transporter | abcT | |
| R13 | actP (A1144G, p.Thr382Ala) | an acetate permease | actP | |
| R13 | phoH (1011nt, 103th A loss) | phosphohydrolase | phoH | |
| R13 | decT (753nt, G233A, p.Thr78Met) | di-trans,poly-cis-decaprenylcistransferase | ||
| R13 | dcaP (1332nt, G395T, p.Pro132Glu) | DcaP-like protein | ||
| R13 | Udp (720nt, T35C, p.Leu 12 Ser) | UDP-2,3-diacylglucosamine diphosphatase | ||
| R28 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R33 | gtr9 (IS5 insertion) | Capsule biosynthesis | gtr9 | |
| R35 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R39 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R54 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R78 | gtrOC3 (IS5 family insertion) | LPS/ LOS biosynthesis | gtrOC3 | |
| gtr9 KO | Markerless knock out the entire gtr9 on the background of ATCC17978 | Capsule biosynthesis | gtr9 | |
| gtrOC3-delta A | Subsititute for the wild type gtr0C3 on the background of ATCC17978 | LPS/ LOS biosynthesis | gtrOC3 | |
| R81 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R83 | No PCR product of gtr9 | Capsule biosynthesis | gtr9 | |
| R86 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R115 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R125 | gtr1 (448th A loss, p. Met 150 Cys fs Ter22) | Glycosyltransferase | gtr1 | |
| R130 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R132 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| R132 | gtr1 (448th A loss, p. Met 150 Cys fs Ter22) | Glycosyltransferase | gtr1 | |
| R134 | gtr9 (transposase insertion) | Capsule biosynthesis | gtr9 | |
| XH198 gtr9 KO | Markerless knock out the entire gtr9 on the background of XH198 | Capsule biosynthesis | gtr9 | |
| XH198 gtrOC3-KO | Markerless knock out the entire gtrOC3 on the background of XH198 | LPS/ LOS biosynthesis | gtrOC3 | |
| R518 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
| R587 | gtrOC3 (535th A loss, p.Asn179Ile fs Ter7) | LPS/ LOS biosynthesis | gtrOC3 | |
Yes: After the transformation of the corresponding plasmid which contains the WT gene, the R variant can get infected by Phab24.
No: After the transformation of the corresponding plasmid which contains the WT gene, the R variant cannot get infected by Phab24.
R1-R80 are Phab24 escape mutants from ATCC17978.
R81-R587 are Phab24 escape variants from XH198.
Based on the NCBI accession number CP018664.1, ATCC17978:
AUO97_06900, gtr9, 828nt
AUO97_07355, abcT, 1932nt
AUO97_05550, actP, 1716nt
AUO97_03485, gtrOC3, 768nt
AUO97_03925, phoH, 1011nt
AUO97_06920, gtr1, 1164nt
AUO97_09800, 2-isopropylmalate synthase, isoS, 1698nt
AUO97_15295, LPS assembly protein LptD, 2439nt
AUO97_06215, long-chain fatty acid--CoA ligase, 1680nt
AUO97_17750, di-trans, poly-cis-decaprenylcistransferase, decT, 753nt
AUO97_18665, UDP-2,3-diacylglucosamine, Udp, 720nt
Interestingly, the phoH of the WT or ATCC17978 used for Sanger sequencing has 1012nt, while the phoH of CP018664.1 has 1011nt, the same as the mutated R variants. Maybe that is the reason why the phoH is annotated as the pseudogene.
To demonstrate that the mutations indeed render a strain non-susceptible, we constructed several plasmids encoding the wildtype genes in the A. baumannii shuttle vector pYMAb2-HygR [24,25]. We then introduced the episomal elements into the phage-resistant (R) mutants. The expression of the wildtype genes of the LOS biosynthesis protein gtrOC3 (e.g. in R1) as well as of gtr9 (e.g. in R7) did restore phage sensitivity, unless both mutations were present in the strain (e.g. strain R5) (Table 1). Some phage-resistant isolates could not be complemented by wildtype genes coding for some of the mutations we observed, e.g. the membrane transport protein abcT (AUO97_07355) phoH (AUO97_03925), or actP (AUO97_05550). This indicates the presence of additional, as of yet unidentified, mutations that confer resistance (Supplemental Table 2).
Due to the obvious complexity of the resistance mechanisms, we decided to focus on gtrOC3 and gtr9. To demonstrate the role of these genes in phage susceptibility, in addition to complementation in trans, we engineered the mutations into the parental A. baumannii strain (ATCC17978), or created knock-out mutants of genes. When mutations were introduced into the gene coding for gtrOC3, the bacterium was rendered “immune” to phage Phab24 (Figure 1(B)). Similarly, a reconstructed mutant in which the gtr9 gene was deleted could not be infected by the phage, clearly demonstrating the role of these two genes in phage susceptibility. Both reconstructed mutants could be infected by Phab24 when the complementing wildtype genes are expressed in trans, from plasmids (Figure 1(B, C)).
Attachment of Phab24 to the surface of phage-resistant mutants
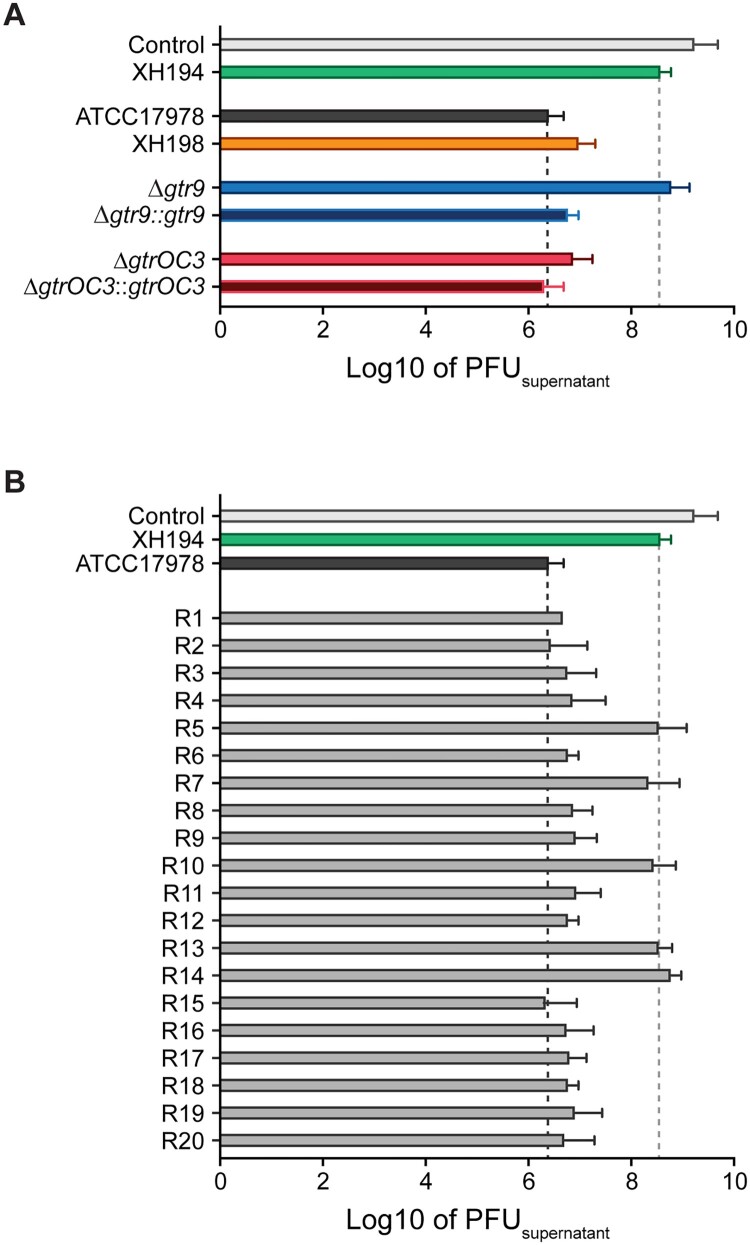
As the mutations found in gtrOC3 and gtr9 code for putative proteins involved in LOS and capsule formation, respectively, we concluded that the surface of the phage-resistant mutants may exhibit modifications compared to the wildtype. Changes on the surface of mutant strains might therefore lead to a reduction in binding of phages to bacteria. We therefore performed binding assays to assess the quantity of phages that bind to the bacterial envelope. To this end, we co-incubated Phab24 with the phage-resistant mutants and controls for 20 minutes and subsequently determined the phage titre left in the supernatant (Figure 2). The positive control, strain ATCC17978, was able to reduce the free-phage titre by a factor of more than 1000-fold, while the negative controls (no bacteria or A. baumannii strain XH194 resistant to Phab24 infection) resulted in only a minor reduction in the number of phage particles in the supernatant. Interestingly, Phab24 bound ATCC17978 with an apparent binding affinity higher than XH198. The gtr9 knockout on the backbone of ATCC17978 showed reduced bacteriophage binding, similar to the negative controls. Expectedly, upon complementation with the wildtype gtr9 gene in trans, binding of Phab24 was restored to a level comparable to parental ATCC17978. Interestingly, the gtrOC3 knockout and its derivative complemented with wildtype gtrOC3 both bound Phab24 avidly just like the parental ATCC17978 strain. In summary, phage Phab24 is able to bind to most phage-resistant strains, with the exception of gtr9 mutants, and those with double mutations in gtr9 and gtrOC3, suggesting the bacterial capsule is the primary receptor used by the phage (Figure 2(B)).
Figure 2.
Attachment assay of Phab24 to bacteria. Titre of free phages detected in media after incubation with (A) reference strain ATCC17978, colistin resistant derivate XH198, the gtr9 and gtrOC3 genetically engineered strains (Δ), and their complementations (::); and (B) Phab24 phage-resistant colonies, “R”, derived from ATCC17978. Control: phage Phab24 incubated without bacteria. XH194: bacterial strain that is colistin resistant and is not infected by Phab24.
The bacterial envelope is altered in both the LOS (gtrOC3) and capsule (gtr9) mutants
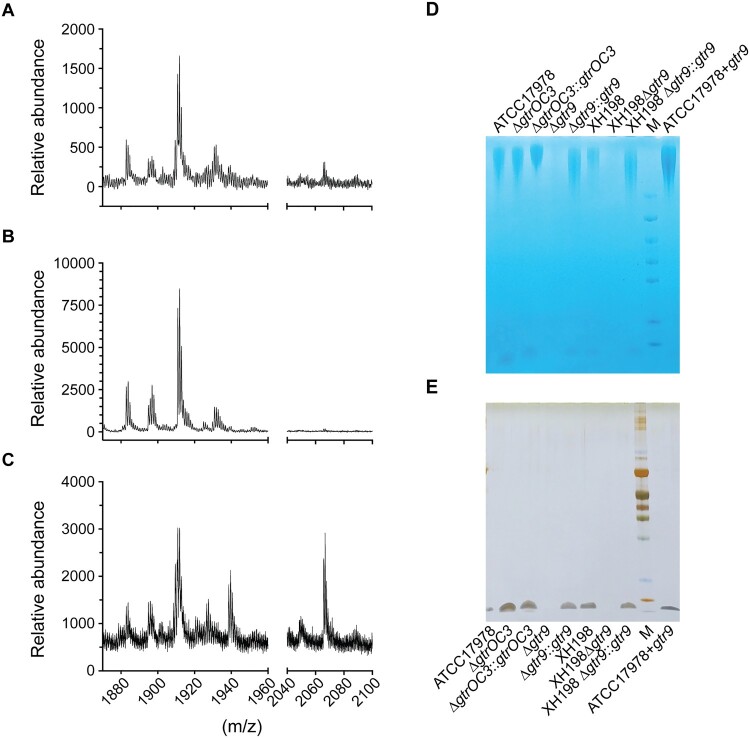
LPS, or LOS in the case of A. baumannii, often serves as a co-receptor in phage binding [26]. As a disruption in the gene coding for a LOS biosynthesis protein (gtrOC3) was observed, we determined if the mutations in the isolated strains lead to a change in LOS composition. Mass spectrometry of isolated lipid A, obtained using the hot aqueous phenol extraction method, was performed to compare the reconstructed mutants (on the ATCC17978 backbone) with the ATCC17978 control (Figure 3(B and A), respectively). In addition, we analysed samples of the plasmid-complemented strain that allows the expression of gtrOC3 in trans. Previously, it was established that the mass/charge (m/z) ratio of A. baumannii lipid A is featured as a prominent peak at 1,910, which was identified as a singly deprotonated lipid A structure that contains two phosphate groups and seven acyl chains (i.e. diphosphoryl hepta-acylated lipid A). In our experiments, this peak was observed in all samples (Figure 3(A–C)). While the mass spectrum of the KO strain (Figure 3(b)) shows molecules with m/z values of 1910 and smaller, the reference spectrum (Figure 3(A)) exhibits several additional small peaks larger than 2,000. These higher molecular weight peaks are more prominent in the plasmid-complemented strain (Figure 3(C)). While the identification of molecules that lead to the occurrence of these peaks with higher m/z values is still outstanding, they possibly represent modified Lipid A molecules. As these peaks are clearly absent from the baseline spectrum recorded in the KO strain sample (Figure 3(B)), it may be reasonable to conclude that the disruption in gtrOC3 results in a modification in the bacterial surface structure. The gene disruption might ultimately prevent the incorporation of one or more types of modified LOS molecules into the bacterial envelope which—when absent—abolish Phab24 phage binding [15,27].
Figure 3.
Composition of the bacterial envelope in parental and phage-resistant A. baumannii. (A–C) Mass spectrometry (positive mode) analysis of lipid A isolated using aqueous phenol extraction from (A) Wildtype strain ATCC17978, (B) Phab24 resistant, genetically introduced reconstructed gtrOC3 gene mutant on ATCC17978, (C) Plasmid complemented Phab24 resistant strain. (D and E) SDS-PAGE gel of isolated capsular polysaccharides stained with (D) Alcian blue, which allows the detection of acidic polysaccharides, and (E) silver staining, which detects lipids and proteins/peptides.
As gtr9 is found in the K locus, and therefore is involved in the formation of the bacterial capsule, we isolated the oligosaccharides of both the reconstructed gtr9 mutant and the parental strain ATCC17978, and compared them using mass spectrometry. Perhaps unexpectedly, we did not observe any additional presence or absence of peaks in the spectra across 180–3200 m/z (Supplemental Figures 1, 2, 3). While ratio and intensity varied for some peaks, there is no indication of the absence of specific polysaccharides, or the presence of others, in the gtr9 mutant. Because the relative quantity of the polysaccharides cannot be firmly established through mass spectrometry, we attempted to determine such differences using SDS PAGE gels, which allow a separation of molecules based on size, while also allowing a quantitative analysis. Here, we observed that the genetically engineered gtr9 mutants of ATCC17978 or of XH198 exhibited a massive reduction in material on the gel compared to the complemented strains (and the reference strains) where gtr9 was expressed in trans (Figure 3(D, E)). A gel with Alcian blue, which stains acidic polysaccharides, shows that the gtr9 mutants of both ATCC17978 and XH198 contain almost undetectable amounts of material, while the plasmid-complemented strains are similar to the wildtype level. In addition to large molecular weight bands, we also observe small molecule components which are stained by Alcian blue (Figure 3(D)), but also by silver ions (Figure 3(E)). As silver staining allows the sensitive detection of lipids and proteins but not of the saccharides (and polypeptides can be excluded due to the preparation which includes a Proteinase K digestion step), the smaller molecules possibly indicate the presence of lipid-saccharide conjugates in the samples, which again, are absent in the gtr9 mutants. Although the mass spectrometry data did not indicate any changes in saccharide composition, the quantitative method of size-separated oligosaccharides on gels suggests that the bacterial envelope surface of the gtr9 mutant is different from that of the A. baumannii reference strain.
Changes in cell morphology and capsule formation in phage-escape mutants
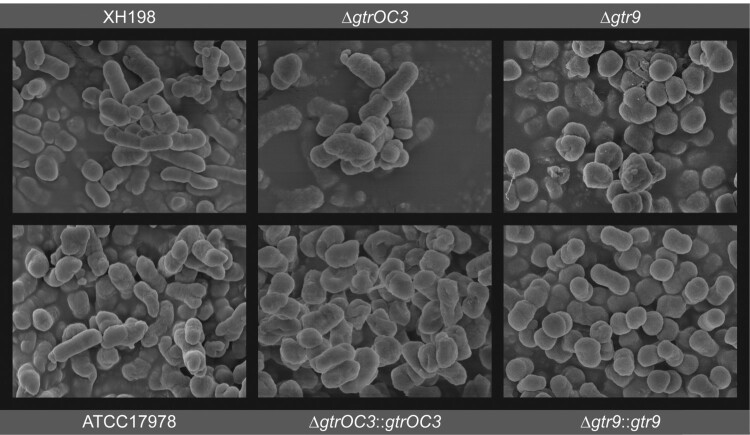
On the molecular level, we could confirm that the composition of the cell envelope is different in the gtrOC3 mutant while we found no indication for a qualitative change in the gtr9 mutant, although the mutant appears to produce substantially less exopolysaccharides. However, we pursued further evidence before conclusively asserting that the surface structure of the cells is altered by the mutations. We first employed Transmission electron microscopy on thin sections of resin-embedded cells with inconclusive results (Supplemental Figure 4). We then used Scanning Electron Microscopy where, both ATCC17978 and XH198 cells, appear similarly smooth and rod-shaped. A similar morphology could be observed in the case of cells containing the gtrOC3 mutant; when complemented in trans by the functional gene, a slightly more “shrivelled”—possibly desiccated—structure was observed. A very even, smooth surface was seen when the gtr9 mutant was complemented with the functional gene in trans while the surface of the gtr9 mutant itself appeared less smooth. In addition, the cells of the gtr9 mutant appear more rounded and adherent to each other, forming clusters (Figure 4). We also observed the formation of mucoid-like strings in preparations of the gtr9 mutant which were absent in all other samples (not shown).
Figure 4.
Surface structure of bacterial cell envelope. Scanning Electron Microscopy. ΔgtrOC3: knock-out of gtrOC3 gene in the ATCC17978. ΔgtrOC3::gtrOC3: knock-out of gtrOC3 gene which was complemented with the wildtype gtrOC3 gene in trans. Δgtr9: knock-out of gtr9 gene in the ATCC17978. Δgtr9::gtr9: knock-out of gtr9 gene which was complemented with the wildtype gtr9 gene in trans.
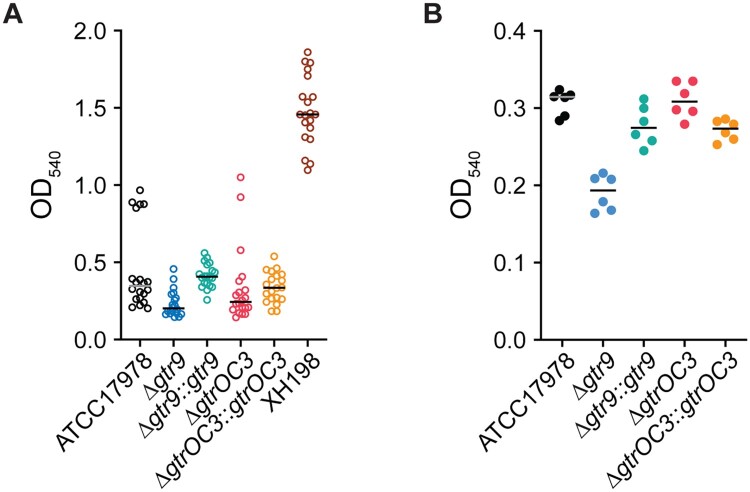
The observed clustering and appearance of “slime” on the surface of the gtr9 mutant may be caused by altered exopolysaccharide production and/or biofilm formation. We therefore assessed the production of these materials by growing bacteria on a solid surface for three days, washing them off, and testing the capacity of the produced material to retain the crystal violet dye. Here, we observed that dye retention, which is commonly used to determine the extent of biofilm formation in the lab setting, was most pronounced in the colistin resistant strain XH198, but remained low in all other strains (Figure 5(A)). Compared to the reference strain ATCC17978, the gtr9 mutant showed reduced biofilm formation (∼42% reduction compared to ATCC17978), while plasmid complementation led to a slight increase in biofilm (∼117% of that of ATCC17978) (Figure 5(A)). In the case of the gtrOC3 mutant, no significant difference in biofilm formation between the complemented or non-complemented strains was observed. Thus, the “mucoid” appearance and aggregation observed in SEM preparations of the cells in the gtr9 mutant cannot be explained by the formation of biofilms. Interestingly, we observed the formation of highly fragile membranous structures in the multi-well plates for this mutant that, however, did not retain the dye, and were washed away easily (Supplemental Figure 5A). In addition to the formation of biofilms we also employed crystal violet staining of capsules of planktonic cells. Biofilms are usually formed when bacteria are incubated for a long duration and allowed to sediment and form clusters. If cells are incubated for shorter times with shaking, biofilm formation does not occur. It is reasonable to assume that the amount of dye that is retained by the capsule correlates with the quantity of material present for the dye to embed in (i.e. more dye is retained by thicker, more extensive capsules) and/ or the density of the packing of the capsule material (i.e. released quicker if the material is less compact). The ATCC17978 strain retained substantially more dye compared to the gtr9 mutant while a complementation results in the same levels observed for the reference strain (Figure 5(B)). One interpretation is that the gtr9 mutant strain can absorb less dye due to a smaller capsule which would correlate with the results of the capsular material separated on SDS-PAGE gels (Figure 3). The gtrOC3 mutant and the complemented strain show levels similar to ATCC17978.
Figure 5.
Biofilm formation and capsular stain. (A) Biofilm formation assessed by the ability to retain crystal violet (CV) dye. Strains were incubated for 72 hours in multi well plates without shaking. (B) Capsule stain of planktonic cells incubated for 8 hours with shaking. Cells were stained with CV to determine the ability of the bacterial capsule to retain the dye. Ethanol was used to destain the capsule and release the CV dye which was then detected by measuring absorbance at 540nm.
Capsule (gtr9) and LOS (gtrOC3) biosynthesis mutants are less virulent in vivo
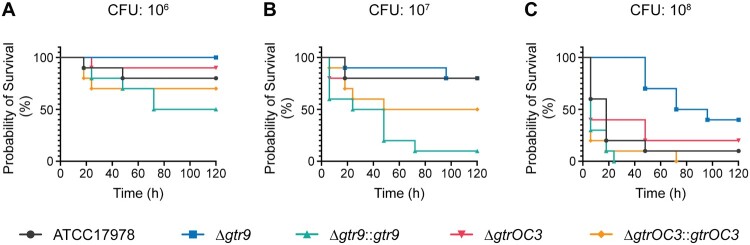
The extended bacterial surface structure, which includes capsules and LPS/LOS molecules, often contributes to bacterial virulence [28–31]. We therefore investigated how the genes that conferred phage-resistance would impact the virulence of the phage-escape mutants. To this end, we used the insect larva model Galleria mellonella, to assess the virulence of the reference strains (ATCC17978, XH198), compared to the single gene mutant strains (gtr9, gtrOC3), and those complemented in trans. The mutation in gtrOC3 impacts the virulence of the strain only to a small extent, with slightly increased survival rates compared to ATCC17978 (Figure 6). In contrast, the gtr9 KO strain had a significantly reduced virulence, demonstrating the importance of gtr9 on the pathogenicity of A. baumannii. When the gtr9 mutant strain was complemented with a plasmid expressing the wildtype gene under a constitutive promoter, the virulence was significantly increased compared to ATCC17978, possibly due to a higher expression level. Similarly, albeit to a lesser extent, virulence of the strain with the gtrOC3 mutation increased when the wildtype gene was expressed in trans.
Figure 6.
In vivo virulence tests of A. baumannii strains and the Phab24 resistant isolates. Survival of G. melonella larvae over 120 h after injection with (A) 106 colony forming units (CFU), (B) 107 CFU, and (C) 108 CFU of A. baumannii strains. Each group consisted of 10 larvae. Shown is a representative experiment of 4 independent repeats.
Phage-resistance mutations in gtr9 decrease colistin resistance
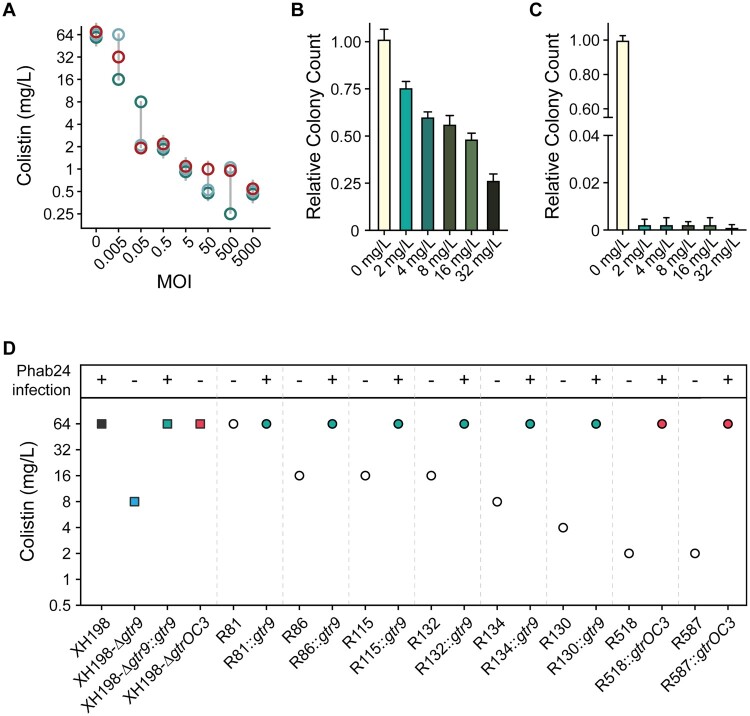
Specific alterations in outer membrane composition are the molecular basis for colistin resistance. We found that phage Phab24 binds to surface exposed molecules of ATCC17978 and XH198, and resistance is mediated by altered envelope structures. To answer the question if phage- and antibiotic-resistance mechanisms impact each other, we performed experiments testing the combination of Phab24 with colistin against our strains. During phage therapy, antibiotics are often used in combination with therapeutic phages, as synergistic effects of phage-antibiotic combinations have often been observed [32,33]. In our case, increasing concentrations of phage reduced the apparent MIC (Minimum Inhibitory Concentration) of colistin (Figure 7(A)). One explanation for this observation is that phage-resistant mutants show a higher sensitivity to colistin. To address this hypothesis, we investigated if phage-resistant bacteria show higher sensitivity to colistin, testing colony survival of XH198 cultures grown in the absence of colistin but in the absence or presence of Phab24 (Multiplicity of Infection of 1) (Figure 7(B, C), respectively). The number of colony forming units (CFUs) were subsequently determined on media with different amounts of colistin. Interestingly, we observed that the number of CFU decreases with increasing colistin concentration, regardless of whether or not the bacteria were co-incubated with Phab24. In the absence of Phab24, the ratio of bacterial colonies dropped by ∼1/4 to ∼3/4 when colistin was present compared to the number of colonies that grew on plates without colistin (Figure 7(B)). However, in the presence of Phab24, the ratio of CFUs was reduced to less than 0.01% compared to the count when colistin was absent (Figure 7(C)). This observation might indicate that the selection for phage-resistance leads to the mutations which in turn increase sensitivity to colistin.
Figure 7.
Colistin and Phage resistance. (A) Synergy of colistin with/without phage Phab24. Co-incubation of different numbers of Phab24 (MOI) at different concentrations of colistin (mg/L) at constant cell numbers. Each circle represents an independent experiment. (B and C) Colistin-resistance levels of emerged phage-resistant colonies. Colony count of XH198 incubated for 3 h in the absence of colistin or on colistin plates with different antibiotic concentrations (B) in the absence of phage, and (C) in the presence of Phab24. (D) Colistin MIC of selected Phab24 resistant isolates. Top panel shows if the strain can be infected by Phab24, indicating successful complementation of a mutated gene. The control, strain XH198, exhibits a MIC of 64 mg/L. Phage-resistant isolates displayed reduced levels of resistance varying from 16 to 1 mg/L, except for one strain (R81). The reconstructed gtr9 mutant shows an increased sensitivity to colistin compared to the reference strain XH198, while plasmid complementation with the wildtype gene fully restores the high level of resistance. The genetic deletion of the entire gtrOC3 gene in XH198 does not result in a reduction in colistin resistance. However, strains with mutations that lead to a truncated gtrOC3 gene product increase colistin sensitivity by 32-fold (R518, R587).
To investigate this finding in more detail, we tested the MIC of individual phage-resistant XH198 mutants (Figure 7(D)). Resistance levels varied widely from 64 to 0.5 mg/L (Supplemental Table 3). The correlation between the observed reduction in colistin resistance with phage resistance is far from trivial. Colistin resistance in the parental strain XH198 is mediated by a mutation in gene pmrB (G315D), which remained unchanged (data not shown). Therefore, the molecular basis for increased sensitivity possibly lies in other mutations found in the strains, including those in gtr9 or gtrOC3. Thus, we tested the impact of gtr9 and gtrOC3 on the sensitivity to colistin.
We first tested XH198-derived phage resistant isolates with gtrOC3 mutations, R518 and R587, where we observed a colistin MIC of 2 mg/L. Both strains could be complemented with a plasmid-encoded gtrOC3, which then allows infection by Phab24, increasing the MIC to 64 mg/L (Figure 7(D)). While the strains may have additional mutations, this might be an indication that the gtrOC3 mutation increases colistin sensitivity. However, unexpectedly, the genetic deletion of the entire gtrOC3 gene in XH198 (a knock-out) apparently does not result in a reduction in colistin resistance. We then tested the reconstructed gtr9 mutant on the backbone of XH198, which showed strongly increased sensitivity to colistin, from 64 mg/L to 8 mg/L (Figure 7(D)). The complementation in trans using a plasmid-encoded wildtype gtr9 restored the high resistance level to colistin. Similarly, mutants containing gtr9 mutations exhibiting various degrees of colistin sensitivity, displayed the high level of resistance when complemented e.g. R130 from 4 mg/L (without plasmid), to 64 mg/L (with plasmid).
Discussion
Bacterial phage susceptibility rests on several mechanisms, one of which is binding to receptors and co-receptors. In our work, we have established the role of two genes in permitting the infection of A. baumannii strain ATCC17978 and its colistin-resistant derivative, XH198, by phage Phab24. One gene has a putative function in the biogenesis of A. baumannii LOS, and is found in the type-2 outer core locus (OCL2), the glycosyltransferase gtrOC3 [34]. The second gene, also a glycosyltransferase, gtr9 (found in the K locus), is involved in the biosynthesis of capsular polysaccharides [34].
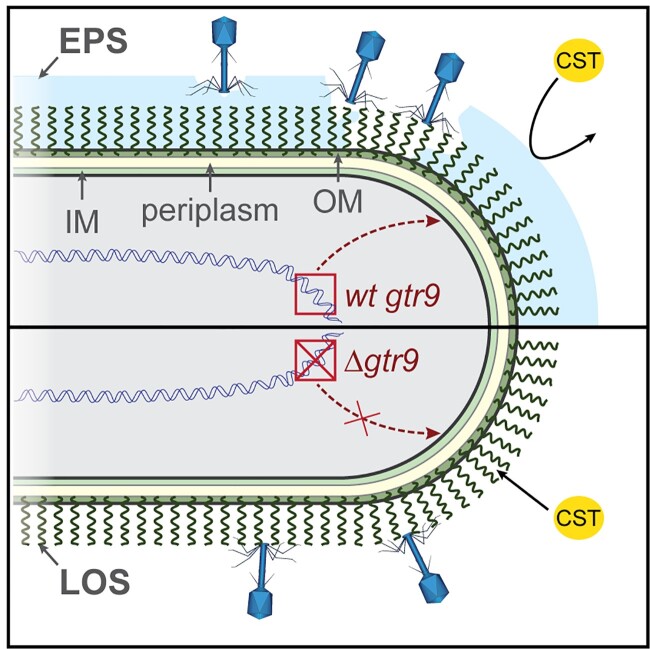
Previous work has shown that capsular molecules can serve as phage receptors [35–38], a finding our study is able to confirm. Phage-resistant isolates that displayed mutations in gtr9 did not permit efficient binding or infection by Phab24. These mutations may either lead to the production of altered surface receptors to which the phage cannot bind, or perhaps abolish the production of the cognate receptor. Surprisingly, we found that binding is unaltered in strains displaying mutations in gtrOC3, yet infection does not occur, which contrasts previous reports [12,39,40]. However, these strains did contain an intact copy of gtr9. Our results suggest a model where Phab24 uses A. baumannii’s capsular polysaccharides, in whose production gtr9 is involved, as a primary receptor, whereas a specific type of LOS molecule, absent in gtrOC3 mutants, acts as a co-receptor, possibly required to then trigger the release of DNA into the bacterium (Figure 8). This simultaneous, or sequential, binding of receptor and co-receptor, essential for the release of DNA into the host, has just been described in E. coli [26] but, to the best of our knowledge, not for A. baumannii-specific phages.
Figure 8.
Model of Phab24 binding to ATCC17978/ XH198 and gtr9 mutants. Phages can bind to the capsule surrounding the cell (top, light blue) as well as to LOS molecules anchored in the outer membrane (OM). In the case of cells in which gtr9 is disrupted or mutated, phage particles may not bind to the cells via the LOS molecules (bottom). When the capsule is missing, colistin (CST) is able to bind to the OM more easily as the absence of a barrier facilitates diffusion to the membrane. IM: inner membrane; EPS: Extracellular polymeric substance.
While a previous study has described phage-resistance caused by capsule loss in A. baumannii [12], our work supports phage resistance being able to emerge by alterations of the bacterial surface structure at two levels: the capsule and the LOS of the outer membrane. In the aforementioned study, capsule-deficient phage-escape mutants of A. baumannii MDR strains showed a much reduced propensity to form biofilms. In our work, we have also observed a decrease in biofilm formation when investigating the gtr9 gene knockout in the colistin sensitive reference strain ATCC17978.
In addition to the gtr gene mutations, we also identified several mutations in genes that were later found not to be responsible for phage resistance. These included the membrane protein abcT, actP and phoH. Genetically engineered mutants were still susceptible to Phab24 while plasmid complementation experiments with abcT, actT, or phoH mutants (containing additional mutations rendering the strains phage-resistant), did not affect infection.
Our study also showed that the two mutations in gtr9 and gtrOC3 which conferred phage-resistance resulted in decreased virulence in our in vivo model. Similar to our findings, it has been shown that phage escape mutants often show attenuated pathogenicity which, in some instances, is directly attributed to the bacterial envelope modifications that serve as the basis for phage resistance [39, 41–44]. Although we investigated only a limited number of colistin resistant A. baumannii strains due to the limited host range of Phab24, increasing evidence indicates that phage-resistance can be accompanied by “trade-off” mechanisms that result in decreased virulence and increased susceptibility to antibiotics [45].
The use of colistin as the last resort for the treatment of MDR infections, has been increasing over the years. The mechanism of action of the antibiotic remains to be fully elucidated, but it has been shown that colistin binds to LPS (LOS) and causes the deterioration of the membrane structure with the antibiotic acting detergent-like, resulting in an osmotic disruption of the cell and ultimately, death [46,47]. However, colistin resistance is on the rise, often mediated by genomic mutations in pmr genes [13]. Other mutations involve the lpx genes that are often identified in in vitro evolution experiments; in contrast to pmrB used in this study, lpx gene mutations can lead to increased sensitivity to vancomycin [48,49]. In clinical practice, patients receiving phage therapy have accessed it for compassionate, or last resort, use. In these patients, therapeutic bacteriophages are commonly deployed in addition to the antibiotic therapy they might be receiving. This is often done despite the fact that the bacteria may be displaying resistance to the used antibiotics in vitro. The rationale behind this approach is the often observed phage-antibiotic synergy; a phenomenon that is also supported by our observations [11,32,50–52]. In our work, we saw a decrease in apparent MIC when colistin was used in combination with phage Phab24. We also confirmed that emerging phage-resistant bacterial clones often exhibited increased levels of antibiotic sensitivity. We hypothesize that the loss of the capsule facilitates the diffusion and ultimately the insertion into the membrane as the absence of the capsule material might allow diffusion of the colistin molecule to the membrane directly without having to cross the capsule barrier (Figure 8). Our findings could potentially be leveraged in the clinical setting, where being able to reduce the doses and duration of colistin treatment a patient receives, could prevent the emergence of its severe side effects [53]. A potential limitation of our study is that we evaluated the impact of phage resistance only for a single colistin resistant bacterial strain. However, our work adds to the mounting evidence that phage resistance often results in fundamental changes in bacteria that can lead to reduced virulence or antibiotic resensitization [38,54,55]. Our work provides experimental data that elucidates a molecular mechanism of phage resistance, resulting in increased colistin susceptibility in A. baumannii, one of the most dangerous MDR pathogens.
Materials & methods
Isolation, purification and the genome of phage Phab24 are described elsewhere [18]. GenBank accession number for Phab24 genome: MZ477002.
DNA genome sequencing and analysis. Genomic DNA was extracted using Bacterial genome DNA isolation kit (Biomed, China), sequenced by Illumina HiSeq platform (150-bp paired reads). Breseq was used to identify single point mutations [21].
Gene knockout or replacement, plasmid-derived complementation were performed as described previously: [24]. All primers for cloning and gene replacement are shown in Supplemental Table 4. Briefly, to construct gene knock out strains, primers were designed to bind a sequence about 1kb upstream and 1kb downstream, respectively, of the target gene in ATCC17978. A PCR fragment was obtained which was then cloned into the suicide plasmid pMo130-HygR by seamless cloning (Gibson assembly) (Biomed Biotech, Beijing, China), transformed into chemically competent DH5α cells (Vazyme, Nanjing, China), and selected on LB plates supplemented with 100 mg/L hygromycin. Sequenced “positive” plasmids were then introduced into A. baumannii strains using electrotransformation. After two crossovers, positive colonies that survived on 10% sucrose plates were screened by colony PCR and finally verified by subsequent Sanger sequencing. For testing complementation, the target genes were amplified via PCR using primers designed using the Gibson assembly principle (seamless cloning) and then inserted into the A. baumannii shuttle vector pYMAb2-HygR, downstream of a constitutional promotor. Primers for confirming mutations identified from the whole genome were designed and used for PCR amplification with subsequent Sanger sequencing to confirm mutations or insertions (Supplemental Table 4).
Determination of bacterial growth rates was performed as described previously [56] with the following modifications for experiments that included phages: An MOI of 5 of a high-titre preparation was added to the culture with a negligible dilution.
Transmission electron microscopy was performed as described previously [47]. Micrographs were obtained with a JEOL JEM1010 at 80 kV. Scanning Electron Microscopy as described here: [24].
Phage adsorption. Adsorption was measured indirectly by quantifying free phage in solution. Overnight bacterial culture was diluted in LB and bacteria at 8×109/mL were incubated with Phab24 at 2×109/mL at 4°C for 20 minutes. Cells were pelleted by centrifugation, before the supernatant was serially diluted and non-adsorbed phages quantified by spot titre.
Laboratory evolution experiment: The soft-agar overlay technique was used to obtain phage resistant colonies which are purified three times by re-streaking, after co-incubation of ATCC17978 (or XH198) with Phab24 (MOI = 1) for 3 hours at 37°C.
Colistin MIC according to the CLSI 2020 antibiogram performance standards (www.clsi.org), was determined by the broth-dilution method as described previously [57].
Lipid A isolation and structural characterization: Lipid A isolation was performed as previously described [58] which was used for MALDI-TOF MS.
Surface polysaccharide extraction were purified by hot aqueous phenol extraction according to a previously described protocol [59].
Biofilm assays were performed as described previously: [60]. Each sample was done five times, with 3 independent experiments.
Crystal Violet (CV) retention assay of planktonic cells: Overnight cultures were diluted in LB media to OD600 = 1. 1 mL of diluted culture was centrifuged and cells were washed with PBS, then resuspended. CV was added (final 0.01% w/v), vortexed and incubated for 10 minutes. Cells were washed 3 times, before being destained with 2 mL of 95% ethanol for 10 minutes. Cell-free supernatant was then transferred into spectroscopic cuvettes and absorbance is measured at 540 nm.
Galleria mellonella infection model. Larvae survival assay was performed as previously described [61]. Ten larvae per group.
Checkerboard Assay was performed as described previously: [32]. 4 × 104 bacteria, with MOI: 5000, 500, 50, 5, 0.5, 0.05, 0.005.
Supplementary Material
Acknowledgements
We thank Mark Toleman (University of Cardiff) for critically reading the manuscript, Jeremy Barr (Monash University) and Nick Scott (University of Melbourne) for helpful discussions. We thank Belinda Loh who has obtained no financial compensation nor salary for her work.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (32011530116).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Leptihn S. Welcome back to the pre-penicillin era. Why we desperately need new strategies in the battle against bacterial pathogens. Infectious Microbes and Diseases. 2019;1(2):33. [Google Scholar]
- 2.Igler C, Rolff J, Regoes R.. Multi-step vs. single-step resistance evolution under different drugs, pharmacokinetics, and treatment regimens. Elife. 2021;10:e64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes EC, Dudas G, Rambaut A, et al. . The evolution of ebola virus: insights from the 2013–2016 epidemic. Nature. 2016;538(7624):193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh B, Chen J, Manohar P, et al. . A Biological inventory of prophages in A. baumannii genomes reveal distinct distributions in classes, length, and genomic positions. Front Microbiol. 2020;11:5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepore C, Silver L, Theuretzbacher U, et al. . The small-molecule antibiotics pipeline: 2014–2018. Nat Rev Drug Discov. 2019;18(10):739. [DOI] [PubMed] [Google Scholar]
- 6.Loh B, Leptihn S.. A call For a multidisciplinary future of phage therapy to combat multi-drug resistant bacterial infections. Infectious Microbes and Diseases. 2020;2(1):1–2. [Google Scholar]
- 7.Gordillo Altamirano FL, Barr JJ.. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 2019;32(2):e00066–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viertel TM, Ritter K, Horz HP.. Viruses versus bacteria–novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemoth. 2014;69(9):2326–2336. [DOI] [PubMed] [Google Scholar]
- 9.Kortright KE, Chan BK, Koff JL, et al. . Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232. [DOI] [PubMed] [Google Scholar]
- 10.Labrie SJ, Samson JE, Moineau S.. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. [DOI] [PubMed] [Google Scholar]
- 11.Tkhilaishvili T, Winkler T, Muller M, et al. . Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;64(1):e00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordillo Altamirano FL, Barr JJ.. Unlocking the next generation of phage therapy: the key is in the receptors. Curr Opin Biotech. 2021;68:115–123. [DOI] [PubMed] [Google Scholar]
- 13.Sun B, Liu H, Jiang Y, et al. . New mutations involved in colistin resistance in Acinetobacter baumannii. Msphere. 2020;5(2):e819–e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu X, Wang N, Li X, et al. . The effect of colistin resistance-Associated mutations on the fitness of Acinetobacter baumannii. Front Microbiol. 2016;7:1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arroyo LA, Herrera CM, Fernandez L, et al. . The pmrCAB operon mediates Polymyxin resistance in Acinetobacter baumannii ATCC 17978 and Clinical Isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Ch. 2011;55(8):3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayol A, Poirel L, Brink A, et al. . Resistance to colistin Associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Ch. 2014;58(8):4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan M, Nhu NTK, Achard MES, et al. . Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemoth. 2017;72(10):2729–2736. [DOI] [PubMed] [Google Scholar]
- 18.Loh B, Wang X, Hua X, et al. . Complete genome sequence of the lytic bacteriophage Phab24, which infects clinical strains of the nosocomial pathogen Acinetobacter baumannii. Microbiol Resour Announc. 2021;10(40):e0066921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tynecki P, Guziński A, Kazimierczak J, et al. (2020). PhageAI-bacteriophage life cycle recognition with machine learning and natural language processing. BioRxiv doi: 10.1101/2020.07.11.198606. [DOI]
- 20.Wielgoss S, Bergmiller T, Bischofberger AM, et al. . Adaptation to parasites and costs of parasite resistance in mutator and nonmutator bacteria. Mol Biol Evol. 2016;33(3):770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deatherage DE, Barrick JE. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq, p 165-188, Engineering and analyzing multicellular systems doi: 10.1007/978-1-4939-0554-6_12. Springer. [DOI] [PMC free article] [PubMed]
- 22.Singh JK, Adams FG, Brown MH.. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front Microbiol. 2019;9:3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenyon JJ, Hall RM.. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. Plos One. 2013;8(4):e62160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Chen T, Yan B, et al. . Dual role of gnaA in antibiotic resistance and virulence in Acinetobacter baumannii. Antimicrob Agents Ch. 2019;63(10):e00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo SC, Yang SP, Lee YT, et al. . Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne bla(OXA-72) in Taiwan. Bmc Infect Dis. 2013;13:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Q, Wang X, Huang H, et al. . Novel host Recognition mechanism of the K1 capsule-specific phage of Escherichia coli: capsular polysaccharide as the first receptor and Lipopolysaccharide as the secondary receptor. J Virol. 2021;95(18):e92021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier MR, Casella LG, Jones JW, et al. . Unique structural modifications Are present in the Lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Ch. 2013;57(10):4831–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talyansky Y, Nielsen TB, Yan J, et al. . Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. Plos Pathog. 2021;17(2):e1009291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding CM, Hennon SW, Feldman MF.. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber BS, Harding CM, Feldman MF.. Pathogenic acinetobacter: from the cell surface to infinity and beyond. J Bacteriol. 2016;198(6):880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua X, Liang Q, Fang L, et al. . Bautype: capsule and Lipopolysaccharide serotype prediction for Acinetobacter baumannii genome. Infectious Microbes and Diseases. 2020;2(1):18–25. [Google Scholar]
- 32.Gu LC, Green SI, Min L, et al. . Phage-antibiotic synergy is driven by a Unique combination of antibacterial mechanism of action and stoichiometry. Mbio. 2020;11(4):e01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan EM, Alkawareek MY, Donnelly RF, et al. . Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol Med Microbiol. 2012;65(2):395–398. [DOI] [PubMed] [Google Scholar]
- 34.Kenyon JJ, Hall RM.. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. Plos One. 2013;8(4):e62160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sørensen MCH, van Alphen LB, Harboe A, et al. . Bacteriophage f336 recognizes the capsular phosphoramidate modification of campylobacter jejuni NCTC11168. J Bacteriol. 2011;193(23):6742–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knirel YA, Shneider MM, Popova AV, et al. . Mechanisms of Acinetobacter baumannii capsular polysaccharide cleavage by phage depolymerases. Biochemistry (Mosc. 2020;85(5):567–574. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Mi Z, Mi L, et al. . Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. Peerj. 2019;7:e6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira H, Costa AR, Konstantinides N, et al. . Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ Microbiol. 2017;19(12):5060–5077. [DOI] [PubMed] [Google Scholar]
- 39.Gordillo Altamirano F, Forsyth JH, Patwa R, et al. . Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol. 2021;6(2):157–161. [DOI] [PubMed] [Google Scholar]
- 40.Hesse S, Rajaure M, Wall E, et al. . Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. Mbio. 2020;11(1):e02530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filippov AA, Sergueev KV, He Y, et al. . Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. Plos One. 2011;6(9):e25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai R, Wang G, Le S, et al. . Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front Microbiol. 2019;10:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oechslin F. Resistance development to bacteriophages occurring during Bacteriophage therapy. Viruses. 2018;10(7):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.León M, Bastías R.. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol. 2015;06:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burmeister AR, Turner PE.. Trading-off and trading-up in the world of bacteria-phage evolution. Curr Biol. 2020;30(19):R1120–R1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zavascki AP, Goldani LZ, Li J, et al. . Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemoth. 2007;60(6):1206–1215. [DOI] [PubMed] [Google Scholar]
- 47.Leptihn S, Har JY, Chen J, et al. . Single molecule resolution of the antimicrobial action of quantum dot-labeled sushi peptide on live bacteria. Bmc Biol. 2009;7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García-Quintanilla M, Carretero-Ledesma M, Moreno-Martínez P, et al. . Lipopolysaccharide loss produces partial colistin dependence and collateral sensitivity to azithromycin, rifampicin and vancomycin in Acinetobacter baumannii. Int J Antimicrob Ag. 2015;46(6):696–702. [DOI] [PubMed] [Google Scholar]
- 49.Gazel D, Tatman Otkun M, Akçalı A.. In vitro activity of methylene blue and eosin methylene blue agar on colistin-resistant A. baumannii: an experimental study. J Med Microbiol. 2019;68(11):1607–1613. [DOI] [PubMed] [Google Scholar]
- 50.Aslam S, Courtwright AM, Koval C, et al. . Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am J Transplant. 2019;19(9):2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson JK, Robinson GL, Pineles LL, et al. . Carbapenem MICs in Escherichia coli and Klebsiella species producing extended-spectrum β-lactamases in critical Care patients from 2001 to 2009. Antimicrob Agents Ch. 2017;61(4):e01718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tagliaferri TL, Jansen M, Horz H.. Fighting pathogenic bacteria on Two fronts: phages and antibiotics as combined strategy. Front Cell Infect Mi. 2019;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spapen H, Jacobs R, Van Gorp V, et al. . Renal and neurological side effects of colistin in critically ill patients. Ann Intensive Care. 2011;1(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangalea MR, Duerkop BA.. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect Immun. 2020;88(7):e00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burmeister AR, Fortier A, Roush C, et al. . Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc Natl Acad Sci U S A. 2020;117(21):11207–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi Y, Hua X, Xu Q, et al. . Mechanism of eravacycline resistance in Acinetobacter baumannii mediated by a deletion mutation in the sensor kinase adeS, leading to elevated expression of the efflux pump AdeABC. Infect Genet Evol. 2020;80(104185):1. [DOI] [PubMed] [Google Scholar]
- 57.M100-S11 . Performance standards for antimicrobial susceptibility testing. Clin Microbiol Newsl. 2001;23(6):49. [Google Scholar]
- 58.Caroff M, Novikov A.. Lipopolysaccharides: structure, function and bacterial identification. OCL. 2020;27(31):1. [Google Scholar]
- 59.Davis JMR, Goldberg JB.. Purification and visualization of Lipopolysaccharide from gram-negative bacteria by hot aqueous-phenol extraction. J Visualized Exp. 2012;63:e3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu L, Shi Y, Xu Q, et al. . Capsule thickness, not biofilm formation, gives rise to mucoid Acinetobacter baumannii phenotypes that are more prevalent in long-term infections: A study of clinical isolates from a Hospital in China. Infect Drug Resist. 2020;13:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richmond GE, Evans LP, Anderson MJ, et al. . The Acinetobacter baumannii Two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio. 2016;7(2):e00430–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.