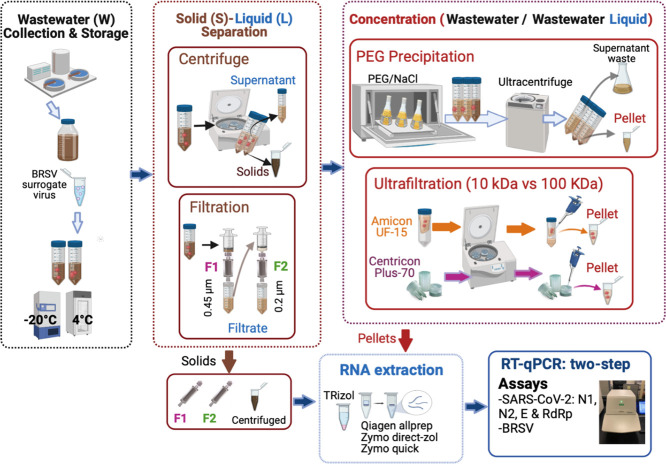
Abstract
In this study, 14 virus concentration protocols based on centrifugation, filtration, polyethylene glycol (PEG) precipitation and ultrafiltration were tested for their efficacy for the quantification of SARS-CoV-2 in wastewater samples. These protocols were paired with four RNA extraction procedures resulting in a combination of 50 unique approaches. Bovine respiratory syncytial virus (BRSV) was used as a process control and seeded in each wastewater sample subjected to all 50 protocols. The recovery of BRSV obtained through the application of 50 unique approaches ranged from <0.03 to 64.7% (±1.6%). Combination of centrifugation as the solid removal step, ultrafiltration (Amicon-UF-15; 100 kDa cut-off; protocol 9) as the primary virus concentration method, and Zymo Quick-RNA extraction kit provided the highest BRSV recovery (64.7 ± 1.6%). To determine the impact of prolonged storage of large wastewater sample volume (900 mL) at −20 °C on enveloped virus decay, the BRSV seeded wastewaters samples were stored at −20 °C up to 110 days and analyzed using the most efficient concentration (protocol 9) and extraction (Zymo Quick-RNA kit) methods. BRSV RNA followed a first-order decay rate (k = 0.04/h with r2 = 0.99) in wastewater. Finally, 21 wastewater influent samples from five wastewater treatment plants (WWTPs) in southern Maryland, USA were analyzed between May to August 2020 to determine SARS-CoV-2 RNA concentrations. SARS-CoV-2 RNA was quantifiable in 17/21 (81%) of the influent wastewater samples with concentration ranging from 1.10 (±0.10) × 104 to 2.38 (±0.16) × 106 gene copies/L. Among the RT-qPCR assays tested, US CDC N1 assay was the most sensitive followed by US CDC N2, E_Sarbeco, and RdRp assays. Data presented in this study may enhance our understanding on the effective concentration and extraction of SARS-CoV-2 from wastewater.
Keywords: Wastewater, Surveillance, SARS-CoV-2, COVID-19, Virus concentration, RT-qPCR
Graphical abstract
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an infectious RNA virus that emerged in Wuhan (Hubei, China) towards the end of 2019 (Zhu et al., 2020). This novel virus induces respiratory illness Coronavirus Disease 2019 (COVID-19) and spread globally causing the World Health Organization (WHO) to declare a pandemic in March 2020. As of 19 November 2021, it has infected about 255 million of people and resulted in more than 5 million deaths globally (WHO, 2021). Hence, the current spread of SARS-CoV-2 is of pressing public health concern with novel variants emerging (Cui et al., 2019; Naqvi et al., 2020).
SARS-CoV-2 RNA is shed by symptomatic and asymptomatic individuals through their feces, sputum, nasopharyngeal secretions, saliva, and urine (Cevik et al., 2021; Collivignarelli et al., 2020; Gao et al., 2020; Tang et al., 2020; Xiao et al., 2020) and deposited in wastewater. SARS-CoV-2 RNA have been detected globally in wastewater samples in about 60 countries (COVIDPoops19, 2021). Wastewater surveillance has the potential to overcome the limitations of clinical testing by screening composite wastewater samples containing SARS-CoV-2 RNA fragments from both symptomatic and asymptomatic individuals (Tang et al., 2020; Xu et al., 2020). Information on the presence or circulation of SARS-CoV-2 at a community level can aid public health officials implementing more targeted approaches to manage the spread of COVID-19 (Prado et al., 2021).
Detection of SARS-CoV-2 in wastewater is a complex process involving several steps such as representative sampling, virus concentration, RNA extraction and detection methods (Kantor et al., 2021). There is no standardized method that has been shown to produce consistent results across laboratories (Pecson et al., 2021). When the concentration of SARS-CoV-2 is high (>6 log10 gene copies (GC/100 mL)) in wastewater, most of the methods are likely to produce positive results (Pecson et al., 2021). However, for trace detection (early in the pandemic, the tailing phase or a community with no known clinical cases), methods must be optimized. Various methods have been used to concentrate SARS-CoV-2 from wastewater samples including polyethylene glycol precipitation (PEG) (Ahmed et al., 2020b; Wu et al., 2020), ultrafiltration (UF) device (Balboa et al., 2020; Medema et al., 2020), adsorption-extraction (Ahmed et al., 2020a; Sherchan et al., 2020), adsorption-precipitation (Randazzo et al., 2020), and ultracentrifugation (Wurtzer et al., 2021). A limited number of studies have investigated the RNA extraction protocols in various combinations with concentration protocols for SARS-CoV-2 detection in wastewater samples (Colosi et al., 2021; Eisen et al., 2020; O'Brien et al., 2021; Pérez-Cataluña et al., 2021; Weidhaas et al., 2021). Furthermore, although investigated for smaller volumes of wastewater samples (Ahmed et al., 2020b; Bivins et al., 2020; Hart and Halden, 2020; Hokajärvi et al., 2021; Wang et al., 2005; Weidhaas et al., 2021; Ye et al., 2016) the impact of storing untreated bulk wastewater samples at −20 and −80 °C are still unclear (Ahmed et al., 2020c).
The aims of this study were: (i) evaluation of 14 virus concentration and four RNA extraction protocols for the effective concentration and extraction of bovine respiratory syncytial virus (BRSV) RNA from BRSV seeded influent wastewater samples; (ii) determine the decay of BRSV in bulk wastewater at −20 °C (iii) determine the sensitivity of four RT-qPCR assays (US CDC N1, US CDC N2, E_Sarbeco and RdRp) for the detection and quantification of SARS-CoV-2 RNA in wastewater; (iv) monitoring of untreated wastewater samples from the influent of five WWTPs in Maryland, USA. BRSV was used as a process control virus in this study. The findings of this study will enable researchers to identify enhanced protocols for the detection and quantification of SARS-CoV-2 in untreated wastewater.
2. Materials and methods
2.1. BRSV stock preparation
Freeze-dried Inforce 3 intranasal cattle vaccine containing attenuated strain of BRSV was obtained from Zoetis (Parsippany, NJ, USA). BRSV was rehydrated with the supplied sterile diluent by following manufacturer's instructions. Multiple vials were rehydrated and pooled to prepare a stock solution for wastewater seeding purpose. Aliquots (1 mL each) were stored at −20 °C for later use. BRSV RNA in stock was 1.05 × (±0.9) 107 GC of BRSV/μL determined using RT-qPCR protocol detailed below in Section 2.9 after the extraction of resuspended BRSV vaccine via Zymo Quick RNA extraction kit (Quick RNA Mini prep, Zymo Research, CA, USA) following manufacturer's instructions (other details in Section 2.7).
2.2. Virus concentration protocols
Bulk wastewater (~3 L) sample was collected in May 2020 from the influent of an urban wastewater treatment plant (WWTP) B (Table 1 ) serving 174,257 people with an influent flow rate of 9.54 × 107 L/day, Maryland, USA. Wastewater was transported on ice to the laboratory and stored at 4 °C for less than 2 h. Aliquots (30–100 mL) from the bulk wastewater sample were seeded with known numbers (1.05 × (±0.9) 107 GC of BRSV/mL of wastewater) and subjected to several virus concentration and extraction protocols (i.e., centrifugation, filtration, PEG precipitation and ultrafiltration (UF)) (Table S1). These four concentration protocols and their variants (14 protocols) were paired with four RNA extraction protocols (as explained in Section 2.7) resulting in a combination of 50 unique protocols (Table 2 ). All concentrated samples and filters were stored at −20 °C and viral RNA was extracted from triplicates samples within 24-h.
Table 1.
Details of five wastewater treatment plants (WWTPs).
| WWTPs | Population | Service area (miles2) | Hospital | Flow (107 × L/day) |
|---|---|---|---|---|
| WWTP A | 55,848 | 13.9 | Medical Center A | 2.48 |
| WWTP B | 174,257 | 102.4 | Medical Center B | 9.54 |
| WWTP C | 241,316 | 126.7 | Medical Center C | 8.41 |
| WWTP D | 201,694 | 46.9 | Medical Center D | 6.12 |
| WWTP E | 5354 | 4.9 | – | 0.33 |
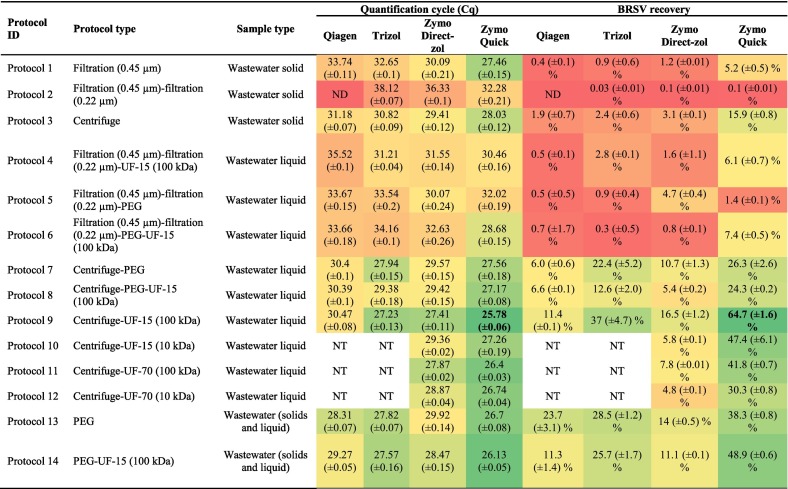
Table 2.
BRSV recovery and quantification cycle (Cq) obtained through each or combination of methods subjected to each of four RNA extraction approaches. Protocols 10–12 were not tested with Qiagen and Trizol RNA extraction approaches.
*NT: not tested. ND: not detected (<LOD) UF-15: Amicon UF-15; UF-70: Centricon UF-70. Recovery percentage show in bold font indicates highest recovery. Colors from red to green indicates highest to lowest values for Cq while for BRSV recoveries, colors from red to green indicates lowest to highest, in other words, meaning worst to best results for both parameters.
2.3. Centrifugation-based protocols
Centrifugation-based protocols (Table S1) were utilized independently (protocol 3) or in combination with PEG or ultrafiltration or both (protocols 7–12). BRSV seeded wastewater samples (100 mL) were mixed vigorously and then transferred into sterile 50 mL falcon tubes. Samples were centrifuged at 3400g in a swinging-bucket rotor for 15 min to pellet solid particles. Each pellet was resuspended in ~1 mL nuclease-free water (Millipore, MA, USA) and stored at −20 °C for RNA extraction (protocol 3). Supernatant was carefully removed and transferred into a new 50-mL tube and concentrated further by either PEG precipitation (protocol 7) or ultrafiltration (protocol 9–12), or a combination of both methods (protocol 8).
2.4. Filtration-based protocols
Sterivex-GP cartridges, with different pore-sizes (0.45 and 0.22 μm) (Millipore, MA, USA) were used in five different protocols (Table S1), namely protocols 1, 2, and 4–6. BRSV seeded wastewater samples (98 mL) were filtered through Sterivex-GP cartridges with 0.45 μm membrane by using a 60-mL sterile syringe (protocol 1). This step was repeated three times to pass through ~33 mL wastewater at a time to obtain ~98 mL filtrate in total. This filtrate was subjected to a second filtration step by passing through a Sterivex-GP cartridge with 0.22 μm filter by using a 60-mL sterile syringe (protocol 2). Approximately, 49–53 mL filtrate from protocol 2 was stored at 4 °C for further concentration within the same day using ultrafiltration (protocol 4), or PEG precipitation (protocol 5), or a combination of both methods (protocol 6). The filters (0.45 and 0.22 μm) were stored at −20 °C until RNA extraction stage (Section 2.7).
2.5. PEG precipitation-based protocols
PEG precipitation was utilized in six different virus concentration protocols (Table S1), namely, protocols 5–8, 13, and 14. PEG 8000 (Fisher Scientific, MA, USA) (8% w/v) and 0.3 M NaCl (Fisher Scientific, MA, USA) were added to each sample (30–60 mL) and shaken at room temperature (20 °C) for 15 min to dissolve PEG and NaCl (Wu et al., 2020). Wastewater samples were then incubated at 4 °C on an orbital shaker at 150 rpm for 12 h. Samples were subsequently centrifuged at 14,000g for 2 h at 4 °C to pellet the virus particles. The supernatant was discarded, and pellets were resuspended in ~1 mL nuclease-free water.
2.6. Ultrafiltration-based protocols
BRSV seeded wastewater samples such as filtrate from filtration-based protocols (protocol 1 and 2), supernatant from centrifugation step (protocol 3), or resuspended pellets from PEG precipitation-based protocols (protocols 6, 8 and 14) were concentrated using two different centrifugal units (Table S1): Amicon Ultra-15 and Centricon Plus-70 (Millipore) with two different molecular weight cut-offs: 10 or 100 kDa by following manufacturer's instructions. A primary reason of ultrafiltration application after PEG precipitation was to further purify the pellet, therefore, 1 mL of pellet coming from PEG-stage was brought to 15 mL with nuclease-free water and the ultrafiltration method was applied. Finally, to have a similar final volume across the protocols, concentrates (300–400 μL) of ultrafiltration-based protocols were brought to 1 mL by adding nuclease-free water the collection cups to rinse and recover as much concentrate as possible.
2.7. Viral RNA extraction
Viral RNA was extracted from an aliquot (250–300 μL) of each concentrated sample (protocols 3–14) using three commercial RNA extraction kits: Qiagen (Allprep Power Viral RNA/DNA, Qiagen, Hilden, Germany), Zymo Quick (Quick RNA Mini prep, Zymo Research, CA, USA), and Zymo Direct-zol (Direct-zol RNA Miniprep Plus, Zymo Research) following the manufacturer's instructions. Furthermore, a reagent-based traditional RNA extraction method, TRIzol-chloroform was also used for viral RNA extraction (Wu et al., 2020).
RNA was extracted from filters (0.45 and 0.22 μm) (protocol 1 and 2) following the same protocols used to extract RNA from liquid concentrated samples, with a slight modification at the initial lysis step. Briefly, the filter inside the Sterivex-GP cartridges was cut into 2 to 4 pieces with a flame sterilized blade. Filter pieces were inserted into 2 mL bead-beating tubes (0.5 mm diameter glass beads, Fisher Scientific). Before bead beating, lysis buffer (750 μL) provided with the RNA extraction kits (i.e., Qiagen, Zymo Quick and Zymo Direct-zol RNA) or a mixture of Trizol (750 μL) and chloroform (150 μL) with Trizol extraction protocol were added into the tubes containing samples. In addition, β-mercaptoethanol (10 μL/mL as the final concentration) was added to the lysis solution used in the Qiagen RNA extraction kit. The samples were then processed on a FastPrep-24 5G bead beater (MP Biomedical, NSW, Australia) for 1 min at 5 m/s. Samples were subsequently centrifuged for 5 min at 16,000g at 4 °C. Approximately, 600 μL of supernatant was then removed and subjected to each RNA extraction protocol. For all methods and samples, RNA was eluted in 100 μL using nuclease-free water and stored at −20 °C. RT-qPCR was performed within 24 h (Boxus et al., 2005).
2.8. Reverse transcription (RT)
RT: complementary DNA (cDNA) was synthesized from RNA using random hexamers (Cat. no: N8080127, Invitrogen) and ProtoScript II reverse transcriptase (M0368, New England Biosciences). Briefly, RNA (4–6 μL) mixed with 50 μM random hexamers (1.5 μL) was incubated at 70 °C for 5 min and 4 °C for 3 min, followed with the addition of 5× ProtoScript II buffer (5 μL), 0.1 M DTT (2.5 μL), ProtoScript II reverse transcriptase (1.25 μL, 200 U/μL), 10 mM dNTP (1.25 μL), RNase Inhibitor (0.5 μL, 40 U/μL), and nuclease-free water to bring the reaction volume to 25 μL. The mixture was then incubated at 42 °C for 1 h and inactivated at 65 °C for 20 min (Wu et al., 2020), then chilled at 4 °C for RT-qPCR done within the same day.
2.9. RT-qPCR assays
BRSV target and standard: one BRSV assay targeting N gene was evaluated in this study. to generate a stock standard for quantification of BRSV RNA, we carried out a PCR reaction using DreamTaq Green PCR Master Mix (Cat. No. K108, ThermoFisher Scientific) and specific PCR primer set designed for this study targeting a region of BRSV N gene and gBlocks (IDTDNA.com) as the BRSV template. gBlocks gene fragments of BRSV N gene (double-stranded DNA, 446 bp) were purchased from IDT. PCR reaction was conducted by using 25 μL of 2× DreamTaq Green PCR Master Mix, 17 μL nuclease-free water, 2 μL of forward and reverse primers (each at 500 nM), and 4 μL of gBlock (1 ng/μL). Both the PCR cycling program and the sequences of PCR primers (forward and reverse) are reported in Table S2. PCR amplicons (318 bp) were then purified with QIAquick PCR Purification Kit (Qiagen, Germany) and used as the stock standard to generate standard curves for quantification of BRSV RNA. The copy numbers (GC) of BRSV N gene in the stock standard, 7.36 × 109 GC/μL, was calculated as described earlier (Ritalahti et al., 2010).
qPCR for BRSV target: a previously published qPCR assay targeting the N gene was used to quantify BRSV in wastewater (Boxus et al., 2005). For quantitation of the BRSV RNA in each sample, standard curves for each run were generated by using 10-fold serial dilutions of BRSV standard (purified PCR product) with copies ranging from 101 to 108 per reaction. Both the thermocycling program and the sequences of qPCR primers and probe are reported in Table S2.
SARS-CoV-2 targets and standard: four SARS-CoV-2 assays (US CDC N1, US CDC N2, E_Sarbeco and RdRp) were evaluated in this study (Corman et al., 2020; Lu et al., 2020). Circular plasmids containing SARS-CoV-2 nucleocapsid gene (N) provided with 2019-nCoV RUO Kit (Cat. No: 10006625, IDT), 2019-nCoV_E (Cat. No: 10006896, IDT), and 2019-nCoV_RdRP (ORF1ab) (Cat. No: 10006897, IDT) were linearized individually using ScaI-HF enzyme (R3122S, New England Biosciences) by following manufacturer's protocol prior their use as a qPCR standard. Standard curves for CDC N1, CDC N2, E_Sarbeco, and RdRp assays were prepared by serial dilutions (10-fold) of linearized plasmid with concentrations ranging from 100 to 105 per reaction.
qPCR for SARS-CoV-2 targets: qPCR reactions were performed in 20 μL with 10 μL TaqMan Fast Advanced Master Mix (Cat. No. 4444557, ThermoFisher Scientific, MA, USA), primers/probe sets specific to each target (final concentrations are given in Table S2) and 2–3 μL of template by following manufacturer's protocol. The qPCR was carried out on a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad, CA, USA). Both the thermocycling program and the sequences of qPCR primers and probe are reported in Table S2.
2.10. Assay limit of detection (ALOD)
To determine, assay limit of detection (ALOD), serially diluted standard curves for each assay were tested with qPCR in triplicates. The lowest GC of diluted standards detected in triplicate assays was accepted as ALOD for each assay. Values above ALODs with quantification cycle values (Cqs) below 40 were accepted as quantifiable and are reported. Viral RNA concentration (GC/L) in wastewater were calculated using Eq. S1.
2.11. Quality controls
To minimize variability between the technical replicates of each concentration and extraction protocol, duplicate samples were concentrated and pooled. Duplicate RNA was extracted from each pooled concentrated sample and again RNA samples were pooled. Wastewater samples were also concentrated to determine background BRSV level, but BRSV could not be detected in any of the wastewater samples. Furthermore, method negative controls (i.e., nuclease-free water) were included for each concentration protocol. Additionally, triplicate method positive controls (i.e., BRSV seeded into nuclease-free water) were concentrated using protocol 7 and extracted with Zymo Quick RNA extraction kit to reveal BRSV recovery from water samples. To account for any contamination during RNA extraction, extraction negative controls were included. No template controls (NTCs) were included in each qPCR. No amplification was observed in any of the negative controls. RT-qPCR assays were performed in triplicate for each sample. Results are reported as the average of triplicate analysis with standard deviations for each sample.
2.12. Wastewater inhibition control
To assess PCR inhibition on BRSV quantification, BRSV was quantified in four qPCR reactions (i.e., reference points): two of those reactions received BRSV cDNA as a template, while the others BRSV cDNA and wastewater sample cDNA as the template during the qPCR reactions. This was only tested on wastewater samples (n = 3) concentrated with protocol 9 only. The Cq cycles of those reactions were compared to assess PCR inhibition. The difference in RT-qPCR Cq values of 1 cycle is used as a threshold for PCR inhibition (Pecson et al., 2021).
2.13. Calculation of BRSV recovery
BRSV recovery was calculated using Eq. (1); briefly, by dividing the BRSV RNA concentration in a given sample quantified after concentration and extractions to the BRSV RNA concentration seeded to the wastewater sample prior to any processing. Efficiency of RNA extraction protocols were evaluated by comparing Cq and recovery values calculated for each concentration method by using Eq. (1) as follows:
| (1) |
2.14. Viral RNA decay rate calculation
To determine the decay rate of viral RNA, the concentrations of BRSV in wastewater (n = 16) were stored determined using RT-qPCR assay. For this purpose, bulk volumes of wastewater (900 mL) were seeded with BRSV and stored in 1 L Nalgene wide-mouth HDPE packaging bottles (ThermoFisher Scientific, MA, USA) at −20 °C for up to 110 days until RT-qPCR analysis of SARS-CoV-2 RNA. At each time points (0, 47, 77, and 110), triplicate samples (30 mL each) were aliquoted from a bottle of wastewater (900 mL) that was sacrificed by thawing at a 4 °C fridge. Protocol 9 and Zymo Quick kit were followed for viral RNA concentration and extraction, respectively. The details of RT-qPCR analysis of BRSV are provided above in the preceding sections. Decay rates were calculated using the Eq. (2) (Li et al., 2021);
| (2) |
where Ct is the measured concentrations of BRSV RNA in wastewater on day 0, 47, 77, and 110, and C0 is the seeded BRSV RNA concentration (day 0) before freezing, and k (day−1) is the decay rate constant. Measured BRSV RNA concentrations were linearized using the natural log (ln) transformation of the normalized concentrations as shown in Eq. (3);
| (3) |
The ln (Ct/C0) values and their associated time points (days) are plotted against each other to find k and the model fitness can be assessed by R 2.
The time required to achieve one-log (90%) reduction (T 90) was calculated using Eq. (4).
| (4) |
2.15. Wastewater sample collection from WWTPs, storage, and analysis for SARS-CoV-2
During method optimization and comparison, composite wastewater influent samples (24-h flow-dependent) were collected from five WWTPs in Maryland during the period of May to August 2020 by operators at the WWTPs. The flow and the population served by each WWTPs and the catchment areas of each WWTP are given in Table 1. Samples were transported on ice to the University of Maryland, MD. Samples (~1 L) were immediately transferred into 1 L Nalgene wide-mouth HDPE bottles and seeded with BRSV 1.05 (±0.9) × 107 GC/mL of wastewater (Bivins et al., 2021; Gonzalez et al., 2020), mixed vigorously and stored at −20 °C for 12–110 days until RT-qPCR analysis of SARS-CoV-2 RNA due to the lengthy method development stage.
All frozen WWTP samples were thawed concurrently at a 4 °C fridge. Triplicate samples from each WWTP were analyzed after virus concentration through protocol 9 followed by RNA extractions using Zymo Quick RNA kit. RT-qPCR assays of SARS-CoV-2 were performed according to the methods described above. In addition, to confirm that BRSV was absent in wastewaters that may induce recovery estimation bias, triplicate samples from each WWTP were analyzed and all samples were negative for BRSV. A total of 21 samples (n = 21/21) were analyzed by using BRSV, and SARS-CoV-2 N1, N2, and RdRP assays, while 20 samples (n = 20/21) were analyzed with E gene assays due to depletion of cDNA.
2.16. Statistical analysis
Statistical analysis was conducted in Microsoft Excel by applying t-tests to two groups of experimental data. To determine statistical significance, two-tailed p values were calculated, and p < 0.05 was accepted as statistically significant.
3. Results
3.1. qPCR performance characteristics and limit of detection (LOD)
Standard curve parameters for BRSV, SARS-CoV-2 CDC N1, CDC N2, and E_Sarbeco are listed in Table S3 together with their ALODs. BRSV ALOD was 44.2 GC/reaction, while ALOD values for CDC N1 and N2 assays were 3 and 5 GC/reaction, respectively, and 12 and 36 GC/reaction for E_Sarbeco and RdRp, respectively. Standard curve parameters (slope and intercept) for RdRp were comparably high and the efficiency was lower (Bivins et al., 2021) which was reflected in SARS-CoV-2 detection rate by this assay as discussed below.
3.2. Comparison of concentration methods and RNA extraction kits
Filtration based (i.e., protocols 1 and 2) protocols resulted in the lowest (p < 0.05) BRSV recoveries (0.03–5.2%) for the four RNA extraction protocols (Fig. S1, Table 2). Similarly, significantly lower (p < 0.05) recoveries were observed in protocols utilizing filtration for solid phase removals (protocols 5 and 6) compared to the recoveries of their replicates analyzed following PEG precipitation-based methods (protocols 13 and 14): 1.4% for protocol 5 vs. 38.3% in protocol 13 and 7.4% in protocol 6 vs. 48.9% in protocol 14. Compared to filtration, significantly greater (p < 0.05) BRSV recoveries (ranging from 1.9% for protocol 3 to 64.7% for protocol 9) were obtained when centrifugation was applied as the solid removal method (as in protocols: 3, 7, 8, 9, 10, 11, and 12). In contrast, among PEG precipitation-based protocols where solids were not removed as in protocol 13 and 14, greater recoveries (ranging from 11.1 to 48.9%) were obtained compared to the results of their replicates analyzed with protocols 6 and 7 (ranging from 0.3 to 26.3%).
Although the solid removal is essential for UF based methods to prevent clogging of the units, overall higher results were obtained UF protocols for the all four RNA extraction methods (Fig. S2, Table 2). Protocols utilizing wastewater liquids (especially the ones using centrifugation as the solid removal method, protocol 7–12) resulted in comparably higher (p < 0.05) BRSV recoveries than the protocols utilizing either the solid parts of the same wastewater sample (protocol 1–3) or wastewater itself (solids and liquid: protocol 12 and 13).
Among the 14 protocols tested for their effectiveness in wastewater viral RNA concentration, protocol 9 provided the highest overall mean BRSV recovery (32.4 ± 24.2%) followed by protocol 13 (PEG precipitation: 26.1 ± 10.1%) and protocol 14 (PEG precipitation followed by UF-15-100 kDa: 24.3 ± 17.8%). Although lower Cq values observed with protocol 14 for all four RNA extraction protocols (Table 2) than protocol 13, higher recoveries albeit not statistically significant (p > 0.05) were obtained through protocol 13 (14.0–38.3%) compared to protocol 14 (11.1–48.9%), indicating Cq values may not always warrant higher recoveries.
Among all four RNA extraction protocols presented Table 2 (also in Fig. S1), Zymo Quick RNA extraction protocol performed as the best with significantly higher (p < 0.005) recoveries ranging between 30.3% and 64.7% followed by Zymo Direct-zol with recoveries ranging between 4.8% and 16.5% for the protocols (protocol 9–12) utilizing UF as the virus concentration methods.
Furthermore, BRSV recovery (0.1%) from BRSV seeded nuclease-free water sample via protocol 9 and Zymo Quick RNA extraction kit (Table S3) was significantly lower (p < 0.05) than the recovery from seeded wastewater sample (64.7%) and the recovery from the rinsate of the inner surface of the falcon tube (4.6%), that was used to centrifuge the BRSV seeded nuclease-free water. This result indicated that the seeded BRSV might have partially attached to inner surface of the plastic falcon tube, which was collected after the centrifuge step by rinsing it with 1 mL nuclease-free water.
Approximate timelines for each protocol are provided in Table S1. Among all protocols, PEG-precipitation methods (protocols 5–8, 13, and 14) takes the longest time (14–16 h.) to concentrate viral RNA, while ultrafiltration methods including solids removal via centrifuge only takes 1–1.5 h.
3.3. Virus decay
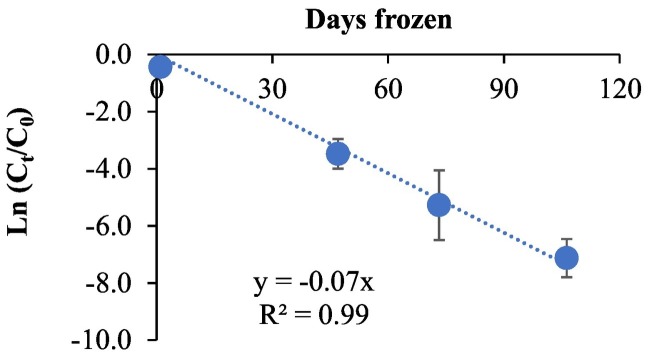
A significant decrease (p < 0.05) in BRSV RNA concentrations was observed over the course of sample storage (110 day), from 8.6 (±0.2) × 108 GC/L (the concentration before freezing) measured at day 0 the beginning of the experiment to 1.9 (±0.4) × 104 GC/L measured on day 110 after freezing. Similarly, the recovery level of BRSV showed a decreasing trend with an increase in storage length at −20 °C, ranging from 0.1 (±0.1) % in samples stored for 110 to 3.3 (±1.3) % in samples stored for 47 days (Fig. 1 ).
Fig. 1.
Average natural log (ln) reduction of BRSV RNA (average ln (Ct/C0)) where C is BRSV RNA concentrations in wastewater samples stored at −20 °C measured on day 0, 47, 77, and 110 and C0 is the seed BRSV RNA concentration (before freezing).
Viral RNA decay in wastewater over time was evaluated and the results follow first-order decay (Li et al., 2021). Hence, the first-order decay rate for BRSV RNA in wastewater at −20 °C (n = 19) was determined as 0.04/h (R 2 of 0.997 and Pearson's r of 0.999), and T 90 was found as 33.2 days by using Eq. (4).
3.4. Method application for the quantification of SARS-CoV-2 RNA in influent WWTP
After demonstrating the ability of each concentration method, protocol 9 (UF-15 with 100 kDa cut-off) was selected as the most efficient virus concentration method and Zymo Quick kit as the RNA extraction kit. BRSV with Cq values ranging between 22.1 and 31.1 was detected in all 16 wastewater samples (100%) seeded with BRSV prior to freezing (Table 3 ). The presence of SARS-CoV-2 RNA was assessed through four SARS-CoV-2 assays (i.e., N1, N2, E, and RdRp) in all 21 WWTP samples. Standard curves for BRSV, SARS-CoV-2 CDC N1, CDC N2, and E_Sarbeco met the Minimum Information for Publication of Quantitative Real-Time PCR (MIQE) guidelines (Bustin et al., 2009) by demonstrating a strong linear correlation within their assay dynamic range (Table S3). A sample was considered positive for SARS-CoV-2 and BRSV assays by RT-qPCR when Cq was below 40 cycles (Bustin et al., 2009; Gonçalves et al., 2021; Wu et al., 2020).
Table 3.
Quantification cycle (Cq) (±standard deviation) values and positivity (detection) rates for SARS-CoV-2 assays (i.e., N1, N2, E_Sarbeco and RdRP) and BRSV measured via protocol 9 (Amicon UF-15-100 kDa centrifugal filters) and extracted with Zymo Quick RNA extraction kit.
| WWTP | Sampling Date | BRSV | N1 | N2 | E_Sarbeco | RdRp |
|---|---|---|---|---|---|---|
| WWTP A | 5/28/20 | 25.7 (±0.2) | 35.3 (±0.3) | ND | ND | ND |
| 6/25/20 | 24.8 (±0.7) | ND | ND | ND | ND | |
| 7/23/20 | 23.5 (±0.3) | 35.2 (±0.4) | ND | ND | ND | |
| 8/27/20 | NA | 32.4 (±0.7) | 34.3 (±0.9) | 33.8 (±0.7) | ND | |
| WWTP B | 5/28/20 | 31.1 (±0.4) | 36.1 (±0.3) | 30.6 (±0.1) | ND | ND |
| 6/25/20 | 27.9 (±0.3) | ND | ND | ND | ND | |
| 7/2/20 | 26.7 (±0.2) | 36.9 (±0.9) | 36.6 (±1.7) | ND | ND | |
| 7/23/20 | 22.1 (±0.1) | ND | ND | ND | ND | |
| 8/27/20 | NA | 35.3 (±0.1) | 36.5 (±0.4) | ND | – | |
| WWTP C | 5/21/20 | 29.3 (±0.2) | ND | ND | ND | ND |
| 6/22/20 | 28.7 (±0.3) | 36.1 (±0.02) | 32.1 (±0.2) | 35.9 (±0.3) | ND | |
| 7/23/20 | 25.5 (±0.1) | 34.4 (±0.3) | ND | ND | ND | |
| 8/27/20 | NA | 27.9 (±0.1) | 28.3 (±0.2) | 29.7 (±0.4) | 38.9 (±0.3) | |
| WWTP D | 5/25/20 | 27.8 (±0.3) | ND | 36.8 (±0.1) | ND | ND |
| 6/22/20 | 24.9 (±0.2) | 33.4 (±0.2) | ND | ND | ND | |
| 7/23/20 | 24.4 (±0.1) | 36.6 (±0.1) | 36.5 (±0) | ND | ND | |
| 8/27/20 | NA | 36.4 (±0.2) | ND | ND | ND | |
| WWTP E | 6/4/20 | 24.5 (±0.02) | 36.2 (±0.3) | 36.5 (±0.2) | 35.6 (±0.2) | ND |
| 6/22/20 | 24.0 (±0.1) | 36.0 (±0.8) | 36.0 (±1.0) | 33.1 (±0.2) | ND | |
| 7/23/20 | 23.6 (±0.1) | 35.1 (±0.2) | 35.3 (±0.2) | 35.5 (±0.03) | ND | |
| 8/27/20 | NA | 34.9 (±1.3) | 35.1 (±0.3) | 34.7 (±0.2) | ND | |
| Detection rate | 100% (n = 16/16) | 76.2% (n = 16/21) | 57.1% (n = 12/21) | 33.3% (n = 7/21) | 5.0% (n = 1/20) | |
“NA”: not applicable since those wastewater samples were not seeded with BRSV and they were only analyzed for background BRSV analysis, which was non-detect; ND: not detected (<LOD). n: number of samples with positive detection over the total number of samples analyzed. “–”: not tested, there was not enough RNA to analyzed for the target.
About 76% (n = 16/21) and 57% (n = 12/21) of the samples tested positive for SARS-CoV-2 with CDC N1 assay (Cqs: 27.9–36.9) and CDC N2 (Cqs: 28.3–36.8), respectively, indicating N1 assay had higher sensitivity than N2. However, the positivity of samples for SARS-CoV-2 RNA was significantly lower when assessed using E (33.3%) and RdRp (5%) assays (Table 3). All samples tested positive with E assay were also tested positive for both N1 and N2 with no significant differences in their Cq values (p > 0.05). Among all samples analyzed, only one wastewater sample (WWTP C sample collected on 8/27/2020) tested positive for all four SARS-CoV-2 assays, which had the lowest Cq for each assay ranging between 27.9 and 38.9, indicating the highest SARS-CoV-2 RNA concentration in that sample.
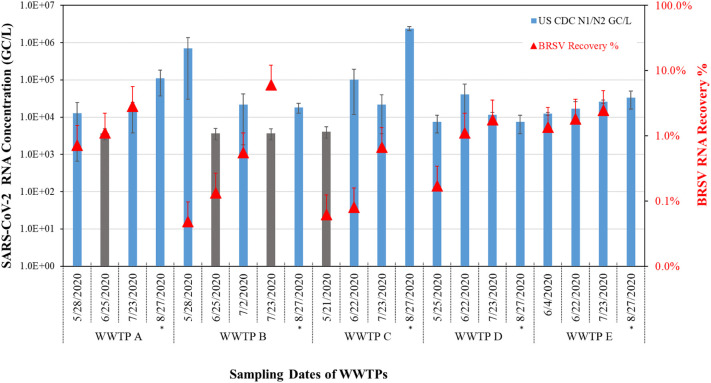
Overall, when combined, the N1 and N2 assays were able to detect SARS-CoV-2 RNA in 81% of the samples (n = 17/21) and despite fluctuation within the sampling period, the overall increases in SARS-CoV-2 RNA were observed in WWTP samples (Fig. 2 ), which ranged between 1.24 × 104 and 2.38 × 106 GC/L. WWTP E, the only plant among the five WWTP without a hospital in sewershed, had an about 5-fold increase in SARS-COV-2 concentration from May to Aug. 2020. However, the presence of a hospital did not seem to be an important factor, as higher concentrations were observed in the samples from the other four WWTPs during the same sampling period.
Fig. 2.
SARS-CoV-2 measurements (mean values of N1 and N2 assays, blue bars) and recovery of surrogate virus (BRSV, red triangles) RNA seeded into wastewater samples from the five WWTPs. Mean of triplicate analysis for each sampling date for each assay were reported with error bars indicating standard deviations of triplicate analysis. SARS-CoV-2 concentrations for samples below assay limit of detection (ALOD) (gray bars) were calculated using 3 GC/reaction. * indicates samples that were not seeded with BRSV for background BRSV analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Comparison of concentration methods and RNA extraction kits
Virus concentration is a critical step especially for the detection of trace level virus RNA (Gonçalves et al., 2021), which can be present during low levels of clinical cases in a community. Review of the literature showed that there is no consensus for solely analyzing the liquid phase (Wu et al., 2020), the solid phase (Peccia et al., 2020) or both phases (Pecson et al., 2021; Ye et al., 2016). The testing protocols in this study were employed to assess the presence of virus in both the solid and liquid phases.
Results of individual analysis performed on both liquid and solid phases of seeded wastewater samples (Fig. S1, Table 2) showed the presence of BRSV RNA in both phases, while significantly higher recovery was obtained from liquid parts of the wastewater samples (i.e., 64.7% via protocol 9) than its associated solid phase (15.9% via protocol 3). Similar results were reported for MHV in an earlier study, where 70% of the virus was present in the liquid part of the wastewater (Ye et al., 2016). In addition, Weidhaas et al. (2021) reported about 90% of the SARS-CoV-2 RNA in the liquid part of the wastewater influent compared to the influent solids.
Solids removal via centrifugation prior to wastewater concentration with PEG precipitation as in protocol 7 and 8 (Table 3, Fig. S1) resulted in about 32–50% lower BRSV recoveries compared to PEG protocols without solids removal as in protocol 13 (38.3%) and 14 (48.9%). This impact was higher when solids were removed by filtration prior to PEG precipitation as in protocol 5 and 6. This was supported by a previous study (Ahmed et al., 2020b), where a pre-filtration step was result in 30% loss of particle-associated viruses.
A recent interlaboratory (32 laboratories across the U.S.) study utilizing 36 standard operating procedures (8 method group) for concentration and quantification of SARS-CoV-2 reported that solid removal prior to SARS-CoV-2 quantification did not have any systematic impact on SARS-CoV-2 concentrations when the results were corrected with the surrogate (i.e., OC43) recovery rates (Pecson et al., 2021). However, the recovery of OC43 (90–95%) of five replicates of the two influent samples from WWTPs in California was one of the highest recoveries as reported for method 2S.6 (which is the protocol 9 developed and described herein) among the 36 procedures (Pecson et al., 2021). This result indicate that solids removal prior to ultrafiltration resulted in a higher concentration of SARS-CoV-2 than the protocols without solids (Pecson et al., 2021). The recovery differences observed for OC43 in the earlier study (Pecson et al., 2021) and for BRSV in this study might be due to behavioral differences of different viruses and the differences in the quantification methods used for different assays. While the analysis and the quantification of OC43 in wastewater samples provided by Pecson et al. (2021) was carried out by following protocol 9 of this study, there is no available information how the stock concentration of OC43 was determined.
Similar to the results obtained by Bivins et al. (2021), in this study, higher BRSV recovery was obtained from wastewater samples compared to the recovery from nuclease-free seeded water. The reason for this observation was linked to the extraction kit being more efficient for wastewater than nuclease-free water (Bivins et al., 2021). However, when combined with the higher recoveries observed in liquid part of the wastewater samples, these results might be due to the hydrophobic nature of the virus. Hence, it is likely that the virus was repelled from the water that is free of colloids/solids to the inner surface as in the falcon tube case or the attracted/attached to the colloidal particles present in wastewater samples as in the case of liquid phase of the wastewater that is hard to be spun down with solids when centrifuged at low forces (3500g).
The lowest recoveries (Fig. S1, Table 3) were obtained by the filtration-based protocols (protocol 1: 0.45 μm and protocol 2: 0.22 μm). Similarly, significantly lower recoveries were observed by Gonzalez et al. (2020) for BCoV and BRSV (4.8% and 6.6%, respectively) through a filtration-based concentration method utilizing electronegative filters (0.45 μm) compared to the recoveries obtained via direct extraction (59% and 75%, respectively).
Accordingly, our results indicated that the type of the RNA extraction method has a significant impact on the results. Gonzalez et al. (2020) obtained higher virus recovery via direct extraction of wastewater liquid (no bead beating step during RNA extraction) compared to results obtained via filtration-based method using electronegative filters (with a bead beating step). One main reason of obtaining lower recoveries via filtration-based methods as observed in other studies (Gonzalez et al., 2020; LaTurner et al., 2021) than via UF or PEG methods could be due to RNA degradation as result of bead beating applied during RNA extraction from filters.
4.2. The effect of molecular weight cut-off values on the efficiency of UF protocols
The differences observed in the recovery rates of Amicon UF-15 (5.8–64.7%) and Centricon UF-70 (4.8–41.8%) were possibly due to the size and design differences between the UF devices (Ikner et al., 2012). Centricon UF-70 has a larger surface area for filtration than the Amicon UF-15 (Ikner et al., 2012), hence, higher loading volumes (i.e. two times the maximum capacity of the filter). Accordingly, more BRSV RNA fragments might have been adsorbed to the membrane due to the van der Waals forces and hydrophobic binding (Ikner et al., 2012).
Regardless of UF type (Amicon or Centricon) and RNA extraction kits (Zymo Direct-zol or Zymo Quick), significantly higher recoveries (p < 0.05) were obtained with 100 kDa cut-off (7.8–64.7%) compared to 10 kDa (4.8–47.4%). Altogether, these results suggest that smaller sized (<100 kDa) inhibitory substances might be retained by UF 10 kDa which were otherwise passed through the 100 kDa membrane (Gonçalves et al., 2021). The difference in RT-qPCR cycle numbers obtained for the wastewater sample extract and the PCR grade water that were spiked with BRSV was found 0.3 (<1 cycle). Hence, it was concluded that PCR inhibition was not present (Pecson et al., 2021). This evaluation was solely performed for samples concentrated with protocol 9 (UF-15 with 100 kDa) and Zymo Quick RNA extraction kit.
Although RT-qPCR inhibition has not yet been reported, UF centrifugal filters can concentrate PCR inhibitors, especially when performed to concentrate viral RNA from large volumes of water samples (Gonçalves et al., 2021). Therefore, it is possible that the protocols that utilized UF units with 10 kDa cut-off (protocol 10 and 12) for sample concentration concentrated inhibitors alongside with virus RNA and, hence, resulted in lower recoveries than their counterparts with 100 kDA cut-off (protocol 9 and 11).
Higher recoveries were observed in this study compared to previous studies (Medema et al., 2020; Ye et al., 2016). For example, the BRSV recoveries found in this study for UF methods with two different cut-off values (10 kDa versus 100 kDa) were comparably higher than that of Ye et al. (2016) obtained for enveloped (28–22% for MHV and 27–9% for Phi6) viruses and non-enveloped (72–39% for MS2 and 110–61% for T3). On the other hand, up to 3.7% of MHV and 2% of MS2 was recovered from the solids (Ye et al., 2016), indicating non-enveloped viruses were more resistant to the methods. Overall results indicated that the UF cut-off value and the selection of RNA extraction kit were the key factors for higher virus recoveries.
4.3. Impact of prolonged storage length on virus decay
Previous studies have provided ample information on the impact of storage temperature and length on decay rates of SARS-CoV-2 RNA or surrogate viruses in wastewater and water (Ahmed et al., 2020b; Bivins et al., 2020; Hart and Halden, 2020; Wang et al., 2005; Weidhaas et al., 2021; Ye et al., 2016). However, the impact of prolonged storage wastewaters in large volumes at −20 °C is still unclear. Therefore, in this study, we evaluated the impact of prolonged storage (up to 100 days) on an enveloped virus, BRSV, in bulk volume (0.9–1 L) of wastewater at −20 °C.
Previously, a wide decay rate ranging between 0.02 and 0.143 h−1 for temperatures ranging between 4 and 35 °C was reported for SARS-CoV and other coronaviruses (Hart and Halden, 2020; Ye et al., 2016). In contrast, about 10-fold lower RNA decay rates (0.002 h−1 at 4 °C and 0.013 h−1 at 37 °C) were reported for SARS-CoV-2, while about 20–1000 fold higher decay rates (0.048–0.059 h−1 for 4–10 °C and 0.142 h−1 for 25 °C) were reported for an enveloped virus (MHV RNA) in untreated wastewater (Ahmed et al., 2020b). In a recent study, with sample volume as 250 mL, a range of decay rate (0.09 to 0.12 h−1) was reported for viral RNA when stored at 4 °C and 10 °C for about 24 h (Weidhaas et al., 2021). RNA was still detectable after 22 h of incubation at 4 °C and 10 °C and after one week at −80 °C with overall reductions at 67%, 86.5 ± 0.5% and 92.4 ± 10.3%, respectively, while it was not detectable after 6 h at 35 °C (Weidhaas et al., 2021), indicating higher decay rates with increase in storage temperatures.
Previous reports indicate that the impact of temperature comprised between 4 °C and at least 40 °C on SARS-CoV-2 RNA detection varies modestly based on the length of the storage. However, the results of this study indicated that prolonged storage of bulk wastewaters especially at −20 °C can have a dramatic negative impact on the concentration of SARS-CoV-2 and similar viruses, hence, result in false negative results. This is especially important for public health when wastewater monitoring used as an epidemiological tool. Compared to this study, however, the storage temperatures of earlier studies (4 °C and 37 °C) were significantly higher and the sample volumes (5–250 mL) were comparably lower than of this study (−20 °C and 900 mL, respectively). Similar to results of this study, the substantially lower MS2 bacteriophage titers were observed for samples stored at 4 °C for a 40-day period than for samples stored at −80 °C. In line with the results of this study, higher viral degradation was observed for samples stored at −20 °C compared to 4 °C and −80 °C, due to the formation of ice crystals, which provokes viral damage (Olson et al., 2004), supporting our results.
4.4. Sensitivity of assays for detectability SARS-CoV-2 RNA in influent samples from WWTP
As observed previously (Pérez-Cataluña et al., 2021; Vogels et al., 2020), CDC N1 assay was the most sensitive assay followed by N2 with SARS-CoV-2 detection in 76% (n = 16/21) and 57% (n = 12/21) of all WWTP samples, respectively. The detection rates were significantly lower with E_Sarbeco (33.33%, n = 7/21) and RdRp (5%, n = 1/20) assays. However, similar to Corman et al. (2020), Gonçalves et al. (2021) also reported higher sensitivity for RdRP (66.7% detection rate) than the E_Sarbeco (40%). The reason for higher detection rates for RdRp than E in the previous studies could be due to the utilization of different probes: both probe 1 and probe 2 reported by Corman et al. (2020) were utilized for RDRp gene in the previous studies, while only probe 2 was utilized in this study. RdRp probe 1 can detect 2019-nCoV, SARS-CoV, and bat-SARS-related CoVs, while probe 2 is specific to 2019-nCoV (SARS-CoV-2), hence, does not detect SARS-CoV (Corman et al., 2020). It is possible that other SARS-CoV viruses could be detected with utilization of the both probes, hence, results in higher quantification values, which can be misleading when only 2019-nCoV is of interest.
The findings of the current study are based on seeding wastewater with BRSV. The behavior of an exogenous control, such as the one used in the current study, compared to an endogenous SARS-CoV-2 shed into wastewater via infected individuals remains uncharacterized. This study included only a single wastewater sample for recovery efficiency, however recovery rates may be variable for different wastewater samples.
5. Conclusion
Based on the research performed in this study, the following conclusions have been drawn:
-
•
Among the 50 approaches, UF-15 with 100 kDa cut-off and Zymo Quick-RNA kit (protocol 9) provided the highest (64.7 ± 1.6%) virus recovery. The main advantages of UF methods are the speed of the protocols (~up to 1 h versus 16 h as in PEG-precipitation protocols) and higher virus recovery despite the relatively modest cost of the filters. The low-speed centrifugation step is still required to remove large debris and solids prior to concentration that may seem to be a limitation, the results indicated improved recovery. In addition, lower virus recovery from solids phase of the samples compared the liquid phase indicates UF method could be an option for the trace level virus detection.
-
•
PCR assay selection, detection, and stability not only in the sewage system but also during the sampling and storage are the vital elements of WBE to produce reliable information. Our results indicated that CDC N1 has the superior sensitivity compared to N2, E_Sarbeco and RdRp.
-
•
For accurate and reliable results, sampling, sample process, storage temperature and length and viral quantification methods should be evaluated and validated. Samples should be transported on ice to laboratories, stored at 4 °C and processed within 2–3 days. At times longer than that, a surrogate virus that has similar characteristics with the virus of interest, is not human pathogen and present in the tested samples, and can be obtained easily should be added to samples (50 mL) at a pre-determined concentration before freezing them away. Further research will be needed to test efficacy of RNA preservations on the integrity of virus RNA under various storage temperatures and length.
CRediT authorship contribution statement
Devrim Kaya: Conceptualization, Methodology, Investigation, Writing-Original Draft, Visualization, Writing-Review and Editing.
Debra Niemeier: Conceptualization, Project Administration, Funding Acquisition.
Warish Ahmed: Validation and Writing-Review and Editing.
Birthe V. Kjellerup: Conceptualization, Methodology, Supervision, Resources, Writing-Review and Editing, Project Administration, Funding Acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could influence the work and results presented in this paper.
Acknowledgements
We would like to thank our collaborator at the Utility for organizing the sampling effort and the operators for collecting and providing wastewater samples. Funding for this study was provided by Dr. Schwartz, Chair of Civil and Environmental Engineering, University of Maryland during the time of the study as well as funding from Dr. Niemeier, Maryland Transportation Institute and Dr. Kjellerup, Department of Civil and Environmental Engineering. Advice and help from Laboratory Manager Marya Anderson made it possible to navigate laboratory activities during the time of research restrictions due to COVID19 during 2020.
Editor: Damià Barceló
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.152033.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. medRxiv. 2020. The fate of SARS-CoV-2 in WWTPs points out the sludge line as a suitable spot for monitoring. (2020.05.25.20112706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxus M., Letellier C., Kerkhofs P. Real time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus. J. Virol. Methods. 2005;125:125. doi: 10.1016/j.jviromet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Prot. 2020;143:196–203. doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosi L.M., Barry K.E., Kotay S.M., Porter M.D., Poulter M.D., Ratliff C., Simmons W., Steinberg L.I., Wilson D.D., Morse R., Zmick P., Mathers A.J. Development of wastewater pooled surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from congregate living settings. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.00433-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVIDPoops19 COVID-19 WBE Collaborative. 2021. https://www.covid19wbec.org/
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A.K.A., Demoliner M., Gularte J.S., Hansen A.W., Schallenberger K., Mallmann L., Hermann B.S., Heldt F.H., de Almeida P.R., Fleck J.D., Spilki F.R. bioRxiv. 2020. Comparison of different kits for SARS-CoV-2 RNA extraction marketed in Brazil. (2020.05.29.122358) [Google Scholar]
- Gao Q.Y., Chen Y.X., Fang J.Y. 2019 novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020;21:125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J., Koritnik T., Mioč V., Trkov M., Bolješič M., Berginc N., Prosenc K., Kotar T., Paragi M. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.143226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki,Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Kantor R., Novitsky V., Carpenter-Azevedo K., Howison M., Manne A., Darpolor J.K., Bobenchik A., Tripathi A., Huard R.C., King E. SARS-CoV-2 variants in Rhode Island. R. I. Med. J. 2021;104(7):16–20. [PubMed] [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Gamas K.R., Terwilliger A., Crosby T., Ali P., Avadhanula V., Santos H.H., Weesner K., Hopkins L., Piedra P.A., Maresso A.W., Stadler L.B. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., Tamin A., Thornburg N.J., Villanueva J.M., Lindstrom S. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1654. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M., Rundell Z.C., Nemec M.D., Langan L.M., Back J.A., Lugo J.N. medRxiv. 2021. A comparison of four commercially available RNA extraction kits for wastewater surveillance of SARS-CoV-2 in a college population. (2021.06.01.21257858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.R., Axler R.P., Hicks R.E. Effects of freezing and storage temperature on MS2 viability. J. Virol. Methods. 2004;122:147–152. doi: 10.1016/j.jviromet.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Omer S.B. medRxiv. 2020. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. (2020.05.19.20105999) [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., Consortium SA-C-I Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Chagas do Vale V.H., RMS Braz, de Andrade JdSR, Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritalahti K.M., Cruz-García C., Padilla-Crespo E., Hatt J.K., Löffler F.E. RNA extraction and cDNA analysis for quantitative assessment of biomarker transcripts in groundwater. 2010. pp. 3671–3685. [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., et al. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child,China. Emerg. Infect. Dis. 2020;26:1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.-E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W., Zhu X., Gu C., Yin J., Wei W., Yao W., Liu C., Li J., Ou G., Wang M., Fang T., Wang G., Qiu Y., Wu H., Chao F., Li J. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People's Liberation Army. Water Sci. Technol. 2005;52:213–221. [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. WHO Coronavirus Disease (COVID-19) Dashboard. [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. medRxiv: the preprint server for health sciences. 2020. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. (2020.06.15.20117747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Ferrier-Rembert A., Frenois-Veyrat G., Mouchel J.M., Boni M., Maday Y., Marechal V., Moulin L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 2021;198 doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material