Abstract
The transmission of Pneumocystis carinii from person to person was studied by detecting P. carinii-specific DNA in prospectively obtained noninvasive deep-nasal-swab samples from a child with a documented P. carinii pneumonia (PCP), his mother, two contact health care workers, and 30 hospital staff members who did not enter the patient's room (controls). Nested-DNA amplification was done by using oligonucleotide primers designed for the gene encoding the mitochondrial large subunit rRNA of rat P. carinii (P. carinii f. sp. carinii) that amplifies all forms of P. carinii and internal primers specific for human P. carinii (f. sp. hominis). P. carinii f. sp. hominis DNA was detected in samples from the patient and all of his contacts versus none of the 30 hospital staff members. The results, as previously shown in murine models of P. carinii pneumonia, document that person-to-person transmission of P. carinii is possible. This observation suggests that immunocompromised patients not on PCP prophylaxis should not enter the room of a patient with PCP, and it also raises the question as to whether healthy contacts can transmit the disease to immunocompromised patients at risk.
Clusters of Pneumocystis carinii pneumonia (PCP) cases have been reported in wards of immunocompromised patients, suggesting the person-to-person transmission of P. carinii (2, 6, 8, 12, 13). This form of transmission is also suggested by the documentation of a rise in P. carinii antibodies in the contacts of patients with P. carinii pneumonia (J. A. Giron, S. Martinez, and P. D. Walzer, Letter, Lancet ii:46, 1982) and by animal models in a controlled environment showing that P. carinii can be transmitted through the airborne route (7). The recent finding that P. carinii DNA can be detected in the room air of patients with PCP (1) led us to hypothesize that P. carinii DNA could be detected with the use of highly sensitive nested-DNA amplification technology in deep-nasal-swab samples from a patient with PCP and his contact health care workers. Hospital staff members who did not enter the patient's room were selected as controls. Our results show that P. carinii DNA can be transiently detected in immunocompetent close contacts of patients with PCP and raises the possibility that such contacts might serve as vectors for transmission of the disease.
MATERIALS AND METHODS
Index patient, contacts, and controls.
An 8-month-old human immunodeficiency virus-negative boy was referred from a rural hospital with a 1-week history of mild cough and a 4-day history of intermittent fever up to 38.5°C. His past medical history revealed recurrent episodes of oral and esophageal candidiasis starting at the age of 3 months. Upon admission the infant presented with extensive white plaques in the oral cavity, mouth breathing, mild nonproductive cough, a respiratory rate of 45/min, and a pulse oximetry of 85% on room air. His lung examination revealed coarse breath sounds with no wheezes, rhonchi, or rales. Chest radiography showed extensive bilateral infiltrates. Laboratory tests showed depressed T-cell counts (white cells, 4,400 /mm3; lymphocytes, 616 /mm3; CD4 166 [27%]; CD8, 215 [35%]; CD4/CD8, 0.77) and normal immunoglobulin levels (immunoglobulin A [IgA], 25 mg/dl; IgG, 720 mg/dl; IgM, 68 mg/dl). The infant was admitted with a diagnosis of pneumonia to a single, standard, hospital room without a controlled environment. The diagnosis of PCP was documented by a Gomori-Grocott stain of the bronchoalveolar lavage fluid performed 2 days after admission, and specific chemotherapy was started (day 1; Fig. 1) with trimethoprim-sulfamethoxazole (20 mg of trimethoprim component per kg per day) given intravenously in four divided doses. He received supplemental oxygen (1 liter/min) via a nasal catheter during the first 2 weeks of hospitalization and subsequently improved without requiring mechanical ventilation. He was discharged from the hospital after 48 days. There were no other patients with P. carinii infection identified in this pediatric hospital during the time of the study. The mother of the patient, the attending physician, and one nurse were prospectively sampled at intervals as depicted in Fig. 1. All of them were considered contacts because they had entered the patient's room for more than 5 min per day and had performed procedures such as physical examination, drawing blood, and taking vital signs with the patient. Thirty members of the hospital staff working on the same floor who did not enter the room were selected as controls in order to document that P. carinii DNA is not normally carried by immunocompetent people. All of these people were apparently healthy, had not received medications with anti-P. carinii activity, and had no evidence of an immunosuppressive disease.
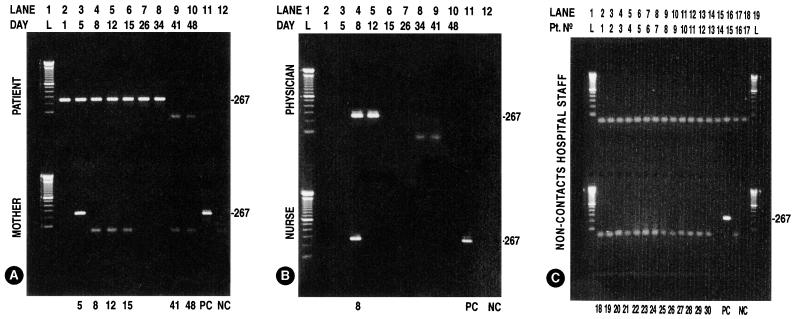
FIG. 1.
P. carinii DNA amplification with nested PCR with primers pAZ102-E and pAZ102-H designed for the P. carinii gene encoding the mitochondrial large subunit rRNA and with primers pAZ102X and pAZ102-Y, which are internal to the first set of primers and specific for P. carinii sp. f. hominis on noninvasive respiratory samples on the index patient and contacts. (A) Top, index patient on days 1 to 34 showing the amplification product of 267 bp (positive) and no amplification product (negative) on days 41 and 48; bottom, mother positive on day 5 and negative on days 8, 12, and 15. (B) Top, attending physician positive on days 8 and 12 and negative on days 34 and 41; bottom, nurse positive on day 8. (C) Thirty noncontact staff members tested on day 34 (all negative). L, Ladder (A and B, lines 1; C, lines 1 and 19); PC, positive control (A and B, lines 11, bottom; C, line 16, bottom); NC, negative control (A and B, lines 12, bottom; C, line 18, bottom).
Sampling procedure and timing.
Because the nasal turbinates are the first natural air filter encountered in humans, we elected to do deep-nasal-swab sampling of the nasal turbinates from persons in the study. A moist sterile cotton-swab was introduced deep into the nose, left there for 30 s, rotated, and withdrawn and placed in a sterile tube containing 300 μl of saline. The first sample of the patient on day 1 was a nasopharyngeal aspirate and subsequent samples on days 5, 8, 12, 15, 26, 34, 41, and 48, as well as all other respiratory samples in this study, were deep nasopharyngeal swabs. The mother was sampled on days 5, 8, 12, and 15, and the attending physician was sampled on days 8, 12, 34, and 41. The nurse was sampled once on day 8, and the 30 controls were also sampled once on day 34 as depicted in Fig. 1. Samples were kept frozen at −20°C until DNA extraction.
DNA amplification.
A volume of approximately 200 μl of saline was examined for P. carinii by DNA amplification as previously described (14, 15). Briefly, the samples were digested with proteinase K (20 mg ml−1) at 60°C in the presence of 10 mM EDTA and 0.5% sodium dodecyl sulfate. Total DNA was purified and concentrated by using the Pharma-Gen Clean Up system (Pharma-Gen, Madrid, Spain) and then recovered in a volume of 50 μl.
For DNA amplification, the P. carinii f. sp. carinii oligonucleotide primers pAZ102-E (5′-GATGGCTGTTTCCAAGCCCA-3′) and pAZ102-H (5′-GTGTACGTTGCAAAGTACTC-3′) were used. Amplification was carried out at 94°C for 1.0 min, 55°C for 1.0 min, and 72°C for 2.0 min for 40 cycles. In order to achieve a greater level of sensitivity and specificity, primers pAZ102-X (5′-GTGAAATACAAATCGGACTAGG-3′) and pAZ102-Y (5′-TCACTTAATATTAATTGGGGAGC-3′), which are internal to the first set of primers and specific for P. carinii f. sp. hominis, were used in a second round of PCR (14, 15). Thirty-five cycles of amplification were carried out under the same conditions as for the external primers. The PCR products were separated on 2.0% agarose gels and visualized with ethidium bromide. A P. carinii DNA amplification product of 267 bp was detected. Negative controls with no added template DNA were included after each sample to monitor for cross-contamination. A sample of human-derived P. carinii DNA was used as a positive control in each experiment. All manipulations during DNA extraction and amplification were performed in laminar flow cabinets by using disposable pipettes, tubes, and reagent aliquots to avoid contamination. DNA amplification was performed twice with each sample.
Antibody determinations.
Antibody titer determinations were done in two 2-week interval serum samples from the patient and his contacts by comparing enzyme-linked immunosorbent assay (ELISA) readings obtained by using, respectively, a crude sonicate of P. carinii-infected mouse lungs (ODIML) and a crude sonicate of normal P. carinii noninfected mouse lungs (ODNIML) as solid-phase antigen, as previously described (4, 5). Briefly, P. carinii-infected and noninfected mouse lungs were sonicated, and the supernatant was then clarified by centrifugation and microfiltration (0.45 μm, pore size), diluted to a protein concentration of ∼10 μg/ml in carbonate buffer, and used to coat microtiter wells in Linbro flat-bottom microtiter plates (Flow Laboratories, McLean, Va.). Plates were incubated tightly covered in a moist chamber at 37°C for 90 min to allow the antigen to adhere. Coated wells were blocked with 5% commercial nonfat dry milk in phosphate-buffered saline (PBS) for 1 h at 37°C or overnight at 4°C. Sera were diluted 1:50 in PBS containing 0.05% Tween 20 (PBS-T; Sigma, St. Louis, Mo.), and 50 μl of each serum sample per well was incubated overnight at 4°C. Plates were then washed three times with PBS-T, and bound antibody was detected by using goat anti-human IgG or IgM conjugated to alkaline phosphatase (Jackson Immunoresearch Laboratories, West Grove, Pa.) diluted 1:5,000 in PBS-T. After a 4-h incubation period at 37°C, the plates were washed four times with PBS-T and developed by the addition of p-nitrophenyl phosphate (Sigma) at a concentration of 1 mg/ml in diethanolamine buffer. This ELISA was previously standardized by using a variety of monoclonal and polyclonal antibodies known to be specific for P. carinii. Irrelevant monoclonal antibodies give optical densities of <0.05 in this ELISA. Samples were labeled as positive if the optical densities if the ODIML values were ≥0.15 and the ODNIML values were <0.15, possible positive if the ODIML values were 0.11 to 0.14 and the ODNIML values were <0.05, uninterpretable if the OD value in both was >0.15, and negative if the ODIML and the ODNIML values were <0.10.
RESULTS
Fifty nasopharyngeal swab samples from the patient (n = 9), the mother (n = 6), the attending nurse (n = 1), the attending physician (n = 4), and the 30 controls were analyzed by DNA amplification as depicted in Fig. 1. Of 11 samples taken from the three contact individuals, 4 were positive for P. carinii DNA. The 30 hospital personnel (controls) who had not entered the patient's room were sampled once, and none of these individuals was positive for P. carinii DNA. None of the contacts developed symptoms suggestive of P. carinii disease during the period of the study, and their antibody titers remained stable. Antibody titers (ODIML/ODNIML) obtained in the patient showed 0.44/0.17 to be uninterpretable in the first serum sample drawn on day 1 and positive on the second serum sample on day 39 (0.18/0.05). Serum titers from the patient contacts were positive in the first (day 11) and second (day 29) samples in the mother (0.50/0.10 and 0.62/0.12) and in the attending physician on days 12 and 29 (0.47/0.14 and 0.42/0.13). The nurse was possibly positive in the first reading on day 12 (0.30/0.15) and positive in the second reading on day 29 (0.26/0.09). Sera from the controls for these days were not available.
DISCUSSION
The detection of P. carinii f. sp. hominis DNA in deep-nares swab samples from health care workers in contact with a patient with PCP suggests the air spread of P. carinii from the patient and inhalation by his contacts, supporting the concept of airborne person-to-person transmission of P. carinii (1, 2, 6–8, 12, 13; Giron et al., Letter). Previous attempts to document person-to-person transmission of P. carinii by amplifying P. carinii sequences in induced sputum and oropharyngeal washing samples from people in contact with PCP patients have not detected evidence of P. carinii DNA (9, 10). In this study, we elected to sample the nasal cavities of the patient, his contacts, and the control group because the nasal turbinates constitute the first air breath filter in the respiratory system. We believe that deep nasal swabs or nasopharyngeal aspirates should be preferred as a research tool for detecting P. carinii DNA in individuals who don't have clinical evidence of PCP. Whether the finding of P. carinii DNA in the nares of the contacts of this patient with PCP indicates the presence of infectious material and whether the contacts could be a vector for disease transmission remains to be determined. Unfortunately, the finding of P. carinii DNA does not provide information about the presence of cysts or trophozoites or whether they are viable if present. The lack of a culture system for P. carinii makes it difficult to study the viability or infectious potential of this organism outside its host. Documenting an eventual role, if any, for transiently colonized immunocompetent individuals as transmitters of the disease warrants further research, and recommendations such as the use of a mask or respiratory isolation precautions for health care workers who see multiple patients, with or without P. carinii infection, cannot be derived from our findings. Standard P. carinii stains such as Gomori-Grocott methenamine silver and immunofluorescence stains were not done in addition to DNA amplification because of the poor sensitivity of these stains in nasopharyngeal samples. P. carinii genotyping was not available. However, documenting eventual differences in P. carinii organisms from the patient and his contacts by sequence analysis of PCR products would be difficult to interpret since different P. carinii genotypes have been identified in a same host, suggesting that one patient can transmit more than one P. carinii genotype (3).
P. carinii was cleared from the nares of the mother and the physician in relation to the clearing of P. carinii from the patient samples. Therefore, the presence of P. carinii DNA in immunocompetent contact individuals was self-limited and transitory and might have been the result of continuous inhalation of P. carinii particles, indicating surface (nares) contamination and carriage rather than an active infectious process.
All of the 30 immunocompetent health care personnel who had not entered the patient's room tested negative by PCR, suggesting that the transmission of P. carinii is not common among healthy adults unless they are exposed to a patient with PCP (H. S. Oz and W. T. Hughes, American Lung Association Conference and Symposium, abstr. 7, 1999). No other patients with PCP were diagnosed in this pediatric hospital during the time of the study.
Given the index patient's young age (8 months), the antibody titers determined in the crude ELISA format is likely an indication that the patient mounted an immune response during a primary infection with P. carinii. All adult contacts were also antibody positive. However, they did not experience a rise in antibody titers after P. carinii DNA was detected in their nares. This lack of serological response is more consistent with transient colonization of the contacts rather than with an active infectious process. Adults normally exhibit an antibody response to the major antigenic determinants present in the P. carinii wall (11).
In summary, this study provides molecular evidence that airborne person-to-person transmission of P. carinii sp. f. hominis is possible, thus suggesting that patients with PCP should not come into close contact with immunocompromised persons. Immunocompromised patients who are not receiving PCP prophylaxis should not enter the room of a patient with PCP. The data show that the immunocompetent contacts of patients with PCP can acquire transient colonization by the organism. A potential role, if any, of immunocompetent hosts transiently colonized with P. carinii as vectors of the disease warrants further investigation.
ACKNOWLEDGMENTS
This work was supported in part by FONDECYT research grant 1960940, Fondo Nacional de Desarrollo Científico y Tecnológico, Santiago, Chile, and by St. Jude International Outreach Program and American Lebanese Syrian Associated Charities, St. Jude Children's Research Hospital, Memphis, Tenn.
We thank the members of the Infectious Diseases Unit, Luis Calvo Mackenna Hospital, Santiago, Chile, for their interest and participation in this study and Veronique Carrat for excellent technical assistance.
REFERENCES
- 1.Bartlett M S, Vermund S H, Jacobs R, Durant P J, Shaw M M, Smith J W, Tang X, Lu J-J, Li B, Jin S, Lee C-H. Detection of Pneumocystis carinii DNA in air samples: likely environmental risk to susceptible persons. J Clin Microbiol. 1997;35:2511–2513. doi: 10.1128/jcm.35.10.2511-2513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chave J-P, David S, Wauters J-P, Van Meele G, Francioli P. Transmission of Pneumocystis carinii from AIDS patients to other immunosuppressed patients: a cluster of Pneumocystis carinii pneumonia in renal transplant recipients. AIDS. 1991;5:927–932. doi: 10.1097/00002030-199108000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Cushion M T, Orr S, Arnold J A. Interactions between 2 Pneumocystis populations within the same host. J Eukaryot Microbiol. 1998;44:9S. doi: 10.1111/j.1550-7408.1997.tb05739.x. [DOI] [PubMed] [Google Scholar]
- 4.Gigliotti F, Harmsen A. Pneumocystis carinii host origin defines the antibody specificity and protective response induced by immunization. J Infect Dis. 1997;176:1322–1326. doi: 10.1086/514128. [DOI] [PubMed] [Google Scholar]
- 5.Gigliotti F, Garvy B A, Haidaris C G, Harmsen A G. Recognition of Pneumocystis carinii antigens by local antibody-secreting cells following resolution of P. carinii pneumonia in mice. J Infect Dis. 1998;178:235–242. doi: 10.1086/515607. [DOI] [PubMed] [Google Scholar]
- 6.Helweg-Larsen J, Tsolaki A G, Miller R F, Lundgren B, Wakefield A E. Clusters of Pneumocystis carinii pneumonia: analysis of person-to-person transmission by genotyping. QJM. 1998;91:813–820. doi: 10.1093/qjmed/91.12.813. [DOI] [PubMed] [Google Scholar]
- 7.Hughes W T. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs J L, Libby D M, Winters R A, Gelmont D M, Fried E D, Hartman B J, Laurence J. A cluster of Pneumocystis carinii pneumonia in adults without predisposing illnesses. N Engl J Med. 1991;324:246–250. doi: 10.1056/NEJM199101243240407. [DOI] [PubMed] [Google Scholar]
- 9.Lidman C, Olsson M, Björkman A, Elvin K. No evidence of nosocomial Pneumocystis carinii infection via health care personnel. Scand J Infect Dis. 1997;29:63–64. doi: 10.3109/00365549709008666. [DOI] [PubMed] [Google Scholar]
- 10.Lundgren B, Elvin K, Rothman L P, Ljungström I, Lidman C, Lundgren J D. Transmission of Pneumocystis carinii from patients to hospital staff. Thorax. 1997;52:422–424. doi: 10.1136/thx.52.5.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei Q, Turner R E, Sorial V, Klivington D, Angus C W, Kovacs J A. Characterization of major surface glycoprotein genes of human Pneumocystis carinii and high-level expression of a conserved region. Infect Immun. 1998;66:4268–4273. doi: 10.1128/iai.66.9.4268-4273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruebush T K, Weinstein R A, Baehner R L, Wolf D, Bartlett M, Gonzales-Crussi F, Sulzer A J, Schultz M G. An outbreak of Pneumocystis pneumonia in children with acute lymphocytic leukemia. Am J Dis Child. 1978;132:143–148. doi: 10.1001/archpedi.1978.02120270041009. [DOI] [PubMed] [Google Scholar]
- 13.Singer C, Armstrong D, Rosen P P, Schottenfeld D. Pneumocystis carinii pneumonia: a cluster of eleven cases. Ann Intern Med. 1975;82:772–777. doi: 10.7326/0003-4819-82-6-722. [DOI] [PubMed] [Google Scholar]
- 14.Tsolaki A G, Beckers P, Wakefield A E. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotypic similarity with contemporary isolates. J Clin Microbiol. 1998;36:90–93. doi: 10.1128/jcm.36.1.90-93.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakefield A E. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]