Abstract
Highly detailed steered molecular dynamics simulations are performed on differently glycosylated receptor binding domains of the severe acute respiratory syndrome coronavirus-2 spike protein. The binding strength and the binding range increase with glycosylation. The interaction energy rises very quickly when pulling the proteins apart and only slowly drops at larger distances. We see a catch-slip-type behavior whereby interactions during pulling break and are taken over by new interactions forming. The dominant interaction mode is hydrogen bonds, but Lennard-Jones and electrostatic interactions are relevant as well.
Graphical abstract

Significance
Glycosylation of the receptor binding domain of the spike protein of SARS-CoV-2 as well as the ACE2 receptor leads to stronger and longer ranged binding interactions between the proteins. Particularly at shorter distances, the interactions are between residues of the proteins themselves, whereas at larger distances these interactions are mediated by the glycans.
Introduction
As of July 2021, more than 182 million people globally have been confirmed to be infected with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which causes coronavirus disease 2019. This zoonotic pandemic has disrupted society and spurred a wide range of scientific endeavors to improve our knowledge of coronaviruses and address the crisis. As the disease spreads and in order to prepare for potential future events, there is a critical need to understand the interaction of the virus with proteins involved in infection and immune clearance or with proteins used as potential countermeasures or for the purpose of improved tests. Here, we study the interactions between SARS-CoV-2 and the human receptor responsible for binding using a molecular dynamics approach and validate it experimentally.
The SARS-CoV-2 spike (S) protein is a major structural protein and is therefore involved in many interactions. Through the receptor binding domain (RBD), S binds to the human angiotensin-converting enzyme 2 (hACE2 or ACE2) receptor on the cell surface and initiates infection. Significant effort has been directed at understanding this interaction both experimentally and computationally (1, 2, 3, 4, 5, 6, 7). Such studies are critical for the development of more efficient tests and therapeutics, including vaccines.
Viral structural proteins such as S are often glycosylated to help pathogens evade the host immune system, modulate access to proteases, and enhance cellular attachment through modification of protein structure and/or direct participation at the virus-host interface (8, 9, 10, 11, 12, 13, 14). Furthermore, many mammalian viruses use glycans on cell-surface glycoproteins or glycolipids as receptors (15). Despite the important role of glycans in virus-host interactions, the glycans themselves are often only partially resolved in experimental structures generated from experimental techniques such as cryoelectron microscopy (16). Computational modeling of these glycans is therefore helpful in predicting their behavior and structural contributions.
S is a trimer in which each monomer is expected to be highly glycosylated with 22 N-linked glycosylation sequons and 4 O-linked predicted glycosylation sites (17). Only 16 N-linked glycosylation sites were observed in a cryoelectron microscopy map of S produced in HEK293F cells (18). A study by Watanabe et al. (16) determined site-specific glycoform analysis of full-length trimeric S protein made in HEK293F cells. In another study of S, glycosylation patterns including O glycosylation were determined (19). In a similar vein, it has recently been argued that glycosylation can exert influence postvaccination and for vaccine resistance (20). Yet the influence of glycosylation on the S-ACE2 interaction has been studied to a lesser extent (21,22). We address this gap in knowledge in the present study to reveal how glycans modulate the interaction of S with ACE2.
We expect that, as both S and ACE2 are glycosylated, the interaction is possibly modulated by the glycans. Few computational studies explicitly take the glycosylation of the receptor and/or the virus into account (23, 24, 25, 26). This is true in general, as glycosylation has only very recently become a stronger focus in simulations (27, 28, 29, 30, 31). One previous study addressed the free energy of binding between the RBD and ACE2, including the impact of protein glycosylation (32). However, previous studies were limited to a single simple glycan model and did not examine interactions of glycans or the influence of different complex glycan distributions beyond pulling force and protein contacts. Additional studies have shown experimentally and computationally that the RBD and ACE2 have different binding strengths and dissociation rates when they are glycosylated versus non-glycosylated (33,34). However, previous computational efforts often used simpler models for the glycans. We previously developed a fully glycosylated model for the SARS-CoV-2 RBD and ACE2 proteins with different glycosylation patterns (2). We extend this model here to explore how a combination of complex glycans affect the energy and duration of binding. This is particularly important to improve rapid tests in which viral antigens may be made in a variety of hosts with different glycan distributions.
In our previous study, we modeled ACE2 combined with the Fc domain as a therapeutic decoy. The extracellular domain of ACE2 was fused with the Fc region of human immunoglobulin (IgG1) (7). The fusion ACE2 to the Fc domain of IgG1 has several advantages as a therapeutic decoy, as it increases circulatory half-life and facilitates purification through the use of the common protein A affinity chromatography platform. This served to neutralize the S protein on the virus and block the S protein’s binding to cellular ACE2 for virus entry. ACE2-Fc was also modeled with plant glycosylation patterns. Because of the anticipated demand for high-speed production of the recombinant ACE2-Fc, plant-based transient expression systems are well suited for rapid production. Plant cells can readily produce glycoproteins with either native, plant glycosylation (35) or modified human-like glycoforms through genetic manipulation (36). We simulated two plant glycovariants of ACE2-Fc in our previous work: variant 1 was targeted for endoplasmic reticulum (ER) retention with high-mannose glycoforms, and variant 2 was targeted for secretion with plant complex glycoforms. As heterologous glycoproteins can be retained in the ER by adding a C-terminal H/KDEL tag and the formation of Man8GlcNAc2 (Man8) N-glycans is typical for H/KDEL-tagging (37), variant 1 was fully glycosylated with MAN8 glycans. Variant 2 was fully glycosylated with GlcNAc2XylFucMan3GlcNAc2 (GnGnXF3), which is a standard plant glycoform, and the S protein fragment was glycosylated with ANaF6 (2). Fig. 1 shows the glycans used in our systems. In our previous study, we simulated the influence of the two glycoforms on the interaction of S protein and the specific recombinant ACE2-Fc fusion protein. We expect that the glycosylation influence is not restricted to the fusion proteins. In this study, we focus on the contribution of these different glycosylation patterns on the protein-protein interactions via hydrogen bonding and interaction energies and determine the corresponding free energies.
Figure 1.
Glycans used in the simulations, adapted from previous work (2), with linkages of interest in MAN8 and GnGnXF3 glycans for dynamic analysis.
Materials and methods
Simulation
Binding between the receptor binding domain of spike and ACE2 receptor was determined using steered molecular dynamics, also known as the pulling of proteins (38). The starting atomic coordinates for all pulling systems were taken from the final 75 ns configurations of our previous study (2). In that study, two sequence variants of ACE2-Fc were used to model the interaction between ACE2-FC and SARS-CoV-2 RBD. Variant 1 (AFM8/SpFr) contained a C-terminal SEKDEL tag, which is used for ER-retained proteins to express high-mannose glycoforms and variant 2 (AFGG/SpFr), which does not contain the SEKDEL tag and expresses standard plant glycoforms. ACE2-B0AT1 and ACE2-B0AT1/SpFr structures were obtained from the Protein Data Bank. These structures had been determined using cryoelectron microscopy (PDB: 6M18 and PDB: 6M17 [39]). These structures were fused to the Fc domain (PDB: 3SGJ [40]). The Zn2+ coordinating residues and water were taken from structure PDB: 1R42 (41) in the case of variant 1 ACE2. Variant 2 has two mutations that prevent Zn2+ coordination. The presence of zinc in protein structures is still actively being studied to determine its role in adjusting binding specificity (42,43). It has been demonstrated that Zn2+ plays a role in stabilizing some protein structures and can aid in the formation of biological oligomers (42,43). The final frame of the 75 ns trajectories for both ACE2-Fc/SpFr variants was selected, and proteins were trimmed at residue 780 ALA (Fig. 2) to make the pulling simulations a manageable 851 residues with glycans and 780 residues without glycans for AFM8/SpFr and 845 residues with glycans and 780 residues without glycans for AFGG/SpFr. Because the system changed, the force-field files had to be regenerated using AmberTools (44), as described previously (2). Briefly, the molecules were trimmed and glycans were removed, then Man8 glycans were reattached to the truncated variant 1 of ACE2, GnGnXF3 to the truncated variant 2 of ACE2, and ANaF6 to the SpFr in both variants using Glycam.org (45). The coordinating Zn2+ was reattached to truncated and glycosylated variant 1 using MCPB.py (46). Special care was taken to align the shortened original coordinates and the newly generated force field. Truncations from variant 1 and variant 2 that remained aglycosylated for both ACE2 and RBD were also studied to compare the influence of glycosylation on binding. The truncated systems were named A1FrM8/SpFr, A1Fr/SpFr, A2FrGG/SpFr, and A2Fr/SpFr, respectively. All amino acid sequences are available in the Supporting material, and all S-S bridges are retained in our simulations.
Figure 2.
Schematics of generating the different initial structures. The full recombinantly made Ace2-FC systems on the left are used in the BLI experiments for determining binding affinities to the RBD, whereas the four truncated systems on the right containing only a fragment of ACE2 are modeled in simulations. From top to bottom, the truncated systems correspond to (a) A1FrM8/SpFr and A1Fr/SpFr, (b) A2FrGG/SpFr, and A2Fr/SpFr.
After the initial structures and corresponding force fields were generated, the proteins were rotated so that the pulling direction was along one of the principal axes, and the simulation boxes were expanded to 10 × 10 × 26 nm for A1FrM8/SpFr and A1Fr/SpFr and 10 × 10 × 30 nm for A2FrGG/SpFr and A2Fr/SpFr so that the spike RBD fragments did not experience interactions with the ACE2 fragments across periodic boundaries during pulling. Then the new box was solvated with 80,271 water molecules and 24 Na+ as counter ions for A1FrM8/SpFr, 80,764 waters and 23 Na+ cations for A1Fr/SpFr, 93,541 waters and 26 Na+ cations for A2FrGG/SpFr, and 93,989 waters and 25 Na+ cations for A2Fr/SpFr. Energy minimizations were performed until the convergence criteria were met (emtol = 1000 kJ/mol/nm), followed by a 100 ps constant volume (NVT) (dt = 2 fs, T = 310 K) and a 100 ps constant pressure (NPT) (dt = 2 fs, T = 310 K, P = 1 atm) to equilibrate the systems. All simulations for equilibration were performed at 310 K and 1 atm with the Velocity Rescale thermostat (47) and Parrinello-Rahman barostat (48). All water bonds were constrained using SETTLE (49), and all other bonds were constrained using LINCS (50). Box expansion, solvation, and equilibration were performed using the Gromacs suite version 2019.1 (51).
Pulling simulations were then performed to study the free energy of binding as well as the structural arrangement of the separating proteins during interaction. For both variants, the ACE2 fragment was set to be immobile but deformable, whereas the spike RBD fragment (also flexible) was pulled away from the ACE2 fragment. Pull simulations were performed under NPT conditions using a 2 fs timestep, a pull coordination spring constant of 1000 kJ/mol/nm2, a Nose-Hoover thermostat (52) at 310 K, and a Parrinello-Rahman barostat at 1 atm.
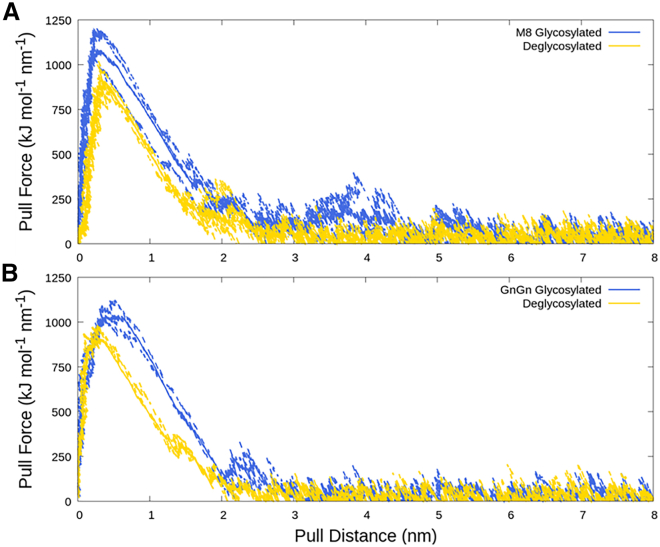
A total of 36 pulling simulations were performed at three different pulling rates (1, 5, and 10 nm/ns) on the four truncated structures using Gromacs version 2019.1 (51). Each structure was pulled at each rate three times for sampling purposes. The starting configuration was the same for each independent run, but the random seed for the velocities in each run was randomly assigned, resulting in independent behaviors. This approach clearly generated independent runs as seen in Fig. 3. Systems were pulled over a distance of 8 nm until full separation (no interaction) was achieved (see Fig. 3).
Figure 3.
Traces of pull force versus pull distance. (A) Man8 glycosylated A1FrM8/SpFr and aglycosylated structure A1Fr/SpFr. (B) GnGnXF glycosylated A2FrGG/SpFr and aglycosylated structure A2Fr/SpFr. Blue lines correspond to glycosylated structures and gold lines to deglycosylated structures. Dashed lines are individual replicas, and solid lines are averages.
Hydrogen bonds were analyzed using the built-in Gromacs bond command (51) with a default cutoff distance of 3.5 Å. This command was used to generate the hydrogen bonds and Lennard-Jones contacts as a function of time as well as a hydrogen bond interaction bitmap and corresponding index file of the different interactions. The hydrogen bonding interaction bitmap was recreated in Python using matplotlib (53) in order to add labels for donor acceptor pairs and calculate the percent occupancy of hydrogen bonds across the simulation (script information available in Supporting material). Short range Lennard-Jones and Coulombic interaction energies were calculated from the Gromacs .edr file by specifying energy groups on the ACE2 and RBD using the gmx energy command (54).
Experiments
Protein deglycosylation
ACE2-Fc (AC2-H5257; Acro Biosystems, Newark, DE) and RBD (40592-V08B; Sino Biological, Chesterbrook, PA) deglycosylation was performed using Remove-iT PNGase F (Bio-Rad, Hercules, CA). Samples with PNGase F were incubated at 310 K for 5 h. PNGase F was then removed by incubating the samples in chitin magnetic beads according to the manufacturer’s instructions (New England Biolabs, Ipswich, MA). Deglycosylation of proteins was confirmed via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Laemmli sample buffer (8 μL; Bio-Rad) and 2 μL β-mercaptoethanol (Bio-Rad) were added to 30 μL sample. Samples were heated at 368 K for 5 min, then run on Mini-PROTEAN TGX Stain-Free Precast Gels (Bio-Rad) at 200 V for 36 min. Gels were imaged using a ChemiDoc Imaging System (Bio-Rad).
Biolayer interferometry
Anti-hIgG-Fc biosensors (FortéBio, Fremont, CA) were used to immobilize ACE2-Fc by immersing the biosensors in solution containing 100 nM ACE2-Fc for 10 min. The Octet RED384 was used to obtain response measurements for protein association and dissociation. Two-fold serial dilutions of RBD were tested, from 250 to 7.81 nM. Data were collected for 60 s for the baseline, 400 s for association, and 800 s for dissociation. The experiment was performed at 299 K.
FortéBio Data Analysis Software version 8.1.0.53 was used for data processing and analysis. From the raw data, reference well values were subtracted, the y axes were aligned to baseline, interstep correction was applied for alignment to dissociation, and Savitzky-Golay filtering (55) was used for smoothing. Using a 1:1 binding model, steady-state analysis was performed on the response average from 390 to 395 s. From the binding affinities of glycosylated and deglycosylated ACE2-Fc and RBD, the change in binding energy following deglycosylation of ACE2-Fc and RBD was calculated as
Results
Fig. 3 presents the pull force as a function of the pull distance between the ACE2 fragments and RBD for different glycosylation states at a 1 nm/ns pulling rate. The pull distances are calculated on the basis of the centers of mass for the ACE2 fragments and RBD but normalized to start from 0 nm to highlight differences between configurations. Plots of pull force versus pull distance for higher pulling rates can be seen in the Supporting material (Fig. S1). Fundamentally, we see that for all conditions under study, there is an immediate sharp increase in force when pulling the two proteins away from each other, indicating strong local binding between the ACE2 binding domain and RBD. After going through a peak in force, the force drops off at increasing distance but with a clearly smaller slope than the initial increase. As expected, the pull force increases with pulling rate (Fig. S1, blue, orange, and green lines) such that the lowest force is most relevant for comparison with experiments. Importantly, for the same fragment the peak force is clearly higher, by ∼250 kJ/mol/nm at 1 nm/ns, with glycosylation than without. This indicates an overall stronger binding of the glycoproteins than their aglycosylated counterparts for both types of glycosylation simulated. Additionally, the force curves are much broader for the glycosylated structures compared with the aglycosylated ones, indicating that the presence of glycans extends the range for binding in addition to strengthening it. Also, the force is longer ranged (only at larger distances does it reach zero), which indicates that the glycans that extend away from the proteins contribute to the binding at longer distances. As shown in Fig. 3, A and B, the aglycosylated structures return to baseline at roughly 2.5 nm of pulling distance. Importantly, the glycosylated structures in Fig. 3, A and B have an extended window of pulling force of 2–3 nm for A1FrM8/SpFr and a smaller difference of roughly 1 nm for A1FrGG/SpFr compared with their aglycosylated counterparts. This indicates that both Man8 and GnGnXF glycans increase binding strength and binding range, but the type of glycan affects both the strength and interaction distance of the specific binding.
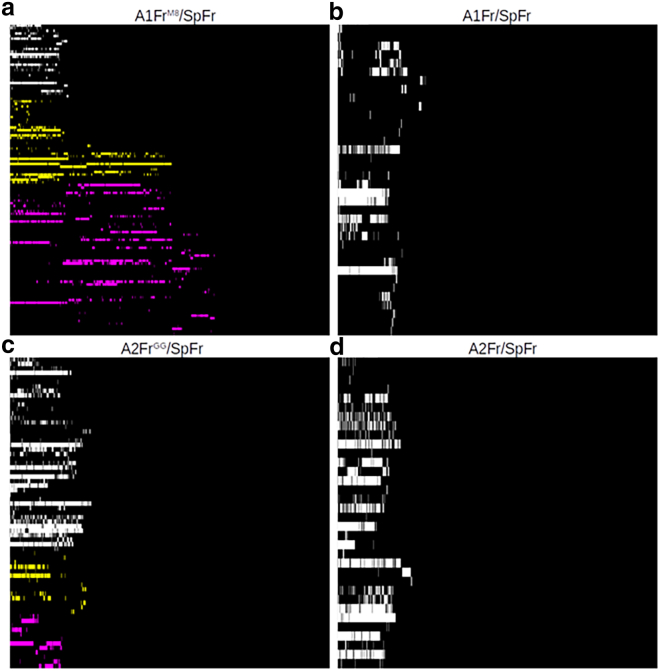
To further characterize the extension of binding interactions, Fig. 4 shows hydrogen bonding interaction maps between the ACE2 and RBD proteins. Fig. 4, a and c are for A1FrM8/SpFr and A2FrGG/SpFr respectively, while Fig. 4, b and d are the corresponding aglycosylated versions. (Full-scale images with donor:acceptor pairs labeled are available in Figs. S2–S5) The y axis contains information about the donor and acceptor pair for the hydrogen bond, and the x axis corresponds to simulation time. Interaction types are colored and sorted according to the interaction type: protein-protein interactions are colored in white, protein-glycan interactions in yellow, and glycan-glycan interactions in magenta. Hydrogen bonding is clearly a major interaction mode between proteins. It is interesting that in A1FrM8/SpFr (Fig. 4 a), the predominant interactions involve glycans directly, while for A2FrGG/SpFr (Fig. 4 c), the predominant interactions are protein-protein interactions, which are indirectly strengthened by glycosylation. This indirect protein-protein strengthening is most clearly seen when comparing occupancy calculated from these heatmaps, as shown in the tables in Figs. 5 and S6–S9. There are multiple binding regimes as a function of time for the two glycosylated structures; this is more pronounced in the A1FrM8/SpFr case. This behavior manifests because of the original active hydrogen bonds in the complex releasing, but other hydrogen bonds catch and eventually release at larger distances before complete unbinding is seen. This catch-slip behavior is particularly attributable to the glycans, as the H-bonds present at longer distance are particularly ones involving glycans, either protein-glycan or direct glycan-glycan bonding. Both non-glycosylated structures shown in Fig. 4, b and d express maps of similar protein-protein interactions, though the A2Fr/SpFr shown in Fig. 4 d contains many more interactions, as indicated by the increased number of rows.
Figure 4.
Hydrogen bond interactions versus simulation time. (a) Man8 glycosylated A1FrM8/SpFr. (b) Aglycosylated structure A1Fr/SpFr. (c) GnGnXF3 glycosylated A2FrGG/SpFr. (d) Aglycosylated structure A2Fr/SpFr. Colors indicate interaction type: white, protein-protein; yellow, protein-glycan; magenta, glycan-glycan.
Figure 5.
Top five hydrogen bond donor:acceptor pairs and occupancy. (a) Man8 glycosylated A1FrM8/SpFr. (b) Aglycosylated structure A1Fr/SpFr. (c) GnGnXF3 glycosylated A2FrGG/SpFr. (d) Aglycosylated structure A2Fr/SpFr. Table colors indicate interaction type: white, protein-protein; yellow, protein-glycan; magenta, glycan-glycan. On the four configurations, residues highlighted with blue indicate donors and those highlighted with pink indicate acceptors.
Fig. 5 shows the configurations in which RBD with and without ANaF6 started to be pulled away from the ACE2 fragment for the four different systems. The top five hydrogen bonds by occupancy (i.e., the fraction of time a given hydrogen bond is active) and their corresponding donor:acceptor pairs are highlighted. (The top 25 hydrogen bonds by occupancy for the four different configurations are available in Figs. S6–S9.) A1FrM8/SpFr clearly shows that the predominant interactions are between the RBD glycan and ACE2 glycan and between the RBD glycan and the ACE2 protein, while for A2FrGG/SpFr, the predominant interactions are between the protein backbones. It is also interesting to note that the predominant interactions in A2FrGG/SpFr are the protein-protein interactions. The strongest glycan interaction for A2FrGG/SpFr are not found until hydrogen bond #9 ranked by occupancy (Fig. S8), while the top 3 hydrogen bonds ranked by occupancy involve glycans for A1FrM8/SpFr. A1FrM8/SpFr also clearly shows a different starting orientation than A2FrGG/SpFr, with minor changes in ACE2 structure and obvious rotation in the RBD with direct glycan-glycan interaction. These minor structural and orientational differences are also seen in the aglycosylated structures. Interacting groups for the hydrogen bonding shown follow AMBER nomenclature (56). The first letter corresponds to element with subsequent letters and numbers being linkage bookkeeping. For example, N, NZ, and NE2 all refer to nitrogen with different linkages, while O and its variants refer to oxygen.
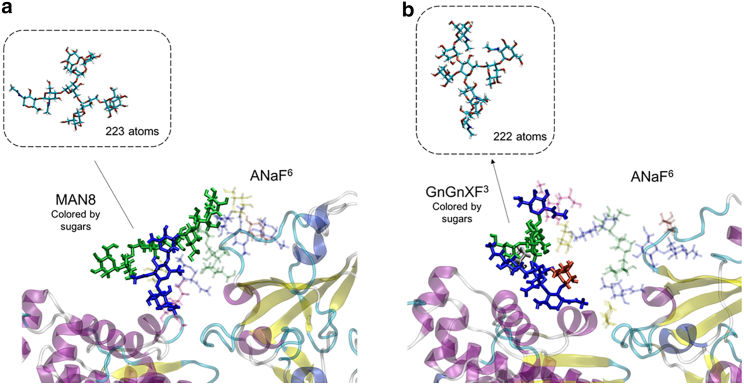
Fig. 6 shows how the different structures of MAN8 and GnGnXF3 affect the hydrogen bonding regime. Although MAN8 and GnGnXF3 have similar size (223 vs. 222 atoms), their shapes are very different. MAN8 is relatively flatter compared with GnGnXF3, making it bend less flexibly. Therefore, when MAN8 is close to ANaF6, they interact in a side-by-side fashion, whereas when GnGnXF3 is close to ANaF6, they interact in a head-to-head fashion, forming fewer hydrogen bonds than the MAN8/GnGnXF3 pair.
Figure 6.
Different structures and hydrogen bonding regimes of MAN8 and GnGnXF3 when interacting with ANaF6 on RBD. (a) MAN8 that interacts with ANaF6. (b) GnGnXF3 that interacts with ANaF6. Insets: shape and size of the MAN8 and GnGnXF3 without bending towards ANaF6. The glycans attached to proteins were colored by different sugars: blue, GlcNAc; green, mannose; yellow, galactose; red, fucose; silver, xylose; purple, Neu5Ac.
An autocorrelation function (ACF) analysis was performed for the angles and dihedrals of interest in both glycosylations, MAN8 and GnGnXF3, to further study the flexibility of the different glycans. These different flexibilities might be able to explain some of the emerging hydrogen bonding patterns. The angles and dihedrals chosen for the analysis are the ones between sugars (i.e., at the linkages). Fig. 1 shows the linkages of interest; the angles and dihedrals at linkage beta4_1, beta4_2, and alpha6 of the glycans at the six glycosylation sites on the ACE2 fragment in A1FrM8/SpFr and A2FrGG/SpFr at positions N219, N256, N269, N488, N598, and N712 were studied. We specifically focused on glycans at N488 for both systems, as it interacts with ANaF6 on RBD. To improve statistics, trajectories from the previous 75 ns runs (2) were used for the ACF analysis. Fig. 7 shows the angle and dihedral motions for both MAN8 and GnGnXF3 at glycosylation sites N219, N269, and N488. ACF results for glycans at all six sites are available in Figs. S10 and S11. Glycans on sites N219 and N269 show typical ACF behaviors of all glycans that do not directly interact with ANaF6 on RBD. Comparing the angle motion with dihedral motion for both glycans, ACFAngle decreases significantly, whereas ACFDihedral decreases slowly, indicating that angle motions are more favored for glycans, and dihedral motions are constrained (alpha6 at N269 in MAN8 is the only exception in which two motions are similarly favored). Comparing ACF of the different linkages, ACF of linkage alpha6 decreases much faster than the two beta4 linkages, indicating that linkage alpha6, which is the linkage to the branches, is the most flexible linkage. Comparing ACF of MAN8 and GnGnXF3, ACFAngle and ACFDihedral of MAN8 decrease either at similar rate or slower than those of GnGnXF3, with very few exceptions (angle: N219_beta4-2, N598_beta4-2 [Fig. S10]; dihedral: N219_alpha6, N256_beta4-1 [Fig. S11]), indicating that MAN8 is generally less flexible than GnGnXF3 for the angle and dihedral motions at linkage beta4_1, beta4_2, and alpha6. The glycans at N488 are the ones interacting with ANaF6 on RBD. All angle motions and dihedral motions of MAN8 at N488 are less flexible than for GnGnXF3, which further proves that side-by-side hydrogen bonding fashion with ANaF6 is favored by MAN8, resulting in more hydrogen bonds between glycans before pulling, whereas a head-to-head arrangement is favored by GnGnXF3, resulting in fewer hydrogen bonds between glycans before pulling. In addition, the angle motions of glycans at N488 are more constrained than those of glycans at N219, and the dihedral motion of glycans at N488 are more constrained than those of glycans at N269, indicating that glycans at N488 are generally constrained because they are connected to the protein on one end, and interacting with ANaF6 on the other end.
Figure 7.
Autocorrelation function analysis of angles and dihedrals at linkage beta4_1, beta4_2, and alpha6 for MAN8 and GnGnXF3 at ACE2 fragment glycosylation sites in semilog lots. Glycans at N219 (a and b) and at N269 (c and d) show typical behaviors and glycan at N488 (e and f) are the ones directly interacting with ANaF6 on RBD. Dashed lines are the dynamic motions of MAN8, and solid lines are the dynamic motions of GnGnXF3.
In addition to hydrogen bonding, we find that electrostatic and Lennard-Jones interactions contribute to the binding between ACE2 and RBD. These interactions are plotted in Fig. 8 with subplots a–d corresponding to the same variants as before. The y axis corresponds to the interaction energy between the ACE2 and RBD groups with the yellow line corresponding to Coulombic interactions and blue to short-range Lennard-Jones energies. Interestingly, it appears that at very short distances, the electrostatic interaction is more important (more negative interaction potential) than the Lennard-Jones interaction; this reverses at intermediate distances (1–2 nm from close contact), where the two lines cross for most of the systems. In some cases, there is a recrossing before the lines essentially merge and the interaction dies out. The glycosylated systems show a similar extension in interaction energies as in the hydrogen bonds, roughly 2–3 nm for the A1FrM8/SpFr and 1 nm for A2FrGG/SpFr. A1 variants demonstrate a differently shaped interaction curve than A2 variants for both glycosylated and aglycosylated systems, this can also be attributable to difference in starting orientation and zinc coordination.
Figure 8.
Lennard-Jones and electrostatic energies. (a) Man8 glycosylated A1FrM8/SpFr. (b) Aglycosylated structure A1Fr/SpFr. (c) GnGnXF glycosylated A2FrGG/SpFr. (d) Aglycosylated structure A2Fr/SpFr.
Visual inspection of the starting configurations of the two systems shows a difference in RBD alignment in the binding pocket. To evaluate if this difference was due to a rocking motion of the RBD or was caused by differences in the glycans, a principal-component analysis was performed on the trajectories from our previous study (2) to determine the dominant motions of the RBD. Results of the principal-component analysis are presented in Figs. 9 and S12–S15. Fig. 9, A and B show still structures with arrows indicating direction of projected motion from the dominant principal component. Corresponding video files are available in the Supporting materials along with time dependence and pairwise plots of principal components (Figs. S12–S15). Fig. 9 A shows that the motion of the spike fragment from A1FrM8/SpFr is a scissoring between helices and oscillation of the turn at the top of the structure. Fig. 9 B shows a similar motion, but the oscillation of the turn is missing because of the formation of a helix at that site. This structural change comes from the stable structure after 75 ns simulation due to differences between the glycans and ACE2 interaction. Fig. 9 C and D show cumulative variance versus the number of principal components for A1FrM8/SpFr and A2FrGG/SpFr, respectively. This clearly shows that most of the variance is explained by the first principal component (∼90% and ∼96% for A1FrM8/SpFr and A2FrGG/SpFr, respectively).
Figure 9.
First principal component (PC1) projected motion and cumulative variance. (A) PC1 projected motion for A1FrM8/SpFr. (B) PC1 projected motion for A2FrGG/SpFr. (C) Principal component cumulative variance A1FrM8/SpFr. (D) Principal component cumulative variance A2FrGG/SpFr. Arrows indicate contraction.
To determine whether changes in binding affinity due to deglycosylation can be observed experimentally, we performed biolayer interferometry using ACE2-Fc and RBD with and without removal of N-glycans. Biolayer interferometry is an optical technique that measures biomolecular interactions by detecting changes in the interference pattern of reflected light from a surface before and after binding (57). The response is measured as a shift in wavelength in units of nanometers. Fig. 10 a shows that deglycosylation of proteins via PNGase F treatment results in slightly lower bands on an SDS-PAGE gel, as expected from the smaller protein sizes following glycan removal. We then performed biolayer interferometry on ACE2-Fc and RBD, which are either both deglycosylated or glycosylated (Fig. 10, b–d). To do this, ACE2-Fc was immobilized onto a biosensor using the Fc tag and placed in a solution containing the RBD analyte. Steady-state analysis was performed on the response using a 1:1 Langmuir binding model, where the response indicates the shift in interference patterns caused by analyte binding (Fig. 10 d). Glycosylated ACE2-Fc and RBD have a binding affinity, KD, of 30 nM, which is similar to values reported by other groups (34,58). Deglycosylation of ACE2-Fc and RBD results in a 2- to 3-fold increase in binding affinity to 77 nM. From the increase in binding affinity, the magnitude of the binding energy decreases by 2.3 kJ/mol following removal of N-linked glycans. This is consistent with our simulation results that predicts that less pulling force is required to break the protein interactions after deglycosylation.
Figure 10.
Biolayer interferometry on glycosylated and deglycosylated ACE2-Fc and RBD. (a) SDS-PAGE on ACE2-Fc and RBD with and without PNGase F treatment. A total of 1 µg of protein is loaded onto each lane. Subscript D indicates deglycosylated proteins, and subscript G indicates glycosylated proteins. (b–d) Biolayer interferometry response for (b) deglycosylated ACE2-Fc and RBD, (c) glycosylated ACE2-Fc and RBD, and (d) glycosylated ACE2-Fc and RBD without glyco buffer 2 and incubation at 37°C. Red lines are the fits to the raw data shown in blue, brown, and green, respectively. (e) Quantification of the equilibrium dissociation constants (KD) for ACE2-Fc and RBD with and without deglycosylation. Control samples (Glycosylated ACE2-Fc + RBD) were prepared by adding glyco buffer 2 (50 mM sodium phosphate), but no PNGase F, and incubating at 37°C for 5 h. Additional control samples (Control ACE2-Fc + RBD) were prepared without glyco buffer 2 and without incubation at 37°C. Error bars represent standard error. ∗p < 0.05. ns, not significant (p > 0.05). Probability values were calculated using one-way ANOVA followed by Tukey’s test.
Discussion
Detailed mechanistic studies of binding interaction events can improve our understanding of how specific changes to proteins affect binding strength. Differences in binding dissociation rate could have implications in infectivity (59, 60, 61). Viral protein and host receptor interactions are complex because of the interplay among interaction types, different degrees of motion during a binding event, and the role of glycans in shielding or strengthening receptor binding. SARS-CoV-2 spike protein and ACE2 interactions are no different. Understanding the implications of different glycans on the binding behavior of spike could prove useful as more variants emerge with potentially different glycosylation patterns. Recent studies have shown experimentally and computationally that the ACE2 and RBD of coronavirus spike fragments have different binding strengths and dissociation rates when they are glycosylated versus non-glycosylated (33,34).
Previous computational efforts focused on the binding difference between SARS-CoV-1 and SARS-CoV-2 with glycan interactions modeled by a generic pentasaccharide (32). Their analysis focused on the difference in binding strengths and protein contacts between RBDCoV1 and RBDCoV2. Our results are in alignment with this trend of stronger interactions caused by the glycans but go further in the analysis of the mechanisms behind this stronger interaction and evaluate more realistic glycan models.
First, our results clearly show that the glycans result in stronger and longer ranged interactions that get extended by a catch-slip mechanism between the glycans (i.e., a hydrogen bond breaks and another one at larger distance takes its place). This catch-slip behavior is clearly seen in the hydrogen bonding maps shown in Fig. 4. The catch-slip behavior is a result of the original hydrogen bond interactions that are present relaxing and then reforming later. Analysis of A1FrM8/SpFr in Fig. 4 a clearly shows the relaxation and reformation of glycan contributed hydrogen bonds. This behavior can be attributed to the increased flexibility of the glycans, which increases the ability of these late-stage hydrogen bonds to form because of both increased contacts and increased ability to extend through solution. The different structures of MAN8 and GnGnXF3 also contribute to the different hydrogen bond interactions between an ACE2 glycan and RBD glycan. The flatter MAN8 allows more hydrogen bonds between MAN8 and ANaF6, therefore causing more glycan-glycan and glycan-protein interactions during pulling for A1FrM8/SpFr than for A2FrGG/SpFr. Angle and dihedral motions are less flexible for MAN8 than for GnGnXF3, especially for the MAN8 and GnGnXF3 glycans that directly interact with ANaF6, proving that MAN8 is more constrained by the hydrogen bonds between MAN8 and ANaF6. The hydrogen bond map of A2FrGG/SpFr in Fig. 4 c shows that there is a present, but less pronounced, hydrogen bond formation between the glycans. The distance extension is seen clearly in the pull force versus center of mass distances (Fig. 3) as well as the interaction energies versus center of mass distances (Fig. 8), where the glycosylated structures have their interaction distance extended by as much as 2 nm. This extension can be clearly attributed to the glycans when compared against the hydrogen bond map in Fig. 4.
Second, an analysis of hydrogen bond occupancy elucidates that the glycans not only result in secondary binding motifs but also strengthen and extend the existing protein-protein interactions. This is most clearly seen in the percentage occupancy numbers for the A2FrGG/SpFr structure, with an increase of several percent in most of the top hydrogen bonds. This trend is also present in A1FrM8/SpFr when looking at the top protein-protein interactions, such as RBD-GLY167:ACE2-LYS519, showing an increase of more than 3%. This strengthening of the protein-protein hydrogen bonds may be a result of the extra stabilization in the RBD structure provided by the glycan. That the glycans strengthen the interactions is consistent with our biolayer interferometry results. A frequent interaction point of interest is the N-glycosylation site ASN90 on ACE2 and GLN409 and THR415 of the spike RBD. Our results suggest a strong interaction in a nearby site ACE2-TYR249 (equivalent to TYR83 in standard numbering) and RBD-ASN152 (equivalent to ASN 487) for all variants studied. This interaction agrees with previous results suggesting a long interaction at this site due to the flexibility of the RBD loop (32). It is interesting to note that this interaction is seemingly not affected by the glycan as it pertains to percentage occupancy.
It is necessary to comment on the difference in starting orientation of the RBD and the ACE2 between the two different starting truncations. By taking the final structure of the simulations from our previous study, it was possible that this resulted in a lower probability starting orientation. A principal-component analysis was performed (Fig. 10) to verify that the starting structures were truly the dominant orientation from our previous paper and not just an unlucky snapshot of a less favorable state. These results show that the dominant motion from the highest principal component is scissoring of helices and oscillation of a turn and not the rocking of the spike fragment. This suggests that the structure was stable in the ACE2 binding pocket and that the difference in starting structure is due to the differences between glycosylation and the effect of Zn2+ on the stability of ACE2. Fig. 10, a and b clearly show the structural changes resulting from these interactions. These structural changes result in differences in the interaction behavior as seen by a slight 1 nm extension of interaction energies as shown in Fig. 10, b and d.
Conclusion
We have expanded on our previously developed model of fully glycosylated ACE2-Fc and SARS-CoV-2 spike protein fragments through investigation of the binding strength and role of glycosylation on binding between these groups. This investigation provides further evidence that binding between SARS-CoV-2 spike and ACE2 receptor is aided by the glycosylation on each protein. We found that for multiple complex glycan types, the interactions between RBD and ACE2 were strengthened and longer ranged. Protein-protein interactions were extended because of the increased stability provided by the glycans, and binding strength is affected by a catch-slip behavior between the glycans. These computational results were corroborated by experimental evidence that the magnitude of the binding energy is decreased for deglycosylated proteins. Further work in analyzing the larger fragments of spike will be necessary for a more realistic model of RBD stability in order to address effects of mutations.
Acknowledgments
Y.H. and R.F. were partially supported by the National Science Foundation under grant CBET 1911267. S.A.M. and S.J. were partially supported by a COVID-19 Research Accelerator Funding Track award by the UC Davis Office of Research (https://covid19research.ucdavis.edu/tags/craft). B.S.H. was partially supported by the Laboratory Directed Research and Development program of Lawrence Livermore National Laboratory, under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344. K.A.M. and S.N. were partially supported by NASA Space Technology Research (award NNX17AJ31G) and by the Translational Research Institute through NASA (grant NNX16AO69A). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of NASA or the Translational Research Institute for Space Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Editor: Margaret Cheung.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.12.002.
Supporting material
References
- 1.Liu Z., Xiao X., et al. Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardi A., Huang Y., et al. Faller R. Development and simulation of fully glycosylated molecular models of ACE2-Fc fusion proteins and their interaction with the SARS-CoV-2 spike protein binding domain. PLoS One. 2020;15:e0237295. doi: 10.1371/journal.pone.0237295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan K.K., Dorosky D., et al. Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson G., Buzko O., et al. Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 doi: 10.1101/2021.01.13.426558. 2021.2001.2013.426558. [DOI] [Google Scholar]
- 5.Mehdipour A.R., Hummer G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2100425118. e2100425118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prajapat M., Shekhar N., et al. Medhi B. Virtual screening and molecular dynamics study of approved drugs as inhibitors of spike protein S1 domain and ACE2 interaction in SARS-CoV-2. J. Mol. Graph. Model. 2020;101:107716. doi: 10.1016/j.jmgm.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson A.M., Wysocki J., Batlle D. Interaction of SARS-CoV-2 and other coronavirus with ACE (angiotensin-converting enzyme)-2 as their main receptor. Hypertension. 2020;76:1339–1349. doi: 10.1161/HYPERTENSIONAHA.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigerust D.J., Shepherd V.L. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtsubo K., Marth J.D. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Phillips M., Nudelman E., et al. Paulson J. ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science. 1990;250:1130–1132. doi: 10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- 11.Sperandio M., Gleissner C.A., Ley K. Glycosylation in immune cell trafficking. Immunol. Rev. 2009;230:97–113. doi: 10.1111/j.1600-065X.2009.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y., Bowden T.A., et al. Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee N., Mukhopadhyay S. Viral glycoproteins: biological role and application in diagnosis. Virusdisease. 2016;27:1–11. doi: 10.1007/s13337-015-0293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson A.J., de Vries R.P., Paulson J.C. Virus recognition of glycan receptors. Curr. Opin. Virol. 2019;34:117–129. doi: 10.1016/j.coviro.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe Y., Allen J.D., et al. Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020:eabb9983. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen K.G., Rambaut A., et al. Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls A.C., Park Y.-J., et al. Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanda M., Morrison L., Goldman R. N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021;93:2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernández A. Glycosylation of SARS-CoV-2 steers evolutionary outcomes in the postvaccination phase. ACS Pharmacol. Transl. Sci. 2021;4:410–412. doi: 10.1021/acsptsci.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen J.D., Watanabe Y., et al. Crispin M. Subtle influence of ACE2 glycan processing on SARS-CoV-2 recognition. J. Mol. Biol. 2021;433:166762. doi: 10.1016/j.jmb.2020.166762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen K., Chakraborty S., et al. Gnanakaran S. Exploring the role of glycans in the interaction of SARS-CoV-2 RBD and human receptor ACE2. Viruses. 2021;13:927. doi: 10.3390/v13050927. https://www.mdpi.com/1999-4915/13/5/927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sztain T., Ahn S.-H., et al. Amaro R.E. A glycan gate controls opening of the SARS-CoV-2 spike protein. bioRxiv. 2021 doi: 10.1101/2021.02.15.431212. 2021.2002.2015.431212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo H., Park S.-J., et al. Im W. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y.K., Cao Y., et al. Im W. Structure, dynamics, receptor binding, and antibody binding of the fully glycosylated full-length SARS-CoV-2 spike protein in a viral membrane. J. Chem. Theor. Comput. 2021;17:2479–2487. doi: 10.1021/acs.jctc.0c01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casalino L., Gaieb Z., et al. Amaro R.E. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vuorio J., Vattulainen I., Martinez-Seara H. Atomistic fingerprint of hyaluronan–CD44 binding. PLoS Comput. Biol. 2017;13:e1005663. doi: 10.1371/journal.pcbi.1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuorio J., Škerlová J., et al. Martinez-Seara H. N-Glycosylation can selectively block or foster different receptor–ligand binding modes. Sci. Rep. 2021;11:5239. doi: 10.1038/s41598-021-84569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernardi A., Kirschner K.N., Faller R. Structural analysis of human glycoprotein butyrylcholinesterase using atomistic molecular dynamics: the importance of glycosylation site ASN241. PLoS One. 2017;12:e0187994. doi: 10.1371/journal.pone.0187994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong Y., Karuppanan K., et al. Nandi S. Effects of N-glycosylation on the structure, function, and stability of a plant-made Fc-fusion anthrax decoy protein. Front. Plant Sci. 2019;10 doi: 10.3389/fpls.2019.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.S., Qi Y., Im W. Effects of N-glycosylation on protein conformation and dynamics: protein Data Bank analysis and molecular dynamics simulation study. Sci. Rep. 2015;5:8926. doi: 10.1038/srep08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao W., Dong C., et al. Zhang X.F. Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J. 2021;120:1011–1019. doi: 10.1016/j.bpj.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang J., Ye G., et al. Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Petitjean S.J.L., et al. Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomord V., Chamberlain P., et al. Faye L. Biopharmaceutical production in plants: problems, solutions and opportunities. Trends Biotechnol. 2005;23:559–565. doi: 10.1016/j.tibtech.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Castilho A., Neumann L., et al. Steinkellner H. Generation of biologically active multi-sialylated recombinant human EPOFc in plants. PLoS One. 2013;8:e54836. doi: 10.1371/journal.pone.0054836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Meyer T., Depicker A. Trafficking of endoplasmic reticulum-retained recombinant proteins is unpredictable in Arabidopsis thaliana. Front. Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izrailev S., Stepaniants S., et al. Schulten K. In: Computational Molecular Dynamics: Challenges, Methods, Ideas. Deuflhard P., Hermans J., et al.Skeel R.D., editors. Springer; 1998. Steered molecular dynamics. [Google Scholar]
- 39.Yan R., Zhang Y., et al. Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrara C., Grau S., et al. Benz J. Unique carbohydrate–carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. U S A. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towler P., Staker B., et al. Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis ∗. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padjasek M., Kocyła A., et al. Krężel A. Structural zinc binding sites shaped for greater works: structure-function relations in classical zinc finger, hook and clasp domains. J. Inorg. Biochem. 2020;204:110955. doi: 10.1016/j.jinorgbio.2019.110955. [DOI] [PubMed] [Google Scholar]
- 43.Gamsjaeger R., Liew C.K., et al. Mackay J.P. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Case D.A., Betz R.M., et al. Kollman P.A. University of California; 2016. AMBER 2016. [Google Scholar]
- 45.Woods Group. 2005-2020. 2020. http://glycam.org
- 46.Bernardi A. MCPB_Glycam_merge. 2020. https://github.com/austenb28/MCPB_Glycam_merge
- 47.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 48.Parinello M., Rahman A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982;76:2662–2666. [Google Scholar]
- 49.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. doi: 10.1002/jcc.540130805. [DOI] [Google Scholar]
- 50.Hess B., Bekker H., et al. Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 51.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 52.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. doi: 10.1063/1.447334. [DOI] [Google Scholar]
- 53.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 54.Reinhardt M., Grubmüller H. GROMACS implementation of free energy calculations with non-pairwise Variationally derived Intermediates. Comput. Phys. Commun. 2021;264:107931. doi: 10.1016/j.cpc.2021.107931. [DOI] [Google Scholar]
- 55.Savitzky A., Golay M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964;36:1627–1639. doi: 10.1021/ac60214a047. [DOI] [Google Scholar]
- 56.Ponder J.W., Case D.A. Advances in Protein Chemistry. Academic Press; 2003. Force fields for protein simulations; pp. 27–85. [DOI] [PubMed] [Google Scholar]
- 57.Abdiche Y., Malashock D., et al. Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 58.Bayarri-Olmos R., Rosbjerg A., et al. Skjoedt M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozono S., Zhang Y., et al. Tokunaga K. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 2021;12:848. doi: 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu H., Zhang Q., et al. Zhang G. The basis of a more contagious 501Y.V1 variant of SARS-CoV-2. Cell Res. 2021;31:720–722. doi: 10.1038/s41422-021-00496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steckbeck J.D., Orlov I., et al. Cole K.S. Kinetic rates of antibody binding correlate with neutralization sensitivity of variant simian immunodeficiency virus strains. J. Virol. 2005;79:12311–12320. doi: 10.1128/JVI.79.19.12311-12320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.