Abstract
Purpose
To investigate the clinical findings in Chinese patients diagnosed with familial exudative vitreoretinopathy (FEVR) and carrying pathogenic mutations.
Methods
One hundred twenty unrelated patients with FEVR were enrolled in this study. Genomic DNA and ophthalmic examinations were collected from all the patients and their available relatives. Targeted next-generation sequencing was performed to detect mutations. In silico programs were used to evaluate the pathogenicity of all the mutations.
Results
Eighty identified mutations were found in 81 unrelated patients (31/81 in LRP5, 25/81 in FZD4, 12/81 in TSPAN12, 8/81 in NDP, 4/81 in KIF11, and 1/81 in ZNF408). Among those mutations, 53 were novel (23/35 in LRP5, 15/21 in FZD4, 8/11 in TSPAN12, 3/8 in NDP, 3/4 in KIF11, 1/1 in ZNF408). Patients with LRP5, FZD4, TSPAN12, or NDP mutations were mainly classified into stage 4 and stage 5 and one-half of patients with KIF11 mutations were in stage 4. In addition, all the patients in NDP group were found to have bilateral symmetry in FEVR stage.
Conclusions
Our results present profound phenotypic variability and a wide mutation spectrum of FEVR in the Chinese population, which could be useful for a precise and comprehensive genetic diagnosis for patients with FEVR in the future.
Keywords: familial exudative vitreoretinopathy, inheritance, ocular manifestation, retinal vascular development
Familial exudative vitreoretinopathy (FEVR) is an inherited retinal disease characterized by incomplete development of retinal vessel and abnormal neovascularization, which was first reported by Criswick and Schepens in 1969.1 The retinal vascular anomalies in FEVR could result in several secondary changes, including fibrovascular proliferation, vitreous hemorrhage, retinal folding, vitreoretinal traction, macular dragging, and partial or total retinal detachment. The clinical manifestation of FEVR varies widely, ranging from no visual impairment to total blindness, and the severity of ocular symptoms could differ within one family harboring the same mutation or between bilateral eyes of the same individual.2
FEVR is clinically and genetically heterogeneous, and it has three inherited forms: autosomal dominant, autosomal recessive, and X-linked recessive. To date, variants in several genes have been identified as causative for FEVR development, including FZD4, LRP5, NDP, TSPAN12, KIF11, and ZNF408. The proteins encoded by the first four genes have been reported to link to the Norrin/β-catenin signaling pathway, which is crucial for the retinal vascular formation during eye development.3,4 KIF11 is a kinesin family member motor protein localized to spindle microtubules, which is involved in mitotic progression, and its lack could result in severely stunted growth of the retinal vessels.5 The ZNF408 protein belongs to the family of zinc fingers, and the ZNF408 mutation leads to retinal vascularization defects and abnormal trunk vascularization in the zebrafish model.6
To date, several studies have attempted to explore the genotype–phenotype correlations in Chinese patients with FEVR. Despite the complex relationship, some trends for FEVR have been reported. Wang et al.7 observed that nearly one-half of patients with FZD4 mutations showed stage 5 and more than one-half of them displayed asymmetric symptoms, indicating a large-scale degree of phenotypic severity in FZD4 mutation. Rao et al.8 found that patients with LRP5 mutations exhibited broader phenotypic spectrum varying from stage 2 to stage 5, and all patients with NDP or truncating mutations were in stage 4 or worse. In addition, patients with FEVR with NDP, TSPAN12, or KIF11 mutations seemed to have symmetrical retinopathy bilaterally, but a greater frequency of asymmetry was found in patients with LRP5 and FZD4 mutations.9 Interestingly, Li et al.10 reported that the phenotype in patients with digenic variants tended to be worse than that with monogenic variants of FEVR-associated genes.
Here, we screened for pathogenic mutations in six genes (FZD4, LRP5, NDP, TSPAN12, KIF11, and ZNF408) in 120 unrelated Chinese patients with FEVR, and then investigated and analyzed their ocular manifestations according to the mutation spectrum.
Methods
Patients
We recruited 120 patients with FEVR from the Department of Ophthalmology at Peking University People's Hospital. Written informed consent was obtained from all participants or parents on behalf of child participants. The collection of clinical and genetic data was approved by the patients and their families. This study was approved by the ethical committee of Peking University People's Hospital and adhered to the tenets of the Declaration of Helsinki. The diagnosis of FEVR was based on the clinical criteria described previously.11 Patients with a history of premature birth, systemic abnormalities, and ocular trauma were excluded.
Clinical Examinations
Comprehensive ophthalmic examinations, such as indirect ophthalmoscopy, color fundus photography, fluorescein fundus angiography, ocular B-scan ultrasound examinations, and optical coherence tomography, were obtained from all patients and their family members where available. Data for sex, age, and family history were recorded, and ocular symptoms and medical history were reviewed. The severities of the affected eyes were assessed following the clinical classification of FEVR as previously described.12
Molecular Analysis
Peripheral blood samples were drawn from all patients and their available relatives, and genomic DNA was extracted using the QIAamp DNA Blood Kit (Qiagen, Germany) and fragmented to generate 350 to 400 bp products. DNA fragments were amplified by PCR and allowed to hybridize with DNA capture probes, which were designed for the targeted genes. The DNA products were eluted and amplified again, and targeted next‑generation sequencing was performed using the Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA). A custom-inherited retinal diseases panel based on targeted exome capture technology was previously established and covered the known FEVR genes: FZD4, LRP5, NDP, TSPAN12, KIF11, and ZNF408.
The possible pathogenicity of missense variants was further estimated by using SIFT (http://sift.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), and Mutation Taster (http://www.mutationtaster.org) online algorithms and via evolutionary conservation analysis.
The Genome Aggregation database (http://gnomad-sg.org/) and the Exome Aggregation Consortium database (http://exac.broadinstitute.org/faq) were used to evaluate the minor allele frequencies in study participants. In addition, identified variants were also evaluated regarding pathogenicity according to the standards and guidelines of American College of Medical Genetics and Genomics.13
Analysis of FEVR-Related Studies
We collected the FEVR-related literature on Chinese patients to analyze the spectrum of FZD4, LRP5, NDP, TSPAN12, KIF11, and ZNF408 genes. This review process was performed using PubMed and Web of Science for an extensive search. Additionally, related articles and their references were screened to obtain additional and potentially eligible information.
Results
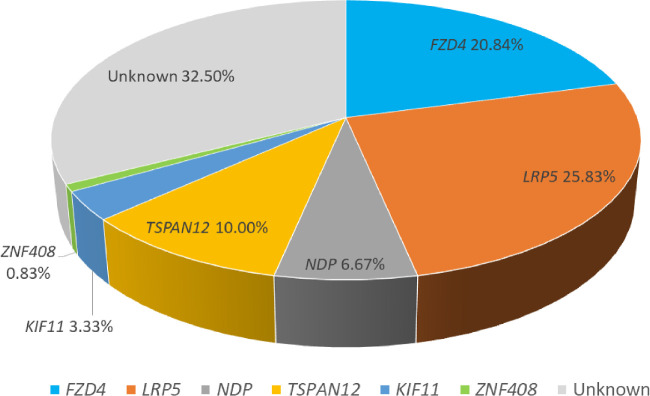
In this study, 81 of 120 patients with FEVR were detected to harbor mutations; 21 were females and 60 were males. The average patients age was 10.64 years (range, 4–40 years). Among the patients carrying mutations (Fig. 1), LRP5 accounted for the greatest proportion (31/120 [25.83%]), followed by FZD4 (25/120 [20.83%]), TSPAN12 (12/120 [10.00%]), NDP (8/120 [6.67%]), KIF11 (4/120 [3.33%]), and ZNF408 (1/120 [0.83%]). In addition, we retrospectively reviewed reports on Chinese patients with FEVR in the past 5 years,8,9,14–17 focusing on the mutation spectrum shown in Supplementary Table S1 (number of genes, ≥3). The largest cohort of patients with FEVR collected and screened for genetic analysis by a single clinic in the Chinese literature was in 2018. The age of patients with FEVR ranged from 0 to 56 years, and the percentage of patients with detected variants ranged from 23.00% to 67.40%. Among these domestic reports, the frequency of LRP5 mutations ranged from 10.00% to 25.93%, FZD4 mutations ranged from 6.45% to 21.35%, TSPAN12 mutations ranged from 3.23% to 12.90%, NDP mutations ranged from 4.11% to 9.68%, and KIF11 mutations ranged from 1.61% to 6.74%. One study found the ZNF408 mutations were identified in 1.80% of patients with FEVR.
Figure 1.
Mutation spectrum of FZD4, LRP4, NDP, TSPAN12, KIF11, and ZNF408 in 120 Chinese patients with FEVR.
Mutations in the FZD4 Gene
We found 21 mutations in FZD4 gene from the 25 probands. Of these mutations, six were previously reported2,18–22 and 15 were novel. The mutations included 15 missense mutations, 2 nonsense mutations, and 4 frameshift mutations (Table 1). The most frequently encountered mutations in FZD4 was c.205C>T; p.H69Y, detected in five patients (5/25 [20.00%]), followed by the two mutations c.1282_1285delGACA; p.D428Sfs*2 and c.313A>G; p.M105V, each found in three (3/25 [12.00%]) and two patients (2/25 [8.00%]) respectively. In addition, two mutations occurred in the signal sequence portion (Supplementary Fig. S1), three were located in the cysteine-rich domain, one was located in upstream of the first transmembrane domain, 14 were located in the seven transmembrane domains (TMDs), and one was located in KTXXXW motif of the intracellular domain (ICD).
Table 1.
Identified Variants in FZD4 Gene of Patients With FEVR
| Patient ID | Sex | Age | Stage (OD/OS) | Gene | Nucleotide Changes | Protein Changes | SIFT | PP-2 | MT | ACMG | ExAC | gnomAD | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P01 | F | 9 | 4/4 | FZD4 | c.40_49delCCCGGGGGCG | p.P14Sfs*44 | – | – | DC | P | – | 0.00000426 | Reported |
| P02 | F | 7 | 4/5 | FZD4 | c.42_43delCG | p.G16Rfs*113 | – | – | DC | P | – | – | Novel |
| FZD4 | c.1300A>G | p.M434V | D | PbD | DC | LP | – | – | Novel | ||||
| P03 | M | 10 | 4/4 | FZD4 | c.205C>T | p.H69Y | D | B | DC | U | 0.00056200 | 0.00022400 | Reported |
| P04 | F | 16 | 1/1 | ||||||||||
| P05 | M | 9 | 4/3 | ||||||||||
| P06 | M | 12 | 2/2 | ||||||||||
| P07 | M | 23 | 4/1 | ||||||||||
| P08 | M | 14 | 4/2 | FZD4 | c.313A>G | p.M105V | T | PsD | DC | LP | 0.00001670 | 0.00002400 | Reported |
| P09 | M | 16 | 4/4 | ||||||||||
| P10 | M | 15 | 4/4 | FZD4 | c.341T>C | p.I114T | D | PsD | DC | LP | – | – | Reported |
| P11 | M | 16 | 4/4 | FZD4 | c.631T>C | p.Y211H | D | B | DC | U | – | 0.00002390 | Novel |
| FZD4 | c.1030A>C | p.S344R | D | PbD | DC | U | – | – | Novel | ||||
| FZD4 | c.1031G>C | p.S344T | D | PbD | DC | U | – | – | Novel | ||||
| P12 | M | 12 | 2/2 | FZD4 | c.684C>A | p.S228R | D | PsD | DC | LP | – | – | Novel |
| P13 | M | 5 | Normal eye/4 | FZD4 | c.686T>C | p.L229P | D | PbD | DC | LP | – | – | Reported |
| P14 | F | 6 | 4/4 | FZD4 | c.694A>G | p.I232V | T | B | DC | U | – | – | Novel |
| P15 | M | 10 | 4/5 | FZD4 | c.733T>A | p.S245T | T | B | DC | U | – | – | Novel |
| P16 | F | 11 | 1/3 | FZD4 | c.807T>G | p.Y269* | – | – | DC | LP | – | – | Novel |
| P17 | M | 8 | 5/5 | FZD4 | c.877A>G | p.I293V | T | B | DC | LP | – | – | Novel |
| P18 | F | 16 | 4/4 | FZD4 | c.983T>C | p.F328S | D | PbD | DC | U | – | – | Novel |
| P19 | M | 10 | 3/4 | FZD4 | c.1282_1285delGACA | p.D428Sfs*2 | – | – | DC | P | 0.00000824 | 0.00001060 | Reported |
| P20 | M | 9 | 5/5 | ||||||||||
| P21 | F | 11 | Artificial eye/5 | ||||||||||
| P22 | M | 9 | 5/1 | FZD4 | c.1328T>C | p.L443P | D | PbD | DC | U | – | – | Novel |
| P23 | M | 12 | 5/2 | FZD4 | c.1387delG | p.A463Hfs*17 | – | – | DC | LP | – | – | Novel |
| P24 | F | 11 | 1/5 | FZD4 | c.1482G>A | p.W494* | – | – | DC | LP | – | – | Novel |
| P25 | M | 16 | 1/5 | FZD4 | c.1502T>C | p.L501P | D | PbD | DC | U | – | – | Novel |
ACMG, American College of Medical Genetics and Genomics; B, benign; D, damaging; DC, disease causing; LP, likely pathogenic; MT, mutation taster; PbD, probably damaging; Pm, polymorphism; PP-2, Polyphen-2; P, pathogenic; PsD, possibly damaging; T, tolerated; U, uncertain.
Mutations in the LRP5 Gene
We identified 35 mutations of LRP5 gene in the 31 probands; 12 mutations had been reported,8,15,23–28 and 23 mutations were newly detected. The LRP5 mutations included 27 missense mutations, 3 nonsense mutations, 2 splicing mutations, 2 duplications, and 1 frameshift mutation (Table 2). The duplication c.55_60dupCTGCTG; p.19_20dulLL was detected in three patients (3/31 [9.68%]), three mutations c.58_60dupCTG; p.20dupL, c.1330C>T; p.R444C and c.4643G>T; p.C1548F, were each found in two patients (2/31 [6.67%]), it is noteworthy that the minor allele frequencies of c.58_60dupCTG; p.20dupL was 0.153 (>10%) in ExAC, and it was classified as a benign mutation. There were nine patients who harbored two heterozygous mutations in LRP5 gene, one mutation was inherited from the mother and the other one from the father. In addition, 2 mutations were in the signal peptide, 26 mutations occurred in the 4 tandem YWTD-type β-propeller (BP) domains (six in BP1, 12 in BP2, five in BP3, and three in BP4), two were located in the LDLR type A domains, and three were located in the ICD (Supplementary Fig. S2).
Table 2.
Identified Variants in LRP5 Gene of Patients With FEVR
| Patient ID | Sex | Age | Stage (OD/OS) | Gene | Nucleotide Changes | Protein Changes | SIFT | PP-2 | MT | ACMG | ExAC | gnomAD | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P29 | M | 40 | 4/4 | LRP5 | c.55_60dupCTGCTG | p.19_20dupLL | – | – | Pm | U | 0.00218000 | 0.00125000 | Reported |
| P30 | M | 8 | 2/4 | ||||||||||
| P26* | F | 10 | 4/4 | LRP5 | c.55_60dupCTGCTG | p.19_20dupLL | – | – | Pm | U | 0.00218000 | 0.00125000 | Reported |
| LRP5 | c.1294T>G | p.W432G | D | PbD | DC | LP | – | – | Novel | ||||
| P27 | M | 13 | 2/4 | LRP5 | c.58_60dupCTG | p.20dupL | – | – | Pm | B | 0.15300000 | 0.08750000 | Reported |
| P28† | M | 7 | 2/2 | LRP5 | c.58_60dupCTG | p.20dupL | – | – | Pm | B | 0.15300000 | 0.08750000 | Reported |
| LRP5 | c.3246C>G | p.Y1082* | – | – | DC | LP | – | – | Novel | ||||
| P31 | M | 9 | 5/5 | LRP5 | c.107T>C | p.L36P | D | PbD | DC | LP | – | – | Novel |
| P32 | M | 8 | 5/5 | LRP5 | c.235T>G | p.W79G | D | PbD | DC | LP | – | – | Novel |
| P33 | M | 5 | 3/4 | LRP5 | c.280C>A | p.Q94K | T | B | DC | U | – | – | Novel |
| P34 | M | 13 | 4/4 | LRP5 | c.542T>C | p.M181T | D | PbD | DC | LP | – | – | Novel |
| P35‡ | M | 26 | 5/5 | LRP5 | c.676G>A | p.G226S | D | PbD | DC | LP | – | – | Novel |
| LRP5 | c.1123G>A | p.A375T | D | PsD | DC | LP | 0.00002480 | 0.00000796 | Reported | ||||
| P36 | F | 10 | 4/4 | LRP5 | c.1057C>T | p.R353W | D | PbD | DC | LP | 0.00001670 | 0.00000399 | Novel |
| P37§ | F | 10 | 5/5 | LRP5 | c.1133T>C | p.I378T | D | PsD | DC | LP | – | – | Novel |
| LRP5 | c.2422G>A | p.D808N | D | PsD | DC | LP | 0.00000832 | 0.00000399 | Novel | ||||
| P38‖ | M | 7 | 4/5 | LRP5 | c.1145C>T | p.P382L | D | PbD | DC | P | – | 0.00000398 | Reported |
| LRP5 | c.1364C>T | p.S455L | T | PbD | DC | P | – | 0.00000401 | Reported | ||||
| P39¶ | M | 9 | 5/5 | LRP5 | c.1148T>C | p.L383P | D | PsD | DC | LP | – | – | Novel |
| LRP5 | c.4105_4106delAT | p.M1369Vfs*2 | – | – | DC | LP | 0.00000826 | 0.00000399 | Novel | ||||
| P40 | M | 15 | 4/4 | LRP5 | c.1199C>T | p.A400V | T | B | DC | U | 0.00029800 | 0.00013800 | Novel |
| P41 | M | 8 | 4/5 | LRP5 | c.1330C>T | p.R444C | D | PsD | DC | LP | 0.00002500 | 0.00000657 | Reported |
| P42 | M | 10 | 3/4 | ||||||||||
| P43# | F | 10 | 4/4 | LRP5 | c.1349G>A | p.R450H | D | PbD | DC | P | – | 0.00000400 | Reported |
| LRP5 | c.3245A>G | p.Y1082C | D | PbD | DC | LP | 0.00008340 | 0.00007480 | Novel | ||||
| P44 | M | 8 | 5/5 | LRP5 | c.1385G>A | p.R462Q | D | PbD | DC | U | 0.00000848 | 0.00000804 | Novel |
| P45 | M | 7 | 3/2 | LRP5 | c.2237G>A | p.R746Q | D | PbD | DC | LP | 0.00003320 | 0.00000657 | Reported |
| P46** | M | 6 | 4/4 | LRP5 | c.2488T>C | p.S830P | D | PbD | DC | LP | – | Novel | |
| LRP5 | c.479C>T | p.P160L | D | PbD | DC | LP | 0.00000848 | 0.00000413 | Novel | ||||
| P62 | F | 7 | 1/1 | LRP5 | c.2512C>T | p.R838W | D | PbD | DC | U | 0.00000828 | 0.00000803 | Novel |
| P47 | M | 7 | 4/4 | LRP5 | c.2555C>T | p.T852M | D | PbD | DC | LP | – | 0.00001310 | Reported |
| P48 | F | 8 | 2/5 | LRP5 | c.3237–2A>G | – | – | – | – | U | – | – | Novel |
| P49 | M | 7 | 4/Normal eye | LRP5 | c.3361A>G | p.N1121D | T | B | DC | LP | 0.00066500 | 0.00039500 | Reported |
| P50 | M | 7 | 2/1 | LRP5 | c.3901G>A | p.A1301T | T | B | Pm | U | 0.00018500 | 0.00013100 | Novel |
| P51 | M | 14 | 4/4 | LRP5 | c.4488+5G>C | – | – | – | – | U | – | – | Novel |
| P52 | M | 11 | 5/5 | LRP5 | c.4600C>T | p.R1534* | – | – | DC | P | – | – | Novel |
| P53, | M | 11 | 4/4 | LRP5 | c.4643G>T | p.C1548F | D | PbD | DC | LP | 0.00028400 | 0.00019700 | Reported |
| P54 | M | 10 | 2/4 | ||||||||||
| P55†† | F | 10 | 4/4 | LRP5 | c.4670G>A | p.W1557* | – | – | DC | P | – | – | Novel |
| LRP5 | c.1378G>A | p.E460K | D | PbD | DC | P | – | 0.00000657 | Reported |
ACMG, American College of Medical Genetics and Genomics; B, benign; D, damaging; DC, disease causing; LP, likely pathogenic; MT, mutation taster; PbD, probably damaging; Pm, polymorphism; PP-2, Polyphen-2; P, pathogenic; PsD, possibly damaging; T, tolerated; U, uncertain.
P26 inherited the mutation c.55_60dupCTGCTG; p.19_20dupLL from her father, who had no signs of FEVR, the c.1294T>G; p.W432G was inherited from her mother, whose information was unavailable.
P28 inherited the mutation c.58_60dupCTG; p.20dupLL from his father, and the mutation c.3246C>G; p.Y1082* was inherited from his mother; neither parent had signs of FEVR.
P35 inherited the mutation c.676G>A; p.G226S from his father, and inherited the mutation c.1123G>A; p.A375T from his mother; neither parent had signs of FEVR.
P37 inherited the mutation c.1133T>C; p.I378T from her father, and inherited the mutation c.2422G>A; p.D808N from her mother, neither parents had signs of FEVR.
P38 inherited the mutation c.1364C>T; p.S455L from his father, and inherited the mutation c.1145C>T; p.P382L from his mother; neither parent had signs of FEVR.
P39 inherited the mutation c.4105_4106delAT; p.M1369Vfs*2 from his father, who had no signs of FEVR, the c.1148T>C; p.L383P was inherited from his mother, who had stage 1 of FEVR.
P43 inherited the mutation c.1349G>A; p.R450H from her father, and inherited the mutation c.3245A>G; p.Y1082C from her mother; neither parent had signs of FEVR.
P46 inherited the mutation c.479C>T; p.P160L from his father, and inherited the mutation c.2488T>C; p.S830P from his mother; neither parent had signs of FEVR.
P55 inherited the mutation c.1378G>A; p.E460K from her father, and inherited the mutation c.4670G>A; p.W1557* from her mother; neither parent had signs of FEVR.
Mutations in the NDP Gene
We detected eight mutations of NDP gene in the eight probands. Among these mutations, five were previously reported15,29–31 and three were novel. The NDP mutations included three missense mutations, four nonsense mutations, and one deletion (Table 3). Only one mutation c.22G>C; p.A8P was located in the signal peptide, the other seven mutations were located in the C-terminal cysteine knot domain (Supplementary Fig. S3).
Table 3.
Identified Variants in NDP, TSPAN12, KIF11, and ZNF408 Genes of Patients With FEVR
| Patient ID | Sex | Age | Stage (OD/OS) | Gene | Nucleotide Changes | Protein Changes | SIFT | PP-2 | MT | ACMG | ExAC | gnomAD | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P56 | M | 22 | 4/4 | NDP | c.22G>C | p.A8P | D | PbD | DC | LP | – | – | Reported |
| P57 | M | 4 | 5/5 | NDP | c.239_241delCGT | p.80_81delSF | – | – | DC | LP | – | – | Novel |
| P58 | M | 4 | 4/4 | NDP | c.268C>T | p.R90C | D | PbD | DC | LP | – | – | Reported |
| P59 | M | 4 | 4/4 | NDP | c.325C>T | p.R109* | – | – | DC | P | – | – | Reported |
| P60 | M | 6 | 5/5 | NDP | c.343C>T | p.R115* | – | – | DC | P | – | – | Reported |
| P61 | M | 9 | 5/5 | NDP | c.358T>G | p.Y120D | D | PbD | DC | U | – | – | Novel |
| P63 | M | 9 | 5/5 | NDP | c.384C>A | p.128C* | – | – | DC | P | – | – | Reported |
| P64 | M | 12 | 2/2 | NDP | c.388G>T | p.E130* | – | – | DC | LP | – | – | Novel |
| P65 | M | 12 | 5/5 | TSPAN12 | c.95delC | p.S32Lfs*4 | – | – | DC | LP | – | – | Novel |
| P66 | F | 33 | 2/1 | TSPAN12 | c.194C>T | p.P65L | T | B | DC | LP | – | 0.00000658 | Reported |
| P67 | F | 10 | 4/4 | TSPAN12 | c.232G>A | p.G78R | D | PbD | DC | LP | – | – | Novel |
| P68 | M | 10 | 4/4 | TSPAN12 | c.352G>T | p.E118* | – | – | DC | P | – | – | Reported |
| P69 | F | 10 | 4/Normal eye | TSPAN12 | c.361–2A>G | – | – | – | – | P | – | – | Novel |
| P70 | M | 8 | 5/5 | TSPAN12 | c.559C>A | p.P187T | T | B | DC | U | – | – | Novel |
| P71 | M | 12 | 5/5 | TSPAN12 | c.617G>T | p.C206F | D | PbD | DC | U | – | – | Novel |
| P72 | F | 13 | 1/4 | TSPAN12 | c.689T>A | p.I230N | D | PbD | DC | LP | – | – | Novel |
| P73 | M | 4 | 4/4 | TSPAN12 | c.689T>C | p.I230T | D | PbD | DC | U | 0.00002470 | 0.00002630 | Novel |
| P74 | F | 6 | 4/4 | TSPAN12 | c.738G>A | p.W246* | – | – | DC | LP | – | – | Novel |
| P75, | M | 16 | 1/4 | TSPAN12 | c.765G>T | p.P255P | T | – | Pm | B | 0.80300000 | 0.00000658 | Reported |
| P76 | F | 6 | 2/1 | ||||||||||
| P77 | M | 4 | 4/4 | KIF11 | c.308+1G>A | – | – | – | – | P | – | – | Reported |
| P78 | M | 8 | 2/2 | KIF11 | c.388–2A>G | – | – | – | – | U | – | – | Novel |
| P79 | M | 4 | 4/4 | KIF11 | c.1349_1353delGTAAA | p.C450Ffs*2 | – | – | DC | P | – | – | Novel |
| P80 | M | 7 | 2/3 | KIF11 | c.2268–4A>G | – | – | – | – | U | – | 0.00000657 | Novel |
| P81 | M | 5 | 3/4 | ZNF408 | c.1102C>A | p.L368I | D | PbD | DC | U | – | – | Novel |
ACMG, American College of Medical Genetics and Genomics; B, benign; D, damaging; DC, disease causing; LP, likely pathogenic; MT, mutation taster; PbD, probably damaging; Pm, polymorphism; PP-2, Polyphen-2; P, pathogenic; PsD, possibly damaging; T, tolerated; U, uncertain.
Mutations in the TSPAN12 Gene
We identified 11 mutations of the TSPAN12 gene in the 12 probands; three mutations had been reported14,15,32 and nine mutations were novel. There were six missense mutations, two nonsense mutations, one frameshift mutation, and one splicing mutation in TSPAN12 (Table 3). The mutation c.765G>T; p.P255P, which was detected in P75 and P76, had a minor allele frequency of 0.803 (>10%) in ExAC and was known as a common single nucleotide polymorphism without pathogenicity/ Additionally, five mutations were located in the TMDs, three mutations were located in the large extracellular loop (ECL-2), and two mutations occurred in the ICD (Supplementary Fig. S4).
Mutations in the KIF11 Gene
We found one reported splicing mutation c.308+1G>A,15 two novel splicing mutations, c.388–2A>G and c.2268–4A>G, and one novel frameshift mutation c.1349_1353delGTAAA; p.C450Ffs*2 in KIF11 gene from four probands (Table 3). The frameshift mutation was located in the downstream of kinesin motor domain, which is responsible for proper function of the KIF11 protein.
Mutation in the ZNF408 Gene
There is only one novel missense ZNF408 mutation, c.1102C>A; p.L368I, detected in a 5-year-old boy, who displayed bilateral retinal detachment, and he inherited this heterozygous mutation from his mother. The mutated 368 amino acid, which located in the first zinc finger of ZNF408 protein, was conserved among various vertebrates (Supplementary Fig. S5) and predicted to be pathogenic by three in silico programs (Table 3).
Clinical Presentation of Patients With Identified Mutations
Table 4 shows the FEVR stage of patients with mutations in six genes. Most of patients in FZD4 (21/25, 84.00%), LRP5 (27/31, 87.10%), NDP (7/8, 87.50%), or TSPAN12 (10/12, 83.33%) group had severe retinopathy (stages 4–5), and patients with FZD4 mutations presented relatively wider phenotypes that varied from stage 1 to stage 5. One-half of the patients (2/4 [50.00%]) with KIF11 mutations and one patient carrying ZNF408 mutation had in stage 4 disease.
Table 4.
Patients at Five Different Stages of FEVR in Different Gene Groups
| Stage* | Total | FZD4 | LRP5 | NDP | TSPAN12 | KIF11 | ZNF48 | Number With Identified Mutation |
|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 2 (50.00%) |
| 2 | 11 | 2 | 2 | 1 | 2 | 1 | 0 | 8 (72.73%) |
| 3 | 4 | 1 | 1 | 0 | 0 | 1 | 0 | 3 (75.00%) |
| 4 | 56 | 12 | 17 | 3 | 7 | 2 | 1 | 42 (75.00%) |
| 5 | 45 | 9 | 10 | 4 | 3 | 0 | 0 | 26 (57.78%) |
| Total | 120 | 25 | 31 | 8 | 12 | 4 | 1 | 81 (67.50%) |
The severity of patients was determined by the highest stage of FEVR in either eye.
Nearly one-half of patients in FZD4 (12/25 [48.00%]) group presented symmetry of staging (Table 4), while more than one-half of patients with LRP5 (20/31 [64.52%]), TSPAN12 (7/12 [58.33%]), or KIF11 (3/4 [75.00%]) mutations displayed bilateral symmetry. It is noteworthy that all patients with NDP (8/8 [100.00%]) mutations showed symmetrical FEVR stage between eyes. Only unilateral FEVR was found in three patients carrying FZD4, LRP5, and TSPAN12 mutation, accounting for 3.70% (3/81) of all the cases with identified mutations.
Discussion
The current study comprehensively screened six known genes (FZD4, LRP5, NDP, TSPAN12, KIF11, and ZNF408) in 120 unrelated patients with FEVR, and uncovered 78 of 120 patients (65.00%), except for three cases carrying the benign variants; this percentage in our study is similar to that of that of Wang et al.,9 who reported that 67.5% of probands had genetic confirmed FEVR.
We found that up to 76 patients (63.33%) were detected to harbor mutations in Norrin/β-catenin signaling genes, including FZD4, LRP5, NDP, and TSPAN12; the proteins encoded by these genes are found to had intense interaction with each other, and mechanisms for these cooperative proteins in retinal vascular formation have been explored in recent years.3,4,33 Norrin acts as a ligand, it could bind to FZD4 receptor or LRP5 coreceptor specifically and form a ternary complex; this process is meditated by TSPAN12 selectively. Then, a downstream β-catenin signaling is initiated, the increasing cytoplasmic β-catenin is translocated to the nucleus and interacts with T-cell factor or lymphoid enhancing factor, resulting in RNA transcription and elongation consequently. In this study, we found that patients with FZD4, LRP5, NDP, and TSPAN12 mutations were mainly classified into stages 4 and 5 (Table 4), suggesting that Chinese patients with FEVR carrying mutations in the Norrin/β-catenin signaling genes tended to exhibit severe retinopathy.
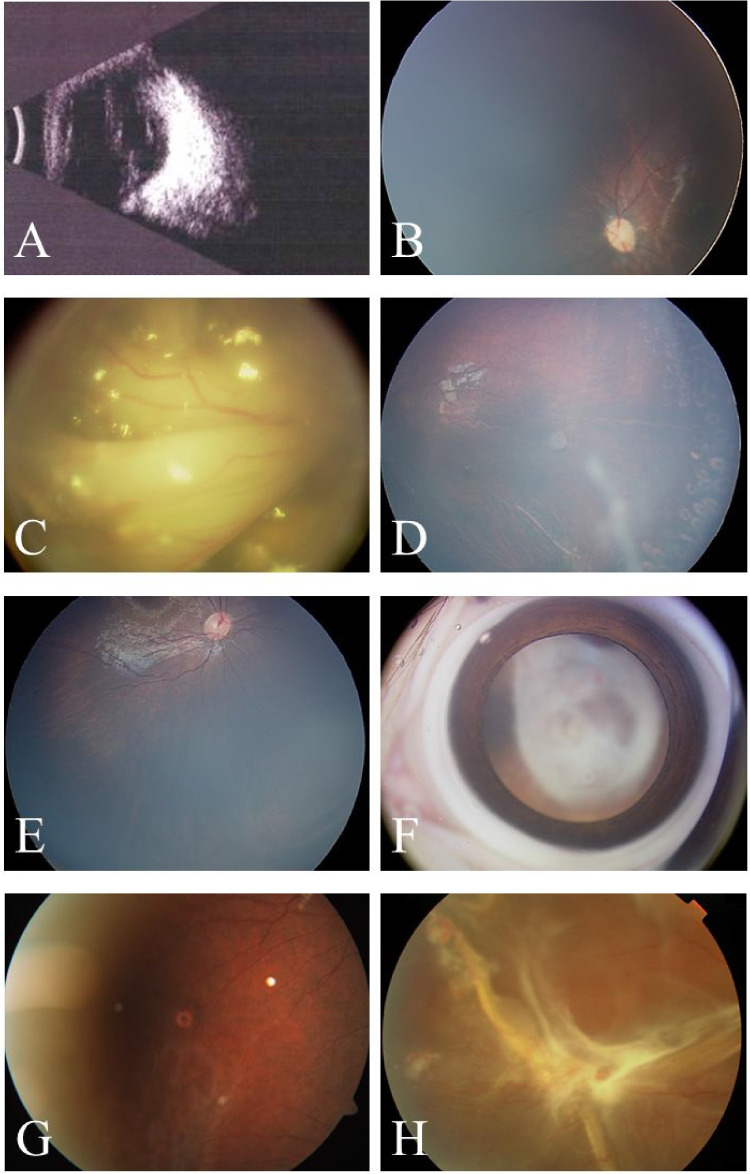
FZD4 works as the receptor for Wnt or Norrin, and it mainly consists of signal sequence, cysteine-rich domain, seven TMDs, and ICD, which many FZD4 mutations occur in these functional areas. Compared with the previous studies for FZD4-mutated patients from Northern America,34–37 the detection rates of identified mutations in each area of FZD4 protein was different. Our study found that mutations located in TMDs accounting for 66.67%, whereas the cysteine-rich domain was the most frequently mutated domain in the Northern American population (48.00%), suggesting that the genotypic spectrum for FZD4 gene in Chinese patients might differ from that of other populations. We also noticed that three FZD4 mutations, including c.205C>T; p.H69Y, c.1282_1285delGACA; p.D428Sfs*2, and c.313A>G; p.M105V, had been reported as a hotspot in several studies.7,8,15,36,38 There were five, three, and two patients of this study harboring the three mutations, but the reason for their high frequency in FZD4 mutations remained unclear. In addition, four patients carrying mutations c.1328T>C; p.L443P, c.1387delG; p.A463Hfs*17, c.1482G>A; p.W494*, and c.1502T>C; p.L501P, which were located in a highly conserved motif (KTXXXW) and its upstream, showed a difference of three or more grades between bilateral eyes (Fig. 2). The KTXXXW motif was important for the activation of canonical Wnt signaling pathway, membrane relocalization, and phosphorylation of Dishevelled,39,40 although we were unable to determine how the four mutations affect their phenotypes. This finding indicated that FZD4 mutations, which were considered to cause abnormality of KTXXXW, might result in asymmetric severe retinopathy of patients with FEVR.
Figure 2.
Ophthalmic examinations of four probands who carried four FZD4 mutations c.1328T>C; p.L443P, c.1387delG; p.A463Hfs*17, c.1482G>A; p.W494*, and c.1502T>C; p.L501P. (A, B) Ocular B-scan ultrasound and color fundus photography (CFP) showed the stage 5 of right eye and the stage 1 of left eye in P22 respectively. (C, D) CFP showed the stage 5 of right eye and the stage 2 of left eye in P23. (E, F) CFP showed the stage 1 of right eye and the stage 5 of left eye in P24. (G, H) CFP showed the stage 1 of right eye and the stage 5 of left eye in P25.
LRP5 belongs to the low-density lipoprotein receptor family and interacts with FZD4 synergistically to bind Norrin/Wnt ligands, forming a functional complex to trigger the Norrin/β-catenin or the Wnt signaling pathway. There were 35 mutations in LRP5 identified from 31 patients, accounting for the greatest proportion in our study. This result is consistent with those of Rao et al.8 and Li et al.15 However, some studies have reported the greatest involvement in the FZD4 gene.7,9,14,16 Considering the cooperative relationship between FZD4 and LRP5 proteins, a greater number of cases was required to validate whether FZD4 or LRP5 accounts for the most frequently gene of FEVR in Chinese population.
Norrin is a secreted signaling factor with the characteristics of autocrine or paracrine, and is constitutively expressed by Müller cells of the retina.41 This protein contains a signal peptide and a highly conserved C-terminal cysteine knot domain. The known mutation c.22G>C; p.A8P detected in P56 affected the directing localization for Norrin; the other eight mutations were in the C-terminal cysteine knot and might impact its function. In our study, all patients carrying mutations in NDP had bilateral symmetry, and most of them (7/8 [87.50%]) had stage 4 or stage 5 disease, highlighting the higher frequency of symmetric severe retinopathy in NDP phenotypes. This finding is similar to that of Wang et al.,9 who suggested that patients with NDP mutations might be likely to exhibited symmetrical and severe disease stage between eyes.
The TSPAN12 gene encodes a 305 amino acid protein, which is the member of the Tetraspanin family. TSPAN12 is involved in the retinal vascular development by promoting the Norrin/β-catenin but not the Wnt/β-catenin signaling pathway.4 In our study, 6 of 11 mutations in TSPAN12 were located in the second extracellular loop (ECL-2) and its upstream, and this extracellular loop is responsible for TSPAN12 interacting with FZD4/Norrin and meditating the FZD4 ligand selectivity.33 Therefore, these TSPAN12 mutations were considered to affect its function in the activation of Norrin/β-catenin signaling. In addition, the ratio of female-to-male patients in the TSPAN12 group (6:6) was relatively higher than that of the FZD4 (8:17), LRP5 (7:24), or NDP (0:8) groups, indicating a high percentage of female individuals among the patients with TSPAN12 mutations.
KIF11 and ZNF408 are newly recognized FEVR-associated genes, which are required for mitotic spindle assembly and high affinity DNA binding, respectively,5,6 and the mechanisms for KIF11 or ZNF408 mutations causing abnormal retinal vascularization were independent of Norrin/β-catenin signaling pathway. Here, we detected four KIF11 mutations and one ZNF408 mutation from five patients, which each accounted for 3.33% and 0.83% of the cohort, respectively. To date, only a small number of mutations in KIF11 and ZNF408 has been reported from patients with FEVR, and the roles of these two proteins in retinal vascular development require further investigation to uncover the involvement of KIF11 or ZNF408 in the pathogenesis of FEVR. In a domestic study of 389 Chinese patients, 8 patients were detected to carry 9 ZNF408 mutations,15 which might be the largest sample size of patients with FEVR with ZNF408 mutations. The ZNF408 protein, which was altered in P81, is composed of 10 Zinc fingers domains with different and still unclear cellular functions; variants in different domains could affect the interaction of ZNF408 with specific targets, thus leading to the dysregulation of different target genes and subsequently are underlying either FEVR or retinitis pigmentosa.42 Owing to the lack of evidence supporting the pathogenicity of mutation c.1102C>A; p.L368I in FEVR, our study tentatively classified it as a variant with unknown significance.
In conclusion, our study expanded the mutation spectrum of Chinese patients with FEVR, thus contributing to knowledge of genotype-phenotype relationship in this inherited ocular disease. Owing to the limited number of patients in this study, establishing a comprehensive database with more patient sources is required to further explore the etiology of FEVR, which is helpful for clinical and genetic counselling of FEVR or other retinal vascular diseases.
Supplementary Material
Acknowledgments
Supported by the National Natural Science Foundation of China Grant (81470649, 81670870), the National Key Research and Development Program of China (2020YFC2008200), and the Science and Technology Innovation Project of Chinese Academy of Medical Sciences (2019-RC-HL-019).
Disclosure: T. Tao, None; N. Xu, None; J. Li, None; H. Li, None; J. Qu, None; H. Yin, None; J. Liang, None; M. Zhao, None; X. Li, None; L. Huang, None
References
- 1. Criswick VG, Schepens CL.. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969; 68: 578–594. [DOI] [PubMed] [Google Scholar]
- 2. Tian T, Chen C, Zhang X, Zhang Q, Zhao P.. Clinical and genetic features of familial exudative vitreoretinopathy with only-unilateral abnormalities in a Chinese cohort. JAMA Ophthalmol. 2019; 137: 1054–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu Q, Wang Y, Dabdoub A, et al.. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004; 116: 883–895. [DOI] [PubMed] [Google Scholar]
- 4. Junge HJ, Yang S, Burton JB, et al.. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009; 139: 299–311. [DOI] [PubMed] [Google Scholar]
- 5. Wang Y, Smallwood PM, Williams J, Nathans J.. A mouse model for kinesin family member 11 (Kif11)-associated familial exudative vitreoretinopathy. Hum Mol Genet. 2020; 29: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collin RW, Nikopoulos K, Dona M, et al.. ZNF408 is mutated in familial exudative vitreoretinopathy and is crucial for the development of zebrafish retinal vasculature. Proc Natl Acad Sci USA. 2013; 110: 9856–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang S, Zhang X, Hu Y, et al.. Clinical and genetical features of probands and affected family members with familial exudative vitreoretinopathy in a large Chinese cohort. Br J Ophthalmol. 2021; 105: 83–86. [DOI] [PubMed] [Google Scholar]
- 8. Rao FQ, Cai XB, Cheng FF, et al.. Mutations in LRP5,FZD4, TSPAN12, NDP, ZNF408, or KIF11 genes account for 38.7% of Chinese patients with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2017; 58: 2623–2629. [DOI] [PubMed] [Google Scholar]
- 9. Wang Z, Chen C, Sun L, et al.. Symmetry of folds in FEVR: A genotype-phenotype correlation study. Exp Eye Res. 2019; 186: 107720. [DOI] [PubMed] [Google Scholar]
- 10. Li Y, Peng J, Li J, et al.. The characteristics of digenic familial exudative vitreoretinopathy. Graefes Arch Clin Exp Ophthalmol. 2018; 256: 2149–2156. [DOI] [PubMed] [Google Scholar]
- 11. Ranchod TM, Ho LY, Drenser KA, Capone A Jr., Trese MT. Clinical presentation of familial exudative vitreoretinopathy. Ophthalmology. 2011; 118: 2070–2075. [DOI] [PubMed] [Google Scholar]
- 12. Kashani AH, Brown KT, Chang E, et al.. Diversity of retinal vascular anomalies in patients with familial exudative vitreoretinopathy. Ophthalmology. 2014; 121: 2220–2227. [DOI] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S, et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C, Sun L, Li S, et al.. The spectrum of genetic mutations in patients with asymptomatic mild familial exudative vitreoretinopathy. Exp Eye Res. 2020; 192: 107941. [DOI] [PubMed] [Google Scholar]
- 15. Li JK, Li Y, Zhang X, et al.. Spectrum of variants in 389 Chinese probands with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2018; 59: 5368–5381. [DOI] [PubMed] [Google Scholar]
- 16. Tang M, Sun L, Hu A, et al.. Mutation spectrum of the LRP5, NDP, and TSPAN12 genes in Chinese patients with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2017; 58: 5949–5957. [DOI] [PubMed] [Google Scholar]
- 17. Chen C, Wang Z, Sun L, et al.. Next-generation sequencing in the familial exudative vitreoretinopathy-associated rhegmatogenous retinal detachment. Invest Ophthalmol Vis Sci. 2019; 60: 2659–2666. [DOI] [PubMed] [Google Scholar]
- 18. Jia LY, Li XX, Yu WZ, Zeng WT, Liang C.. Novel frizzled-4 gene mutations in Chinese patients with familial exudative vitreoretinopathy. Arch Ophthalmol. 2010; 128: 1341–1349. [DOI] [PubMed] [Google Scholar]
- 19. Omoto S, Hayashi T, Kitahara K, Takeuchi T, Ueoka Y.. Autosomal dominant familial exudative vitreoretinopathy in two Japanese families with FZD4 mutations (H69Y and C181R). Ophthalmic Genet. 2004; 25: 81–90. [DOI] [PubMed] [Google Scholar]
- 20. Kondo H, Hayashi H, Oshima K, Tahira T, Hayashi K.. Frizzled 4 gene (FZD4) mutations in patients with familial exudative vitreoretinopathy with variable expressivity. Br J Ophthalmol. 2003; 87: 1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robitaille JM, Wallace K, Zheng B, et al.. Phenotypic overlap of familial exudative vitreoretinopathy (FEVR) with persistent fetal vasculature (PFV) caused by FZD4 mutations in two distinct pedigrees. Ophthalmic Genet. 2009; 30: 23–30. [DOI] [PubMed] [Google Scholar]
- 22. Nikopoulos K, Venselaar H, Collin RW, et al.. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010; 31: 656–666. [DOI] [PubMed] [Google Scholar]
- 23. Qin M, Hayashi H, Oshima K, et al.. Complexity of the genotype-phenotype correlation in familial exudative vitreoretinopathy with mutations in the LRP5 and/or FZD4 genes. Hum Mutat. 2005; 26: 104–112. [DOI] [PubMed] [Google Scholar]
- 24. Fei P, Zhang Q, Huang L, et al.. Identification of two novel LRP5 mutations in families with familial exudative vitreoretinopathy. Mol Vis. 2014; 20: 395–409. [PMC free article] [PubMed] [Google Scholar]
- 25. Qin M, Kondo H, Tahira T, Hayashi K.. Moderate reduction of Norrin signaling activity associated with the causative missense mutations identified in patients with familial exudative vitreoretinopathy. Hum Genet. 2008; 122: 615–623. [DOI] [PubMed] [Google Scholar]
- 26. Crabbe P, Balemans W, Willaert A, et al.. Missense mutations in LRP5 are not a common cause of idiopathic osteoporosis in adult men. J Bone Miner Res. 2005; 20: 1951–1959. [DOI] [PubMed] [Google Scholar]
- 27. Van Wesenbeeck L, Cleiren E, Gram J, et al.. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003; 72: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chung BD, Kayserili H, Ai M, et al.. A mutation in the signal sequence of LRP5 in a family with an osteoporosis-pseudoglioma syndrome (OPPG)-like phenotype indicates a novel disease mechanism for trinucleotide repeats. Hum Mutat. 2009; 30: 641–648. [DOI] [PubMed] [Google Scholar]
- 29. Royer G, Hanein S, Raclin V, et al.. NDP gene mutations in 14 French families with Norrie disease. Hum Mutat. 2003; 22: 499. [DOI] [PubMed] [Google Scholar]
- 30. Schuback DE, Chen ZY, Craig IW, Breakefield XO, Sims KB.. Mutations in the Norrie disease gene. Hum Mutat. 1995; 5: 285–292. [DOI] [PubMed] [Google Scholar]
- 31. Liu D, Hu Z, Peng Y, et al.. A novel nonsense mutation in the NDP gene in a Chinese family with Norrie disease. Mol Vis. 2010; 16: 2653–2658. [PMC free article] [PubMed] [Google Scholar]
- 32. Kondo H, Kusaka S, Yoshinaga A, et al.. Genetic variants of FZD4 and LRP5 genes in patients with advanced retinopathy of prematurity. Mol Vis. 2013; 19: 476–485. [PMC free article] [PubMed] [Google Scholar]
- 33. Lai MB, Zhang C, Shi J, et al.. TSPAN12 Is a Norrin co-receptor that amplifies Frizzled4 ligand selectivity and signaling. Cell Rep. 2017; 19: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salvo J, Lyubasyuk V, Xu M, et al.. Next-generation sequencing and novel variant determination in a cohort of 92 familial exudative vitreoretinopathy patients. Invest Ophthalmol Vis Sci. 2015; 56: 1937–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drenser KA, Dailey W, Vinekar A, et al.. Clinical presentation and genetic correlation of patients with mutations affecting the FZD4 gene. Arch Ophthalmol. 2009; 127: 1649–1654. [DOI] [PubMed] [Google Scholar]
- 36. Dailey WA, Gryc W, Garg PG, Drenser KA.. Frizzled-4 variations associated with retinopathy and intrauterine growth retardation: a potential marker for prematurity and retinopathy. Ophthalmology. 2015; 122: 1917–1923. [DOI] [PubMed] [Google Scholar]
- 37. Robitaille JM, Zheng B, Wallace K, et al.. The role of Frizzled-4 mutations in familial exudative vitreoretinopathy and Coats disease. Br J Ophthalmol. 2011; 95: 574–579. [DOI] [PubMed] [Google Scholar]
- 38. Seo SH, Yu YS, Park SW, et al.. Molecular characterization of FZD4, LRP5, and TSPAN12 in familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2015; 56: 5143–5151. [DOI] [PubMed] [Google Scholar]
- 39. Umbhauer M, Djiane A, Goisset C, et al.. The C-terminal cytoplasmic Lys-thr-X-X-X-Trp motif in frizzled receptors mediates Wnt/beta-catenin signalling. Embo J. 2000; 19: 4944–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wong HC, Bourdelas A, Krauss A, et al.. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell. 2003; 12: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ohlmann A, Tamm ER.. Norrin: molecular and functional properties of an angiogenic and neuroprotective growth factor. Prog Retin Eye Res. 2012; 31: 243–257. [DOI] [PubMed] [Google Scholar]
- 42. Avila-Fernandez A, Perez-Carro R, Corton M, et al.. Whole-exome sequencing reveals ZNF408 as a new gene associated with autosomal recessive retinitis pigmentosa with vitreal alterations. Hum Mol Genet. 2015; 24: 4037–4048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.