Abstract
The outbreak of COVID-19 pandemics highlighted the need of sensitive, selective, and easy-to-handle biosensing devices. In the contemporary scenario, point-of-care devices for mass testing and infection mapping within a population have proven themselves as of primordial importance. Here, we introduce a graphene-based Electrical-Electrochemical Vertical Device (EEVD) point-of-care biosensor, strategically engineered for serologic COVID-19 diagnosis. EEVD uses serologic IgG quantifications on SARS-CoV-2 Receptor Binding Domain (RBD) bioconjugate immobilized onto device surface. EEVD combines graphene basal plane with high charge carrier mobility, high conductivity, low intrinsic resistance, and interfacial sensitivity to capacitance alterations. EEVD application was carried out in real human serum samples. Since EEVD is a miniaturized device, it requires just 40 μL of sample for a point-of-care COVID-19 infections detection. When compared to serologic assays such ELISA and other immunochromatographic methods, EEVD presents some advantages such as time of analyses (15 min), sample preparation, and a LOD of 1.0 pg mL-1. We glimpse that EEVD meets the principles of robustness and accuracy, desirable analytic parameters for assays destined to pandemics control strategies.
Keywords: SARS-CoV-2, COVID-19, Serologic detections, Graphene, IgG, Biosensor
Graphical abstract
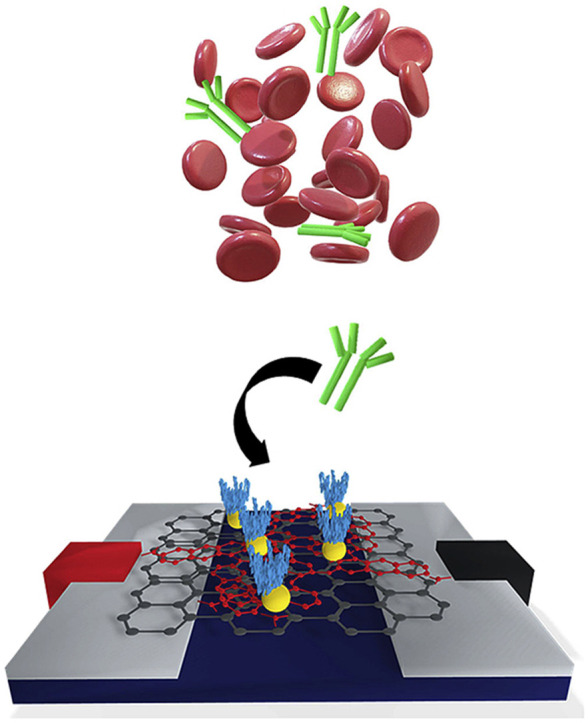
1. Introduction
The outbreak of COVID-19 pandemics shocked all nations throughout the world due to the high transmittivity of its causative virus, SARS-CoV-2. It can be spread out through airborne, droplet, contact, fecal-oral, fomite and bloodborne, and once infected, a person can be either symptomatic or asymptomatic (Chaibun et al., 2021). The severeness of COVID-19 symptoms and high rate of hospitalizations and fatalities (Mattioli et al., 2020) concerned the World Health Organization (WHO), as well as leaderships of more than 200 countries to which SARS-CoV-2 migrated (WHO, 2020). A global mobilization has been established aiming the control of COVID-19 infections, by the adoption of mass immunization through vaccines, social distancing measurements, use of face masks, and mass testing (Raffle et al., 2020).
Understanding the SARS-CoV-2 structure and its functional proteins are of relevant importance to the development of vaccines, medication, and sensible tests for diagnosis. The Spike protein of SARS-CoV-2 (S protein), a structural protein responsible for interacting and invading external receptors, is one of the most studied SARS-CoV-2 encoded genes (Lan et al., 2020; Mattioli et al., 2020). The S protein is divided into two subunits, S1 and S2, and present a furin cleavage in the S1–S2 boundary that can improve its infection capacity in host cells (Lan et al., 2020). To interact with human receptor cells, interactions between S protein and ACE2 (Angiotensin Conversion Enzyme) are established (Lan et al., 2020). S protein uses its Receptor Binding Domain (RBD), localized in the S1 subunit, the main interaction site of SARS-CoV-2 virus (Mattioli et al., 2020). It is reported that RBD domain is used as a highly specific immunotarget of human antibodies produced in response to SARS-CoV-2 infection (Premkumar et al., 2020). The RBD structure has been used as a potential therapeutic target for immunization strategies, as vaccines (Chen et al., 2020; Yang et al., 2020), and protein target and/or immunogen for the development of methodologies aiming either viral or serologic detections (Li et al., 2020; Seo et al., 2020; Yakoh et al., 2021).
COVID-19 diagnoses have been proposed by using RBD as immobilized immunogen for serologic detections of human antibodies produced in response to SARS-CoV-2 infections (Li et al., 2020; Mattioli et al., 2020; Zeng et al., 2020). IgG antibodies are one of the most bioanalytes for the development of SARS-CoV-2 serologic detections, due to its higher stability (Liu and May 2012), higher specificity in comparison to other immunoglobulins (Charles A Janeway et al., 2001), and its long-term presence in human organism after the infection (Sun et al., 2020). Devices based on IgG detection by immunochromatographic methodologies are commonly used strategies for fast serologic detections of human antibody (Isho et al., 2020; Theel et al., 2020). Despite of the well-known advantages of immunochromatographic tests, as low-cost, rapidness of fabrication and easy-to-handle operation, immunochromatographic assays have been frequently associated with unsatisfactory limit of detection (LOD) (Zhang et al., 2020), low repeatability, and poor sensitivity (Ragavendar and Anmol, 2012). Functional nanomaterials are frequently employed as biodevice modifiers in order to improve the analytical features. For instance, gold nanoparticles (AuNP) are employed in biosensors due to size-dependent color, biocompatibility, and high extinction coefficients (Qing et al., 2020b). Due to its optical properties and relatively low cost, AuNP are extensively applied in colorimetric and serological detection devices (Qing et al., 2020a). Furthermore, AuNP use for biosensing and bioelectronics construction with integration between biological units expands its applications for different biosensing architectures, mainly the electrochemical ones (Guo and Wang, 2007; Pingarrón et al., 2008; Qing et al., 2020b). Recently, a large number of electrochemical biosensing devices relying on the application of AuNP as modifiers has been proposed (Akbari Nakhjavani et al., 2019; Filik and Aslıhan Avan, 2020; Prasad et al., 2020; Suresh et al., 2018).
Electrochemical devices have also been widely studied and applied for rapid and sensitive serologic detections through IgG quantifications (Kudr et al., 2021; Mattioli et al., 2020). There are different materials that can be used as a main constituent of these devices, as glassy carbon, screen printed carbon electrodes, pristine graphene, and graphene derivatives (e.g., oxidized graphene). Graphene has attracted much attention for biosensors design due to its high biocompatibility and chemical stability (Jiang et al., 2020; Roy et al., 2017) Graphene-based devices are promising and fit many of the biosensing requirements owing to its expressive sensitivity, ability to reach lower LODs, rapidness, simplicity of operation, and possibility to perform biofunctionalization to improve selectivity (Ali et al., 2021; Hashemi et al., 2021; Mattioli et al., 2021; Torrente-Rodríguez et al., 2020; Yakoh et al., 2021). For instance, serologic detections of IgG and IgM antibodies through RBD SARS-CoV-2 antigen, as well as other SARS-CoV-2 genes using graphene-based electrochemical devices have been reported (Torrente-Rodríguez et al., 2020). The so called “reduced graphene oxide” as a coating material for 3D electrodes aiming COVID-19 antibodies sensing in a microfluidic assay have also been reported (Ali et al., 2021). These devices frequently involve functionalization procedures, as pristine graphene present sluggish electron transfer kinetics (Brownson et al., 2014; Macedo et al., 2019). As a consequence, this leads to a long time of electrodes preparation, and with structures that are not well defined, as is the case of oxidized graphene (Zhu et al., 2010).
Applying pristine graphene in bioelectronics and electrochemical biosensing is interesting due to its high basal plane conductivity, high charge carrier mobility and low intrinsic resistivity (Jiang et al., 2020; Macedo et al., 2019). On the other hand, graphene in field-effect devices configuration (e.g., G-FET) involves not only functionalization procedures to improve selectivity, but also the use of a Ag/AgCl non-polarizable reference electrode as gate electrode for data collection (Reddy et al., 2012). The use of a reference electrode as a gate causes imprecision on potential values reading, affecting the reliability of the proposed detection method (Mattioli et al., 2021). Despite of these issues, graphene field-effect transistors for serological COVID-19 detections has not been reported in literature up to our knowledge.
Here, we propose the application of a Electrical Electrochemical Vertical Device (EEVD), a graphene-based device based on hybrid electrical and electrochemical working principles (Mattioli et al., 2021), for COVID-19 diagnosis through IgG detections. Unlike a conventional G-FET, EEVD uses the vertical electron transfer occurring perpendicularly to the graphene plane, while the electronic current flows through the graphene van der Waals (vdW) heterojunctions. Recent study reported that EEVD can be10 times more sensitive than traditional graphene field-effect transistors, reaching a degree of zepto-molar for the limit of detection (Mattioli et al., 2021). The detection signal is the variation of interfacial potential of the device’s surface, obtained by open circuit potential (OCP) measurements. We performed IgG detections based on antigen-antibodies interactions through immobilized SARS-CoV-2 RBD bioconjugates onto a graphene-based van der Waals heterojunction capable of attaching RBD bioconjugates labels without damaging graphene sp2 structure. This heterojunction was made by electrodeposition of poly-neutral red (PNR), leading to a graphene-PNR (G-PNR) surface. The variation of OCP signal is expected to be caused by the establishment of the specific antigen-antibody interactions.
2. Materials and methods
2.1. Reagents and materials
Sulfuric acid, ethanol, isopropanol, monobasic sodium phosphate (NaH2PO4) and potassium phosphate were acquired from Synth®, Brazil. Hydrochloric acid (37% v/v), toluene, polystyrene (MW ∼192,000 g mol-1), tetrachloroauric acid trihydrate, ethanolamine, Tween 20, potassium chloride, neutral red dye and bovine serum albumin protein were bought from Sigma Aldrich®. Acetone was purchased from Chemis®, Brazil, and hydrogen peroxide (30–32%, v/v) was acquired from Vetec®, Brazil. Monoclonal IgG human antibodies (IgG1 type) from SARS-CoV-2 (ab273073) were obtained from Abcam®, USA. Human IgM antibody was acquired from Rheabiotech, Brazil. Bilirubin oxidase was purchased from Amano, Japan. For graphene devices confection, p-doped Si/SiO2 ( = 90 nm) were purchased from Graphene Supermarket®, USA. CVD monolayer graphene was acquired from Graphenea®, Spain. Metallic spots for electrical contact films deposition of Ti and Pt were obtained from Electron Microscopy Sciences®, USA. All aqueous solutions were made using deionized water with resistivity >18 MΩ cm.
2.2. AuNP synthesis
Colloidal AuNP were synthetized through reduction of Au3+ by citrate ions, according to the Turkevich methodology (Turkevich et al., 1951). Briefly, 20 mL of AuCl3 3H2O (1.0 mmol L-1) aqueous solution was heated to its boiling point under vigorous stirring. Then, 2.0 mL of sodium citrate (1.0%, w/v) was added to the boiling solution, and the mixture was kept under vigorous stirring at boiling point until to reach dark-red color. The resulting AuNP suspension was then rapidly cooled at ice bath under light protection. The final suspension was stored at 4 °C and protected from light to avoid nanoparticle agglomeration.
2.3. Bioconjugate synthesis
Bioconjugate of RBD with AuNP was strategically adopted in order to ensure an efficient modification of EEVD’s G-PNR interface with RBD, as PNR and AuNP present a well-established adsorptive interaction (Mazar et al., 2017; Prakash et al., 2018). Therefore, AuNP based bioconjugates are expected to adsorb effectively on G-PNR EEVD interface. Prior to this, we expressed RBD of SARS-CoV-2 as follows. Firstly, we obtained the clones of the genes encoding for spike S1 RBD in the expression vector pET28a by chemical synthesis from Twist Bioscience (USA). RBD was produced by the E. coli expression system. The bacteria pellet was resuspended in lysis buffer (50 mM phosphate buffer pH 8.0, containing 200 mM NaCl, 1 mM DTT, and 1 mM PMSF and 1 mg mL-1 of egg lysozyme). RBD samples was subjected to chromatography and gel filtration on a Superdex 200 (16/600). RBD was then characterized by slot blot, UV–Vis, Micro-FTIR and circular dichroism. AuNP/RBD bioconjugates were prepared according to procedures reported elsewhere (de Oliveira et al., 2018; Li et al., 2020). For this, 10 μL of RBD (1.0 mg mL-1) was added to 1.1 mL of colloidal AuNP, prior containing 100 μL of borate buffer (0.1 mol L-1, pH 8.5). After incubation during 20 min, 100 μL of BSA (5%) in deionized water was added, and kept under incubation for 20 min. The resulting mixture was homogenized and centrifuged at 10,000 rpm and 4 °C for 20 min. After discarding the supernatant, a washing step with PBS (0.01 mol L-1) and Tween 20 was performed. The final mixture was also centrifuged at 10,000 rpm and 4 °C during 20 min and resuspended in BSA (3%) solution in PBS (0.01 mol L-1).
2.4. UV–Vis analyses
All proteinaceous samples were diluted 50 × from the original solutions used in bioconjugate synthesis. Final bioconjugate AuNP/RBD samples were diluted 10 ×. All spectra were presented with normalized absorbance values from 0 to 1 for maximum absorbance to clarify the data analysis.
2.5. Transmission electron microscopy (TEM)
TEM images were obtained for diluted AuNP and AuNP/RBD suspensions using a JEOL JEM-2100 microscope with a 200 kV electron beam. Diluted samples were sonicated for 30 min and drop cast onto a copper grid with thin carbon film (200 mesh, Electron Microscope Sciences, USA) and let to dry in vacuum at room temperature.
2.6. EEVDs
The Si/SiO2 chips were cleaned with piranha solution (3 H2SO4: 1 H2O2) prior to the electrical contacts deposition. For this, Ti and Pt thin films of 10 and 20 nm, respectively, were subsequently deposited at Si/SiO2 substrates through sputtering deposition at Brazilian Nanotechnology National Laboratory (LNNano) at the Brazilian Center for Research in Energy and Materials (CNPEM). For CVD monolayer graphene transfer to Si/SiO2 chips, the polymer-mediated graphene transferring process was adopted (Hassan et al., 2021; Macedo et al., 2018). Briefly, a drop of polystyrene in toluene was deposited onto small-area pieces of CVD monolayer graphene on Cu foil and let under 70 °C for ∼10 min. Then, the Cu foil was removed by etching in an aqueous solution containing HCl and H2O2 (1.4 and 0.5 mol L-1, respectively). After graphene transfer onto Si/SiO2 EEVDs substrates, a pre-annealing step in air at 90 °C was adopted. Next, the polystyrene layer was removed though toluene washing steps, and a thermal annealing under Ar atmosphere at 585 °C was finally performed. The success of this procedure was evaluated by optical images collected for each EEVD. Figure S1 present a schematic representation of these steps.
2.7. EEVD modification and characterization
Prior to EEVDs use, an electrochemical etching in HCl (0.1 mol L-1) procedure (Figure S3) was performed for removing Cu traces remaining from EEVDs confection (Iost et al., 2014). Pristine graphene EEVDs were modified with poly-neutral red (PNR) by electropolymerization by cyclic voltammetry, as is further described in “Electrochemical experiments” session. The modification of G-PNR EEVDs with AuNP/RBD bioconjugate was performed by drop-casting of 20 μL of the final suspension. AuNP/RBD was let to adsorb on G-PNR interface for 30 min. Next, 10 μL of ethanolamine (5.0 mmol L-1) aqueous solution (pH 8.0) was employed for blockage of extra RBD sites of AuNP/RBD (Raoof et al., 2013). This step was adopted in order to avoid non-specific interactions between these biomolecules and any other matrix component.
2.8. Micro-FTIR experiments
Micro-FTIR characterizations were performed in a Vertex 70v Fourier transform infrared spectrometer coupled to an IR microscope Hyperion 3000 (Bruker). Chemical 2D and 3D images of PNR and AuNP/RBD bioconjugates deposited onto graphene interfaces were obtained using a liquid nitrogen-cooled 64 × 64 Focal Plane Array (FPA) detector. To provide enough reflectivity for the IR beam (Brunner et al., 1997) experiments were performed in a thin Au layer substrate instead of conventional Si/SiO2 substrates. Images were collected over a 200 × 200 μm2 area. Experiments were performed at room temperature and room atmosphere, with 128 scans acquisition at 8 cm-1 resolutions. 2D and 3D chemical map of AuNP/RBD bioconjugate onto G-PNR surface was made according to amide I (1648 cm-1) and amide II (1540 cm-1) bands integration. All 3D contour plots have absorbance intensity in z-axis.
2.9. Raman spectroscopy
Raman spectra were acquired for bare graphene and G-PNR EEVD surfaces (for the latter surface, after electropolymerization and water evaporation in room temperature), from 1000 to 3000 cm-1 under air atmosphere and room temperature by a LabRAM HR Evolution spectrometer (Horiba Scientific) with an HeNe laser of 633 nm wavelength with an acquisition of 64 spectra and an aquisition time of 4s.
2.10. Electrochemical experiments
Electrochemical measurements of all EEVDs were acquired by a PGSTAT 128N potentiostat/galvanostat (Metrohm). Cyclic voltammetry of all electrodes which were performed in PBS (0.01 mol L-1) were made from −1.0 to 1.0 V (vs. Ag/AgClsat), at scan rate (ν) 50 mV s-1. Electropolymerization of PNR at pristine graphene EEVDs interfaces were performed by cyclic voltammetry from −1.0 to 1.2 V (vs. Ag/AgClsat) at ν = 50 mV s-1 for 2 scans, and from −1.0 to 0.5 V (vs. Ag/AgClsat), ν = 50 mV s-1 for 15 scans (Pauliukaite et al., 2007; Pauliukaite and Brett, 2008). Electrochemical impedance spectroscopy (EIS) experiments of graphene, G-PNR, G-PNR-AuNP/RBD and G-PNR-AuNP/RBD after IgG interaction were performed. All plots were obtained from 1 × 105 to 0.1 Hz, DCpot (direct current potential) equals to OCP (open-circuit potential) of each interface with an amplitude of 5 mV s-1 in PBS (0.01 mol L-1, pH 7.4).
2.11. Electrical-electrochemical experiments
Hybrid Electrical-Electrochemical (EE) experiments of all electrodes were performed in a PGSTAT 128N potentiostat/galvanostat (Metrohm). An Ag/AgClsat electrode was applied as reference electrode, connected to potentiostat RE terminal. Both WE and CE terminals were connected to Ti/Pt electrical contacts in a short-circuit-like configuration. The applied potential range for each EE experiment was defined after the determination of the OCP of each interface. All experiments were performed in a drop of ∼40 μL of PBS (0.01 mol L-1) supporting electrolyte. IgG and RBD interactions experiments were performed by the drop-casting of 20 μL IgG (1.0 μg mL-1) in PBS (0.01 mol L-1, pH 7.4) target analyte in a G-PNR-AuNP/RBD EEVD. All curves were obtained in PBS (0.01 mol L-1, pH 7.4); ν = 5 mV s-1. Further details on EEVDs fabrication, theory and working principles can be find elsewhere (Mattioli et al., 2021; Mattioli and Crespilho, 2020)
2.12. Analytical quantifications by EEVDs
For detection studies, calibration curves for IgG detections by G-PNR-AuNP/RBD EEVDs were obtained by hybrid I ds vs. V ds electrical-electrochemical experiments, being I ds = CE to WE terminals current and V ds = CE to WE terminals potential. The LOD of the studied EEVD was evaluated considering the standard deviation (SD) of the lowest concentration sample that could be detected, relying on signal-to-noise ratio approaches recommended by IUPAC (Brunetti B, 2015; Inczedy et al., 1998). The collection and use of human sera was approved by the Ethical Committee number 30178220.3.000.0068 of diagnostic proposal analysis at Brazilian platform CNEP (National Council of Ethical and Clinical Research). The diagnosis of SARS-CoV-2 were PCR-based, Roche and Abbott diagnostic kit. The sera from uninfected humans were taken before June 2019 (Pro-blood foundation, Brazil).
2.13. ELISA Comparative method for IgG positive human sera samples
In house plates were immobilized with RBD (10 μg mL-1) in carbonate buffer (0.1 mol L-1, pH 9.4) overnight incubation at 4 °C. The plates were empty and dried at 37 °C. The patient serum sample were diluted 1:10 in PBS containing BSA (0.05%) and milk casein (2%) for titration across the plate to a final volume of 100 μL/well. The plates were incubated for 30 min, 1 h at 37 °C. At the end of the incubation period, the plates were washed with PBS-T and incubated with 100 μL/well of commercial anti-IgG secondary (RheaBiotech, Brazil) for 30 min and 1 h at 37 °C. The unbound conjugate will be removed by washing with PBS-T. The enzymatic activity by addition of TMB (3,3',5,5'-tetramethylbenzidine, Scienco, Brazil) ready to use was measured after incubating each well with 100 μL of TMB solution. After 15 min at 25 °C the reaction was stopped by adding 100 μL of HCl (1.0 mol L-1) to each well. The absorbance was read at 450 nm in a TECAN sunrise conventional ELISA reader.
3. Results and discussions
3.1. PNR-modified graphene EEVDs studies
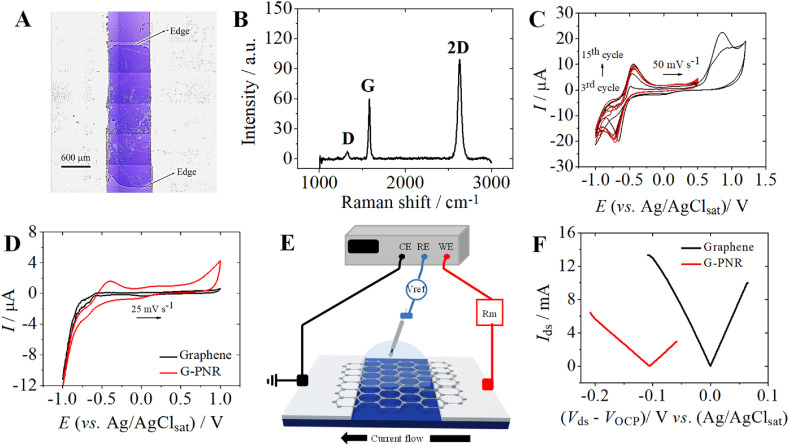
After EEVDs confection (Fig. 1 A), the monolayer features of our transferred pristine graphene onto EEVD Si/SiO2 chips were evaluated by Raman spectroscopy (Fig. 1B). As expected, the obtained Raman spectra presented only D, G, and 2D bands, typical of monolayer graphene structures (Malard et al., 2009), at 1327, 1583, and 2631 cm-1, respectively. I D/I G ratio is usually interpreted as an indicative of sp2 integrity along the monolayer graphene sheet, for which a value equal to 0 refers to an ideal non-defective graphene (Liu et al., 2013). For our monolayer graphene, a I D/I G ratio of 0.35 was obtained and found adequate for considering its features as of a low-defective graphene, taking into account the inevitable defects formation during wet transfer steps (Bleu et al., 2019). Moreover, I 2D/I G ratio is used to obtain insights on the number of layers for graphene-based materials with acceptable accuracy (Bleu et al., 2019). I 2D/I G ratios equal to 2 are obtained for ideal non-defective monolayer graphene. However, there is an agreement in literature for considering monolayer graphene characteristics with ratios ranging from 1.3 up to 2 (Bleu et al., 2019). As our graphene presented an I 2D/I G ratio of 1.5, we inferred that in fact most of its monolayer pristine graphene features could be preserved during transferring procedures.
Fig. 1.
G-PNR EEVDs. A) Optical image of a pristine graphene EEVD after confection; B) Raman spectrum for pristine graphene EEVD (graphene on Si/SiO2); C) Cyclic voltammograms of PNR electropolymerization under Ar atmosphere in a graphene large area (∼0.1 cm2) 2D electrode in PBS (0.01 mol L-1, pH 7.4), KNO3 (0.5 mol L-1), and neutral red monomer (2.0 mmol L-1), from – 1.0 V to +1.0 V (vs. Ag/AgClsat) for 2 scans and from – 1.0 V to +0.5 V (vs. Ag/AgClsat) for 15 scans; ν = 50 mV s-1; D) Cyclic voltammograms of graphene interface in bare PBS (0.01 mol L-1, pH 7.4) before PNR functionalization (black) and after PNR functionalization (red), leading to G-PNR interface; E) Schematic representation of hybrid EE experiments with graphene-based EEVDs; F) Pristine graphene (black) and G-PNR (red) EE Idsvs. Vds curves in PBS (0.01 mol L-1, pH 7.4). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Sequentially, electropolymerization of PNR onto graphene was performed by cyclic voltammetry (Fig. 3 C). It was firstly studied in a large-area graphene electrode (Figure S4A), under inert atmosphere, according to an electrochemical methodology described elsewhere (Pauliukaite et al., 2007; Pauliukaite and Brett, 2008). PNR was chosen due to its ability to adsorb on gold nanoparticles-based bioconjugates with high affinity, useful for our next analytical studies (Mazar et al., 2017; Prakash et al., 2018). Typical redox peaks are observed due to irreversible monomer oxidation at +0.86 V (vs. Ag/AgClsat) and the redox pair interconversion neutral red – leuco-neutral red at −0.585 V (vs. Ag/AgClsat) (Pauliukaite et al., 2007). After this procedure, the formation of G-PNR interface (Figure S4B) and its stability is electrochemically observed in Fig. 3D. Although the reversibility of neutral red – leuco-neutral red redox pair is affected, the presence of oxidation peak concerning this redox pair at c.a. + 0.6 V is interpreted as indicative of a successful and stable electropolymerization.
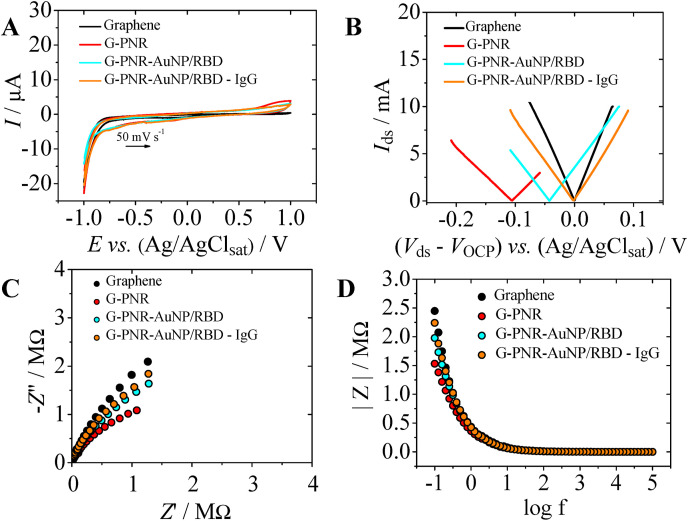
Fig. 3.
Electrochemical and EE studies for IgG interaction with G-PNR-AuNP/RBD surface. A) Cyclic voltammograms for bare graphene (black), G-PNR (red), G-PNR-AuNP/RBD (cyan) and G-PNR-AuNP/RBD after interaction with human IgG (1.0 μg mL-1) (orange) in PBS (0.01 mol L-1, pH 7.4), ν = 50 mV s-1; B) Hybrid Idsvs. Vds EE curves for bare graphene (black), G-PNR (red), G-PNR-AuNP/RBD (cyan) and G-PNR-AuNP/RBD after interaction with human IgG (1.0 μg mL-1) (orange) in PBS (0.01 mol L-1, pH 7.4), ν = 5 mV s-1; C) Nyquist plots for bare graphene (black), G-PNR (red), G-PNR-AuNP/RBD (cyan) and G-PNR-AuNP/RBD after interaction with human IgG (1.0 μg mL-1) (orange) interfaces in PBS (0.01 mol L-1, pH 7.4), from 1 × 105 to 0.1 Hz, amplitude = 5 mV and DCpot = OCP of each interface; D) Respective impedance modulus plots for each interface of item C). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Then, hybrid EE I ds vs. V ds curves were obtained for graphene and G-PNR interfaces (Fig. 1E and F). For this, CE and WE terminals are directly connected through G-PNR basal plane, allowing current flow with high charge carrier mobility and low intrinsic resistivity (Vieira et al., 2016; Wang et al., 2012). Moreover, in this configuration, it is possible to monitor interfacial OCP displacements (ΔVOCP) depending on the modification of the graphene interface (Mattioli et al., 2021). The OCP shifts are correlated to capacitive alterations of the EEVD interface and they are observed at more negative potentials due to PNR adsorption. As, EE experiments are performed in a potential range without the occurrence of PNR faradaic processes, we expected that only capacitive alterations would be manifested in G-PNR interfaces during these studies. As already discussed in our previous work (Mattioli et al., 2021), this shift is also an indicative of charge carrier insertion onto graphene, resulting in n-doping. This occurs probably due to the establishment of weak van der Waals interactions between graphene sp2 lattice and neutral red sp2 electronic density (Georgakilas et al., 2016) and nitrogen atoms of PNR structure. This coupling can be confirmed by G band displacements in Raman spectroscopy data (Figure S5). Despite this, our Raman spectroscopy characterizations of G-PNR could not be useful to corroborate hybrid EE doping insights, as expected due to the creation of some point defects during voltammetric cycling. (See Section 4 of SI for further discussion). Further interfacial capacitive features of graphene and G-PNR EEVDs interfaces were also investigated and confirmed by EIS measurements. A detailed discussion is presented in Section 5 of SI. Therefore, we conclude that G-PNR interfaces would not present any faradaic process, but only capacitive features to be considered during further bioanalytical IgG quantifications by EE studies.
3.2. AuNP/RBD and G-PNR-AuNP/RBD EEVDs surface characterization
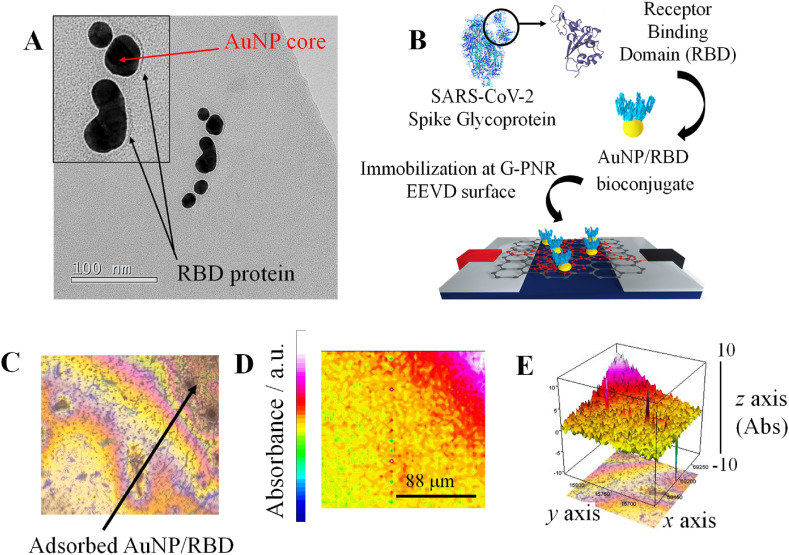
G-PNR EEVDs was modified with RBD protein by non-covalent functionalization with AuNP-based bioconjugates, aiming IgG detections through antigen-antibody interaction. It is known that AuNP-based bioconjugates with proteins are established through electrostatic interactions by chemisorption mechanisms (Busch et al., 2019). UV-Vis spectra were collected after each step of the procedure (Figure S8). Transmission electron microscopy (TEM) images of AuNP/RBD bioconjugate are in accordance with UV–Vis results (See Section S7 for further information). As shown in Fig. 2A, the coating material on AuNP characterizes RBD protein, indicating that the protein effectively interacts with AuNP (Busch et al., 2019). AuNP/RBD bioconjugate was drop-casted onto G-PNR EEVDs as illustrated in Fig. 2B. The distribution of AuNP/RBD onto G-PNR was evaluated by micro-FTIR 2D and 3D chemical mapping of AuNP/RBD spatial distribution.
Fig. 2.
AuNP/RBD characterization and adsorption onto G-PNR. A) TEM micrograph of AuNP/RBD bioconjugate; B) Schematic representation on AuNP/RBD immobilization methodology onto G-PNR; C) Optical image of G-PNR-AuNP/RBD surface in thin layer Au substrate; D) 2D chemical mapping of AuNP/RBD distribution onto G-PNR by proteinaceous RBD amide-I band monitoring. Scales in Fig. 2C and D are the same; E) The respective 3D chemical mapping.
Optical image of G-PNR-AuNP/RBD interface onto Au substrate shown in Fig. 2C indicates that most of AuNP/RBD proteinaceous bioconjugate have a tendency to accumulate on G-PNR edges while aqueous solvent is dried due to coffee-ring effects, typical in protein-based coatings (Devineau et al., 2016; Sedenho et al., 2021). From 2D and 3D special distribution of AuNP/RBD (Fig. 2D and E, respectively), it is observed that the homogeneity of AuNP/RBD distribution is associated to the homogeneity of PNR electropolymerization onto graphene. Fresh solution containing the precursors of electropolymerization was always used. In order to ensure a homogeneous AuNP/RBD immobilization over G-PNR surface, solvent evaporation in drop-casting procedures should be carried out at room temperature, to avoid extensive coffee-ring formation.
3.3. G-PNR-AuNP/RBD interaction with IgG
To understand the expected capacitance alterations at G-PNR-AuNP/RBD interface due to IgG interaction with immobilized RBD sites, EE and electrochemical experiments of the specific antigen-antibody interaction with IgG (1.0 μg mL-1) as target were carried out. Electrochemical behavior of G-PNR-AuNP/RBD was first investigated by cyclic voltammetry (Fig. 3A). Neither the adsorption of AuNP/RBD bioconjugate nor its interaction with human IgG (1.0 μg mL-1) interferes on PNR redox behavior and only capacitive changes are observed. This was already expected, as both bioconjugates do not present redox-active sites (from −1.0V to 1.0V). Therefore, EE I ds vs. V ds curves for these interfaces were obtained within a potential range with no faradaic process occurring (Fig. 3B). The immobilization of AuNP/RBD bioconjugate onto G-PNR leads to OCP value shifts to more positive values in relation to G-PNR interface OCP potential value, probably due to G-PNR sp2 electronic density displacement towards AuNP/protein bioconjugate. These shifts occasioned by charge insertion/removal and electronic density variations in EEVDs are correlated to capacitive changes that lead to OCP potential displacements (Mattioli et al., 2021). Similar to what is observed for Graphene Field-Effect Transistors (GFET), these OCP shifts can be interpreted in a similar way as it is done for the charge neutrality point (CNP). The interaction of IgG with G-PNR-AuNP/RBD interface could be monitored by ΔVOCP shifts, and in this case, the specific RBD-IgG interaction led to ΔVOCP = 0.0437 V compared to the G-PNR-AuNP/RBD initial potential. The shift of OCP potential to more positive values (p-doping) due to specific IgG interaction is coherent to the presence of a positive net charge, as the type of the human IgG antibody (IgG1) used in this work present an isoelectric point (pI) that mainly ranges from 7.9 to 8.6 (Yang et al., 2019). Based on these results, it was concluded that G-PNR-AuNP/RBD EEVDs interface can be sensitive to IgG interaction and detections through ΔVOCP potential displacements.
In order to confirm the capacitive nature of the above-mentioned interfaces and the IgG interaction, EIS data were collected in a non-electroactive electrolyte medium (Fig. 3C and D). Capacitive alterations in accordance to EE data in Fig. 3B can be also seen due to each subsequent modification of EEVD interface based on Nyquist plots inclination changes. The inclination of G-PNR-AuNP/RBD Nyquist plot in Fig. 3C is close to the bare graphene one, indicating that the employed subsequent modifications led to an increase in total interfacial capacitance of the EEVD (Fig. 3D), until it reaches values similar to the bare graphene one (Mattioli et al., 2021). The predominance of capacitive features G-PNR-AuNP/RBD interface can be confirmed. It was also possible to fit a best circuit to describe G-PNR-AuNP-RBD impedance features, which was the same obtained for G-PNR (Figure S7B). These insights can be corroborated by the impedance modulus plots of Fig. 3D as well, based on the absence of peaks through the entire swept frequency range. EIS data corroborated to all ΔVOCP potential displacements occurred due to capacitive alterations at each EEVD interface and, therefore, IgG detections in different concentrations may be performed based on these principles.
3.4. COVID-19 diagnosis through IgG quantifications by G-PNR-AuNP/RBD EEVDs
We proceeded with the development of an analytical quantification method for serological diagnosis related to COVID-19 disease, based on total interfacial capacitance alterations on the EEVD interface due to specific RBD-IgG interaction (Fig. 4 A). For this, ΔVOCP were monitored as a function of IgG concentration, as indicated in Equation (1).
| (1) |
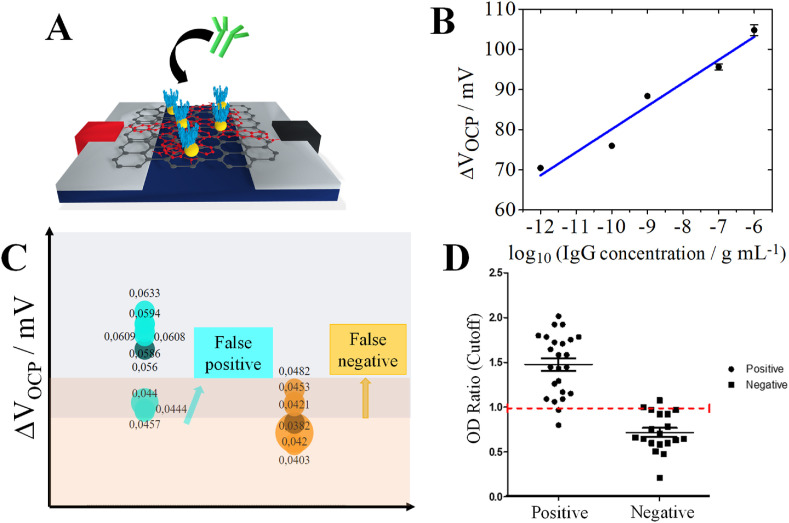
Fig. 4.
IgG detections for COVID-19 diagnosis through EEVD. A) Schematic representation of G-PNR-AuNP/RBD interface detecting IgG antibodies; B) Calibration curve for IgG detections from 1.0 pg mL-1 to 1.0 μg mL-1 concentrations in PBS (0.01 mol L-1, pH 7.4) as support electrolyte by hybrid Idsvs. Vds EE experiments at 5 mV s-1; C) Distribution of OCP displacement values for n = 9 for positive and negative IgG detections in diluted patient serum samples by G-PNR-AuNP/RBD EEVD; D) Analysis via ELISA dimerized intact RBD in samples from 20 positive and negative patients.
In this Equation, corresponds to the interfacial OCP potential of G-PNR-AuNP/RBD interface after adsorption and interaction with IgG and corresponds to the G-PNR-AuNP/RBD interfacial OCP potential before interaction with IgG. All detections were performed in a miniaturized configuration by using a drop of 40 μL of electrolyte as reactional medium. A calibration curve shows an unprecedent wide linear range of IgG concentrations, from 10-12 up to 10-7 mol L-1. Despite of the absence of COVID-19-related IgG detections by GFETs, the extent of this dynamic linear range is wider than the obtained for IgG COVID-19 diagnosis through electrochemical methods reported elsewhere (Torrente-Rodríguez et al., 2020). The EEVD sensitivity obtained for IgG quantifications based on the slope of calibration curve was of 5.8 mV/decade. A LOD of 1.0 pg mL-1 was obtained. The LOD was evaluated according to IUPAC recommendations, by using the standard deviation of the lowest concentration sample to estimate the signal-to-noise ratio of our device, with 99% of confidence level (Brunetti B, 2015; Inczedy et al., 1998). Interestingly, to the best of our knowledge, there has, so far, one of the lowest LODs already reported in literature, where EEVD performance is compared to other serological assays presented in Table 1 . This can be related to the high sensitivity of the EEVD interface to its chemical environment and to the EE detection method through interfacial capacitive alterations. Due to the similarities of the EEVDs to GFETs, a comparison between these two kinds of devices is of relevant interest. However, no FET or GFET was reported in literature for RBD-based IgG detections. The reached LOD value by G-PNR-AuNP/RBD, as well as its sensitivity were compared to the values obtained for general immunoglobulin detections by FETs. For example, G-PNR-AuNP/RBD LOD was comparable to the obtained by Vu and co-authors using a GFET (1.0 pg mL-1) (Vu et al., 2021), and remarkably lower than the reached by Minamiki and co-authors applying an OFET (0.62 μg mL-1) (Minamiki et al., 2014). The sensitivity of G-PNR-AuNP/RBD obtained by the slope of the calibration curve could not be compared to the values reported by different serological assays due to incompatibility of units.
Table 1.
Comparison of several serological detection methods of IgG antibodies produced in response to SARS-CoV-2 infection.
| Detection method | Device components | LOD | Sensitivity | Accuracy/κ coefficient | Reference |
|---|---|---|---|---|---|
| LFIA | Paper strip/colloidal AuNP | Not informed | 69.1% | κ = 0.612 | Wen et al. (2020) |
| ELISA | NovaLisa SARS-CoV-2 test | 0.18 a.u. | 94.9% | Not informed | Tré-Hardy et al. (2020) |
| Electrochemical | COVID-19 ePAD/SWV | 0.96 ng mL-1 | 100% | κ = 0.881 | Yakoh et al. (2021) |
| SERS/LFIA | SERS-based LFIA/Ag@SiO2 | 1.0 pg mL-1 | Not informed | 100% | Liu et al. (2021) |
| Electrochemical | 3D nanoprinted electrodes/rGO, Spike S1 subunit | 2.8 × 10-15 mol L-1 | 1.0 × 10-12 mol L-1 | Not informed | Ali et al. (2021) |
| LFIA | Paper strip/Selenium NPs bioconjugate | 5.0 ng mL-1 | 93.33% | Not informed | Wang et al. (2020) |
| LISA | N protein | 0.4 pg mL-1 | 71% | Not informed | Liang et al. (2021) |
| Colorimetric | Epitope functionalized AuNPs | 3.2 nmol L-1 | 83% | Not informed | Lew et al. (2021) |
| Hybrid EE detections | G-PNR-AuNP/RBD EEVDs | 1.0 pg mL-1 | 5.8 mV/decade | 61.1% | This work |
ePAD = electrochemical paper based analytical device; SWV = Square Wave Voltammetry; rGO = reduced graphene oxide; LFIA = Lateral Flow Immunochromatographic Assay; SERS = Surface Enhanced Raman Scattering; LISA = Luciferase Immunosorbent Assay.
Although the EEVDs reported here for the diagnosis of COVID-19 have a great positive impact in the area of biosensors using graphene monolayers, it is necessary to emphasize that other works using GFETs sensors for different proteins achieved remarkable sensitivity. Just to exemplify among several studies, Danielson and collaborators reached 14 mV/decade for streptavidin (Danielson et al., 2020). Our value of 5.8 mV/decade can be correlated to the low sensitivity of G-PNR interface to the IgG-AuNP/RBD complex formation due to the establishment of antigen-antibody interaction. This value may improve replacing PNR by other non-covalent modifier, capable of stronger interactions with graphene basal plane through more effective heterojunction formation, as, for example, ferrocene derivatives or PBASE for direct IgG immobilization. Consequently, stronger capacitive alterations would be observed with the addition of each target analyte concentration. Despite of this, IgG detections by G-PNR-AuNP/RBD could be performed in a short time of analyses. Each EE experimental run can be performed within 5 min. This feature is considerably advantageous especially in relation to LFIAs and ELISA assays reported in Table 1, which experimental runs usually take more than 15–20 min. Furthermore, other assays reported in this table require extensive device preparations with nanoparticles functionalization and expensive electrode printings, making the proposed methodologies less suitable for fast and simple serologic COVID-19 mass-testing of a population.
To further evaluate the analytical performance of our G-PNR-AuNP/RBD EEVD, IgG detections were performed in human sera with n = 9 replicates, for proof-of-concept purposes (Fig. 4C). Mean ΔVOCP potential shifts for positive and negative IgG detections were, respectively, 0.056 ± 0.009 V and 0.042 ± 0.003 V. We used the value of OCP potential displacements that was possible to discriminate the healthy from the infected patient with COVID-19 as cut-off values. The estimated SARS-CoV-2 IgG cut-off was 40 mV. Nonetheless, some quantifications could not be differentiated between a positive or negative result according to the obtained ΔV value. A false-positive region and a false-negative region of ΔV range between 43 and 48 mV could be estimated by our results, probably due to bioconjugates removal during washing steps and EEVD surface harm. Consequently, from a total of 9 samples studied for positive results, 3 of them appeared in the false-positive region. From all 9 samples studied for negative results, 4 of them appeared in the false-negative region. These problems can be overcome by increasing the population size of samples to be analyzed by our EEVD. Despite these issues and the relatively small population size, the obtained results indicated that the proposed EEVD can be a suitable device for point-of-care serologic diagnosis.
Lastly, we estimated possible interferents for IgG detections by G-PNR-AuNP/RBD in human sera. We evaluated human IgM, BSA and bilirubin oxidase enzyme (Mattioli et al., 2020). Experiments in 0.01 mol L-1 PBS pH 7.4 were performed and ΔVOCP shifts obtained for each studied biomolecule (1.0 μg mL-1 concentration) was compared to the ΔVOCP shift presented by the detection of 1.0 μg mL-1 IgG. The obtained results are illustrated in Figure S10. It is noticeable that human IgM presented the higher interference, as bilirubin oxidase and BSA did not interfere significantly. This tendency was expected as human IgM also has immunogenic activity against SARS-CoV-2 viral infections and can bind to RBD through antigen-antibody interactions. As BSA and bilirubin oxidase does not bind to RBD through this mechanism, low interference from these proteins was already expected. Despite of the interference for the proposed IgG detection method, these results gave us insights on future improvements, by considering IgM as another target analyte for total antibody serologic detections aiming COVID-19 diagnosis. This strategy is similar to what is already used by a number of immunoassays in literature (Carter et al., 2020; Imai et al., 2020; Montesinos et al., 2020; Wang et al., 2020).
Positive and negative tests with EEVD were compared with ELISA, based on RBD and human IgG specific interactions. (Fig. 4D). Nine replicates (n = 9) already evaluated by EEVDs were added in a n = 20 population, in order to minimize relative errors and measurement uncertainty (Biswas and Kumar Saha, 2015). Likewise, ELISA quantifications also presented false results, indicating that this may be originated from non-specific interactions that still happen in extra RBD active sites. Apart from this, all human sera samples with true-positive results (6 from a total of 9 samples) by EEVD recovered positive results by ELISA. True-negative COVID-19 diagnosis returned by IgG detections (5 of a total of 9 samples) by EEVDs also presented negative results by ELISA. Based on these results and validation methodologies from Brazilian Society of Laboratorial Medicine, Brazilian Society of Clinical Analyzes and Brazilian Society of Clinical Pathology (ABRAMED, 2021), it was possible to estimate a accuracy of 61.1% and sensitivity of 66.7% by considering a division of true positive and true negative samples by total of analyzed samples. The sensitivity obtained for our serological evaluation in real samples is lower than the values reported in literature, as presented in Table 1. However, it is expected that both sensitivity and accuracy values can be improved for analyses with larger populations. (Mattioli and Crespilho, 2020) Although false-positive and false-negative results were presented in both assays, it is possible to infer that a satisfactory accuracy could be achieved by employing EEVD in our proof-of-concept studies in real human serum samples.
4. Conclusions
A graphene-based EEVD was developed aiming serologic COVID-19 diagnosis applications. For EEVD fabrication, non-covalent functionalization with PNR was adopted in order to allow AuNP/RBD bioconjugate proper adsorption onto the device interface, without harming graphene sp2 lattice. EEVD working principles relied on the current passage through graphene basal plane with high charge carrier mobility. Monitoring interfacial potential (i.e., OCP) displacement with the adsorption of each EEVD modifier was made similarly to the procedures usually adopted in GFETs studies. The specific interaction of adsorbed RBD with IgG showed to be dependent on the IgG concentration, and therefore, an analytical calibration curve could be obtained. A LOD of 1.0 pg mL-1 was reached with a sensitivity of 5.8 mV per decade, within a wide linear dynamic range of IgG concentrations varying from 10-12 to 10-7 g mL-1. These results give us insights on the possibility of using our proposed devices for trace IgG detections, combined with their miniaturized features for rapid point-of-care purposes. Human blood serum samples were evaluated for positive and negative diagnosis of COVID-19 infection based on IgG quantifications, as a proof-of-concept of our device performance in real samples analyses. Although false-positive and false-negative results were obtained, our results showed a satisfactory accuracy of 61.1%, and a sensitivity of 66.7%. EEVD can be considered as promising alternative for COVID-19 serologic detections in large populations, since the device showed good sensitivity, low LOD, rapidness of analyses, robustness and accuracy. All these are desirable analytic parameters for assays destined to pandemics control strategies.
Author contributions
FNC and IAM conceived the original project. FNC, IAM, KRC, LJAM, GCS and MNO designed the experimental procedures. MNO, IT, PMV, SCF, ERM and GMP prepared human real samples. IAM, KRC, LJAM, GCS, MNO, IT, PMV, SCF, ERM and GMP collected the data. IAM, KRC, LJAM, GCS, MNO, IT, PMV, SCF, ERM, GMP, ECS and FNC analyzed the data. This manuscript was written and revised with contributions by all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the funding agencies: Sao Paulo Research Foundation (FAPESP) for all the financial support under the grants: 17/20493-2 (L.J.A.M.), 18/11071-0 (I.A.M.), 20/04796-8 (G.C.S.), 20/06752-8 and 20/05030-1 (M.N.O.) and 18/22214-6; 19/15333-1 e 19/12053-8 (F.N.C.); Coordinating Agency for Advanced Training of Graduate Personnel (CAPES), Metabolomics and Diagnostic Research Network of Covid-19 (MeDiCo Network), CAPES-Brazil grant number: 88881.504532/2020-01 and for the fellowship grant 88887.511448/2020-00 (K.R.C.); and National Council of Scientific and Technological Development (CNPq). Authors are grateful for Professor Edson Leite's immeasurable effort to Brazilian science during the pandemic, and thankful to Rui Murer for all electrical contacts deposition at LNNano, and CNPEM-Brazil for their facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113866.
Abbreviations
- EEVD
electrical-electrochemical vertical device
- IgG
Immunoglobulin G
- GFET
graphene field-effect transistor
- EE
Electrical-Electrochemical
- PNR
poly-neutral red
- CNP
charge neutrality point
- OCP
open-circuit potential
- EIS
electrochemical impedance spectroscopy
- CVD
carbon vapor deposition
- RBD
receptor binding domain
- LOD
limit of detection
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abramed, Sbac, Cbdl, Sbpc/Ml, 2021.
- Akbari Nakhjavani S., Afsharan H., Khalilzadeh B., Ghahremani M.H., Carrara S., Omidi Y. Biosens. Bioelectron. 2019;141:111439. doi: 10.1016/j.bios.2019.111439. [DOI] [PubMed] [Google Scholar]
- Ali M.A., Hu C., Jahan S., Yuan B., Saleh M.S., Ju E., Gao S.J., Panat R. Adv. Mater. 2021;33:1–15. doi: 10.1002/adma.202006647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Kumar Saha M. Immunochem. Immunopathol. 2015:1–5. 01. [Google Scholar]
- Bleu Y., Bourquard F., Loir A.S., Barnier V., Garrelie F., Donnet C. J. Raman Spectrosc. 2019;50:1630–1641. [Google Scholar]
- Brownson D.A.C., Varey S.A., Hussain F., Haigh S.J., Banks C.E. Nanoscale. 2014;6:1607–1621. doi: 10.1039/c3nr05643k. [DOI] [PubMed] [Google Scholar]
- Brunetti B D.E. Pharm. Anal. Acta. 2015 06. [Google Scholar]
- Brunner H., Mayer U., Hoffmann H. Appl. Spectrosc. 1997;51:209–217. [Google Scholar]
- Busch R.T., Karim F., Weis J., Sun Y., Zhao C., Vasquez E.S. ACS Omega. 2019;4:15269–15279. doi: 10.1021/acsomega.9b02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles A Janeway J., Travers P., Walport M., Shlomchik M.J. Immunobiol. Immune Syst. Heal. Dis. 2001;1–9 [Google Scholar]
- Chen W.H., Hotez P.J., Bottazzi M.E. Hum. Vaccines Immunother. 2020;16:1239–1242. doi: 10.1080/21645515.2020.1740560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson E., Sontakke V.A., Porkovich A.J., Wang Z., Kumar P., Ziadi Z., Yokobayashi Y., Sowwan M. Sensor. Actuator. B Chem. 2020;320:128432. [Google Scholar]
- de Oliveira T.R., Martucci D.H., Faria R.C. Sensor. Actuator. B Chem. 2018;255:684–691. [Google Scholar]
- Devineau S., Anyfantakis M., Marichal L., Kiger L., Morel M., Rudiuk S., Baigl D. J. Am. Chem. Soc. 2016;138:11623–11632. doi: 10.1021/jacs.6b04833. [DOI] [PubMed] [Google Scholar]
- Filik H., Aslıhan Avan A. Microchem. J. 2020;158:105242. [Google Scholar]
- Georgakilas V., Tiwari J.N., Kemp K.C., Perman J.A., Bourlinos A.B., Kim K.S., Zboril R. Chem. Rev. 2016;116:5464–5519. doi: 10.1021/acs.chemrev.5b00620. [DOI] [PubMed] [Google Scholar]
- Guo S., Wang E. Anal. Chim. Acta. 2007;598:181–192. doi: 10.1016/j.aca.2007.07.054. [DOI] [PubMed] [Google Scholar]
- Hashemi S.A., Bahrani S., Mousavi S.M., Omidifar N., Behbahan N.G.G., Arjmand M., Ramakrishna S., Lankarani K.B., Moghadami M., Shokripour M., Firoozsani M., Chiang W.-H. J. Electroanal. Chem. 2021;894:115341. doi: 10.1016/j.jelechem.2021.115341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A., Macedo L.J.A., Mattioli I.A., Rubira R.J.G., Constantino C.J.L., Amorim R.G., Lima F.C.D.A., Crespilho F.N. Electrochim. Acta. 2021;376:138025. [Google Scholar]
- Imai K., Tabata S., Ikeda M., Noguchi S., Kitagawa Y., Matuoka M., Miyoshi K., Tarumoto N., Sakai J., Ito T., Maesaki S., Tamura K., Maeda T. J. Clin. Virol. 2020;128:104393. doi: 10.1016/j.jcv.2020.104393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inczedy J., Lengyel T., Ure A.M. 1998. International Union of Pure and Applied Chemistry. [Google Scholar]
- Iost R.M., Crespilho F.N., Zuccaro L., Yu H.K., Wodtke A.M., Kern K., Balasubramanian K. ChemElectroChem. 2014;1:2070–2074. [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., Pu A., Christie-Holmes N., Gervais C., Ceccarelli D., Samavarchi-Tehrani P., Guvenc F., Budylowski P., Li A., Paterson A., Yun Y.F., Marin L.M., Caldwell L., Wrana J.L., Colwill K., Sicheri F., Mubareka S., Gray-Owen S.D., Drews S.J., Siqueira W.L., Barrios-Rodiles M., Ostrowski M., Rini J.M., Durocher Y., McGeer A.J., Gommerman J.L., Gingras A.C. Sci. Immunol. 2020;5:1–14. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Feng B., Xu J., Qing T., Zhang P., Qing Z. Biosens. Bioelectron. 2020;166:112471. doi: 10.1016/j.bios.2020.112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudr J., Michalek P., Ilieva L., Adam V., Zitka O. TrAC Trends Anal. Chem. (Reference Ed.) 2021;136:116192. doi: 10.1016/j.trac.2021.116192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. 2020. Nature. [DOI] [PubMed] [Google Scholar]
- Lew T.T.S., Aung K.M.M., Ow S.Y., Amrun S.N., Sutarlie L., Ng L.F.P., Su X. ACS Nano. 2021 doi: 10.1021/acsnano.1c04091. [DOI] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Yan H., Huang L., Zhao J., Wang H., Kang M., Wan Z., Shui J., Tang S. J. Virol. Methods. 2021;292:114141. doi: 10.1016/j.jviromet.2021.114141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Sensor. Actuator. B Chem. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., May K. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li Q., Zou Y., Qian Q., Jin Y., Li G., Jiang K., Fan S. Nano Lett. 2013;13:6170–6175. doi: 10.1021/nl4035048. [DOI] [PubMed] [Google Scholar]
- Macedo L.J.A., Iost R.M., Hassan A., Balasubramanian K., Crespilho F.N. ChemElectroChem. 2019;6:31–59. [Google Scholar]
- Macedo L.J.A., Lima F.C.D.A., Amorim R.G., Freitas R.O., Yadav A., Iost R.M., Balasubramanian K., Crespilho F.N. Nanoscale. 2018;10:15048–15057. doi: 10.1039/c8nr03893g. [DOI] [PubMed] [Google Scholar]
- Malard L.M., Pimenta M.A., Dresselhaus G., Dresselhaus M.S. Phys. Rep. 2009;473:51–87. [Google Scholar]
- Mattioli Isabela A., Crespilho F.N. 2020. CC-PI-2020–0101. [Google Scholar]
- Mattioli Isabela Alteia, Crespilho F.N. An. Acad. Bras. Cienc. 2020:1–3. doi: 10.1590/0001-3765202020201208. [DOI] [PubMed] [Google Scholar]
- Mattioli I.A., Hassan A., Oliveira O.N., Crespilho F.N. ACS Sens. 2020 doi: 10.1021/acssensors.0c01382. [DOI] [PubMed] [Google Scholar]
- Mattioli I.A., Hassan A., Sanches N.M., Vieira N.C.S., Crespilho F.N. Biosens. Bioelectron. 2021;175:112851. doi: 10.1016/j.bios.2020.112851. [DOI] [PubMed] [Google Scholar]
- Mazar F.M., Alijanianzadeh M., Molaeirad A., Heydari P. Process Biochem. 2017;56:71–80. [Google Scholar]
- Minamiki T., Minami T., Kurita R., Niwa O., Wakida S.I., Fukuda K., Kumaki D., Tokito S. Materials (Basel) 2014;6:6843–6852. doi: 10.3390/ma7096843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos I., Gruson D., Kabamba B., Dahma H., Wijngaert S. Van Den, Reza S., Carbone V., Vandenberg O., Gulbis B., Wolff F., Rodriguez-Villalobos H. J. Clin. Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauliukaite R., Brett C.M.A. Electroanalysis. 2008;20:1275–1285. [Google Scholar]
- Pauliukaite R., Ghica M.E., Barsan M., Brett C.M.A. J. Solid State Electrochem. 2007;11:899–908. [Google Scholar]
- Pingarrón J.M., Yáñez-Sedeño P., González-Cortés A. Electrochim. Acta. 2008;53:5848–5866. [Google Scholar]
- Prakash A., Pathrose B.P., Mathew S., Nampoori V.P.N., Radhakrishnan P., Mujeeb A. Opt. Mater. (Amst). 2018;79:237–242. [Google Scholar]
- Prasad K.S., Cao X., Gao N., Jin Q., Sanjay S.T., Henao-Pabon G., Li X.J. Sensor. Actuator. B Chem. 2020;305:127516. doi: 10.1016/j.snb.2019.127516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A.J., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C.E., Weimer E., Scherer E.M., Rouphael N., Edupuganti S., Weiskopf D., Tse L.V., Hou Y.J., Margolis D., Sette A., Collins M.H., Schmitz J., Baric R.S., de Silva A.M. Sci. Immunol. 2020;5:1–10. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Z., Li Yacheng, Li Younan, Luo G., Hu J., Zou Z., Lei Y., Liu J., Yang R. Microchim. Acta. 2020;187 doi: 10.1007/s00604-020-04486-2. [DOI] [PubMed] [Google Scholar]
- Qing Z., Luo G., Xing S., Zou Z., Lei Y., Liu J., Yang R. Angew. Chem. Int. Ed. 2020;59:14044–14048. doi: 10.1002/anie.202003964. [DOI] [PubMed] [Google Scholar]
- Raffle A.E., Pollock A.M., Harding-edgar L. BMJ. 2020:10–11. doi: 10.1136/bmj.m3262. [DOI] [PubMed] [Google Scholar]
- Ragavendar M.S., Anmol C.M. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS. 2012:2408–2411. doi: 10.1109/EMBC.2012.6346449. [DOI] [PubMed] [Google Scholar]
- Raoof M., Jans K., Bryce G., Ebrahim S., Lagae L., Witvrouw A. Microelectron. Eng. 2013;111:421–424. [Google Scholar]
- Reddy D., Register L.F., Carpenter G.D., Banerjee S.K. J. Phys. D. Appl. Phys. 2012;45 [Google Scholar]
- Roy E., Patra S., Tiwari A., Madhuri R., Sharma P.K. Biosens. Bioelectron. 2017;89:234–248. doi: 10.1016/j.bios.2016.02.056. [DOI] [PubMed] [Google Scholar]
- Sedenho G.C., Hassan A., Macedo L.J.A., Crespilho F.N. J. Power Sources. 2021;482:229035. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S. Il. ACS Nano. 2020 [Google Scholar]
- Sun B., Feng Y., Mo X., Zheng P., Wang Q., Li P., Peng P., Liu X., Chen Z., Huang H., Zhang Fan, Luo W., Niu X., Hu P., Wang L., Peng H., Huang Z., Feng L., Li Feng, Zhang Fuchun, Li Fang, Zhong N., Chen L. Emerg. Microb. Infect. 2020:1–36. doi: 10.1080/22221751.2020.1762515. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh L., Brahman P.K., Reddy K.R., Bondili J.S. Enzym. Microb. Technol. 2018;112:43–51. doi: 10.1016/j.enzmictec.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Theel E.S., Harring J., Hilgart H., Granger D. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tré-Hardy M., Wilmet A., Beukinga I., Favresse J., Dogné J.M., Douxfils J., Blairon L. J. Med. Virol. 2020;93:803–811. doi: 10.1002/jmv.26303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkevich J., Stevenson P.C., Hillier J. Discuss. Faraday Soc. 1951;55:55–75. [Google Scholar]
- Vieira N.C.S., Borme J., Machado G., Cerqueira F., Freitas P.P., Zucolotto V., Peres N.M.R., Alpuim P. J. Phys. Condens. Matter. 2016;28 doi: 10.1088/0953-8984/28/8/085302. [DOI] [PubMed] [Google Scholar]
- Vu C.A., Pan P.H., Yang Y.S., Chan H.W.H., Kumada Y., Chen W.Y. Sensors (Switzerland) 2021;21:1–12. doi: 10.3390/s21020650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.H., Jin Z., Kim K.K., Hilmer A.J., Paulus G.L.C., Shih C.J., Ham M.H., Sanchez-Yamagishi J.D., Watanabe K., Taniguchi T., Kong J., Jarillo-Herrero P., Strano M.S. Nat. Chem. 2012;4:724–732. doi: 10.1038/nchem.1421. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zheng Z., Hu H., Zhou Q., Liu W., Li X., Liu Z., Wang Y., Ma Y. Lab Chip. 2020;20:4255–4261. doi: 10.1039/d0lc00828a. [DOI] [PubMed] [Google Scholar]
- Wen T., Huang C., Shi F.J., Zeng X.Y., Lu T., Ding S.N., Jiao Y.J. Analyst. 2020;145:5345–5352. doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- Who . 2020. Situation by WHO Region. [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:11912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Kroe-Barrett R., Singh S., Laue T. Antibodies. 2019;8:24. doi: 10.3390/antib8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Jingyun, Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang Guanpeng, Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang Jinliang, Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.S., Dai L., Wu M., Li G., Wang Guangyu, Peng Y., Qian Z., Huang C., Lau J.Y.N., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Zeng L., Li Y., Liu J., Guo L., Wang Z., Xu X., Song S., Hao C., Liu L., Xin M., Xu C. Mater. Chem. Front. 2020 [Google Scholar]
- Zhang Y., Liu X., Wang L., Yang H., Zhang X., Zhu C., Wang W., Yan L., Li B. Sci. Rep. 2020;10:9604. doi: 10.1038/s41598-020-66456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Murali S., Cai W., Li X., Suk J.W., Potts J.R., Ruoff R.S. Adv. Mater. 2010;22:3906–3924. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.