Abstract
With rapid transmission, the coronavirus disease 2019 (COVID-19) has led to over three million deaths worldwide, posing significant societal challenges. Understanding the spatial patterns of patient visits and detecting local cluster centers are crucial to controlling disease outbreaks. We analyze COVID-19 contact tracing data collected from Seoul, which provide a unique opportunity to understand the mechanism of patient visit occurrence. Analyzing contact tracing data is challenging because patient visits show strong clustering patterns, while cluster centers may have complex interaction behavior. Cluster centers attract each other at mid-range distances because other cluster centers are likely to appear in nearby regions. At the same time, they repel each other at too small distances to avoid merging. To account for such behaviors, we develop a novel interaction Neyman–Scott process that regards the observed patient visit events as offsprings generated from a parent cluster center. Inference for such models is challenging since the likelihood involves intractable normalizing functions. To address this issue, we embed an auxiliary variable algorithm into our Markov chain Monte Carlo. We fit our model to several simulated and real data examples under different outbreak scenarios and show that our method can describe the spatial patterns of patient visits well. We also provide useful visualizations that can inform public health interventions for infectious diseases, such as social distancing.
Keywords: Infectious disease, Doubly-intractable distributions, Cluster point process, Bayesian hierarchical model, Markov chain Monte Carlo
1. Introduction
Caused by the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the coronavirus disease 2019 (COVID-19) was first reported in December 2019 in Wuhan, Hubei province, China (WHO, 2020). By May 2021, there have been about 150 million confirmed cases of COVID-19, with more than three million deaths. The disease spreads more quickly than influenza, mainly through close contact with infected people (CDC, 2020). Human-to-human transmission is most common for COVID-19, primarily via respiratory droplets and aerosols from an infected person. Thus, contact tracing, which records the travel history of patients, can be an effective way to gain insights into disease prevalence. Spatial point process models for contact tracing data can be useful to study the probabilistic mechanism of the spread of diseases. Here, we propose a new point process model that can provide useful epidemiological information such as a warning system for local hotspots.
We analyze the locations visited by confirmed patients in Seoul, South Korea. We refer to a “confirmed patient” as a person who tested positive through the polymerase chain reaction (PCR) test, which is considered to be the standard procedure to identify COVID-19 patients because of its high accuracy. The visited locations are first identified through patient interviews and then verified through other measures. The data set contains the thorough contact histories of confirmed patients, collected through a rigorous contact tracing protocol (Park et al., 2020), and hence provides a unique source for point process modeling. We describe the details of the contact tracing protocol in Section 2.
We regard each visit in the contact tracing data as an event in the point process model. Since such events are spatially clustered, a natural way to model this is the Neyman–Scott point process (Neyman and Scott, 1952), which considers observed events as offsprings generated around unobserved parents (cluster centers). In the epidemiological context, we can regard a parent point as a cluster center of disease and offsprings as the corresponding clusters (i.e., patient visits).
Several computational methods have been developed for inference for point processes, including a minimum contrast method (Diggle, 2013) and pseudolikelihood approximation (Guan, 2006, Diggle et al., 2010). However, such approaches are sensitive to the choice of tuning parameters and may not be accurate in the presence of strong spatial dependence among points. Through simulation studies, Mrkvička et al. (2014) report that Bayesian estimation is the most precise for the Neyman–Scott point process. Furthermore, we can easily adjust the priors based on epidemiological knowledge. Therefore, the Bayesian framework can be practical for fitting hierarchical point process models.
A simple Neyman–Scott process has limited applicability here because it assumes an independent parent process. Local cluster centers (parents) may have some interaction behavior, leading to an intractable likelihood function, which is common in many spatial point process models. Several extensions for Neyman–Scott processes have been developed. Waagepetersen, 2007, Mrkvička et al., 2014, Mrkvička and Soubeyrand, 2017 study inference for inhomogeneous Neyman–Scott point processes. Yau and Loh (2012) consider a special case where the parent process exhibits repulsion. Albert-Green et al. (2019) generalize the Neyman–Scott process by allowing parents to follow the log-Gaussian Cox process (Møller et al., 1998). However, these existing models may not be appropriate for COVID-19 data because of the complex interactions of cluster centers. Unobserved local cluster centers attract each other because other cluster centers are likely to appear in nearby communities.
However, an interaction point process only with pairwise attraction is not well defined in general (Moller and Waagepetersen, 2003) due to clumping behavior; the integral of the intensity of the process becomes infinite because adding a new point always increases intensity in the nearby area, which in turn encourages the generation of another point. This creates a loop of positive feedback, leading to infinite intensity in the target area. Therefore, more sophisticated models are often used to consider both attractive and repulsive interactions (Geyer, 1999, Goldstein et al., 2015). By adapting these models, we allow local cluster centers to attract each other at mid-range distances, while make them repel each other at too small distances for a valid model specification.
In this manuscript, we propose a novel parametric approach that exhibits interaction between local clusters. This new approach can infer model parameters that provide guidelines for controlling the spread of disease. Our model allows us to detect unobserved cluster centers, thereby providing a practical warning system for coronavirus hotspots. Recently, Goldstein et al. (2015) proposed an attraction–repulsion point process model by extending the Strauss process (Strauss, 1975) that only allows repulsion behavior. Russell et al. (2016) use such a process to model different types of interactions among animals, including herding behavior and collision avoidance. Adopting this idea, we incorporate the attraction–repulsion (Goldstein et al., 2015) process into the parent process. Our model includes an intractable normalizing function, posing computational and inferential challenges. To address this, we use a novel auxiliary variable Markov chain Monte Carlo (MCMC) (Møller et al., 2006, Liang, 2010) that can avoid the direct evaluation of intractable normalizing functions.
The remainder of this paper is organized as follows. In Section 2, we describe the COVID-19 contact tracing data in Seoul, South Korea. In Section 3, we introduce a new interaction Neyman–Scott process model and discuss its computational and inferential challenges. We describe our MCMC algorithm for this model and provide implementation details. In Section 4, we apply our methods to both simulated and real COVID-19 data sets. We show that our methods can detect the sources of cluster centers and describe the spatial patterns of patient visits. Furthermore, we provide a disease risk map that can give important epidemiological interpretations. In Section 5, we conclude the paper with a discussion and summary.
2. Data description and exploratory analysis
In this section, we provide the background of our contact tracing data and summarize the results from the exploratory data analysis.
2.1. COVID-19 contact tracing data
We study the COVID-19 contacting tracing data in Seoul, which disclose the detailed locations visited by confirmed patients. When a patient is confirmed to be infected by a screening clinic through a PCR test, he or she is immediately transported to a hospital dedicated to treating infectious disease. Once patients are admitted, they are interviewed by an epidemiological investigator, and their visited location history is thoroughly recorded. For asymptomatic cases, the travel history since a possible exposure to an infection source is recorded. For symptomatic cases, the travel history from two days before the symptoms’ onset is recorded. The collected tracing information is anonymized and disclosed for two weeks on local government websites and then archived. This code follows the “Response Guidelines to Prevent the Spread of COVID-19 (for local government)” (KDCA, 2020). The contact tracing data sets used in this study were collected directly from these websites.
Table 1 shows the locations visited by multiple confirmed patients. For example, the first row in Table 1 indicates that a patient was confirmed as infected by a screening clinic on March 7. However, the confirmation dates for the other patients are different because Table 1 provides records from multiple patients. Note that we do not use any personal identification information, including the patient ID in the record for each visited location, to avoid possible privacy violations. Fig. 1 illustrates a graphical representation of the visited locations in Table 1. To study the spatial patterns of patient visits, we regard each visit (coordinate) as a realization of a spatial point process.
Table 1.
Example locations visited by confirmed patients. The X and Y coordinates are the World Geodetic System 1984 (WGS84) coordinates.
| Date time | Address | Description | X | Y |
|---|---|---|---|---|
| 0307 | Chunhodaero 145, Seoul | Screening Clinic | 959371 | 1952879 |
| 0302 | Gyungheedaero 7-1,Seoul | Café | 960469 | 1954862 |
| 0302 | HanChunro 58, Seoul | House | 962047 | 1956113 |
| 0301 | Hoegiro 19, Seoul | Café | 960541 | 1954855 |
| 0301 | Hoegiro 18, Seoul | Restaurant | 960500 | 1954704 |
| 0301 | Imunro 37, Seoul | Café | 960812 | 1954692 |
| 0301 | HanChunro 58, Seoul | House | 962047 | 1956113 |
| 0229 | Shinimunro 40, Seoul | Hospital | 961735 | 1956027 |
| 0229 | Shinimunro 24, Seoul | Pharmacy | 961718 | 1956025 |
| 0229 | Hoegiro 21, Seoul | Restaurant | 960558 | 1954887 |
| 0229 | Imunro 37, Seoul | Café | 960812 | 1954692 |
| 0229 | Hwigyeongro 2, Seoul | Café | 961181 | 1955099 |
| 0229 | HanChunro 58, Seoul | House | 962047 | 1956113 |
Fig. 1.
An illustration of the locations visited by confirmed patients within the Dongdaemun-gu area (an east center district of Seoul). Different colors indicate different confirmed patients. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In this paper, at any given time point, we fit a point process model based on the contact tracing data accumulated over the last two weeks. According to the basic data analysis for the early stages of the COVID-19 spread in South Korea and China, two weeks is the mean period of recovery from infection (Ki, 2020, Choi and Ki, 2020). Two weeks is also the period for epidemic investigations conducted by the local health authority for all confirmed cases. Therefore, when we construct a warning system for COVID-19 at a certain time, modeling patient visits in the last two weeks would be the most useful. In particular, we focus on three different scenarios that end on March 19th, April 2nd, and April 15th, which can be considered as severe, moderate, and mild outbreak cases, respectively, in terms of overall visit numbers. Our goal here is to examine how our model works and provide important epidemiological insights for these different scenarios with various disease spreading patterns.
2.2. Exploratory data analysis
Spatial point processes provide a natural solution to model the spatial patterns of the locations visited by confirmed patients. Here, we provide the motivation for a new interaction point process model with an exploratory data analysis. Let be the realization of the point process over the bounded spatial domain . The pair correlation function (PCF) is useful for exploring point process observations (Stoyan and Stoyan, 1994). The PCF is defined as
where is the area of the spatial domain. Here, Ripley’s is the expected density of the points within distance . Under complete spatial randomness, , which results in . indicates that points have a tendency to cluster at a distance (attraction), while indicates that points tend to remain apart at a distance (repulsion).
Fig. 2 illustrates the spatial pattern in the severe case period (ending on March 19th) and its PCF. We observe that patient visits are spatially clustered (). Therefore, a point process model for the data should capture such behavior. The Neyman–Scott process (Neyman and Scott, 1952) is widely used to study spatially aggregated point patterns. Furthermore, the Neyman–Scott process can detect cluster centers, which can be useful for identifying local hotpots. Consider the Neyman–Scott process , where is the offspring and is the parent (cluster centers). Given parent process , offspring follows an independent Poisson process with intensity . Here, controls the spread of offsprings around their parent and determines the expected number of offsprings per each cluster. With the Gaussian kernel with mean and variance , is called the Thomas process (Thomas, 1949). The basic Neyman–Scott process models the unobserved parent process as a simple Poisson process.
Fig. 2.
The left panel shows the locations visited by patients (March 6–March 19, 2020) within the Seoul area. The right panel shows the estimated PCF from the observation. The red dahsed line indicates the theoretical PCF under complete spatial randomness. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The Neyman–Scott process is appropriate for modeling COVID-19 data because the locations visited by confirmed patients, , are clustered around the unobserved center . From this, we can detect the local cluster centers of COVID-19. However, may also have complex spatial interactions. Naturally, cluster centers tend to clump together because other cluster centers are likely to exist in a nearby region. However, a purely attractive pairwise interaction process is not well defined (Moller and Waagepetersen, 2003) in general because of the degeneracy issue; when a new point is added to an existing set of points, the probability of having another point in the nearby area will increase. This creates a loop of positive feedback and the process will create an infinite number of points; as a result, a process only with pairwise attraction will have an infinite integrated intensity over the study area. To prevent this phenomenon, incorporating repelling behavior at too small in the model is necessary. The basic Neyman–Scott process cannot describe such behavior because the basic model considers an independent Poisson process for . In the following section, we add another layer of complexity to the basic Neyman–Scott process. This new model can provide epidemiological interpretations for understanding local clusters of COVID-19.
3. An interaction Neyman–Scott point process model
In this section, we propose a new Neyman–Scott process by incorporating interaction behavior into . This new parametric approach can detect unobserved cluster centers of COVID-19 and describe their patterns. Based on this, we can provide a warning system for higher-risk regions.
3.1. Model
Consider the realization of Neyman–Scott point process in the bounded plane (Seoul domain). This indicates the observed locations visited by confirmed patients. Let be their unobserved parent process. In our context, each is the center of the kernel that captures the local intensity of point processes. The kernel centers tend to remain apart (repulsion) at small distances to avoid the degeneracy issue. At mid-range distances, they tend to clump together (attraction) because other cluster centers are likely to appear in nearby areas. Cluster centers become independent at sufficiently large distances. To describe such patterns, we model the parent in the Neyman–Scott process as an attraction–repulsion interaction point process (Goldstein et al., 2015). The locations of the unobserved center of the parent process are modeled as
| (1) |
where denotes the Euclidean distance between two parent points and . is a normalizing function that makes as a valid probability density function. The interaction function can be defined as
| (2) |
Here, are set to make the interaction function continuously differentiable (Goldstein et al., 2015). The interaction function is determined by the distance between two parent points, and . There are three parameters in this model, : controls the overall intensity of the parent process and control the shape of the interaction function . provides the peak value of , whereas provides the location of the peak value. Examples of under different parameter values are illustrated in Fig. 3.
Fig. 3.
Illustration of the attraction–repulsion interaction function in Goldstein et al. (2015). The left panel shows the effect of changing the peak value parameter . The right panel shows the effect of changing the location parameter .
When the distance between local cluster centers becomes too small, is less than 1, which means and show repulsion behavior. On the other hand, as becomes larger, is increased, which means local cluster centers show attraction behavior at mid-range distances. Such attractions become weaker as the distance between cluster centers becomes larger. From this, (1) can describe the attraction–repulsion behavior of local cluster centers.
Let be the density of the patient visits with respect to the unit Poisson point process in . Given the local cluster centers (parents), the locations visited by confirmed patients can be modeled as
| (3) |
We use a symmetric Gaussian kernel with a center at each and variance . This results in a higher intensity of patient visits around the cluster centers. In our context, controls the expected number of confirmed patients for each and controls the width of cluster events activity.
3.2. Computational and inferential challenges
The proposed model in the previous section has a hierarchical structure. Unobserved local cluster centers (parent process) follow the spatial interaction process defined in (1), (2). Observed patient visits (offsprings) are distributed around the unobserved parents with Gaussian kernels. The Bayesian framework is useful for such hierarchical models, as it can easily quantify the model parameters’ uncertainty using MCMC. With priors , the joint posterior distribution is
| (4) |
Although (4) is a natural way to construct a joint posterior distribution, there are computational and inferential challenges to fitting such a model due to intractable normalizing functions. The model for cluster centers in (1) can be written as
| (5) |
Here, the calculation of normalizing function requires integration over spatial domain , which is infeasible. Inference for such models is challenging because an intractable is a function of the model parameters. The resulting posterior (4) is referred to as a doubly-intractable distribution (Murray et al., 2006).
Several computational methods have been proposed to address this issue. By assuming the conditional independence of points, Besag (1974) proposed pseudolikelihood that does not have intractable normalizing functions. Due to its convenience, pseudolikelihood approximations are often used in many point process applications (e.g., Diggle et al., 2010, Tamayo-Uria et al., 2014). However, it is well known that such approximations are unreliable when there is strong dependence among points (Mrkvička et al., 2014). Geyer and Thompson (1992) propose theoretically justified Monte Carlo maximum likelihood methods, which maximize a Monte Carlo approximation to the likelihood. However, this has limited applicability because the algorithm requires gradients for , which is not available for interaction point process models such as the one defined above. Bayesian approaches can be an alternative for such cases. Several Bayesian methods have been proposed for doubly-intractable distribution (see Park and Haran (2018) for a comprehensive review). Among current approaches, Park and Haran (2018) report that double Metropolis–Hastings (DMH) is the most practical method for point process models. DMH can avoid the calculation of and alleviate memory issues, which can be problematic for adaptive algorithms (Atchade et al., 2008, Liang et al., 2016). In the following section, we formulate a DMH that can carry out Bayesian inference for an interaction Neyman–Scott point process model.
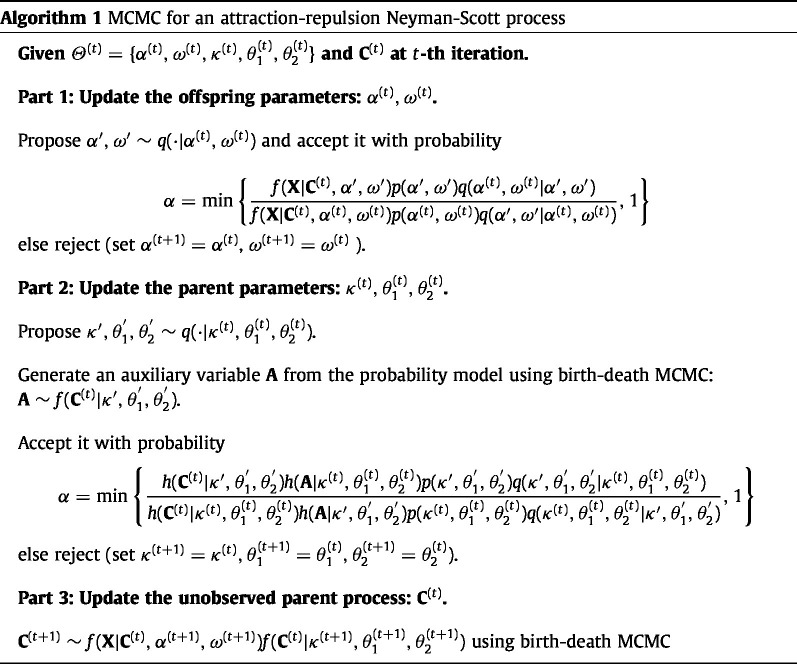
3.3. Markov chain Monte Carlo
Here, we describe an MCMC algorithm for the attraction–repulsion Neyman–Scott point process model. Consider the model parameters and latent parent process in (4) at the th iteration. We update the parameters successively. The offspring parameters can be updated from
with a joint prior density . We can update parent parameters from
with a joint prior density (see Section 4.1 for how we choose the priors in our problem). Compared with updating the offspring parameters, updating the parent parameters is challenging because of the intractable normalizing function in (5). Intractable leads to the intractable acceptance probability in MCMC as
| (6) |
for the proposed parameters , and . To avoid the direct evaluation of the intractable normalizing functions in , we incorporate double Metropolis–Hastings (DMH) (Liang, 2010) into our MCMC algorithm. Instead of evaluating the intractable normalizing functions in , the DMH sampler generates an auxiliary variable from (1) via a birth–death MCMC (Moller and Waagepetersen, 2003). We provide the details of the birth–death MCMC in the supplementary material. For given a proposed parent parameters (), the birth–death MCMC iteration is repeated times to generate an auxiliary variable . Theoretically, we can simulate exactly as ; of course, should be finite in practice. We observe that the acceptance rate of each birth–death iteration is 0.87 on average, which is natural because we propose to change only a single point (birth, death, or move). In DMH (Liang, 2010), the last state of the resulting birth–death MCMC (i.e., th realization) is regarded as an exact draw from . Following the recommendation in Liang (2010) and Park and Haran (2018), we set to be proportional to the dimension of the parent process , which is about in our applications. We show that this choice of can generate reasonably accurate Bayesian inference in simulated and real data examples. Note that the auxiliary variable is from the same probability model as and can be regarded as synthetic point process data. The acceptance probability of DMH is now given as
| (7) |
This does not include intractable terms because the introduction of modifies the original densities in (6) by multiplying into the numerator and into the denominator. If the simulated auxiliary variable resembles the parent process , the proposed parent parameters () are likely to be accepted. Therefore, the realistic simulation of is important for accepting the parent parameters. However, the auxiliary variable does not affect the acceptance of the other components (offspring parameters, ) because the conditional posterior distributions of the other components are not dependent on intractable .
Finally, we obtain from
using a birth–death MCMC. Note that stationary distribution of the birth–death MCMC algorithm for updating is different from that of generating the auxiliary variable in DMH. The stationary distribution for generating the auxiliary variable follows (1) (see the supplementary material for details). Our MCMC algorithm is summarized in Algorithm 1.
4. Application
In this section, we apply our approach to simulated and real data examples. The computer code for this is implemented in R and C++ using the Rcpp and RcppArmadillo packages (Eddelbuettel et al., 2011).
4.1. Prior specification
To complete the posterior specification we now explicitly define the prior distributions. In the Neyman–Scott process, there is a strong dependence between and . The same observed pattern can be regarded as the points generated from a small number of parents, with each having a large number of offsprings (large , large ), or they may come from a large number of parents, with each having a small number of offsprings (small , small ). To avoid such identifiability issues, we need to set the upper and lower bounds for them using uniform priors (Møller and Waagepetersen, 2007, Kopeckỳ and Mrkvička, 2016).
Here, we focus on modeling cluster centers at the Dong scale (the smallest administrative district). To this end, we set the lower and upper bounds for based on the average size of a Dong. This encourages that patient visits from different Dongs come from different cluster centers. Therefore, we set the range of to . We assume that each cluster center (parent) generates at least 3 and at most 30 patient visits (offsprings) in a Dong on average. This choice of priors can avoid identifiability issues in the Neyman–Scott point process while maintaining interpretability and modeling flexibility.
The prior range of needs to be determined in consideration of the overall intensity and offspring intensity . The overall intensity is around , computed by dividing the average number of patient visits within two weeks by the area of Seoul . The average number of patient visits is computed over about 16 weeks (02/20/2020-06/11/2020). Since controls for the intensity of the parent process and controls for the average number of offsprings per each parent, should be around the overall intensity . By specifying the prior of to be uniform with the range , belongs to the range so that the resulting range can include the overall intensity .
Similar to , we set a uniform prior for whose upper and lower bounds are proportional to the average size of a Dong. We use a uniform prior with the range so that gives the location of the peak value within a Dong. To allow for attraction, we set the prior of to be uniform with the range , as in Goldstein et al. (2015).
4.2. Simulated data
We now conduct a simulation study to validate our method. We first simulate the parent process from (1) with the true parent parameters and . We then simulate the offsprings from (3) centered on each parent point with the true offspring parameters and . We consider three different scenarios here: In Scenario 1, we emulate COVID-19 data with a severe outbreak. This has numerous local cluster centers () and each of them also generates a large number of offsprings. This results in patient visits in total. In Scenario 2, we consider a moderate outbreak. This has fewer cluster centers () and each generates a moderate number of offsprings; the simulated patient visits consisted of points. Finally, in Scenario 3, we consider a mild outbreak in which we have the smallest number of cluster centers () and patient visits ().
Table 2 indicates that the true parameter values used in all the scenarios are estimated with reasonable accuracy. We also compare the true and recovered parent points in Fig. 4. We obtain the recovered parent points from the last iteration of the MCMC algorithm (i.e., in Algorithm 1). We observe that our method can detect cluster centers well as there are good agreements between the true and fitted parent points in all the scenarios.
Table 2.
Inference results for simulated data sets. 100,000 MCMC samples are generated, discarding the first 50,000 as burn-in. Posterior mean estimates are reported for each parameter. The highest posterior density (HPD) is calculated using the coda package.
| Scenario 1 | |||||
| Truth | 6 | 360 | 1.2 | 1.5 | 600 |
| Estimates | 5.70 | 370.34 | 1.13 | 1.85 | 626.81 |
| 95%HPD | (4.79, 6.62) | (356.27, 390.39) | (0.39, 2.05) | (1.25, 2.67) | (500.02, 798.47) |
| Scenario 2 | |||||
| Truth | 5 | 400 | 1.0 | 1.5 | 650 |
| Estimates | 5.58 | 400.02 | 1.18 | 1.40 | 684.59 |
| 95%HPD | (4.72, 6.52) | (373.65, 427.80) | (0.60, 1.79) | (1.00, 1.93) | (500.02, 937.61) |
| Scenario 3 | |||||
| Truth | 4 | 440 | 0.5 | 1.5 | 700 |
| Estimates | 4.12 | 423.73 | 0.75 | 1.64 | 716.71 |
| 95%HPD | (3.20, 5.02) | (387.68, 461.51) | (0.34, 1.22) | (1.03, 2.36) | (500.03, 941.41) |
Fig. 4.
Comparisons between the true (circle) and recovered (triangle) parent points for the different scenarios.
We repeat the simulation 100 times for each scenario to investigate whether the credible intervals can appropriately cover the true parameters. Table 3 indicates that coverage of the parameters in the different scenarios is reasonably close to the 95% nominal rate. Note that obtaining an exact frequentist coverage of Bayesian credible intervals can be problem-specific. It might largely depend on the choice of priors or can only be achieved with an increasing sample size. A full-fledged study of these properties for an interaction Neyman–Scott process would be an interesting research direction.
Table 3.
Coverage for the simulated data sets under the different scenario.
| Coverage | |||||
|---|---|---|---|---|---|
| Scenario 1 | 0.91 | 0.97 | 0.99 | 0.98 | 0.99 |
| Scenario 2 | 0.89 | 0.90 | 1.00 | 0.95 | 1.00 |
| Scenario 3 | 0.92 | 0.90 | 0.99 | 1.00 | 1.00 |
4.3. COVID-19 contact tracing data analysis
Parameter interpretation.
We now apply our method to the real observational data described in Section 2.1. Algorithm 1 takes about 2.5 h for 100,000 MCMC iterations. The parameter estimation results for the time periods that end at March 19th, April 2nd, and April 15th are summarized in Table 4. These three different periods can be regarded as severe, moderate, and mild outbreaks, respectively, in terms of the number of patient visits. The average number of offspring points per each parent () and intensity of the parent process () change as the overall number of patient visits changes; that is, more overall events lead to higher and values. The width of the Gaussian kernel controlled by , on the other hand, stays similar. For April 15th, when the total number of patient visits is relatively small (), takes a lower value as well.
Table 4.
Inference results for real data. 100,000 MCMC samples are generated, discarding the first 50,000 as burn-in.
| Severe | |||||
|---|---|---|---|---|---|
| Estimates | 5.42 | 357.40 | 1.33 | 1.72 | 547.43 |
| 95%HPD | (4.69, 6.11) | (356.26, 359.63) | (0.57, 2.06) | (1.31, 2.21) | (500.00, 638.49) |
| Moderate | |||||
| Estimates | 4.61 | 357.44 | 1.00 | 1.92 | 553.22 |
| 95%HPD | (3.88, 5.25) | (356.26, 359.74) | (0.43, 1.71) | (1.37, 2.58) | (500.02, 646.51) |
| Mild | |||||
| Estimates | 4.08 | 358.55 | 0.56 | 2.08 | 568.40 |
| 95%HPD | (3.24, 4.93) | (356.26, 363.21) | (0.25, 0.88) | (1.34, 2.99) | (500.01, 699.50) |
The estimated values of the remaining two parameters, and describe the interaction between the parent points and show how clusters of the events are distributed in Seoul. The distance for the maximum interaction, determined by , increases as the number of parents reduces, reflecting a sparser distribution of parent events. Nevertheless, from the estimates (which are significantly greater than 1), the parent points in all the periods show clear attraction at the location given by (around 550 m). This implies that different event clusters tend to appear near each other rather than independently. Fig. 5 indicates that parent points are generated nearby the observed locations visited by confirmed patients (i.e., offsprings in the Neyman–Scott process). Note that parents are unobserved points; therefore, we do not have information about the types of places for parents’ coordinates. Instead, we can calculate the closest data point from a parent point and identify the place type in that observed point. We find that several parent points occur nearby points of interest (within 800 m), such as supermarkets, Internet cafes, churches, and subway stations. This highlights the fact that our approach might be suitable for detecting local sources of the spread of disease.
Fig. 5.
Comparisons between the offsprings (circle) and parent points (triangle) for real data examples. Parent points are generated nearby observed visits (offsprings).
Visualization of COVID-19 risk.
From the estimated values of , , and , we can create an intensity map of patient visit events. Since local cluster centers cause rapid community transmission, it is essential to avoid higher risk locations in daily life. Understanding the probabilistic mechanism occurring patient visits and detecting local cluster centers (hotspots) are key to manage disease outbreaks. Let and be the posterior mean of and , respectively. Given the parent points from the last iteration of the MCMC, denoted as , the spatial intensity function for the offspring points is given by
| (8) |
for any . This map can be used to identify high-risk areas where the intensity exceeds a certain threshold set by administrative decision. Fig. 6 illustrates the intensity map and high-risk areas defined as km, which corresponds to roughly one patient per 1.427 km each day. Here we divide by 14 to convert the intensity computed for two weeks into the intensity for one day. The area 1.427 km is the average size of each Dong in Seoul.
Fig. 6.
(Left Column) Intensity map for the patient visit events computed by the offspring intensity function in (8). (Right Column) High risk area with an intensity greater than 1 patient/1 day 1.427 km, roughly meaning one patient per day within a “Dong”.
The hotspots shown in Fig. 6 present different types of spreading events. For example, the hotspots in Guro-gu (bottom left corner of the map) are caused by a group infection from a call center building around March. The event led to multiple detected confirmed cases. On the other hand, the hotspots in Jungrang-gu (top right corner of the map) are mostly caused by a single patient. This patient moved around multiple places in the local area, which resulted in many close contacts. The former gained considerable media attention and people became aware of the potential risk in the community. The latter, on the other hand, did not receive media coverage considering privacy protection and the local community was largely unaware of the risk. Our approach may provide a way to warn local people without revealing information about the specific contagious individual. In summary, the visualization allows authorities to identify contagion risk hotspots and help them set up public health interventions while maintaining privacy protection.
Visualization of risk boundaries.
Our approach can also provide a way to generate “risk boundaries” within a short time period (e.g., one day), assuming that the event distribution within that period can be approximated by a model based on the previous two weeks. Such boundaries are potentially useful for issuing public health advisories. The left column of Fig. 7 shows that the patient visits occurred on April 3rd were mostly concentrated within areas with high intensity , given by the existing parent points (), but there are also points located slightly away from the high intensity areas. However, there are no points that are clearly far away from the existing high intensity areas either. Therefore, these points can be viewed as offspring events from new parents that are created near existing parents.
Fig. 7.
The same intensity map as the left column in Fig. 6 (left column) and the risk boundaries defined as circles, whose centers are the existing sampled parent points and the radii are defined as (right column). The black dots in both columns show the events observed on the day following the last date of each period.
The right column of Fig. 7 shows the risk boundaries that consider both the occurrence of new parent points and the range of offspring densities created by them. Specifically, risk boundaries are represented as the orange circles, whose centers are the existing parent points . The radii are given as , the sum of the distance that maximizes the interaction between the parent points () and one side of the 95% interval for the offspring density (). As Fig. 7 shows, these risk boundaries indicate how far new events can reach given the existing parent points.
5. Discussion
In this manuscript, we proposed a novel interaction Neyman–Scott point process for modeling COVID-19 data. Our model can be used to study the spatial patterns of patient visits and detect cluster centers in local communities by considering location-based interactions. To address the intractable normalizing functions involved in the posterior, we embed DMH (Liang, 2010) into our MCMC algorithm. We apply our approach to COVID-19 data sets pertaining to Seoul, and draw meaningful epidemiological conclusions based on the parameter estimates. Furthermore, we provide easy-to-interpret disease risk maps from the fitted results. Through simulation studies, we show that our method can provide reasonably accurate parameter estimates and detect true cluster centers.
Here, we focus on developing a new cluster point process model by regarding each patient visit as a realization from the process. Our models allow both attraction and repulsion for the cluster centers, which have values for the different modeling forms of interactions in the Neyman–Scott point process. Note that there are other spatial models that can be applied to contact tracing data. Examples include dynamic models for movement tracks of animals (Russell et al., 2016) and spatial gradient methods for modeling the spread of invasive species (Goldstein et al., 2019). Adopting such methods for studying COVID-19 contact tracing data could provide another interesting epidemiological insight. Following the standard Neyman–Scott point process, we model the parent intensity parameter as a constant. One can generalize this by embedding a spatially varying intensity function (i.e., ) into our model. As in the well-established mechanistic point process framework (Diggle, 2013, González et al., 2016), we can represent as a function of various socio-demographic covariates (e.g., population sizes). Adopting such covariates into our model would be an interesting future research avenue.
Our method can be useful for planning public health interventions such as deciding social distancing levels. Social distancing is necessary to prevent the spread of infectious diseases, especially for people in risk groups for severe COVID-19. However, raising the social distancing level could adversely affect the local economy. Therefore, it is essential to measure the degree of COVID-19 risk: how dangerous is the current coronavirus outbreak? Based on our approach, we can provide some guidelines for deciding the appropriate distancing levels to minimize economic damage. We can quantify such risks based on the model parameters. For instance, and estimates provide the expected number of patient visits per cluster center and width of spreading events activity (range of spread). From the estimate, we can expect the probable location of a new cluster center. Using these parameter estimates, disease control authorities can decide the threshold for social distancing levels. Note that such an interpretation is not available from existing cluster point process approaches.
Furthermore, our proposed approach has some advantages considering privacy protection. The disease risk maps allow authorities to alert local communities about disease hotspots without revealing privacy-sensitive information. This is opposite to the current practice in South Korea, where local governments and health authorities release each individual’s travel history; although the names of individuals are not revealed, the released information can be used to infer the identity of each patient. The method and ideas developed in this manuscript are generally applicable to other infectious diseases as well.
Supplementary material
The supplementary material available online provides details of the birth–death MCMC algorithms and trace plots for the model parameters. The source code for the simulation study can be downloaded from https://github.com/jwpark88/InteractPoint.
Acknowledgments
Jaewoo Park was supported by the Yonsei University, South Korea Research Fund of 2020-22-0501 and the National Research Foundation of Korea (NRF-2020R1C1C1A0100386811). Boseung Choi was supported by the National Research Foundation of Korea (2020R1F1A1A01066082). The Ohio Super Computer Center (OSC) provided part of computational resources for this study. The authors are grateful to Tomáš Mrkvička for providing useful sample codes and the anonymous reviewers for their careful reading and valuable comments. The authors are also grateful to Korea Spatial Information & Community Co. Ltd. for their support with data and Sungjae Kim and Eunjin Eom for data collection and organization.
Appendix A. Birth-death MCMC
Birth–death samplers (Moller and Waagepetersen, 2003) are often used to generate point processes given the model parameters. We propose adding a new point (birth), removing an existing point (death), or moving a point with equal probability. To generate an auxiliary variable in DMH, we use a birth–death sampler whose stationary distribution is . Algorithm 2 summarizes this step. In our full MCMC algorithm (Algorithm 1 in the manuscript), we update , given all the model parameters. Here, we use a birth–death sampler whose stationary distribution is . Algorithm 3 summarizes this step.
Appendix B. Trace plots for model parameters
See Fig. B.8.
Fig. B.8.
Trace plots for the real data examples in the order of (rows). Columns indicate severe, moderate, mild cases, respectively. 100,000 MCMC samples are generated.
References
- Albert-Green A., Braun W.J., Dean C.B., Miller C. A hierarchical point process with application to storm cell modelling. Canad. J. Statist. 2019;47(1):46–64. [Google Scholar]
- Atchade Y., Lartillot N., Robert C.P. Bayesian computation for statistical models with intractable normalizing constants. Braz. J. Probab. Stat. 2008;27:416–436. [Google Scholar]
- Besag J. Spatial interaction and the statistical analysis of lattice systems. J. R. Stat. Soc. Ser. B Stat. Methodol. 1974;36:192–236. [Google Scholar]
- CDC J. 2020. How COVID-19 spreads. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/how-covid-spreads.html. [Google Scholar]
- Choi S., Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol. Health. 2020;42 doi: 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P.J. CRC Press; 2013. Statistical Analysis of Spatial and Spatio-Temporal Point Patterns. [Google Scholar]
- Diggle P.J., Kaimi I., Abellana R. Partial-likelihood analysis of spatio-temporal point-process data. Biometrics. 2010;66(2):347–354. doi: 10.1111/j.1541-0420.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Eddelbuettel D., François R., Allaire J., Chambers J., Bates D., Ushey K. Rcpp: Seamless R and C++ integration. J. Stat. Softw. 2011;40(8):1–18. [Google Scholar]
- Geyer C. Chapman and Hall/CRC, Monographs on Statistics and Applied Probability; 1999. Likelihood Inference for Spatial Point Processes. Stochastic Geometry: Likelihood and Computation. [Google Scholar]
- Geyer C.J., Thompson E.A. Constrained Monte Carlo maximum likelihood for dependent data. J. R. Stat. Soc. Ser. B Stat. Methodol. 1992:657–699. [Google Scholar]
- Goldstein J., Haran M., Simeonov I., Fricks J., Chiaromonte F. An attraction-repulsion point process model for respiratory syncytial virus infections. Biometrics. 2015;71(2):376–385. doi: 10.1111/biom.12267. [DOI] [PubMed] [Google Scholar]
- Goldstein J., Park J., Haran M., Liebhold A., Bjørnstad O.N. Quantifying spatio-temporal variation of invasion spread. Proc. R Soc. B. 2019;286(1894) doi: 10.1098/rspb.2018.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J.A., Rodríguez-Cortés F.J., Cronie O., Mateu J. Spatio-temporal point process statistics: a review. Spatial Stat. 2016;18:505–544. [Google Scholar]
- Guan Y. A composite likelihood approach in fitting spatial point process models. J. Amer. Statist. Assoc. 2006;101(476):1502–1512. [Google Scholar]
- KDCA Y. 2020. Responseguidelines to prevent the spread of COVID-19 (local government) http://ncov.mohw.go.kr/en/ [Google Scholar]
- Ki M. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Korea. Epidemiol. Health. 2020;42 doi: 10.4178/epih.e2020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopeckỳ J., Mrkvička T. On the Bayesian estimation for the stationary Neyman-Scott point processes. Appl. Math. 2016;61(4):503–514. [Google Scholar]
- Liang F. A double Metropolis–Hastings sampler for spatial models with intractable normalizing constants. J. Stat. Comput. Simul. 2010;80(9):1007–1022. [Google Scholar]
- Liang F., Jin I.H., Song Q., Liu J.S. An adaptive exchange algorithm for sampling from distributions with intractable normalizing constants. J. Amer. Statist. Assoc. 2016;111:377–393. [Google Scholar]
- Møller J., Pettitt A.N., Reeves R., Berthelsen K.K. An efficient Markov chain Monte Carlo method for distributions with intractable normalising constants. Biometrika. 2006;93(2):451–458. [Google Scholar]
- Møller J., Syversveen A.R., Waagepetersen R.P. Log Gaussian cox processes. Scand. J. Stat. 1998;25(3):451–482. [Google Scholar]
- Moller J., Waagepetersen R.P. CRC Press; 2003. Statistical Inference and Simulation for Spatial Point Processes. [Google Scholar]
- Møller J., Waagepetersen R.P. Modern statistics for spatial point processes. Scand. J. Stat. 2007;34(4):643–684. [Google Scholar]
- Mrkvička T., Muška M., Kubečka J. Two step estimation for Neyman-Scott point process with inhomogeneous cluster centers. Stat. Comput. 2014;24(1):91–100. [Google Scholar]
- Mrkvička T., Soubeyrand S. On parameter estimation for doubly inhomogeneous cluster point processes. Spatial Stat. 2017;20:191–205. [Google Scholar]
- Murray I., Ghahramani Z., MacKay D.J.C. Proceedings of the 22nd Annual Conference on Uncertainty in Artificial Intelligence (UAI-06) AUAI Press; 2006. MCMC for doubly-intractable distributions; pp. 359–366. [Google Scholar]
- Neyman J., Scott E. A theory of the spatial distribution of galaxies. Astrophys. J. 1952;116:144. [Google Scholar]
- Park Y.J., Choe Y.J., Park O., Park S.Y., Kim Y.-M., Kim J., Kweon S., Woo Y., Gwack J., Kim S.S., et al. Contact tracing during coronavirus disease outbreak, South Korea, 2020. Emerg. Infect. Diseases. 2020;26(10):2465–2468. doi: 10.3201/eid2610.201315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Haran M. Bayesian inference in the presence of intractable normalizing functions. J. Amer. Statist. Assoc. 2018;113(523):1372–1390. [Google Scholar]
- Russell J.C., Hanks E.M., Haran M. Dynamic models of animal movement with spatial point process interactions. J. Agric. Biol. Environ. Stat. 2016;21(1):22–40. [Google Scholar]
- Stoyan D., Stoyan H. John Wiley & Sons Inc; 1994. Fractals, Random Shapes, and Point Fields: Methods of Geometrical Statistics. [Google Scholar]
- Strauss D.J. A model for clustering. Biometrika. 1975;62(2):467–475. [Google Scholar]
- Tamayo-Uria I., Mateu J., Diggle P.J. Modelling of the spatio-temporal distribution of rat sightings in an urban environment. Spatial Stat. 2014;9:192–206. [Google Scholar]
- Thomas M. A generalization of Poisson’s binomial limit for use in ecology. Biometrika. 1949;36(1/2):18–25. [PubMed] [Google Scholar]
- Waagepetersen R.P. An estimating function approach to inference for inhomogeneous Neyman–Scott processes. Biometrics. 2007;63(1):252–258. doi: 10.1111/j.1541-0420.2006.00667.x. [DOI] [PubMed] [Google Scholar]
- WHO R.P. 2020. Novel coronavirus - China. https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ [Google Scholar]
- Yau C.Y., Loh J.M. A generalization of the Neyman-Scott process. Statist. Sinica. 2012:1717–1736. [Google Scholar]