Abstract
Mouse hepatitis virus (MHV; m-β-CoV) serves as a useful model for studying the cellular factors involved in neuroinflammation. To understand the role of matrix metalloproteinases (MMPs) in neuroinflammation, brain tissues from m-β-CoV-infected mice were harvested at different days post-infection (d.p.i) and investigated for Mmp expression by RT-qPCR. Mmp-2, -3, -8, -12 showed significant mRNA upregulation peaking with viral replication between 5 and 6 d.p.i. Elevated levels of MMP regulator TIMP-1 are suggestive of a TIMP-1 mediated host antiviral response. Biological network assessment suggested a direct involvement of MMP-3, -8, -14 in facilitating peripheral leukocyte infiltrations. Flow cytometry confirmed the increased presence of NK cells, CD4+ and CD8+ T cells, neutrophils, and MHCII expressing cells in the m-β-CoV infected mice brain. Our study revealed that m-β-CoV upregulated Park7, RelA, Nrf2, and Hmox1 transcripts involved in ROS production and antioxidant pathways, describing the possible nexus between oxidative pathways, MMPs, and TIMP in m-β-CoV-induced neuroinflammation.
Keywords: β-coronavirus (β-CoV), MHV-A59, MMPs, ROS, TIMPs, Neuroinflammation
1. Introduction
The newly emergent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Zhu et al., 2020; Yan et al., 2020; Lu et al., 2020) and previous encounters with SARS-CoV (Peiris et al., 2003; Rota et al., 2003; Drosten et al., 2003) and middle east respiratory syndrome coronavirus (MERS-CoV) (Zaki et al., 2012; Drosten et al., 2013; Hilgenfeld and Peiris, 2013) have led to a renewed interest in coronavirus (CoV) research. CoVs are enveloped positive-sense single-stranded RNA virus (Gorbalenya et al., 2006; Perlman and Netland, 2009)[ICTV 9th report, 2011; https://talk.ictvonline.org/ictv-reports/]. Past studies have demonstrated the requirement of several cellular proteases such as cell surface serine proteases (Hoffmann et al., 2020; Iwata-Yoshikawa et al., 2019; Shulla et al., 2011; Matsuyama et al., 2010), intracellular endosomal cathepsins (Shang et al., 2020; Jaimes et al., 2020; Ou et al., 2020; Kawase et al., 2012; Bosch et al., 2008; Qiu et al., 2006), or extracellular proteases like trypsin (Jaimes et al., 2020; Bertram et al., 2011; Matsuyama et al., 2005) and elastases (Matsuyama et al., 2005; Belouzard et al., 2010; Ami et al., 2008) in priming the virus surface spike (S) glycoprotein for successful entry and infection. However, studies on the role of host proteases in virus-induced inflammation are limited.
Matrix metalloproteinases (MMPs) are a large family of zinc-dependant endopeptidases, of which 23 members are identified in mice (Yong, 2005; Cui et al., 2017; Cauwe and Opdenakker, 2010; Nagase et al., 2006). MMPs are best known for their ability to remodel the extracellular matrix (ECM) proteins. Over the years, the substrate repertoire of MMPs has widened to include cytokines, chemokines, growth factors, and hormones (Cauwe and Opdenakker, 2010; Young et al., 2019; Van Lint and Libert, 2007). By acting upon cytokines or chemokines immobilized on ECM or cell surface, MMPs release soluble effector molecules for a successful inflammatory response and are involved in autoimmune diseases such as multiple sclerosis, systemic lupus erythematosus, and Alzheimer's disease (Cauwe and Opdenakker, 2010; Javaid et al., 2013; Yong et al., 2007; Lee et al., 2019; Zhu et al., 2010; Duits et al., 2015; Wang et al., 2014). The proteolytic activity of MMPs is regulated by endogenously produced inhibitory proteins called tissue inhibitors of metalloproteinases (TIMPs) (Cui et al., 2017; Nagase et al., 2006; Murphy, 2011). So far, identified four TIMP molecules (TIMP-1, -2, -3, and -4) could inhibit all known MMPs. TIMPs bind reversibly to the catalytic subunit of MMPs in a 1:1 stoichiometric ratio and inhibit their function. Studies conducted in the past have shown the involvement of MMPs and TIMPs in CoV infection (Phillips et al., 2017; Zhou et al., 2002, 2005; Savarin et al., 2013; Shi et al., 2021).
Murine hepatitis virus (MHV; m-β-CoV) is a member of the same β-coronavirus genus as SARS-CoV-2, MERS-CoV, and SARS-CoV (Bender and Weiss, 2010; Bergmann et al., 2006) [ICTV 9th report, 2011; https://talk.ictvonline.org/ictv-reports/]. MHV is a widely studied β-coronavirus model. Being a natural pathogen of mice, MHV damages the liver and central nervous system (CNS), resulting in hepatitis and meningoencephalitis during acute inflammation. MHV-induced chronic inflammation is associated with demyelination concurrent with axonal loss (Lavi et al., 1984; Das Sarma, 2010; Shindler et al., 2008; Das Sarma et al., 2008, 2009).
Previous analysis of MMP and TIMP expression in the CNS following infection with neurotropic MHV strain, JHMV showed increased Mmp3, Mmp12, and Timp1 mRNAs, which correlated with high virus replication (Zhou et al., 2002, 2005). Some MHV S can be cleaved by intracellular proprotein convertase like furin, and furin inhibition prevents cell-to-cell fusion in the MHV-A59 strain (Phillips et al., 2017; de Haan et al., 2004). The involvement of endosomal acidification, surface protease TMPRSS2, and metalloproteinases in m-β-CoV infection have also been explored recently (Phillips et al., 2017). Broad-spectrum metalloprotease inhibitor demonstrated the most significant effects on inhibition of in-vitro infection by MHV strains and cell-to-cell fusion without affecting S cleavage (Phillips et al., 2017).
In this study, we investigated the relationship between the induction of MMP/TIMP expression and inflammation in acute infection of mice with MHV strains. Data suggest that TIMP-1 induction during acute infection following MHV-A59 infection could serve as an antiviral host response to regulate MMP activities. We also determined the role of S protein in inflammation using isogenic recombinant strains RSA59 and RSMHV2 that differ only in the spike gene. Here we show that RSA59 and RSMHV2 both induce elevated mRNA expression of Mmp3, Mmp8, and Mmp14 but differ in their induction of Mmp2 and Mmp12. Biological network analysis using Ingenuity Pathway Analysis (IPA) suggested that MMP-3, MMP-8, and MMP-14 aid immune cell infiltration, suggesting a spike-independent role of these MMPs in MHV-induced neuroinflammation. Further, flow cytometry studies confirmed the increased infiltration of peripheral lymphocytic and myeloid cells in the CNS upon m-β-CoV infection.
Virus infection generates reactive oxygen species (ROS) inside the cells, and oxidative stress can mediate inflammation (Sun et al., 2020). Transcriptomic analysis showed RSA59 infection-induced gene transcription of Park7 and RelA during acute inflammation. Park7 is involved in the NADPH oxidase-dependant ROS pathway, while RelA is involved in the NF-κB signaling. In addition, increased mRNA levels of Nrf2 and Hmox1, major antioxidant genes, also reported upon m-β-CoV infection. Our study suggests a paradigm shift in our understanding of virus-induced inflammatory response by highlighting a relationship between m-β-CoV MHV infection-induced oxidative stress and metalloprotease expression during neuroinflammation.
2. Experimental procedures
Reagents and kits. Gelatin, paraformaldehyde (PFA), and polyvinylidene difluoride (PVDF) membranes were obtained from Merck Millipore, USA. High-capacity cDNA reverse transcription kit (Cat:4368814), DyNAmo ColorFlash SYBR Green qPCR kit (Cat: F-416L), Pierce™ BCA protein assay kit (Cat: 23225), and SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Cat: 34580) were purchased from Thermo Fischer Scientific, USA. EDTA-free protease inhibitor cocktail (Cat:11836170001) was from Roche Diagnostics, Germany. Mouse TIMP-1 antibody (Cat: AF980) was obtained from R&D Systems, USA. Mouse MMP-8 antibody (Cat: SAB4501895) was purchased from Sigma-Aldrich, USA. Anti-g actin antibody (Cat: BB-AB0025) was purchased from Bio Bharati Life Science, India. Horseradish peroxidase (HRP)-conjugated secondary IgG antibodies, donkey anti-goat (Cat: 705-035-003), and goat anti-rabbit (Cat: 111-035-003) were purchased from Jackson Immuno Research Laboratories, USA. All other reagents were purchased from Sigma, while pre-designed primers were ordered from IDT, USA.
Ethical approval. The use of animals and related experimental techniques were evaluated and sanctioned by the Institutional Animal Care and Use Committee at the Indian Institute of Science Education and Research Kolkata (AUP no. IISERK/IAEC/AP/2017/16). Experiments were performed strictly adhering to the standards of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India.
Viruses. A naturally occurring demyelinating strain of MHV, MHV-A59 (Lavi et al., 1984), and two recombinant strains, RSA59, and RSMHV2 (Shindler et al., 2008; Das Sarma et al., 2000, 2002), was used to infect mice. RSA59 and RSMHV2 are isogenic recombinant strains of MHV-A59 constructed by targeted RNA recombination and differ only in the spike gene described previously (Shindler et al., 2008; Das Sarma et al., 2000). The recombinant demyelinating RSA59 strain expresses the MHV-A59 spike in the background of the MHV-A59 genome. In contrast, the non-demyelinating RSMHV2 strain express MHV-2 [wild-type non-demyelinating MHV (Das Sarma et al., 2001)] spike protein in the MHV-A59 background. Also, the recombinant strains express enhanced green fluorescence protein (EGFP) for easy in-vitro detection of the virus (Das Sarma et al., 2002).
Inoculation of mice. Four-weeks old, MHV-free, male C57BL/6 mice (Jackson Laboratory, USA) were inoculated intracranially with 50% of LD50 doses of MHV-A59 (2000 PFU), RSA59 (25,000 PFU) or RSMHV2 (100 PFU) respectively and as described previously (Shindler et al., 2008; Das Sarma et al., 2000). Virus inoculum was prepared in a 20 μL volume of phosphate-buffered saline (PBS) plus 0.075% bovine serum albumin (BSA) and injected into the right cerebrum of mice (N = 9). Parallelly, we inoculated three mock-infected control mice with only PBS+0.075% BSA. Daily monitoring of mice for disease signs and symptoms was conducted. Control mice were euthanized at the peak of inflammation between 5 and 6 days post-infection (d.p.i). MHV-infected mice were euthanized at 5–6 (peak of inflammation), 10 (acute-chronic inflammation), and 15 (chronic inflammation) d.p.i. Brain and spinal cord tissue samples were flash-frozen in liquid N2 and preserved at −80 °C until used for RNA studies.
Estimation of viral replication. The efficacy of virus replication in mice was assessed by routine plaque assay, as described previously (Lavi et al., 1984; Kishore et al., 2013; Singh et al., 2019) with modifications. Briefly, on 5–6, 10, and 15 d.p.i, mice were euthanized, and brain tissues (either half) harvested aseptically in 1 mL of isotonic saline containing 0.167% gelatin. Tissues were weighed and stored at −80 °C for titer assay. Brain tissues (300 mg) from control and infected mice were homogenized, and 250 μL of serially diluted supernatant was added onto confluent monolayers of DBT (mouse delayed brain astrocytoma) cells. Following 75 min of viral adsorption, culture media containing 1.4% agarose was overlaid on the monolayers and incubated for 27 h (MHV-A59/RSA59) or 36 h (RSMHV2). Cells were fixed with 4% PFA, followed by crystal violet staining. Clear regions indicative of plaques were counted, and values [log10 pfu/gm tissue] were plotted against post-infection time. Plaque forming unit or PFU was calculated as the number of plaques times dilution factor (DF) per mL per gram of tissue per mL [PFU= (no. of plaques*DF per ml)/(tissue weight in gram per ml)].
Gene expression analysis. Total RNA was extracted from brain tissues of MHV-A59, RSA59, or RSMHV2 infected as well as mock-infected mice using TRIzol reagent (Invitrogen) following the manufacturer's instructions. RNA concentration was measured using a NanoDrop 2000/2000c Spectrophotometer (Thermo Fisher Scientific), and cDNA was prepared with 1 μg of total RNA using a cDNA reverse transcription kit. Quantitative real-time PCR (RT-qPCR) was performed using SYBR Green dye-based assay in a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific) with the following reaction conditions: initial denaturation at 95 °C for 7 min, 40 cycles of 95 °C for 10 s and 60 °C for 30 s, and melting curve analysis at 60 °C for 30 s. Reactions were performed in triplicates (n = 3). Primer sequences are provided in Table 1 . The comparative threshold (ΔΔCT) method was used for relative quantification. The mRNA levels of target genes were normalized with the housekeeping GAPDH gene and represented as the relative fold change values compared to their respective mock-infected controls.
Table 1.
Primer sequence for detection of gene expression by RT-qPCR.
| Gene | Forward primera | Reverse primera |
|---|---|---|
| Gapdhb | GCCCCTTCTGCCGATGC | CTTTCCAGAGGGGCCATCC |
| Mmp2 | CACCTGGTTTCACCCTTTCT | GAGGAGGGGAACCATCACTA |
| Mmp3 | TGGAGATGCTCACTTTGACGA | CCTTGGCTGAGTGGTAGAGTC |
| Mmp8 | TGGCATTCAGACAATCTATGGACCT | CACGGAGTGTGGTAGTAGCATCAA |
| Mmp12 | GGAGCTCACGGAGACTTCAACT | CCTTGAATACCAGGTCCAGGATA |
| Mmp14 | TCGGCCCTCTGTCCCAGATAA | GCCAGAACCATCGCTCCTTGA |
| Timp1 | CCAGAGCCGTCACTTTGCTT | AGGAAAAGTAGACAGTGTTCAGGCTT |
| Timp2 | TTGCAGGAAAGGCAGAAGGAGATG | GCACTCACAGCCCATCTGGTA |
| Timp3 | CTGACAGGGCGCGTGTATGAA | CAACCCAGGTGGTAGCGGTAATTG |
| Timp4 | AGGGTTCGAGAAGGCCAAGGA | CCATCACTGAGAATCTGGCCAGTCA |
| Park7 | AACACACCCACTGGCTAAGG | CTCCACAATGGCTAGTGCAA |
| RelA | CCTTCCTCAGCCATGGTACCTCT | CCCAAGTCTTCATCAGCATCAAACTGC |
| Nfkb2 | CCCTACCCATCCCTCTCC | CCCGCCTAAGGTGAAGAGA |
| Nrf2 | GATCCGAGATATACGCAGGAGAGGTAAGA | GCTCGACAATGTTCTCCAGCTTCC |
| Hmox1 | GCCCCACCAAGTTCAAACAGCTCTA | CTCTGTCAGCATCACCTGCAGC |
Primer sequences given in 5′ to 3′ direction. All primers purchased from IDT, USA. Mmp2, Mmp9 and Park7 primers were purchased from RealTimePrimers.com, USA.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blotting. Brain tissues (30 mg) were harvested from mice following transcardial PBS perfusion and flash-frozen in liquid N2. Tissues were homogenized (thrice for 30s at medium speed using Qiagen TissueRuptor II) and lysed in 500 μL of RIPA buffer [50 mM Tris Base (pH 7.6), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% Sodium deoxycholate] containing protease-cocktail inhibitor and phosphatase inhibitors (1 mM NaVO4 and 10 mM NaF) for 1 h 30 min with intermittent vortex every 15 min. The samples were kept on ice during the entire process. Samples were then centrifuged for 15 min at 13,000 rpm at 4 °C to separate the supernatant. The total protein content in the supernatant was determined with a BCA protein assay kit. For immunoblotting, 20–60 μg of total protein per sample was resolved by SDS-PAGE on a 12% polyacrylamide gel, then transferred onto PVDF membranes using transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol). Membranes were blocked for 1 h at room temperature either in 5% v/v goat serum or skimmed milk prepared in TBST (Tris-buffered saline containing 0.1% v/v Tween 20) and subsequently incubated overnight at 4 °C in polyclonal anti-mouse TIMP-1 or polyclonal anti-mouse MMP-8 antibody at 1:1000 dilution in blocking solution respectively. The membranes were washed in TBST and incubated for 1 h at room temperature with HRP-conjugated donkey anti-goat or goat anti-rabbit secondary IgG antibody. As an internal loading control, γ-actin or β-tubulin was used, and membranes were blocked separately in 5% w/v non-fat skimmed milk in TBST. Polyclonal anti-mouse γ-actin or β-tubulin antibody (1:2000 dilution) and HRP-conjugated goat anti-rabbit secondary IgG antibody (1:10,000 dilution) were used. The blots were washed in TBST, and the immunoreactive bands were visualized using the chemiluminescent HRP substrate. Non-saturated bands were visualized with Syngene G: box Chemidoc system using GENSys Software.
Biological network analyses. The list of MMP (Mmp-2, -3, -8, -12, −14) and TIMP (Timp-1 to −4) genes were loaded into QIAGEN's Ingenuity Pathway Analysis software (IPA®, QIAGEN, USA) to perform biological network analyses. IPA enables the identification of likely modulated pathways based on transcriptional inputs, which molecules may drive the expression patterns seen and the possible downstream consequences.
Flow cytometry analysis. Brain tissues were harvested from mock and virus-infected mice and homogenized in 4 ml of Dulbecco's PBS (pH 7.4) using Tenbroeck tissue homogenizers. Following centrifugation at 450g for 10 min at 4 °C, cell pellets were collected and resuspended in RPMI +25 mM HEPES (pH 7.2), adjusted to 30% Percoll, and underlaid with 1 ml of 70% Percoll. Following centrifugation at 800g for 30 min at 4 °C, cells were collected from the 30%–70% interface, washed with RPMI, and suspended in FACS buffer (0.5% bovine serum albumin in Dulbecco's PBS). Fc receptors were blocked with 1% polyclonal mouse serum. Specific cell types were identified by staining with fluorophore-conjugated MAb (all from BD Biosciences except where otherwise indicated) for 30 min on ice in FACS buffer. Surface markers used were CD45 (clone Ly-5), CD4 (clone GK1.5), CD8 (clone 53–6.7), NK1.1(clone PK136), CD11b (clone M1/70), Ly-6G (clone 1A8), AND MHC II (clone M5/114.15.2). Samples were analyzed using a BD LSRFortesa flow cytometer (BD Biosciences) and FlowJo 10 software (Treestar, Inc., Ashland, OR).
Statistical analysis. Data shown are mean ± standard error mean (SEM) for all graphs. Unpaired student t-test was performed to examine significant differences between the two groups. Multiple comparisons were achieved using ordinary one-way ANOVA, followed by Dunnett's multiple comparison test. A p-value < 0.05 was considered statistically significant.
Availability of data. All the data sets used and analyzed in the current study are available from the corresponding author on request.
3. Results
3.1. Mmp mRNA expression in MHV-A59 infected mice brain
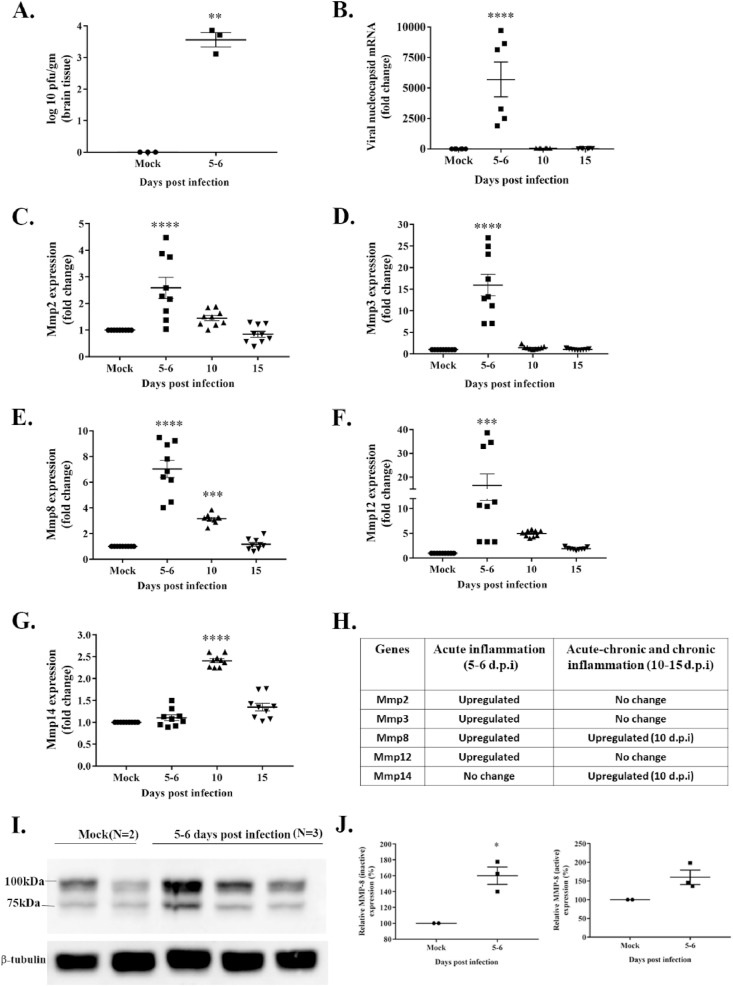
Four weeks old, male C57BL/6 mice inoculated with MHV-A59 (2000 PFU) or mock-infected were euthanized at 5–6 (acute), 10 (acute-chronic), and 15 (chronic) days post-infection (d.p.i), and brains were harvested. Routine plaque assay was performed with serially diluted brain homogenates to determine viral replication. Viral titer was significantly different in infected mice compared to mock between 5 and 6 d.p.i (Fig. 1 A; p < 0.001). and viral plaques were below the detection limit at later time points (data not shown). Total RNA was isolated from mock and virus-infected brain tissues to analyze viral nucleocapsid and Mmp genes through RT-qPCR. Primer sequences are provided in Table 1. Levels of viral nucleocapsid mRNA (Fig. 1, B; p < 0.0001) coincided with viral replication reaching its peak between 5 and 6 d.p.i, marking the acute phase of inflammation. MHV-A59 infected mice exhibit neuroinflammation that peaks by 5–7 d.p.i and is associated with meningitis, encephalitis, perivascular cuffing, and macrophage/microglia nodule formation (Das Sarma, 2010; Shindler et al., 2008). Transcript levels of Mmp2, Mmp3, Mmp8, and Mmp12 were significantly upregulated at 5–6 d.p.i (Fig. 1, C–F; p < 0.001) and coincided with the peak in viral replication and acute neuroinflammation. Additionally, Mmp14 mRNA was significantly upregulated at a later time point, i.e., 10 d.p.i (Fig. 1, G; p < 0.001). Collectively, MHV-A59 infection-induced the upregulation of Mmp2, Mmp3, Mmp8, Mmp12, and Mmp14 (Fig. 1, H). To confirm the increased mRNA levels correlated with protein expression, immunoblotting was performed with whole-brain protein lysate against MMP-8. Presence of both the inactive (∼100 KDa) and active (∼75 KDa) forms of MMP-8 detected in mock and infected brain samples (Fig. 1, I). The protein levels of the inactive (∼100 KDa) MMP-8 were increased at 5–6 d.p.i (Fig. 1, J; p < 0.05). Although the active form (∼75 KDa) demonstrated an increasing trend upon infection, the change was insignificant compared with mock brain lysates (Fig. 1, J).
Fig. 1.
Upregulation of Mmp2, Mmp3, Mmp8, Mmp12, and Mmp14 mRNAs in the brain of MHV-A59 infected mice. Brain tissues from mock- and MHV-A59 (2000 PFU)-infected mice at different days post-infection (d.p.i) were harvested, and viral replication was determined by routine plaque assay. RNA isolated from brain tissues was subjected to cDNA synthesis. An equal amount of cDNA templates was used for RT-qPCR. Gene expression was normalized to GAPDH, and fold-change values were obtained using the ΔΔCt method. A: MHV-A59 titer peaked between 5-6 d.p.i. and was beyond the detectable range at 10 and 15 d.p.i (data not shown). B: Viral nucleocapsid mRNA levels reached the peak at 5–6 d.p.i, similar to viral replication. C–F: Mmp2, Mmp3, Mmp8, and Mmp12 mRNA levels elevated between 5-6 d.p.i coinciding with viral replication peak. G: Membrane-associated MMP-14 mRNA levels reached the peak at a later stage post-infection. H: A list of upregulated Mmps at different stages of inflammation is shown. I: Immunoblot assay performed with 20 μg of total proteins per sample from brain tissue lysate represents the inactive (∼100 kDa) and active (∼75 kDa) forms of MMP-8 from mock and MHV-A59 infected samples at 5–6 d.p.i with 2–3 biological replicates. β-tubulin kept as the internal loading control. J: MHV-A59 infected mice showed significantly upregulated levels of the inactive (∼100 KDa) MMP-8 while the active form (∼75 KDa) of MMP-8 demonstrated an increasing trend upon infection. All data are represented as mean ± SEM. RT-qPCR data is from two independent biological experiments with nine technical replicates. A significant difference between the two groups was compared with the student t-test. Multiple group comparison was made with ordinary one-way ANOVA followed by Dunnet's test. A p-value of <0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
3.2. Induction of TIMP-1 expression following MHV-A59 infection
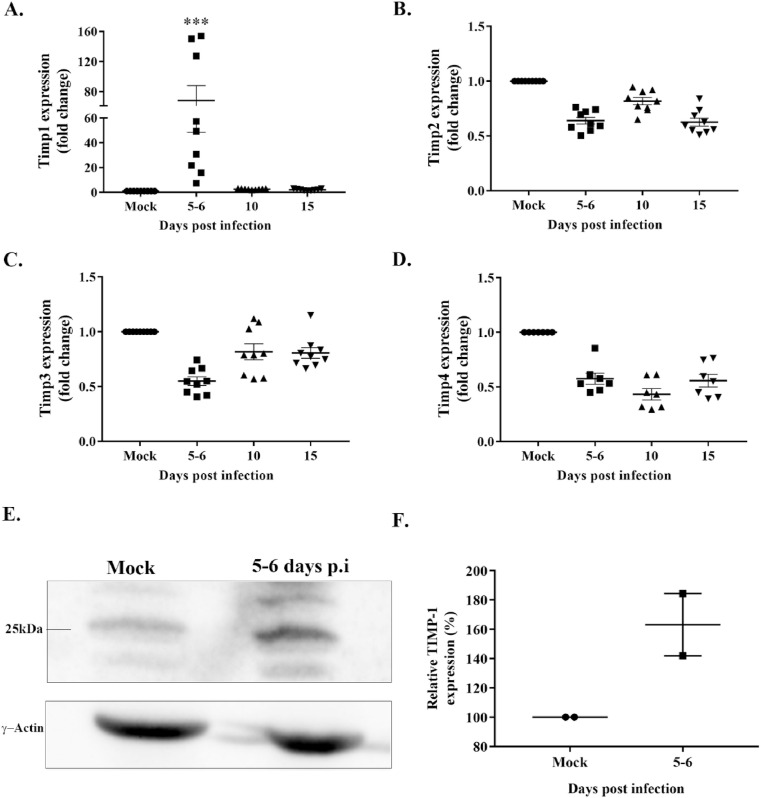
Tissue inhibitors of metalloproteinases or TIMPs are endogenous protein regulators of MMPs (Murphy, 2011). To understand the regulation of MMPs upon MHV-A59 infection, we also considered the mRNA expression of TIMPs. As described above, total RNA from brain samples of mock and MHV-A59 infected mice were subjected to RT-qPCR using specific primers (Table 1) to determine the transcript levels of Timp1, Timp2, Timp3, and Timp4. MHV-A59 infection resulted in significant upregulation of Timp1 mRNA at 5–6 d.p.i (Fig. 2 , A; p < 0.001). Although the transcriptional profile of Timp2, Timp3, and Timp4 shows a downward trend, the biological significance is unknown (Fig. 2B–D). Immunoblotting assay performed with whole-brain protein lysates from mock and MHV-A59 infected mice showed an upward trend in TIMP-1 protein levels upon infection, although the change is not statistically significant (Fig. 2, E-F). Overall, MHV-A59 resulted in the induction of TIMP-1 expression in the brain of infected mice.
Fig. 2.
Preferential upregulation of TIMP-1 following MHV-A59 infection. Brain tissues from mock- and MHV-A59 (2000 PFU)-infected mice at different days post-infection (d.p.i) were harvested and subjected to RT-qPCR and immunoblot assay. A: Data analysis of RT-qPCR revealed a significant upregulation in Timp-1 mRNA levels at 5–6 d.p.i in MHV-A59-infected samples compared with mock-infected samples. B–D: In contrast, Timp-2, -3, and -4 mRNAs exhibit a downward trend post-infection. E: Representative immunoblot from two independent experiments depicts the expression of TIMP-1 (∼25 KDa) and γ-actin (internal loading control) in mock and MHV-A59 infected samples. F: MHV-A59 infected mice demonstrated an increasing trend in TIMP-1 protein levels compared to mock samples. All data are represented as mean ± SEM. RT-qPCR data is from two independent biological experiments with nine technical replicates. A significant difference between the two groups was compared with the student t-test. Ordinary one-way ANOVA followed by Dunnet's test was performed for multiple group comparisons. Statistical significance was considered for p values < 0.05 (***, p < 0.001).
3.3. Similarities in the induction of MMP and TIMP expression following m-β-CoV-RSA59 and m-β-CoV-RSMHV2 infection
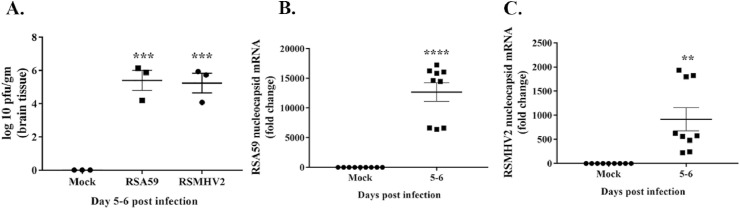
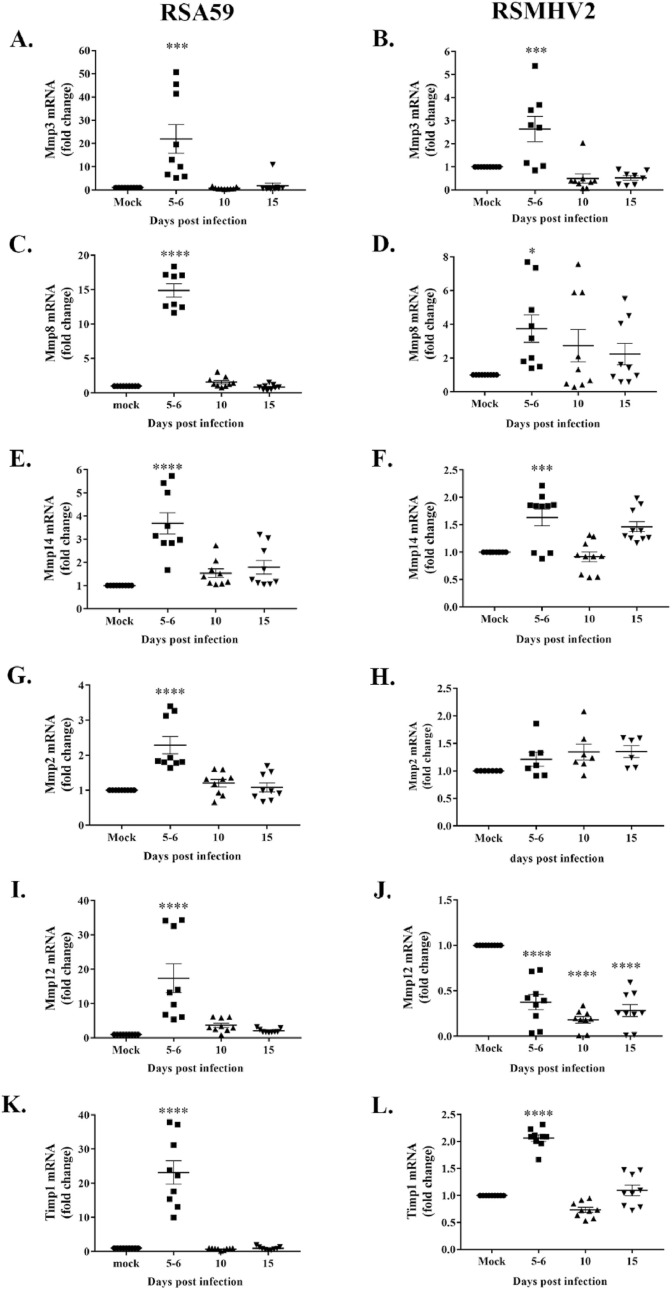
To determine whether the spike (S) protein has any role in inducing Mmp and Timp expression, we employed recombinant m-β-CoV strains RSA59 and RSMHV2. RSA59 and RSMHV2 are isogenic recombinant strains of wild-type MHV-A59 constructed by targeted RNA recombination and differ only in the spike gene described previously (Cauwe and Opdenakker, 2010; Van Lint and Libert, 2007). The recombinant demyelinating RSA59 strain expresses the MHV-A59 spike in the background of the MHV-A59 genome. In contrast, the non-demyelinating RSMHV2 strain express MHV-2 [wild-type non-demyelinating MHV (Javaid et al., 2013)] spike protein in the MHV-A59 background. Histopathological studies demonstrated that RSA59 and RSMHV2 exhibit similarities in CNS inflammation characterized by meningitis and encephalomyelitis involving parenchymal lymphocytic infiltration and microglial nodule formations (Das Sarma, 2010; Shindler et al., 2008; Das Sarma et al., 2008, 2009). However, the recombinant m-β-CoV strains differ in their ability to induce myelin loss. RSA59 induced demyelination and axonal loss during chronic inflammation, whereas no myelin and axonal destruction are evident in RSMHV2 infection (Das Sarma, 2010). We performed RT-qPCR from brain samples of RSA59, RSMHV2, and mock-infected mice euthanized at 5–6, 10, and 15 d.p.i. Brain samples were also harvested for routine plaque assay to determine viral replication. Like the parental MHV-A59 strain, viral titer and nucleocapsid mRNA levels reached a peak by 5–6 d.p.i in both the recombinant strains, as demonstrated in the representative graphs (Fig. 3 A–C). Although we detected no nucleocapsid mRNAs between 10 and 15 d.p.i in RSMHV2, its presence was observed at 10 d.p.i in RSA59 (data not shown). This data corroborates previous findings that demyelinating RSA59 persists in the brain while RSMHV2 does not persist or, if present, nucleocapsid level is significantly low compared with RSA59 (Das Sarma et al., 2000). Also, both the strains showed similar kinetics in Mmp3 and Mmp8 gene expression like their parental MHV-A59. Results from RT-qPCR demonstrated significant upregulation of Mmp3 and Mmp8 mRNA levels upon infection compared with mock-infection at 5–6 d.p.i (Fig. 4 , A-D; p-value varies as <0.05 to <0.0001). In addition, the recombinant m-β-CoVs resulted in an early stage increased transcription of Mmp14 at 5–6 d.p.i (acute inflammation), unlike the delayed (acute-chronic inflammation) expression of Mmp14 in the parental MHV-A59 (Fig. 4, E-F; p-value varies as <0.001 to <0.0001). Moreover, the fold change induction of Mmp3, Mmp8, and Mmp14 differed in the recombinant m-β-CoVs, with higher changes observed upon RSA59 infection. Furthermore, Mmp2 and Mmp12 were differentially altered in the two recombinant strains. RSA59 induced upregulation of Mmp2 (Fig. 4, G; p < 0.0001) and Mmp12 (Fig. 4, I; p < 0.0001) during acute inflammation. In contrast, RSMHV2 infection did not alter Mmp2 mRNA (Fig. 4, H) levels while significantly decreasing transcript levels of Mmp12 (Fig. 4, J; p < 0.0001). Also, both strains significantly increased mRNA levels of Timp1 at 5–6 d.p.i with varying fold change (Fig. 4, K & L; p < 0.0001). Previously, it has been reported that RSA59 and RSMHV2 do not differ significantly in their ability to induce inflammation in the brain (Das Sarma, 2010; Das Sarma et al., 2008, 2009). Therefore, we performed biological pathway analysis to understand the involvement of MMP-3, -8, and -14 in the onset of virus-induced neuroinflammation.
Fig. 3.
Viral replication and nucleocapsid mRNA expression in isogenic recombinant m-β-CoV strains at 5–6 days post-infection. RSA59 and RSMHV2 are isogenic recombinant strains of the wild-type MHV-A59 and differ only in the spike gene. Brain samples from mice infected with half of the LD50 dose of RSA59 (25000 PFU) or RSMHV2 (100 PFU) were harvested at different days post-infection (d.p.i) for routine plaque assay and total RNA extraction. A: Both RSA59 and RSMHV2 showed peak viral replication between 5 and 6 d.p.i. No detectable viral plaques were observed at later time points in a routine plaque assay (data not shown). B–C: RT-qPCR data showed significantly elevated mRNA levels of viral nucleocapsid gene in both RSA59 (B) and RSMHV2 (C) at early d.p.i, coinciding with the levels of viral plaques at the peak of inflammation. Data shown are mean ± SEM from three independent experiments having three technical replicates each. A significant difference between the two groups was compared with the student t-test. Multiple group comparison was made with ordinary one-way ANOVA followed by Dunnet's test. A p-value of <0.05 was considered statistically significant (**, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
Fig. 4.
Elevated brain levels of Mmp3, Mmp8, Mmp14, Timp1 mRNAs at the peak of inflammation and differential induction of Mmp2 and Mmp12 following infection with isogenic recombinant m-β-CoV strains, RSA59 and RSMHV2. Like their parental wild-type MHV-A59 strain, both RSA59 and RSMHV2 induced similar expression patterns of Mmp and Timp genes, as evident from RT-qPCR. A–F: Mmp3, Mmp8, and Mmp14 transcript levels were significantly upregulated between 5-6 d.p.i, coinciding with the peak in the inflammatory response. G–H: Mmp2 gene expression was significantly upregulated at 5–6 d.p.i in RSA59 while remaining unchanged in RSMHV2 post-infection. I–J: Mmp12 showed significantly elevated transcript levels at 5–6 d.p.i in RSA59 infected samples. Levels of Mmp12 mRNA decreased significantly post-infection in RSMHV2. K–L: Additionally, Timp1 mRNA was also upregulated in both the strains and followed a similar expression trend as Mmps during acute inflammation (5–6 d.p.i). RSA59 and RSMHV2 differed in the fold change induction of Mmp3, Mmp8, Mmp14, and Timp1 mRNA levels. Data shown are mean ± SEM from three (A–J) and two (K–L) independent experiments having nine technical replicates. Ordinary one-way ANOVA followed by Dunnet's test was performed for multiple group comparisons. Statistical differences at p < 0.05 was considered significant (*, p < 0.05; ***, p < 0.001; ****, p < 0.0001).
3.4. Classical leukocyte adhesion and diapedesis pathway activated in m-β-CoV-RSA59 and m-β-CoV-RSMHV2 infection
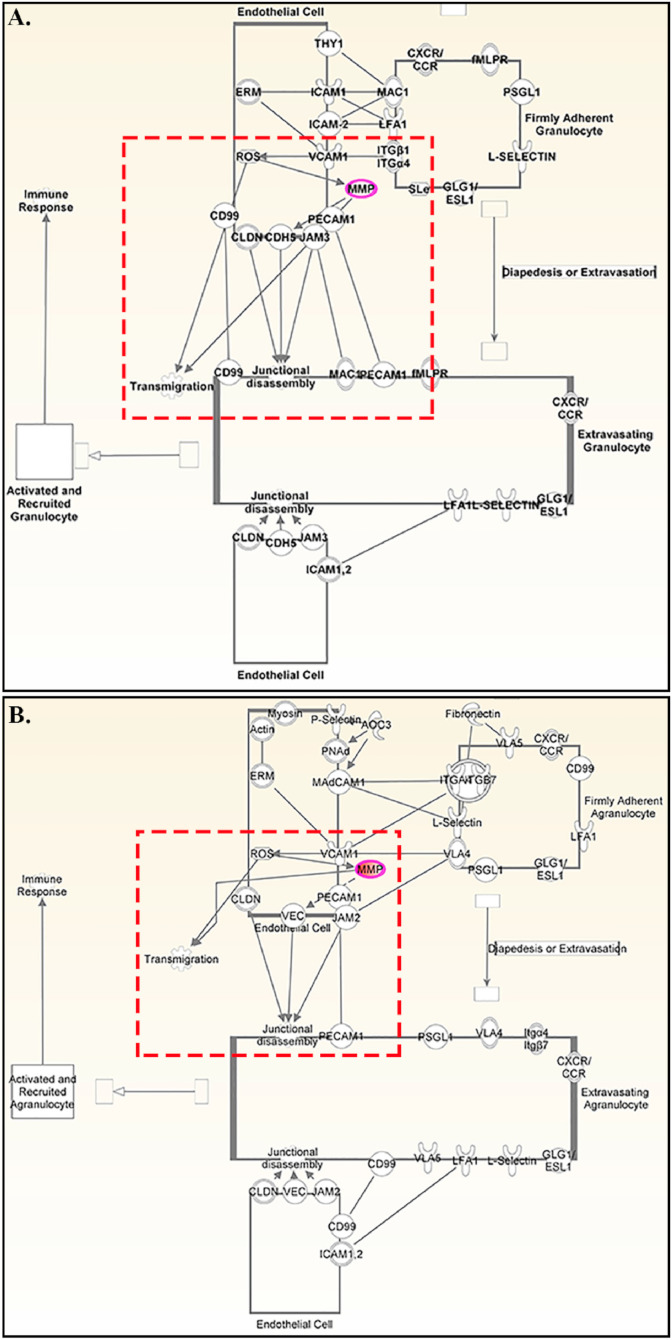
MHV-A59 isogenic recombinant strains RSA59 and RSMHV2 showed similar kinetics in MMP induction and significantly upregulated Mmp3, Mmp8 and Mmp14 during acute inflammation. To understand the significance of upregulated Mmps upon m-β-CoV infection, we performed biological network construction for Mmp3, Mmp8, and Mmp14 genes using QIAGEN's Ingenuity Pathway Analysis (IPA) software. IPA enables the identification of likely modulated pathways based on transcriptional inputs, which molecules may drive the expression patterns seen and the possible downstream consequences. IPA analysis identified that these MMPs could influence several canonical pathways associated with an immune response, such as leukocyte extravasation signaling, granulocyte, and agranulocyte adhesion and diapedesis. IPA revealed that MMPs could facilitate the transmigration of firmly adhered granulocytes (Fig. 5 , A) and agranulocytes (Fig. 5, B) across the endothelial cells in the blood vessel. IPA results suggested a possible relationship between MMPs and junctional proteins like claudin, cadherin, and platelet-derived endothelial cell-adhesion molecules. Collectively, IPA analysis indicates the role of MMP-3, MMP-8, and MMP-14 in peripheral immune cell migration during MHV infection.
Fig. 5.
Network analysis of Mmp3, Mmp8, and Mmp14 in m-β-CoV -RSA59 and m-β-CoV -RSMHV2 infection. The path designer function of the IPA software was used to dissect further the direct and indirect relationships that exist between Mmp genes and inflammatory responses with a focus on immune cell trafficking. Transcript details of Mmp-3, -8, -14, which showed significant upregulation upon RSA59 and RSMHV2 infection during acute inflammation was loaded into the IPA. A–B: Results suggest that Mmps facilitate the transmigration of granulocytes (A) and agranulocytes (B) firmly adhered to endothelial cells in the blood vessel. As indicated through IPA (A–B), MMPs can mediate junctional disassembly between endothelial cells via interacting with adhesion molecules such as claudin (CLDN), cadherin 5 (CDH5), endothelial cell adhesion molecule (PECAM1), and junctional adhesion molecules (JAM). Further, IPA indicates an interactive relationship between reactive oxygen species (ROS) and MMPs during immune cell trafficking. Nodes in the network represent the genes (or their corresponding proteins). The lines connecting the nodes indicate the kind of interaction between the genes (direct is the solid line; indirect is a dotted line). Red dotted rectangles highlight the possible interaction between MMPs and different cellular molecules.
3.5. Immunophenotyping of peripheral immune cell infiltration in the m-β-CoV-RSA59 infected inflamed CNS in the context of MMP expression
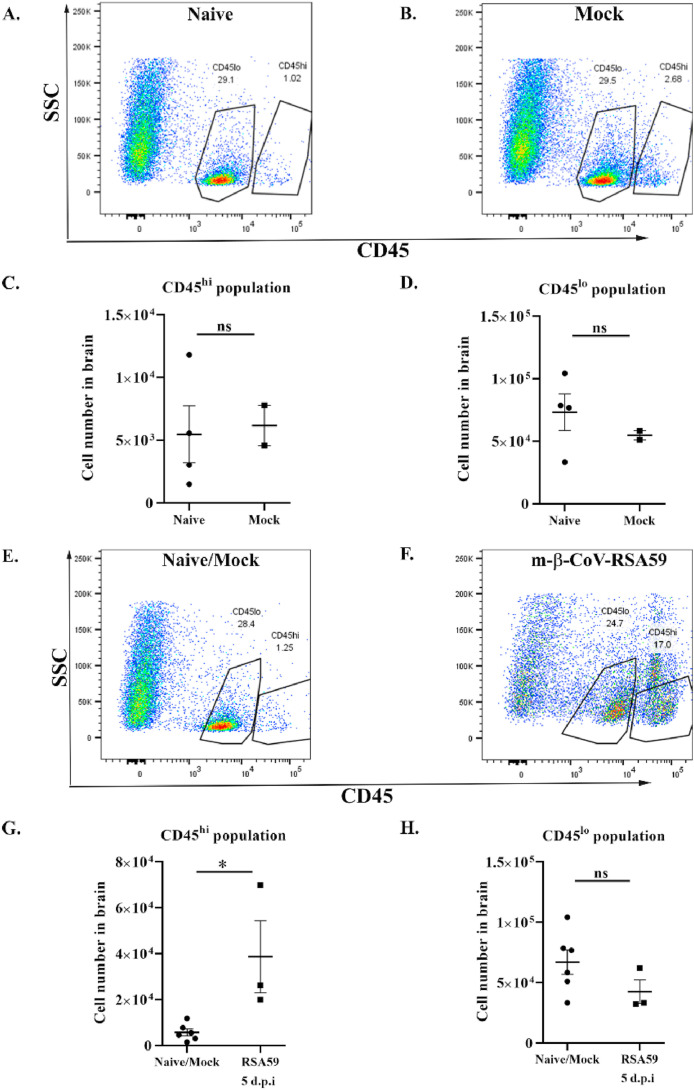
Antiviral host response to murine β-coronaviruses is associated with the recruitment of peripheral lymphocytes and microglial activation in the brain. Recent studies have demonstrated peripheral lymphoid and myeloid cell infiltration time kinetics at different days post-infection, including days 3, 5, 7, 10, 16, and 30 post-infection (Chakravarty et al., 2020; Das Sarma et al., 2020; Savarin and Bergmann, 2018; Butchi et al., 2015). In the context of MMP expression, the current study conducted immunophenotyping at day 5–6 post-infection, which is associated with acute inflammation and elevated MMP mRNA levels. Flow cytometry analysis of whole-brain samples from m-β-CoV-RSA59 and naïve/mock-infected mice was performed to assess the CNS infiltration of immune cells. Gating for CD45 expression was made on live cells and successively subdivided into CD45hi lymphocytes and CD45lo microglia cells.
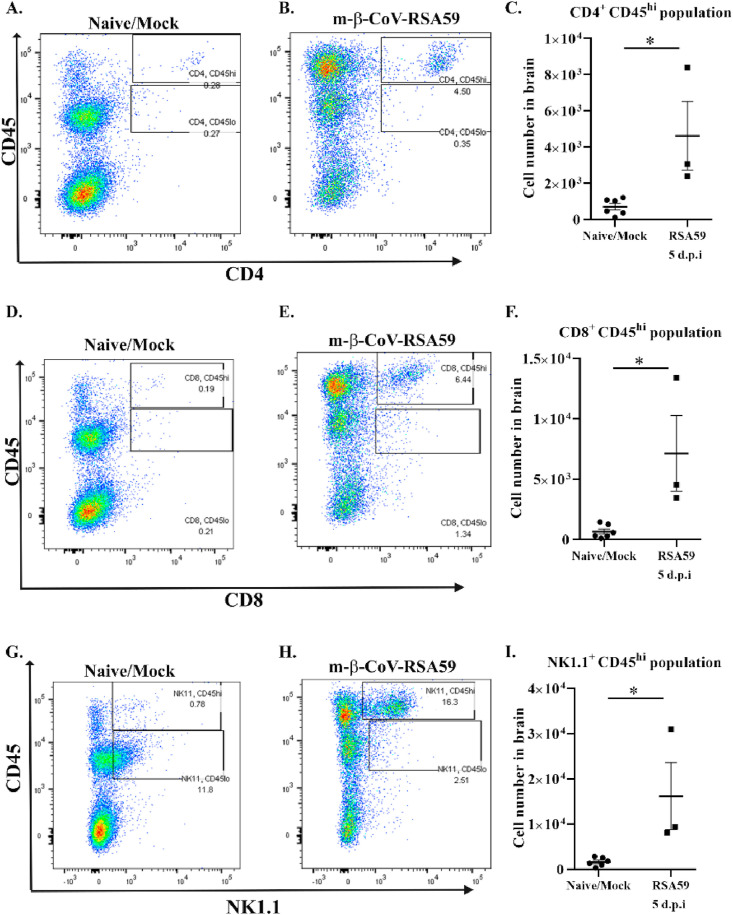
A set of age-matched naïve and mock-infected mice pooled from different experiments were used to compare the increased infiltration of peripheral immune cells. Naïve and mock-infected mice did not show significant differences in the number of CD45hi and CD45lo populations (Fig. 6 A–D). The m-β-CoV-RSA59 infected mice showed higher infiltrating CD45hi in the brain at 5 d.p.i than age-matched naïve and mock-infected mice (Fig. 6, E and G). No marked difference was observed between the groups for the CD45lo population (Fig. 6, F and H). Detection of lymphocytic subsets within CD45hi populations was performed using the surface markers NK1.1 (NK cells), CD4 (TH cells), and CD8 (TC cells). Mice infected with m-β-CoV-RSA59 exhibited a significant increase in all three lymphocyte populations at 5 d.p.i compared with naïve/mock-infected samples (Fig. 7 ).
Fig. 6.
Intracranial infection with m-β-CoV-RSA59 induced higher levels of infiltrating CD45hipopulations in the brain at 5 days post-infection. Brains from age-matched naïve or mock-infected and virus-infected (day 5) mice were harvested for flow cytometric analysis. A–B: Representative flow cytometry plots indicating percentages of CD45hi and CD45lo cells in brains after gating on live cells in age-matched naïve and mock samples. C–D: Age-matched naïve or mock-infected mice did not exhibit any difference in the presence of CD45hi and CD45lo populations. E–F: Representative flow cytometry plots indicating percentages of CD45hi and CD45lo cells in brains after gating on live cells. G–H: The number of cells in the brain are graphically represented for better comparison between naïve/mock and m-β-CoV infected mice. Graphs represent data from three to six biological replicates. Asterix (*) denotes differences that are statistically significant by Student's unpaired t-test analysis (*, p < 0.05).
Fig. 7.
Intracranial infection with m-β-CoV-RSA59 induced higher levels of infiltrating CD4+, CD8+and NK1.1+ T cell subsets within CD45hipopulations in the brain 5 days post-infection. Brains from age-matched naïve or mock-infected and virus-infected (day 5) mice were harvested for flow cytometric analysis. A–C: Representative flow cytometry plots indicating percentages of CD4+ cells within CD45hi population and cell number (C) in brains after gating on live cells. D–F: Representative flow cytometry plots showing percentages of CD8+ cells within CD45hi population and cell number (F) in brains after gating on live cells. G–I: Representative flow cytometry plots indicating percentages of NK1.1+ cells within CD45hi population and cell number (I) in brains after gating on live cells. Graphs represent data from three to six biological replicates. Asterix (*) denotes differences that are statistically significant by Student's unpaired t-test analysis (*, p < 0.05).
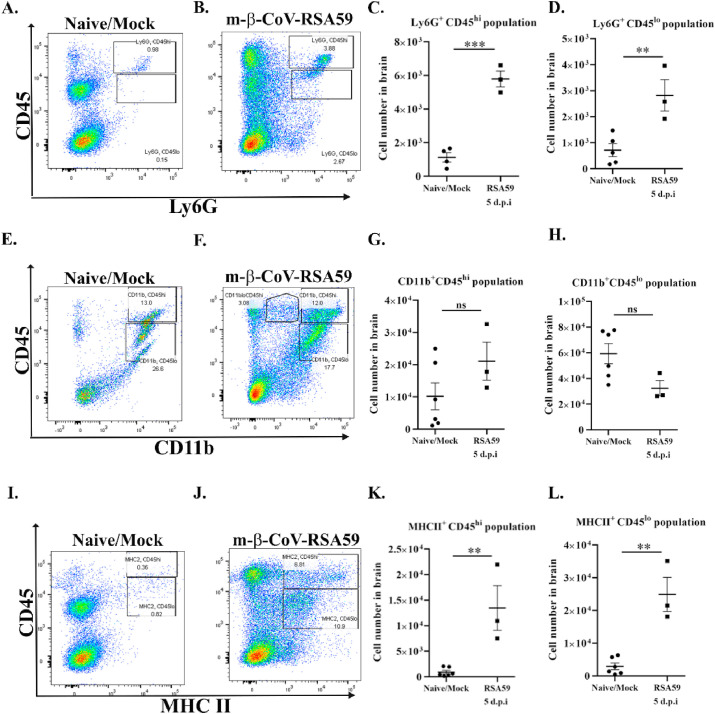
Staining with Ly6G to detect neutrophils revealed a significant increase in the cell number in CD45hi (Fig. 8 A–C) and CD45lo (Fig. 8A and B, D) infiltrating leukocyte populations. While there is a significant infiltration of lymphocytes and neutrophils, no significant changes were observed in the number of CD11b + cell populations. Infection with m-β-CoV-RSA59 did not alter CD11b+ myeloid cells (microglia/monocytes/macrophages) number within CD45hi (Fig. 8E–G) and CD45lo (Fig. 8E and F, H) populations at 5 d.p.i compared with age-matched naïve or mock-infected samples. Further, staining with surface marker, MHC II demonstrated a markedly high level of MHCII + cells within CD45hi (Fig. 8I–K) and CD45lo (Fig. 8I and J, L) population. As expected in the context of MMP expression, m-β-CoV infection induced increased infiltration of peripheral lymphocytes, including natural killer cells and T cells, as well as infiltration of neutrophils. Also, when cells were gated on the CD45 population and looked for MHC II (microglia/monocyte/macrophage activation marker) expression, we observed MHC II expression to be upregulated. Therefore, while CD11b+ cells may not increase in numbers, m-β-CoV may activate microglia/monocytes/macrophages in the inflamed CNS.
Fig. 8.
Intracranial infection with m-β-CoV-RSA59 induced higher infiltrating Ly6G + neutrophils and MHC II+subsets within CD45hiand CD45lopopulations and did not alter CD11b+myeloid cell population by number while in the brain at 5 days post-infection. Brains from naïve or mock-infected (day 15–16) and virus-infected (day 5) mice were harvested for flow cytometric analysis. A–D: Representative flow cytometry plots indicating percentages of Ly6G+ cells within CD45hi/lo populations and cell numbers (C, D) in brains after gating on live cells. E–H: Representative flow cytometry plots indicating percentages of CD11b+ cells within CD45hi/lo populations and cell number (G, H) in brains after gating on live cells. I–L: Representative flow cytometry plots indicating percentages of MHC II + cells within CD45hi/lo populations and cell number (K, L) in brains after gating on live cells. Graphs represent data from three to six biological replicates. Asterix (*) denotes differences that are statistically significant by Student's unpaired t-test analysis (**, p < 0.01; ***, p < 0.001).
3.6. Infection with m-β-CoV-RSA59 induced increased transcription of oxidative and antioxidative pathway genes
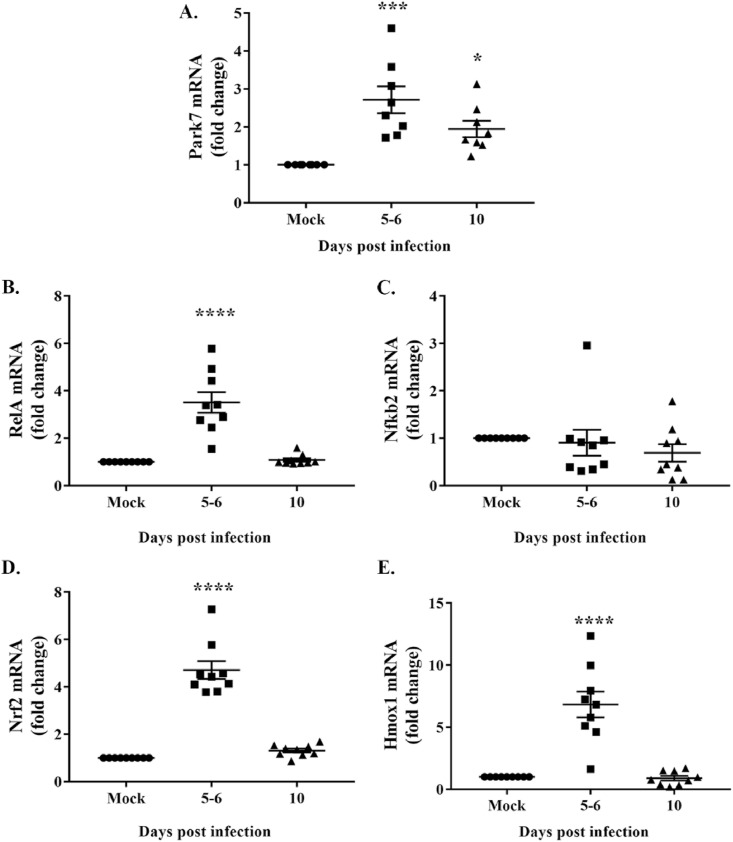
The biological network constructed using IPA indicates a possible interaction between ROS and MMPs during immune cell migration upon m-β-CoV infection. Also, virus infection is known to increase ROS generation (Sun et al., 2020). We examined the effect of RSA59 infection on essential oxidative and antioxidative pathway genes. Brain samples from mice infected with RSA59 and euthanized at 5–6 and 10 d.p.i were harvested for total RNA isolation, followed by cDNA synthesis. Mock-infected samples were used as controls. RT-qPCR was performed using primers (Table 1) specific for genes involved in the oxidative and antioxidative pathways at 5 d.p.i, which coincides with the increased infiltration of peripheral lymphocytes and neutrophils marking the acute inflammatory phase. Transcript levels of the Parkinson's disease 7 (Park7) gene were significantly upregulated following m-β-CoV-RSA59 infection and remained elevated post-infection compared to mock-infected samples (Fig. 9 , A; p varies as <0.05 to <0.001). RelA, an NF-κB subunit, also showed elevated mRNA levels during the acute infection, i.e., 5–6 d.p.i (Fig. 9, B; p < 0.0001). On the contrary, mRNA levels of Nfκb2, a negative regulatory subunit of NF-κB, remained unchanged post-infection (Fig. 9, C). Similar to a recently published paper (Kundu et al., 2021), we also detected significantly high mRNA levels of nuclear factor erythroid 2-related factor 2 (Nrf2) and heme oxygenase-1 (Hmox1) genes during the acute disease phase in the context of MMP expression (Fig. 9, D-E; p < 0.0001). Overall, m-β-CoV-RSA59 infection in mice induced simultaneous transcription of genes such as Park7 and Nfκb involved in the oxidative pathway and antioxidative pathway genes like Nrf2 and Hmox1.
Fig. 9.
Upregulation of gene transcripts involved in oxidative and antioxidative pathways during m-β-CoV-RSA59 infection. Total RNA isolated from brain samples of mice infected with m-β-CoV-RSA59 (25000 PFU) or mock-infected was subjected to cDNA synthesis, and subsequently, RT-qPCR was performed. A: Data analysis revealed elevated mRNA levels of Park7 during acute (5–6 d.p.i) and acute-chronic (10 d.p.i) disease phase. B: Upregulation of mRNA was detected for RelA, a subunit of the NF-kB transcription factor. C: In contrast, no change was observed in the mRNA level of Nfkb2, a negative regulator of NF-kB. D–E: Moreover, m-β-CoV-RSA59 infection also induced increased transcription of antioxidative Nrf2 and Hmox1 genes. Data shown are mean ± SEM from two independent experiments. A significant difference between multiple groups was compared with ordinary one-way ANOVA, followed by Dunnet's test. A p-value of <0.05 was considered statistically significant (*, p < 0.05; ***, p < 0.001; ****, p < 0.0001).
4. Discussion
The current study investigates the regulation of MMPs and their inhibitors during CNS infection with MHV strains. Wild-type MHV-A59 and its isogenic recombinant strains, RSA59 and RSMHV2, were analyzed for their ability to influence MMP and TIMP mRNA expression. Kinetics of Mmp-3, Mmp-8, and Timp-1 mRNA expression, mostly upregulated during the acute disease phase, were similar in both the parental MHV-A59 and recombinant strains. MMP-3 (stromelysin) is produced by leukocytes and endothelial cells and acts on multiple collagen substrates (Cui et al., 2017). Both neutrophils and macrophages produce MMP-8 (neutrophil collagenase), while MMP-14, which is membrane-associated and showed significant upregulation during m-β-CoV infection, promotes the conversion of inactive MMP-2 to its active form. Both MMP-8 and MMP-14 can cleave multiple collagen substrates (Cui et al., 2017). The ability of MMP-3, -8, and -14 to modulate the ECM suggests their potential to assist in the migration of peripheral immune cells. Biological network evaluation using Ingenuity Pathway Analysis (IPA) suggests a possible role of MMP-3,-8, and -14 in granulocyte and agranulocyte transmigration across the endothelial cells by disrupting claudin, cadherin, or junctional adhesion molecules mediated tight junctional assembly. Moreover, it explains the similarity observed in CNS inflammation with both the recombinant strains RSA59 and RSMHV2 despite having differences in their S proteins (Shindler et al., 2008) but similar in the induction of MMP. Past studies associating overproduction of MMPs with increased blood-brain-barrier permeability and, thereby, enhanced inflammation of neurotropic viruses (Wang et al., 2008; Luplertlop et al., 2006), further supports our observation.
Flow cytometric analysis was performed to assess the infiltration of and the types of immune cells in the infected mice brain. Infection with m-β-CoV-RSA59 resulted in markedly high infiltrations of peripheral lymphocytic and myeloid cells, including NK cells, CD4+ T cells, CD8+ T cells, neutrophils, and MHC II expressing antigen-presenting cells. Parenchymal infiltration of peripheral lymphocytes coincided with the elevated levels of MMPs during MHV-A59-induced acute inflammation.
Previous studies involving JHMV strains have shown that MMP-3, MMP-12, and TIMP-1 mRNA expression correlates with viral virulence and viral load (Zhou et al., 2002). Additionally, the role of TIMP-1 in inhibiting protease activity and thereby controlling the differential distribution of T-cell subsets during virus encephalitis was also demonstrated (Zhou et al., 2005). Moreover, TIMP-1 protein expression was found only in CD4+ T-cells and not CD8+ T-cells, suggesting that TIMP-1 regulates differential CNS distribution of T-cells (Zhou et al., 2005). TIMP-1, through its inhibition of protease activity, promotes transient accumulation of CD4+ T cells within the perivascular space and controls the trafficking of T cells into the parenchyma (Zhou et al., 2005). Thus, the dynamic regulation of TIMP and MMP levels is an essential factor determining the onset of inflammation. Overall, infection of mice with MHV serves as a useful model to study acute-inflammatory responses and contributions of the immune system in maintaining homeostasis in the inflamed brain.
We found Timp1 significantly upregulated in the recombinant m-β-CoV strains like the parental MHV-A59. Our study adds further knowledge in understanding the role of TIMP-1. The present work suggests that Timp1 upregulation could be part of a classical host defense mechanism against virus-induced upregulation of different metalloproteases. Whether the downregulation of Timp2, Timp3, and Timp4 is mediated by MHV or a feedback mechanism exists to maintain a balance among different TIMPs is an exciting aspect to address. Moreover, our study further sheds light on the viral-induced oxidative stress pathway and neuroinflammation.
Oxidative stress is a common denominator of cell death and damage, tissue disintegration in any pathogenic infection, and also known to get induced in m-β-CoV infection (Khan et al., 2014). MHV-A59 infection increases ROS production in the retina of infected mice that can be attenuated by treatment with SIRT1 (NAD-dependent deacetylase) activating compound, which induces the expression of mitochondrial enzymes such as SDH and SOD2 and significantly reduces ROS levels (Khan et al., 2014). In our current study, m-β-CoV-RSA59 infection increased transcript levels of Park7, which is involved in oxidative stress. It has been shown that Park7 phosphorylates the p47phox subunit of NADPH oxidase during NADPH oxidase activation, which is crucial for NADPH oxidase-dependant ROS production (Liu et al., 2015). Park7 induced ROS production plays a vital role in initiating inflammation and survivability of mice during LPS-induced sepsis.
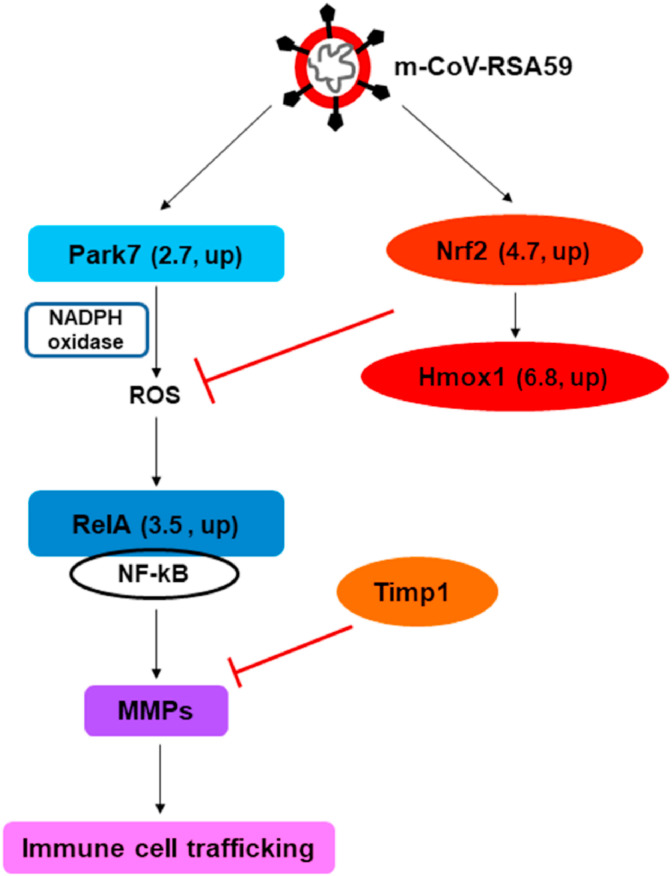
Activation of metalloproteases via oxidative pathways has been demonstrated in the past (Fu et al., 2001; Paquette et al., 2003; Meli et al., 2003). ROS-induced oxidative stress can also activate NF-κB signaling (Lingappan, 2018). The nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) acts as a transcription factor and can induce inflammation-related genes. RelA, a subunit of NF-κB, which gets activated in the canonical pathway via toll-like receptors that recognize pathogenic patterns (Hoffmann et al., 2006), showed increased mRNA levels following m-β-CoV-RSA59 infection. On the other hand, Nfκb2, which acts as both a precursor and suppressor of NF-κB (Hoffmann et al., 2006), demonstrated unchanged mRNA levels upon infection. Previously, it has been documented that NF-κB can induce MMP genes (Wu et al., 2013; Bond et al., 2001). Therefore, our result suggests that Park7 mediated ROS generation could lead to the induction of MMP genes via NF-κB signaling during MHV-induced acute disease (Fig. 10 ).
Fig. 10.
Schematic diagram depicting the possible relationship between ROS, NF-kB, and induction of MMP and TIMP following m-β-CoV-RSA59 infection. m-β-CoV-RSA59 induced the upregulation of Park7 (Parkinson's disease 7) in the brain of virus-infected mice compared with mock samples. Park7 is known to activate NADPH oxidase-dependant ROS production. In the downstream of the ROS pathway lies nuclear factor kappa B or NF-kB. m-β-CoV-RSA59 infection also upregulated the transcripts of RelA, a subunit of NF-kB. The transcription factor NF-kB can induce many inflammation-related genes, including MMPs. Additionally, m-β-CoV-RSA59 infection increased mRNAs of nuclear factor erythroid 2-related factor 2 (Nrf2) and its dependant heme oxygenase-1 (Hmox1) genes that are involved in the antioxidative pathway. The numbers indicate mRNA fold-change and its regulation (up or down) as detected in RT-qPCR in Fig. 6. Further, elevated levels of Timp1 provide for a host homeostatic mechanism to regulate the activities of upregulated MMPs.
We also found that m-β-CoV-RSA59 infection-induced upregulation of Nrf2 and Hmox1 genes. The antioxidative pathway mediated by nuclear factor erythroid 2-related factor 2 (Nrf2) and its dependant heme oxygenase-1 (Hmox1) (Loboda et al., 2016) could therefore play an important role in restoring homeostasis through inhibition of ROS overproduction. One limitation of this study that will be addressed in our future experiments is that the connection between ROS and MMPs has not been validated using inhibitors of ROS as positive controls. Additionally, interactome studies of different pathways and a global unbiased approach such as single-cell RNA-seq and comparing the expression patterns would better uncover new information regarding the activation of the oxidative stress expression responses in the inflamed CNS over time following infection.
While the dependence of β-coronaviruses on cellular proteases for successful entry is widely studied, our findings demonstrate an essential role of cellular proteases in mediating inflammation. Our data suggest that MMP-3, MMP-8, and MMP-14 might play prominent roles in MHV-induced neuroinflammation. Another interesting finding is the existence of a possible nexus between ROS, NF-κB, and MMPs during MHV infection. Further studies are needed to enhance our understanding of the diverse role played by MMPs in inflammation and in developing strategies to interrupt this process, thereby influencing the outcome of CNS inflammatory diseases. Also, it will be interesting to explore the role of MMPs or metalloproteases in general in acute respiratory illness caused by SARS-CoV-2, the causal agent of the COVID-19 pandemic situation.
Funding
This work was supported by a research grant [BT/PR20922/MED/122/37/2016] from the Department of Biotechnology, Ministry of Science and Technology, India.
CRediT authorship contribution statement
Sourodip Sengupta: Conceptualization, designing, and planning of all experiments, performed all the animal experiments, participated in data analysis and data interpretation, drafted the manuscript, revised the manuscript. Sankar Addya: performed the network construction using IPA, data analysis and data interpretation. Diptomit Biswas: performed PCR experiments and its data analysis. Paromita Banerjee: performed immunoblot assays and its data analysis; participated in data analysis and data interpretation, drafted the manuscript, revised the manuscript. Jayasri Das Sarma: Conceptualization, designing, and planning of all experiments, participated in data analysis and data interpretation, drafted the manuscript, critically reviewed the manuscript and supervised all aspects of this work. All authors have read and agreed to the submission of this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the Ministry of Education, India, and the Department of Science & Technology for fellowships to SS and DB, respectively. We also thank the Department of Biological Sciences, IISER-K, for providing the necessary laboratory facilities. We acknowledge the support of the Cancer Genomics Center, Thomas Jefferson University, Philadelphia, USA. The authors thank Patricia Rayman of Lerner Research Institute, Cleveland Clinic, Ohio, USA, for assistance with flow data analysis. The authors thank the IISER-K Animal Facility. The authors also thank the IISER-K SyMeC Lab for providing access to its RT-qPCR facilities.
Data availability
Data will be made available on request.
References
- Ami Y., Nagata N., Shirato K., Watanabe R., Iwata N., Nakagaki K., et al. Co-infection of respiratory bacterium with severe acute respiratory syndrome coronavirus induces an exacerbated pneumonia in mice. Microbiol. Immunol. 2008 Feb;52(2):118–127. doi: 10.1111/j.1348-0421.2008.00011.x. PubMed PMID: 18380809. Pubmed Central PMCID: PMC7168413. Epub 2008/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J. Biol. Chem. 2010 Jul 23;285(30):22758–22763. doi: 10.1074/jbc.M110.103275. PubMed PMID: 20507992. Pubmed Central PMCID: PMC2906266. Epub 2010/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S.J., Weiss S.R. Pathogenesis of murine coronavirus in the central nervous system. J. Neuroimmune Pharmacol.: Offic J. Soc. NeuroImmune Pharmacol. 2010 Sep;5(3):336–354. doi: 10.1007/s11481-010-9202-2. PubMed PMID: 20369302. Pubmed Central PMCID: 2914825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C.C., Lane T.E., Stohlman S.A. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 2006 Feb;4(2):121–132. doi: 10.1038/nrmicro1343. PubMed PMID: 16415928. Pubmed Central PMCID: 7096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011 Dec;85(24):13363–13372. doi: 10.1128/JVI.05300-11. PubMed PMID: 21994442. Pubmed Central PMCID: PMC3233180. Epub 2011/10/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond M., Chase A.J., Baker A.H., Newby A.C. Inhibition of transcription factor NF-kappaB reduces matrix metalloproteinase-1, -3 and -9 production by vascular smooth muscle cells. Cardiovasc. Res. 2001 Jun;50(3):556–565. doi: 10.1016/s0008-6363(01)00220-6. PubMed PMID: 11376631. Epub 2001/05/30. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Bartelink W., Rottier P.J. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008 Sep;82(17):8887–8890. doi: 10.1128/JVI.00415-08. PubMed PMID: 18562523. Pubmed Central PMCID: PMC2519682. Epub 2008/06/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchi N., Kapil P., Puntambekar S., Stohlman S.A., Hinton D.R., Bergmann C.C. Myd88 initiates early innate immune responses and promotes CD4 T cells during coronavirus encephalomyelitis. J. Virol. 2015 Sep;89(18):9299–9312. doi: 10.1128/JVI.01199-15. PubMed PMID: 26136579. Pubmed Central PMCID: 4542380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauwe B., Opdenakker G. Intracellular substrate cleavage: a novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 2010 Oct;45(5):351–423. doi: 10.3109/10409238.2010.501783. PubMed PMID: 20812779. [DOI] [PubMed] [Google Scholar]
- Chakravarty D., Saadi F., Kundu S., Bose A., Khan R., Dine K., et al. CD4 deficiency causes poliomyelitis and axonal blebbing in murine coronavirus-induced neuroinflammation. J. Virol. 2020 Jul 1;94(14) doi: 10.1128/JVI.00548-20. PubMed PMID: 32404525. Pubmed Central PMCID: 7343199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui N., Hu M., Khalil R.A. Biochemical and biological attributes of matrix metalloproteinases. Prog. Molecul. Biol. Translat. Sci. 2017;147:1–73. doi: 10.1016/bs.pmbts.2017.02.005. PubMed PMID: 28413025. Pubmed Central PMCID: 5430303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J. vol. 2010. 2010. (A Mechanism of Virus-Induced Demyelination. Interdisciplinary Perspectives on Infectious Diseases). PubMed PMID: 20652053. Pubmed Central PMCID: 2905936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Fu L., Tsai J.C., Weiss S.R., Lavi E. Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J. Virol. 2000 Oct;74(19):9206–9213. doi: 10.1128/jvi.74.19.9206-9213.2000. PubMed PMID: 10982367. Pubmed Central PMCID: PMC102119. Epub 2000/09/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Fu L., Hingley S.T., Lavi E. Mouse hepatitis virus type-2 infection in mice: an experimental model system of acute meningitis and hepatitis. Exp. Mol. Pathol. 2001 Aug;71(1):1–12. doi: 10.1006/exmp.2001.2378. PubMed PMID: 11502093. Epub 2001/08/15. [DOI] [PubMed] [Google Scholar]
- Das Sarma J., Scheen E., Seo S.H., Koval M., Weiss S.R. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 2002 Oct;8(5):381–391. doi: 10.1080/13550280260422686. PubMed PMID: 12402164. Pubmed Central PMCID: PMC7095158. Epub 2002/10/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Iacono K., Gard L., Marek R., Kenyon L.C., Koval M., et al. Demyelinating and nondemyelinating strains of mouse hepatitis virus differ in their neural cell tropism. J. Virol. 2008 Jun;82(11):5519–5526. doi: 10.1128/JVI.01488-07. PubMed PMID: 18385249. Pubmed Central PMCID: PMC2395180. Epub 2008/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Kenyon L.C., Hingley S.T., Shindler K.S. Mechanisms of primary axonal damage in a viral model of multiple sclerosis. J. Neurosci.: Offic. J. Soc. Neurosci. 2009 Aug 19;29(33):10272–10280. doi: 10.1523/JNEUROSCI.1975-09.2009. PubMed PMID: 19692601. Pubmed Central PMCID: 2747667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Sarma J., Burrows A., Rayman P., Hwang M.H., Kundu S., Sharma N., et al. Ifit2 deficiency restricts microglial activation and leukocyte migration following murine coronavirus (m-CoV) CNS infection. PLoS Pathog. 2020 Nov;16(11) doi: 10.1371/journal.ppat.1009034. PubMed PMID: 33253295. Pubmed Central PMCID: 7738193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Stadler K., Godeke G.J., Bosch B.J., Rottier P.J. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 2004 Jun;78(11):6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. PubMed PMID: 15141003. Pubmed Central PMCID: PMC415802. Epub 2004/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003 May 15;348(20):1967–1976. doi: 10.1056/NEJMoa030747. PubMed PMID: 12690091. [DOI] [PubMed] [Google Scholar]
- Drosten C., Seilmaier M., Corman V.M., Hartmann W., Scheible G., Sack S., et al. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect. Dis. 2013 Sep;13(9):745–751. doi: 10.1016/S1473-3099(13)70154-3. PubMed PMID: 23782859. Pubmed Central PMCID: 7164791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duits F.H., Hernandez-Guillamon M., Montaner J., Goos J.D., Montanola A., Wattjes M.P., et al. Matrix metalloproteinases in alzheimer's disease and concurrent cerebral microbleeds. J. Alzheim. Dis. : JAD. 2015;48(3):711–720. doi: 10.3233/JAD-143186. PubMed PMID: 26402072. [DOI] [PubMed] [Google Scholar]
- Fu X., Kassim S.Y., Parks W.C., Heinecke J.W. Hypochlorous acid oxygenates the cysteine switch domain of pro-matrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J. Biol. Chem. 2001 Nov 2;276(44):41279–41287. doi: 10.1074/jbc.M106958200. PubMed PMID: 11533038. Epub 2001/09/05. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006 Apr;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. PubMed PMID: 16503362. Pubmed Central PMCID: 7114179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013 Oct;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. PubMed PMID: 24012996. Pubmed Central PMCID: 7113673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Natoli G., Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006 Oct 30;25(51):6706–6716. doi: 10.1038/sj.onc.1209933. PubMed PMID: 17072323. Epub 2006/10/31. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Apr 16;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. PubMed PMID: 32142651. Pubmed Central PMCID: PMC7102627. Epub 2020/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019 Mar 15;93(6) doi: 10.1128/JVI.01815-18. PubMed PMID: 30626688. Pubmed Central PMCID: PMC6401451. Epub 2019/01/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience. 2020 Jun 26;23(6) doi: 10.1016/j.isci.2020.101212. PubMed PMID: 32512386. Pubmed Central PMCID: PMC7255728. Epub 2020/06/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaid M.A., Abdallah M.N., Ahmed A.S., Sheikh Z. Matrix metalloproteinases and their pathological upregulation in multiple sclerosis: an overview. Acta Neurol. Belg. 2013 Dec;113(4):381–390. doi: 10.1007/s13760-013-0239-x. PubMed PMID: 24002649. [DOI] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012 Jun;86(12):6537–6545. doi: 10.1128/JVI.00094-12. PubMed PMID: 22496216. Pubmed Central PMCID: PMC3393535. Epub 2012/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R.S., Dine K., Das Sarma J., Shindler K.S. SIRT1 activating compounds reduce oxidative stress mediated neuronal loss in viral induced CNS demyelinating disease. Acta Neuropathol Commun. 2014 Jan 2;2:3. doi: 10.1186/2051-5960-2-3. PubMed PMID: 24383546. Pubmed Central PMCID: PMC3892130. Epub 2014/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore A., Kanaujia A., Nag S., Rostami A.M., Kenyon L.C., Shindler K.S., et al. Different mechanisms of inflammation induced in virus and autoimmune-mediated models of multiple sclerosis in C57BL6 mice. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/589048. PubMed PMID: 24083230. Pubmed Central PMCID: PMC3780522. Epub 2013/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., Saadi F., Sengupta S., Antony G.R., Raveendran V.A., Kumar R., et al. DJ-1-Nrf2 axis is activated upon murine beta-coronavirus infection in the CNS. Brain disorders. 2021 Dec;4 doi: 10.1016/j.dscb.2021.100021. PubMed PMID: 34514445. Pubmed Central PMCID: 8418700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi E., Gilden D.H., Wroblewska Z., Rorke L.B., Weiss S.R. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology. 1984 May;34(5):597–603. doi: 10.1212/wnl.34.5.597. PubMed PMID: 6324031. [DOI] [PubMed] [Google Scholar]
- Lee J.M., Kronbichler A., Park S.J., Kim S.H., Han K.H., Kang H.G., et al. vol. 2019. 2019. (Association between Serum Matrix Metalloproteinase- (MMP-) 3 Levels and Systemic Lupus Erythematosus: A Meta-Analysis. Disease Markers). PubMed PMID: 31396295. Pubmed Central PMCID: 6668546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappan K. NF-kappaB in oxidative stress. Curr Opin Toxicol. 2018 Feb;7:81–86. doi: 10.1016/j.cotox.2017.11.002. PubMed PMID: 29862377. Pubmed Central PMCID: PMC5978768. Epub 2018/06/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wu H., Chen L., Wen Y., Kong X., Gao W.Q. Park7 interacts with p47(phox) to direct NADPH oxidase-dependent ROS production and protect against sepsis. Cell Res. 2015 Jun;25(6):691–706. doi: 10.1038/cr.2015.63. PubMed PMID: 26021615. Pubmed Central PMCID: PMC4456629. Epub 2015/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016 Sep;73(17):3221–3247. doi: 10.1007/s00018-016-2223-0. PubMed PMID: 27100828. Pubmed Central PMCID: PMC4967105. Epub 2016/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.J., Zhao X., Li J., Niu P.H., Yang B., Wu H.L., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb 22;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. PubMed PMID: WOS:000514849400027. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luplertlop N., Misse D., Bray D., Deleuze V., Gonzalez J.P., Leardkamolkarn V., et al. Dengue-virus-infected dendritic cells trigger vascular leakage through metalloproteinase overproduction. EMBO Rep. 2006 Nov;7(11):1176–1181. doi: 10.1038/sj.embor.7400814. PubMed PMID: 17028575. Pubmed Central PMCID: PMC1679776. Epub 2006/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U. S. A. 2005 Aug 30;102(35):12543–12547. doi: 10.1073/pnas.0503203102. PubMed PMID: 16116101. Pubmed Central PMCID: PMC1194915. Epub 2005/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010 Dec;84(24):12658–12664. doi: 10.1128/JVI.01542-10. PubMed PMID: 20926566. Pubmed Central PMCID: PMC3004351. Epub 2010/10/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli D.N., Christen S., Leib S.L. Matrix metalloproteinase-9 in pneumococcal meningitis: activation via an oxidative pathway. J. Infect. Dis. 2003 May 1;187(9):1411–1415. doi: 10.1086/374644. PubMed PMID: 12717622. Epub 2003/04/30. [DOI] [PubMed] [Google Scholar]
- Murphy G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011 Nov 11;12(11):233. doi: 10.1186/gb-2011-12-11-233. PubMed PMID: 22078297. Pubmed Central PMCID: 3334591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H., Visse R., Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006 Feb 15;69(3):562–573. doi: 10.1016/j.cardiores.2005.12.002. PubMed PMID: 16405877. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020 Mar 27;11(1):1620. doi: 10.1038/s41467-020-15562-9. PubMed PMID: 32221306. Pubmed Central PMCID: PMC7100515. Epub 2020/03/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette B., Bisson M., Therriault H., Lemay R., Pare M., Banville P., et al. Activation of matrix metalloproteinase-2 and -9 by 2- and 4-hydroxyestradiol. J. Steroid Biochem. Mol. Biol. 2003 Oct;87(1):65–73. doi: 10.1016/s0960-0760(03)00386-8. PubMed PMID: 14630092. Epub 2003/11/25. [DOI] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003 Apr 19;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. PubMed PMID: 12711465. Pubmed Central PMCID: PMC7112372. Epub 2003/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009 Jun;7(6):439–450. doi: 10.1038/nrmicro2147. PubMed PMID: 19430490. Pubmed Central PMCID: 2830095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.M., Gallagher T., Weiss S.R. Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell-cell fusion. J. Virol. 2017 Apr 15;91(8) doi: 10.1128/JVI.01564-16. PubMed PMID: 28148786. Pubmed Central PMCID: PMC5375694. Epub 2017/02/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., et al. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006 Jun;80(12):5768–5776. doi: 10.1128/JVI.00442-06. PubMed PMID: 16731916. Pubmed Central PMCID: PMC1472567. Epub 2006/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003 May 30;300(5624):1394–1399. doi: 10.1126/science.1085952. PubMed PMID: 12730500. [DOI] [PubMed] [Google Scholar]
- Savarin C., Bergmann C.C. Fine tuning the cytokine storm by IFN and IL-10 following neurotropic coronavirus encephalomyelitis. Front. Immunol. 2018;9:3022. doi: 10.3389/fimmu.2018.03022. PubMed PMID: 30619363. Pubmed Central PMCID: 6306494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarin C., Bergmann C.C., Hinton D.R., Stohlman S.A. MMP-independent role of TIMP-1 at the blood brain barrier during viral encephalomyelitis. ASN neuro. 2013 Nov 26;5(5) doi: 10.1042/AN20130033. PubMed PMID: 24156369. Pubmed Central PMCID: 3840398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020 May 26;117(21):11727–11734. doi: 10.1073/pnas.2003138117. PubMed PMID: 32376634. Pubmed Central PMCID: PMC7260975. Epub 2020/05/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Su M., Shen G., Hu Y., Yi F., Zeng Z., et al. Matrix metalloproteinase 3 as a valuable marker for patients with COVID-19. J. Med. Virol. 2021 Jan;93(1):528–532. doi: 10.1002/jmv.26235. PubMed PMID: 32603484. Pubmed Central PMCID: 7362036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindler K.S., Kenyon L.C., Dutt M., Hingley S.T., Das Sarma J. Experimental optic neuritis induced by a demyelinating strain of mouse hepatitis virus. J. Virol. 2008 Sep;82(17):8882–8886. doi: 10.1128/JVI.00920-08. PubMed PMID: 18579591. Pubmed Central PMCID: PMC2519666. Epub 2008/06/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011 Jan;85(2):873–882. doi: 10.1128/JVI.02062-10. PubMed PMID: 21068237. Pubmed Central PMCID: PMC3020023. Epub 2010/11/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Kishore A., Maity D., Sunanda P., Krishnarjuna B., Vappala S., et al. A proline insertion-deletion in the spike glycoprotein fusion peptide of mouse hepatitis virus strongly alters neuropathology. J. Biol. Chem. 2019 May 17;294(20):8064–8087. doi: 10.1074/jbc.RA118.004418. PubMed PMID: 30824541. Pubmed Central PMCID: PMC6527167. Epub 2019/03/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wang X., Saredy J., Yuan Z., Yang X., Wang H. Innate-adaptive immunity interplay and redox regulation in immune response. Redox Biol. 2020 Oct;37 doi: 10.1016/j.redox.2020.101759. PubMed PMID: 33086106. Pubmed Central PMCID: PMC7575795. Epub 2020/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint P., Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol. 2007 Dec;82(6):1375–1381. doi: 10.1189/jlb.0607338. PubMed PMID: 17709402. [DOI] [PubMed] [Google Scholar]
- Wang P., Dai J., Bai F., Kong K.F., Wong S.J., Montgomery R.R., et al. Matrix metalloproteinase 9 facilitates West Nile virus entry into the brain. J. Virol. 2008 Sep;82(18):8978–8985. doi: 10.1128/JVI.00314-08. PubMed PMID: 18632868. Pubmed Central PMCID: PMC2546894. Epub 2008/07/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.X., Tan M.S., Yu J.T., Tan L. Matrix metalloproteinases and their multiple roles in Alzheimer's disease. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/908636. PubMed PMID: 25050378. Pubmed Central PMCID: 4094696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Huang W., Luo G., Alain L.A. Hypoxia induces connexin 43 dysregulation by modulating matrix metalloproteinases via MAPK signaling. Mol. Cell. Biochem. 2013 Dec;384(1–2):155–162. doi: 10.1007/s11010-013-1793-5. PubMed PMID: 24002703. Pubmed Central PMCID: PMC3825321. Epub 2013/09/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Shin W.I., Pang Y.X., Meng Y., Lai J., You C., et al. The first 75 Days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment. Int. J. Environ. Res. Publ. Health. 2020 Mar 30;17(7) doi: 10.3390/ijerph17072323. PubMed PMID: 32235575. Pubmed Central PMCID: 7177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong V.W. Metalloproteinases: mediators of pathology and regeneration in the CNS. Nat. Rev. Neurosci. 2005 Dec;6(12):931–944. doi: 10.1038/nrn1807. PubMed PMID: 16288297. Epub 2005/11/17. [DOI] [PubMed] [Google Scholar]
- Yong V.W., Zabad R.K., Agrawal S., Goncalves Dasilva A., Metz L.M. Elevation of matrix metalloproteinases (MMPs) in multiple sclerosis and impact of immunomodulators. J. Neurol. Sci. 2007 Aug 15;259(1–2):79–84. doi: 10.1016/j.jns.2006.11.021. PubMed PMID: 17382965. [DOI] [PubMed] [Google Scholar]
- Young D., Das N., Anowai A., Dufour A. Matrix metalloproteases as influencers of the cells' social media. Int. J. Mol. Sci. 2019 Aug 7;(16):20. doi: 10.3390/ijms20163847. PubMed PMID: 31394726. Pubmed Central PMCID: 6720954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012 Nov 8;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. PubMed PMID: 23075143. [DOI] [PubMed] [Google Scholar]
- Zhou J., Stohlman S.A., Atkinson R., Hinton D.R., Marten N.W. Matrix metalloproteinase expression correlates with virulence following neurotropic mouse hepatitis virus infection. J. Virol. 2002 Aug;76(15):7374–7384. doi: 10.1128/JVI.76.15.7374-7384.2002. PubMed PMID: 12097550. Pubmed Central PMCID: PMC136378. Epub 2002/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Marten N.W., Bergmann C.C., Macklin W.B., Hinton D.R., Stohlman S.A. Expression of matrix metalloproteinases and their tissue inhibitor during viral encephalitis. J. Virol. 2005 Apr;79(8):4764–4773. doi: 10.1128/JVI.79.8.4764-4773.2005. PubMed PMID: 15795262. Pubmed Central PMCID: PMC1069551. Epub 2005/03/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.Q., Li T.T., Chen R., Pan H.F., Tao J.H., Li X.P., et al. Elevated serum levels of MMP-2, MMP-3, and MMP-13 in Chinese patients with systemic lupus erythematosus. Scand. J. Rheumatol. 2010;39(5):439–441. doi: 10.3109/03009741003742789. PubMed PMID: 20684736. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 Feb 20;382(8):727–733. doi: 10.1056/NEJMoa2001017. PubMed PMID: 31978945. Pubmed Central PMCID: 7092803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.