Abstract
Key message
This study focused on the key regulatory function of Physcomitrium patens GRAS12 gene underlying an increasing plant complexity, an important step in plant terrestrialization and the evolutionary history of life.
Abstract
The miR171‐GRAS module has been identified as a key player in meristem maintenance in angiosperms. PpGRAS12 is a member of the GRAS family and a validated target for miR171 in Physcomitrium (Physcomitrella) patens. Here we show a regulatory function of miR171 at the gametophytic vegetative growth stage and targeted deletion of the PpGRAS12 gene adversely affects sporophyte production since fewer sporophytes were produced in ΔPpGRAS12 knockout lines compared to wild type moss. Furthermore, highly specific and distinct growth arrests were observed in inducible PpGRAS12 overexpression lines at the protonema stage. Prominent phenotypic aberrations including the formation of multiple apical meristems at the gametophytic vegetative stage in response to elevated PpGRAS12 transcript levels were discovered via scanning electron microscopy. The production of multiple buds in the PpGRAS12 overexpression lines similar to ΔPpCLV1a/1b disruption mutants is accompanied by an upregulation of PpCLE and downregulation of PpCLV1, PpAPB, PpNOG1, PpDEK1, PpRPK2 suggesting that PpGRAS12 acts upstream of these genes and negatively regulates the proposed pathway to specify simplex meristem formation. As CLV signaling pathway components are not present in the chlorophytic or charophytic algae and arose with the earliest land plants, we identified a key regulatory function of PpGRAS12 underlying an increasing plant complexity, an important step in plant terrestrialization and the evolutionary history of life.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11103-021-01125-z.
Keywords: GRAS family, CLAVATA, microRNA171, Growth arrest, Meristem regulation, Land plant evolution
Introduction
The plant-specific GRAS genes encode transcriptional regulators that play key roles in plant growth and development. The name of GRAS derives from the first three GRAS proteins identified in A. thaliana, GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GA (RGA) and SCARECROW (SCR) (Di Laurenzio et al. 1996; Peng et al. 1997; Silverstone et al. 1998). Based on the protein sequence and structural characteristics, the GRAS protein family is divided into eleven subfamilies: DELLA, HAIRY MERISTEM (HAM), PHYTOCHROME A SIGNAL TRANSDUCTION1 (PAT1), LATERAL SUPPRESSOR (LAS) & SCARECROW-LIKE 4/7 (SCL4/7), SCARECROW (SCR), SHORT ROOT (SHR), SCARECROW-LIKE 3 (SCL3), LISCL (Ll SCL), Clonorchis sinensis (C. sinesis) GRAS34 (CsGRAS34), Oryza sativa 19 (Os19) and DWARF AND LOW-TILLERING (DLT) (Zhang et al. 2019). GRAS protein subfamilies are known to be involved in various processes of plant growth and development such as gibberellin signal transduction (DELLA), radial root patterning and root growth (SCR and SHR), initiation and formation of axillary meristems (LAS), shoot meristem maintenance (HAM), phytochrome A signal transduction (PAT1 and SCL21), and gametogenesis (LlSCL) (Schumacher et al. 1999; Bolle et al. 2000; Helariutta et al. 2000; Wysocka-Diller et al. 2000; Greb et al. 2003; Morohashi et al. 2003; Engstrom 2012; Park et al. 2013; Torres-Galea et al. 2013). GRAS proteins also appeared to be involved in plant disease resistance and abiotic stress response (Mayrose et al. 2006).
The presence of a miR170/171 binding site within the encoded transcripts is characteristic for most members of the HAM family. MicroRNAs (miRNAs) are a class of non-coding ~ 21 nt RNAs that mediate gene silencing through cleavage of target messenger RNA (mRNA) and/or translational inhibition. miR171 is a conserved miRNA family that exists in all major land plant groups including bryophytes (Axtell and Bowman 2008) and plays critical roles in regulating plant growth and development through repressing expression of SCARECROW-LIKE (SCL) transcripts. In P. patens, two members of the GRAS family (Pp1s205_1V6.1/Pp3c12_10V3.1 and Pp1s130_63V6.1/ Pp3c7_1494V3.1) are validated targets of miR171 (Axtell et al. 2007; Hiss et al. 2017). Three members of the HAM subfamily in A. thaliana [AtSCL6-II (Atg45160), AtSCL6-III (At3g60630) and AtSCL6-IV (At4g00150) also known as the HAM (HAIRY MERISTEM) or LOM (LOST MERISTEM)] are reported targets of miR171 (Llave et al. 2002) and play an important role in shoot apical meristem maintenance and axillary meristem formation, polar organization and chlorophyll synthesis (Schulze et al. 2010; Wang et al. 2010). In A. thaliana LOM1 and LOM2 were shown to stimulate cell differentiation at the periphery of shoot meristems and to assist to maintain their polar organization (Schulze et al. 2010). Furthermore AtHAM1, AtHAM2 and AtHAM3 genes are not only essential for shoot apical meristem maintenance, but also play an important role in the maintenance of root indeterminacy (Engstrom et al. 2011). The Petunia HAM gene promotes shoot indeterminacy by an undefined non-cell-autonomous signaling mechanism (Engstrom et al. 2011). Tomato (Solanum lycopersicum) encodes three HAM homologs which are guided for cleavage by miR171 (Hendelman et al. 2016) and their silencing led to over-proliferation of cells in the periphery of the meristems. SlHAM genes not only function in meristem maintenance, but also play minor roles in the morphogenesis of a simple leaf that is determinate in tomato (Hendelman et al. 2016).
Apical meristems are a built by a specialized group of cells that principally reside at the tips of roots and shoots. Maintenance and programming of the meristematic cells are crucial steps for the cell division, shoot, and root branching. Any misregulation of the meristematic cells may result in perturbation and disorder in cell division, shoot, and root branching. Both shoot and root meristems are generated during embryogenesis, but do not contribute to the construction of the embryo and are activated once the seedling germinates (Doerner 2003). Following germination, the plant undergoes several developmental phases and shoot meristems change their identity in the course of these phase changes. In contrast, no identity alterations occur in root meristems during development. In A. thaliana the shoot meristem identity alteration appears as leaves during the initial vegetative growth, leaves and axillary meristems during the transition to flowering, and floral meristems and bracts by the inflorescence meristem during reproductive growth (Doerner 2003). Shoot apical meristems (SAMs) are responsible for developing the above-ground parts of the plant, such as stems, leaves, and flowers, while the under-ground parts of plants including root systems are generated by root apical meristems (Barton and Poethig 1993). The shoot apical meristem comprises a small bank of densely cytoplasmic, undifferentiated, dividing cells (Barton and Poethig 1993). Based on striking features including the ability of proliferation, regeneration into a new meristem after damage and the aptitude to produce a variety of differentiated cell types, meristem cells can be classified as stem cells (Sussex 1952; Potten and Loeffler 1990). Eight types of stem cells were reported to be formed in P. patens during its life cycle (Kofuji and Hasebe 2014). The common ancestor of land plants was haplontic and generated stem cells only in the gametophytic generation. Other types of body fragments in moss, such as the protonema and rhizoid filaments, leafy-shoot and thalloid gametophores, and gametangia were formed during land plant evolution by the divergence of stem cells in the gametophytic generation. Stem cells follow different morphological and anatomical patterns among land plants. The subapical cells of caulonemal filaments branch to form more filaments and three‐faced buds, which develop into leafy stems, called gametophores (Cove et al. 2009). While stem cells in shoot and roots of angiosperms and gymnosperms are multiple cells, in P. patens (protonema, gametophore, leaf, rhizoid, and sporophyte), stem cells are a single-cell (Kofuji and Hasebe 2014).
In plants, several transcription factors (TFs) were shown to be involved in meristem maintenance. Recessive mutations in the WUSCHEL (WUS) gene lead to an interruption in A. thaliana shoot meristem maintenance (Laux et al. 1996). The defect is restricted to shoot and floral meristems and persists at all developmental stages. In A. thaliana, KANADI1 (KAN1), KANADI2 (KAN2), ASYMMETRIC LEAVES 2 (AS2), and YABBY3 (YAB3) encode differentiation promoting TFs. WUS regulates KAN1, KAN2, AS2, and YAB3 genes via direct binding to their regulatory regions to represses their expression (Yadav et al. 2013).
Another important signaling pathway involving CLAVATA (CLV) and WUS that controls stem cell maintenance via an auto-regulatory negative-feedback loop was first reported in A. thaliana (Schoof et al. 2000). WUS initially acts as an activator of CLV3, which further binds with the receptor kinase CLV1 and the receptor-like protein CLV2/CORYNE (CRN) complex and negatively regulates its own expression. The A. thaliana jabba-1D (jba-1D) mutant was reported to show multiple enlarged shoot meristems (Williams et al. 2005). Furthermore, jba-1D exhibits radicalized leaves, reduced gynoecia, and vascular defects. High WUS expression levels are detected in mutants since the jba-1D meristem phenotypes require a dramatic increase in WUS expression levels. Furthermore, overexpression of miR166g, which targets and reduces PHABULOSA, PHAVOLUTA and CORONA (CNA)/ATHB15 transcript levels, is essential for the development of jba-1D meristem phenotypes. Williams et al. (2005) described the indirect involvement of miRNAs in controlling meristem formation via regulation of WUS expression. In addition to the WUS-CLV pathway, the ERECTA pathway, a leucine rich repeat receptor‐like kinase signaling pathway, represents an independent route that controls inflorescence architecture by regulating WUS expression (Mandel et al. 2014). Mutations of the eukaryotic translation initiation factor 3 subunit H, eIF3h, resulted in the formation of an enlarged shoot apical meristem in A. thaliana (Zhou et al. 2014). Unlike A. thaliana, homologs of CLV2 and CRN are missing in P. patens, but CLV3-like (CLE) peptides activate an intracellular MAPK signaling cascade through membrane localized CLV1 or RPK2 receptor kinases (Whitewoods et al. 2018). P. patens harbors two CLV1 genes (PpCLV1a-b), a single gene encoding an RPK2 receptor kinase (PpRPK2), one NO GAMETOPHORES 1 gene (PpNOG1), four AINTEGUMENTA, PLETHORA and BABY BOOM genes (PpAPB1-4), one DEFECTIVE KERNEL 1 (PpDEK1) and seven CLE (PpCLE1-7) genes and disruption of any of these genes adversely affects 2D to 3D transition in moss (Perroud et al. 2014; Demko et al. 2014; Johansen et al. 2016; Moody et al. 2018; Whitewoods et al. 2018). Analyses in PpcleAmiR1-3, PpcleAmiR4-7, ΔPprpk2, ΔPpDEK1and ΔPpCLV1a/1b mutants showed division plane misorientation and/or defects in gametophore cell division and all of these mutant lines generated multiple/supernumerary buds, and hence defects in gametophore development (Perroud et al. 2014; Demko et al. 2014; Johansen et al. 2016; Whitewoods et al. 2018; Moody 2019).
Even though miR171 and its targets are known to play an important role in plant development and growth and are conserved within all major land plants, its regulatory functions in bryophytes have not been studied yet. In this study we show that PpGRAS12 (Pp1s205_1V6.1/Pp3c12_10V3.1), a member of the GRAS family and a validated target for miR171 plays an important role in simplex meristem regulation. We provide evidence that PpGRAS12 negatively regulates PpCLV1a-b, PpAPBs, PpNOG1, PpDEK1, PpRPK2 expression and causes the formation of multiple/supernumerary buds at the gametophytic vegetative stage in P. patens.
Material and methods
Plant material and growth conditions
All experiments were performed with Physcomitrium (Physcomitrella) patens ssp. patens (Hedwig) ecotype ‘Gransden 2004’ cultured under standard growth conditions as described by Reski and Abel (1985).
Phenotypic analysis
Phenotypic analysis was performed by adjusting protonema cultures to an equal density of 100 mg/L dry weight and 5 µL of the adjusted cultures were spotted onto standard solid medium {250 mg L−1 KH2PO4, 250 mg L−1 KCl, 250 mg L−1 MgSO4 × 7H2O, 1 g L−1 Ca(NO3)2 × 4H2O and 12.5 mg L FeSO4 × 7H2O, pH 5.8 with micro-elements (ME) [H3BO3, MnSO4, ZnSO4, KI, Na2MoO4 × 2H2O, CuSO4, Co(NO3)2] and 12 g L−1 purified agar; Oxoid, Thermo Scientific, Waltham, MA, USA} or solid medium supplemented with 2 µM ß-estradiol (Sigma-Aldrich, St. Louis, USA) that was used as inducing agent for the PpGRAS12 overexpression lines (PpGRAS12-iOE). Moss was grown in a growth cabinet under long day conditions (16 h light:8 h dark photocycle, 22 °C) at 100 µmol photons m−2 s−1. For the analysis of phenotypic changes at the leafy gametophore stage the inducer was directly applied onto colonies from transgenic lines as well as WT controls. Pictures of plants were taken by a Nikon stereoscopic microscope (C-DSD230, Minato, Japan).
Subcellular localization of PpGRAS12
The complete PpGRAS12 coding sequence was amplified by PCR from genomic DNA with the primers PpGRAS12::C_F and PpGRAS12::C_R containing SalI and BglII restriction sites (Table S1). The PCR product was digested with SalI and BglII and cloned into the SalI- and BglII-sites of a modified pMAV4 plasmid (Martin et al. 2009), where the GFP reporter gene was replaced with a citrine coding sequence. The citrine coding sequence was C-terminally fused in frame to the PpGRAS12 coding sequence and confirmed by sequencing. The resulting construct was transiently transfected into P. patens protoplasts following standard procedures (Frank et al. 2005). Nuclei were stained by the addition of 2.5 mg/mL of 4′,6-diamidino-phenylindole (DAPI, Sigma-Aldrich, USA). Fluorescence microscopy was performed using an inverted Leica TCS SP5 confocal laser scanning microscope (Carl Zeiss, Germany). The excitation/emission wavelengths were 514 nm/520–620 nm for YFP, 358 nm/460–490 nm for DAPI and 633 nm/650–720 nm for chlorophyll. Images were processed and assembled by ImageJ.
Generation of ∆PpGRAS12 mutants
The ∆PpGRAS12 knockout construct was assembled by three-template PCRs and cloned into pJet1.2. These fragments were the 958 bp PpGRAS12-5′UTR, a 2174 bp aph4 selection cassette conferring resistance to hygromycin and a 904 bp PpGRAS12-3′UTR. Primers used to generate these fragments are listed in Supplementary Table 1. The final insert was amplified via three templates PCR and inserted into pJet1.2. The insert was released from the vector backbone by KpnI and NcoI to generate a fragment with 5′ and 3′ homologous ends and transfected into P. patens protoplasts (Frank et al. 2005). Subsequently, transgenic lines were selected on solidified Knop-ME medium containing 25 mg/L hygromycin. 50 plants surviving the selection procedure were directly screened by PCR for the integration of the ∆PpGRAS12 knockout construct into the nuclear DNA. Positive lines were further analyzed by RT-PCR to confirm loss of the PpGRAS12 transcript and subsequently by Southern blot to identify lines that harbor a single insertion at the PpGRAS12 locus and to exclude additional genomic integration sites.
Generation of PpGRAS12-iOE mutants
The complete PpGRAS12 coding sequence harboring six neutral mutations within the miR171 binding site to prevent miR171-directed cleavage was amplified from the mutated version of PpGRAS12 (Fig. 1). The cloning step was performed using the pENTR/D-TOPO cloning kits (Invitrogen, USA). A pair of primers (Supplementary Table 1) was designed to amplify the miR171-resistant PpGRAS12 fragment from the plasmid harboring the mutated version of PpGRAS12 and the amplified PpGRAS12 fragment was cloned into the Gateway pENTR/D-TOPO vector (Invitrogen, USA). The fragment orientation was confirmed by sequencing and the pENTR/D-TOPO vector was cloned into the PpGX8 destination vector containing a hygromycin resistance cassette (Kubo et al. 2013) using the gateway LR Reaction (Invitrogen, USA). The inducible overexpression construct without homologous recombination sites was linearized using PmeI (NEB, USA) and transfected into P. patens protoplasts (Frank et al. 2005). Selection of stable transformants was performed as described above. 50 plants surviving the selection procedure were directly screened by PCR for integration of the PpGRAS12 inducible overexpression construct into the nuclear DNA. Positive lines were further analyzed by RT-PCR and Northern blot to confirm an inducible overexpression of PpGRAS12.
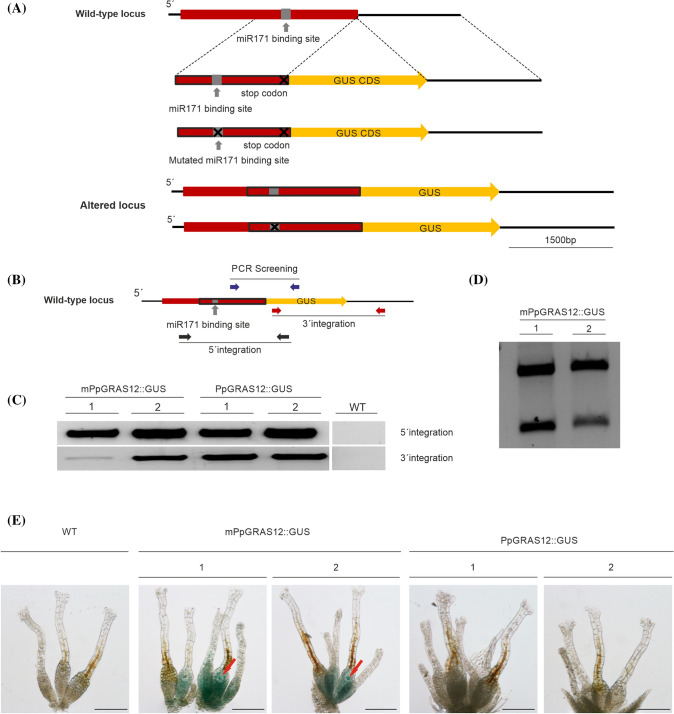
Fig. 1.
Generation of the PpGRAS12::GUS protein fusion reporter lines. a Scheme representing the generation of the PpGRAS12::GUS and mPpGRAS12::GUS fusion reporter constructs. Two variants of GUS fusion reporter constructs were generated and introduced to their cognate genomic locus by means of homologous recombination. The red box indicates the PpGRAS12 coding region. The red box with the black border lines indicates 1482 bp from the coding sequence including the miR171 binding site (native/mutated), which was fused to the GUS coding sequence (yellow box). The PpGRAS12 stop codon was removed and the coding sequence fused to the GUS coding sequence. b Purple, red, and black arrows show the primer pairs sequentially applied for PCR-based analyses of the PpGRAS12::GUS protein fusion reporter lines. c Upper panel: confirmation of 5′ integration of the constructs using black primers. Lower panel: confirmation of 3′ integration of the construct using red primers. d Validation of mPpGRAS12::GUS protein fusion reporter lines by digestion of RT-PCR products with PauI (a PauI restriction site was introduced into the mutated miR171 binding site). e Correspondent blue colors were detected only in the archegonia and egg cells of mPpGRAS12::GUS protein fusion reporter lines. Red arrows: egg cells. Scale bars: 1 mm
Generation of the PpGRAS12::GUS protein fusion reporter lines
The PpGRAS12 coding sequence harboring a mutated or native miR171 binding site was fused to the GUS coding sequence and introduced to their cognate genomic locus by means of homologous recombination. Three sets of primers were used for the generation of PpGRAS12::GUS fusion constructs. The first set of primers was designed to amplify 1482 bp (5′ flanking region of the construct) from the coding sequence including the miR171 binding site, where the SacI restriction site was added to the 5′ end and an EcoRI restriction site was added to the 3′ end. The second set including EcoRI and SalI restriction sites was designed to amplify the GUS coding region. The third set of primers was designed to amplify 1528 bp downstream of the PpGRAS12 coding region including the 3′ UTR (3′ flanking region of the construct), where the SalI restriction site was added to the 5′ end and the KpnI restriction site was added to the 3′ end. All three fragments were digested with EcoRI and SalI, gel purified, ligated, and subsequently cloned into the pJET cloning vector. PpGRAS12::GUS and mPpGRAS12::GUS fusion reporter constructs were released from the pJET backbone by SacI and KpnI digestion and transfected into P. patens protoplasts. 50 plants surviving the selection procedure for each construct were screened by PCR to validate proper 5′ and 3′ integration of the GUS reporter constructs. PCR products from positive lines harboring the transfected constructs were further validated by sequencing.
RNA isolation and qRT-PCR
Plant tissue was homogenized under liquid nitrogen and total RNA was extracted using TRI Reagent according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, USA). RNA was treated with RNase-free DNase I (NEB, USA), reverse transcribed as described Arif et al. (2018) and the synthesized cDNA was used as template for qRT-PCR analysis performed on a CFX96 Real-Time System (Bio-Rad, USA) using SYBR Green mix. The relative expression levels of genes were calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001) using two constitutively expressed PpEf1α (Pp1s7_445V6.1/Pp3c2_6650V3.1) and PpC45, encoding 60S ribosomal protein L21 (Pp1s107_181V6.1/Pp3c13_2360V3.1) as internal controls for normalization. Oligonucleotides used for qRT-PCR analyses are listed in Supplementary Table 1.
Southern blot
Determination of copy numbers of inserted DNA fragments by Southern blot was performed according to QIAGEN Bench Guide (Qiagen, Hilden, Germany). 2 µg of genomic DNA was digested with the indicated restriction enzymes, resolved with PerfectBlue Gel System Mini L Revolution (PEQLAB Biotechnology, Erlangen, Germany) and blotted to a positively charged nylon membrane (Amersham Hybond-N+, GE Healthcare UK Limited, Buckinghamshire, UK). Samples were fixed by UV-crosslinking and hybridized with a [α32P] dCTP radioactively labeled probe which was amplified from selection cassette by PCR (Supplementary Table 1).
RNA gel blot analysis
RNA gel blot analysis using 20 µg of total RNA was performed as described (Khraiwesh et al. 2008). A 721 bp PpGRAS12 coding sequence was amplified with specific primers and used as a probe (Supplementary Table 1).
Scanning electron microscopy
Gametophores from P. patens WT and mutant lines were fixed with 2.5% glutaraldehyde in 75 mM cacodylate buffer containing 2 mM MgCl2. After 4 washing steps with pure buffer, post-fixation was carried out with 1% OsO4 for 90 min. Two washing steps with buffer were followed by washing three times with double-distilled water. After this, the samples were dehydrated in a graded acetone series and critical-point-dried. Finally, the samples were mounted on aluminum stubs and sputter-coated with platinum. Scanning electron microscopy was performed on a Hitachi S-4100 SEM (Hitachi, Japan) at acceleration voltages between 3 and 5 kV.
Results
MiR171 regulates PpGRAS12 expression
The existence of the GRAS domain categorized PpGRAS12 as a member of the GRAS family (Supplementary Fig. 1). PpGRAS12 is a validated target of miR171 (Axtell et al. 2007) and belongs to the HAM subfamily (Chen et al. 2019). Plant miRNAs frequently play a role in defining the spatiotemporal expression of their cognate target mRNAs and the miR171‐GRAS module has been described as a key player in meristem maintenance in A. thaliana (Huang et al. 2017). To study whether miR171 regulates the spatiotemporal expression of PpGRAS12, PpGRAS12::GUS protein fusion reporter lines were generated. To generate these lines, the PpGRAS12 coding sequence harboring a mutated or native miR171 binding site was fused to the GUS coding sequence and introduced to the cognate genomic locus by means of homologous recombination (Fig. 1a). Integrated DNA constructs were detected via PCR screening (purple primers, Fig. 1b) and the precise integration of the PpGRAS12::GUS fusion construct into the genome was confirmed for two independent lines by 5′ (black primers, Fig. 1b) and 3′ (red primers, Fig. 1b) integration PCR (Fig. 1c) and subsequent sequencing of the amplified products. Validation of the transgenic lines that had integrated the mPpGRAS12::GUS (miR171-resistant) construct was performed for two positive lines by subsequent digestion of RT-PCR products with PauI since a PauI recognition site was embedded within the miR171 binding site to generate silent mutations that prevent miR171-mediated cleavage of the PpGRAS12 mRNA (Fig. 1d).
Histochemical GUS staining was performed for both miR171-resistant and miR171-sensitive lines. Correspondent blue color indicating GUS activity and expression of the PpGRAS12 gene was neither observed in the native PpGRAS12::GUS or in the mutated mPpGRAS12::GUS protein fusion reporter lines at the protonema and gametophore stage (Fig. 1e). However, the blue color was only detected in the archegonia and egg cells of mPpGRAS12::GUS protein fusion reporter lines (Fig. 1e) resembling previously determined high levels of native PpGRAS12 expression in the early phase of sporophyte development (Physcomitrium eFP browser; Ortiz-Ramírez et al. 2016). Expression of GUS in archegonia and egg cells of the miR171-resistant lines indicates that miR171 significantly regulates expression of PpGRAS12 in these cells.
Loss of the nuclear-localized PpGRAS12 protein results in reduction of sporophyte production
We performed transient expression of a C-terminal PpGRAS12-citrine protein fusion in P. patens protoplasts and confirmed an expected nuclear localization for the PpGRAS12 protein (Fig. 2a). The citrine fluorescence signals in the transformed protoplasts overlapped with nuclei stained by 4′,6-diamidino-2-phenylindole (DAPI), but we also observed signals in the cytosol. The observed nuclear localization was in agreement with the proposed function of GRAS proteins as transcription factors (Di Laurenzio et al. 1996; Gallagher and Benfey 2009; Heo et al. 2011; Yoshida et al. 2014). The cytoplasmic accumulation of PpGRAS12-citrine fusion protein might be due to a high ectopic expression level or the localization of PpGRAS12 into the nucleus may require the formation of heterodimers and a limited abundance of the dimerizing partner prevents complete targeting of PpGRAS12 to the nucleus and leads to remaining fluorescent signals in the cytoplasm.
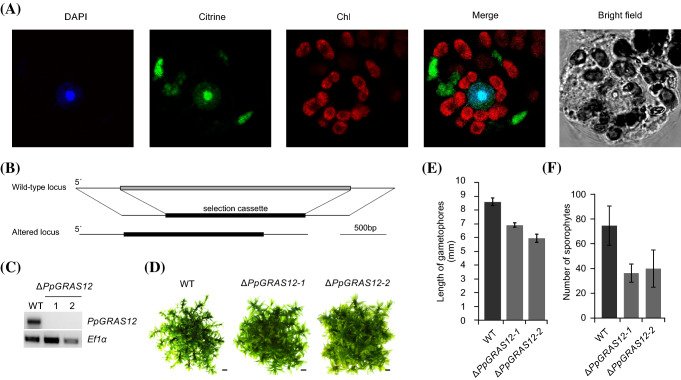
Fig. 2.
Generation and phenotypic analysis of the ΔPpGRAS12 lines. a Subcellular localization of the PpGRAS12::citrine protein fusion in P. patens protoplasts. Pictures were taken 3 days after transfection of the PpGRAS12::citrine fusion construct into P. patens protoplasts. DAPI: DAPI signal. Citrine: citrine signal. Chl: chlorophyll auto-fluorescence. Merge: merged images of citrine and chlorophyll auto-fluorescence. b Scheme depicting the targeted knockout approach of the PpGRAS12 coding sequence. c RT-PCR from cDNA derived from the indicated lines using PpGRAS12-specific primers; note that the two ΔPpGRAS12 mutant lines are null mutants lacking the PpGRAS12 transcript; RT-PCRs performed with primers for the constitutively expressed gene PpEf1α served as a control to monitor successful cDNA synthesis. d Phenotypic analyses of the knockout lines. Initially, a single gametophore from the indicated lines was cultured on standard growth medium and pictures were taken after 45 days of growth under standard growth conditions. Scale bars: 1 mm. e Comparison of the gametophore length in the WT and two independent ∆PpGRAS12 lines. Gametophore length was measured from colonies grown for 45 days under standard growth conditions; error bars represent standard errors (n = 30). f Comparison of the sporophyte numbers in the WT and two independent ∆PpGRAS12 lines; error bars represent standard errors (n = 27)
To analyze the function of PpGRAS12, we generated ∆PpGRAS12 knockout lines by targeted disruption of the corresponding genomic locus via the insertion of an nptII cassette (Fig. 2b). Using homologous recombination, gene targeting was performed and putative gene deletion mutants were first selected by 5′ integration PCR. Six out of 50 plants surviving the selection procedure and that showed proper 5′ integration were further analyzed by RT-PCR to confirm the loss of GRAS12 transcript. Subsequently, two independent knockout lines with a single integration of the gene deletion construct were selected by Southern blot (Supplementary Fig. 2) and were further confirmed by reverse transcriptase PCR (RT-PCR) to be ∆PpGRAS12 null mutants that lack the PpGRAS12 transcript (Fig. 2c). In the primary phase of growth including protonema and budding stage, no distinct differences were observed in the ∆PpGRAS12 lines compared to the WT. Mild phenotypic deviations were observed in the ΔPpGRAS12 lines at the gametophytic growth stage (Fig. 2d) and confirmed by statistical analyses of the gametophore length in the WT and two independent ΔPpGRAS12 lines (Fig. 2e). Further phenotypic analysis revealed that the absence of the PpGRAS12 gene significantly influences the sporophyte production and consequently, fewer sporophytes were produced in the knockout lines compared to the WT (Fig. 2f).
PpGRAS12 overexpression leads to the formation of multiple apical simplex meristems
We observed a mild phenotypic deviation in the ΔPpGRAS12 lines at the gametophytic growth stage (Fig. 2e, f) and prominent phenotypic aberrations in the sporophytic generation (Fig. 2f). To analyze the impact on P. patens growth and development upon PpGRAS12 overexpression, PpGRAS12 inducible overexpression lines (PpGRAS12-iOE lines) were generated. For this, we amplified the PpGRAS12 cDNA and introduced six silent mutations within the miR171 binding site to inhibit miR171-mediated cleavage without affecting the encoded amino acid sequence (Supplementary Fig. 3). The fragment orientation was checked by sequencing and the pENTR/D-TOPO vector was cloned into the PpGX8 destination vector (Kubo et al. 2013). This construct was used for the transfection of P. patens protoplasts. After the selection of regenerating protoplasts on hygromycin-containing medium, a PCR-based screening was performed to identify desired transgenic lines within 50 plants surviving the selection period. The PCR-based screen (Supplementary Table 1) of regenerating lines identified two independent PpGRAS12 overexpression lines (PpGRAS12-iOE). To verify the inducible expression of PpGRAS12, protonema tissue from both independent PpGRAS12-iOE lines was treated for 4 h with 2 μM ß-estradiol that was used in all experiments to induce PpGRAS12 expression. Whereas the untreated PpGRAS12-iOE lines had similar PpGRAS12 transcript levels as the WT control we detected a strong induction of the PpGRAS12 transgene in both PpGRAS12-iOE lines by RNA gel blot (Supplementary Fig. 4). The overexpression of PpGRAS12 in both PpGRAS12-iOE lines was detected in protonema 2 h after the addition of ß-estradiol (Supplementary Fig. 4a). Time course analysis of both PpGRAS12-iOE lines confirmed inducible and increasing PpGRAS12 expression over time (Supplementary Fig. 4b).
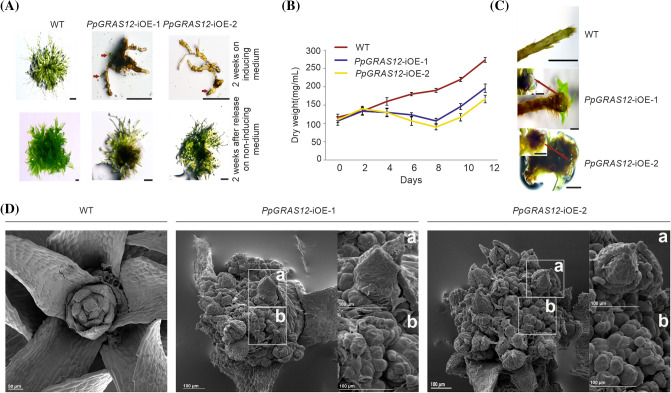
Phenotypic analysis of the PpGRAS12-iOE lines in comparison to WT was performed by adjusting pure protonema cultures to an equal density of 100 mg/L dry weight and 5 µL of the adjusted cultures were spotted onto standard solid growth medium supplemented with 2 µM of ß-estradiol or without inducer. We did not observe any phenotypic differences between WT and both PpGRAS12-iOE lines on standard growth medium without inducer. However, highly specific and distinct growth arrests including a callus-like mass at the gametophore base and strong cell fate and proliferation defects were observed in the PpGRAS12-iOE lines upon the induction at the protonema stage (Fig. 3a, upper panel) whereas the PpGRAS12-iOE lines were able to recover and to return to normal growth and development after release to non-inducing conditions (Fig. 3a, lower panel). To analyze the growth behavior of the PpGRAS12-iOE lines in liquid medium, protonema tissue from WT and both PpGRAS12-iOE lines were transferred into liquid medium supplemented with 2 µM of ß-estradiol and growth of the cultures was monitored by the determination of the dry weight every 2 days. We observed a decrease in the growth rate of both PpGRAS12-iOE lines compared to the WT 2 days after the induction (Fig. 3b). The decrease in the growth rate of both PpGRAS12-iOE lines showed a steep decline until 8 days of growth in the induced medium followed by a slight recovery (Fig. 3c). However, the growth rate in both PpGRAS12-iOE lines remained lower compared to the WT after 12 days of growth in the induced medium. The slight increase after 8 days might be related to the gradual degradation of the inducer. Additionally, the effect of PpGRAS12 induction at later growth stages was investigated using colonies that were grown on solid medium and developed leafy gametophores. For this, 2 µM of ß-estradiol was directly applied onto the colonies of both PpGRAS12-iOE lines as well as WT. Strikingly, atypical enlargement of the gametophore apical stem cells at the shoot apex was observed in both PpGRAS12-iOE lines 7 days after the induction (Fig. 3d). Furthermore, we noticed an abnormal enlarged structure at the tip of both PpGRAS12-iOE lines (Fig. 3d). Further investigation using scanning electron microscopy revealed that the abnormal structure, which was formed in response to an elevated level of PpGRAS12 at the tip zone, is formed by multiple apical simplex meristems (Fig. 3d).
Fig. 3.
Phenotypic analysis of the PpGRAS12-iOE lines. a Equal amounts of protonema tissues from the WT and both PpGRAS12-iOE lines were spotted on standard solid growth medium supplemented with 2 µM ß-estradiol. Upper panel: protonema tissue after growth for 14 days on the medium supplemented with 2 µM ß-estradiol. Lower panel: 14 days after growth on inducing medium protonema tissue was transferred onto standard growth medium without inducer for 2 weeks. Red arrows indicate viable green cells. Scale bars: 1 mm. b PpGRAS12-iOE lines and WT were grown in standard liquid medium. Protonema from the PpGRAS12-iOE lines and WT were induced with 2 µM of ß-estradiol and dry weight of samples was measured every 2 days for a period of 12 days. Error bars indicate mean values ± SE (n = 3). c Formation of abnormal structures at the tip of both PpGRAS12-iOE lines. Scale bar: 1 mm for the WT and 0.5 mm for the mutants. d SEM analysis of PpGRAS12-iOE lines. The formation of supernumerary apical meristems in the PpGRAS12-iOE lines upon the induction with 2 µM of ß-estradiol. Box a: a leafy gametophore that was formed from an individual apical meristem. Box b: enlarged supernumerary apical meristems
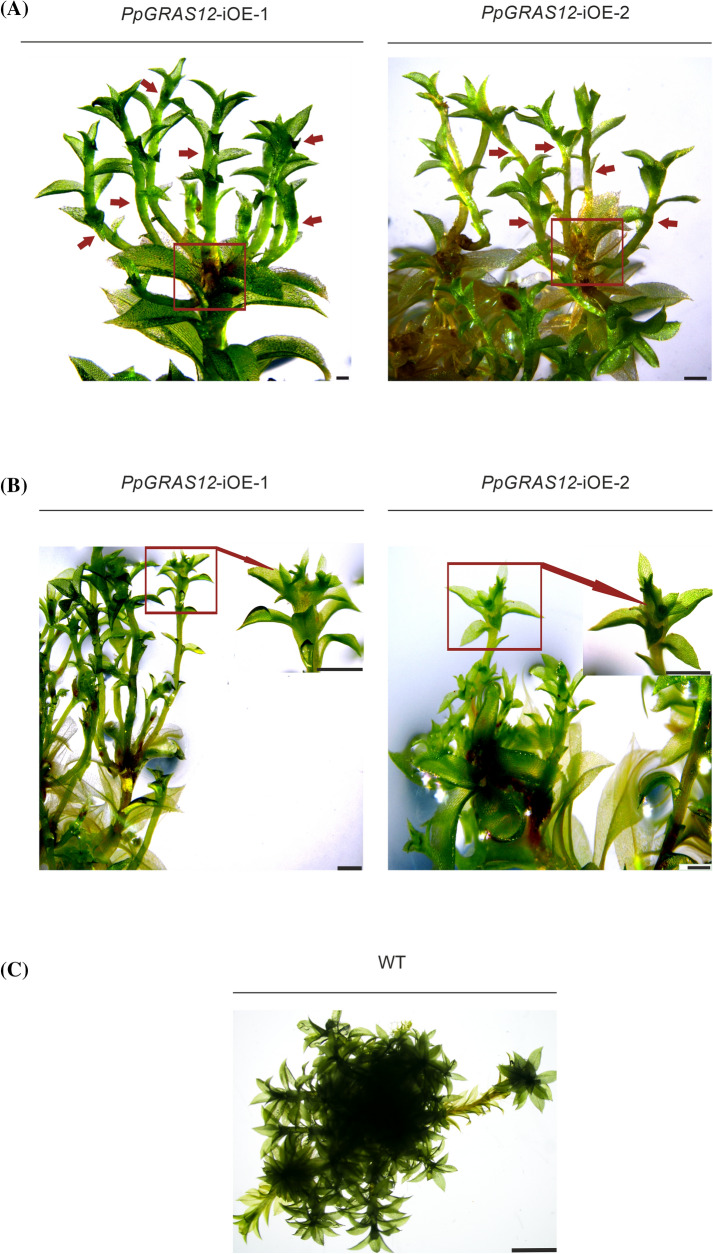
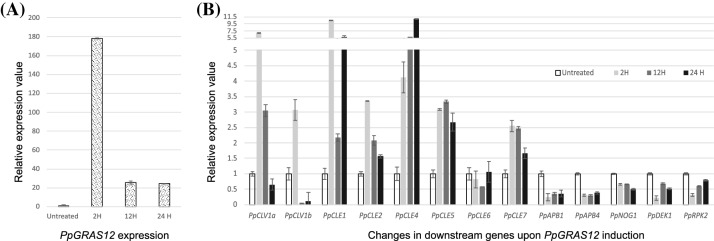
Moreover, after withdrawal of the inducer for 3 weeks causing its dilution and/or degradation individual apical cells were able to undergo normal developmental progression and developed into leafy gametophores (Figs. 3d, 4a). However, the developmental progression of PpGRAS12-iOE lines was still significantly delayed compared to WT. Furthermore, if a new gametophore, which has previously emerged from an individual apical simplex meristem, once more was exposed to the inducer, multiple apical simplex meristems were formed over again from the tip (Fig. 4b). A. thaliana CLV1 was previously reported to play an important role in maintaining meristem identity and controlling meristem size (Clark et al. 1993) since the A. thaliana clv1 mutant develops enlarged shoot apical meristems (Clark et al. 1995; Shinohara and Matsubayashi 2015). To investigate whether the formation of multiple apical meristms upon PpGRAS12 overexpression might be caused by reduced levels of the P. patens CLV1a and CLV1b homologs as well as altered expression levels of additional genes that act in the signaling cascade we analyzed their expression levels in one representative PpGRAS12-iOE line by qRT-PCR 2 h, 12 h and 24 h after the induction of PpGRAS12 in gametophores. Since we did not detect differential expression of the analyzed genes between WT and the uninduced PpGRAS12-iOE line, the uninduced PpGRAS12-iOE line was used as a control. Even though an over 175-fold increase in the PpGRAS12 expression level was observed 2 h after induction, the transcript levels dropped to an approximately 25-fold increase and remained constant after 24 h of induction (Fig. 5a). 24 h after induction, all CLE genes, PpCLE1, PpCLE2, PpCLE4, PpCLE5, PpCLE6 and PpCLE7, were upregulated upon PpGRAS12 induction (Fig. 5b). PpCLV1 expression analysis showed downregulation of both PpCLV1a and PpCLV1b homologs in response to the elevated PpGRAS12 levels (Fig. 5b). Statistically significant differences for each gene were analyzed by one-way ANOVA and other than PpCLE6, changes in relative expression levels were significantly different (p < 0.05). Furthermore, qRT-PCR analysis also revealed the downregulation of PpAPB1, PpAPB4, PpNOG1, PpDEK1 and PpRPK2 upon PpGRAS12 induction. The downregulation of both PpCLV1a and PpCLV1b as well as reduced transcript levels of additional genes acting downstream might explain the formation of multiple simplex meristems similar to ΔPpCLV1a/1b, Pprpk2 and ΔPpDEK1 disruption mutants.
Fig. 4.
Supernumerary gametophore formation from multiple apical meristems in the induced PpGRAS12-iOE lines. a Supernumerary gametophores were formed from multiple apical meristems in the PpGRAS12-iOE lines upon PpGRAS12 induction. The red box shows the development of multiple gametophores from apical meristems in the PpGRAS12-iOE lines. Red arrows indicate a single gametophore. Pictures were taken 75 days after the induction; scale bar: 1 mm. b Renewal of induction resulted in the formation of multiple apical meristems and consequently, the formation of multiple gametophores on the top of previous gametophores. The red arrow shows multiple gametophores. Pictures were taken, 12 days after the renewal of induction; scale bar: 1 mm. c Representative WT gametophores grown on standard solid growth medium; scale bar: 100 µm
Fig. 5.
The relative expression levels of PpGRAS12 and other members of MAPK signaling pathway in the WT and PpGRAS12-iOE lines. a The relative expression levels of PpGRAS12 2 h, 12 h and 24 h after induction. b The relative expression levels of PpCLV1a, PpCLV1b, PpCLE1, PpCLE2, PpCLE4, PpCLE5, PpCLE6, PpCLE7, PpAPB1, PpAPB4, PpNOG1, PpDEK1 and PpRPK2 2 h, 12 h and 24 h after induction of the PpGRAS12 gene. Plants were grown on standard solid growth medium for 4 weeks, induced for 2 h, 12 h and 24 h and RNA from gametophore tissue was used for qRT-PCR. Relative gene expression levels were normalized to PpEf1a and PpC45 and transcription rates in the uninduced PpGRAS12-iOE line were set to 1. Error bars indicate mean values ± SE (3 biological replicates with three technical replicates each). Statistically significant differences for each gene were analyzed by one-way ANOVA (p < 0.05)
Discussion
PpGRAS12 is a member of the GRAS family (Pysh et al. 1999; Tian et al. 2004; Hirsch and Oldroyd 2009) harboring a conserved order of the characteristic GRAS motifs. Here, we show that PpGRAS12 is nuclear-localized, which is in agreement with the proposed function of GRAS proteins as transcription factors (Di Laurenzio et al. 1996; Gallagher and Benfey 2009; Heo et al. 2011).
The presence of the miR170/171 binding site is a characteristic feature for most members of the HAM families. A. thaliana orthologs of Petunia HAM were shown to be targets of miR170/171 (Llave et al. 2002) and they were shown to be involved in meristem regulation and the CLV-WUS pathway (Zhou et al. 2018) that controls stem cell maintenance via an auto-regulatory negative-feedback loop (Schoof et al. 2000). HAM and WUS share collective targets in vivo and their physical interaction is vital in driving downstream transcriptional programs and promoting shoot stem cell proliferation (Zhou et al. 2015). AtGPR23, AtTIT2;2, and AtTPL are reported as collective targets of HAM and WUS and they are noticeably affected when WUS and HAM interact (Zhou et al. 2015). Analysis of the PpGRAS12::GUS protein fusion reporter lines showed a regulatory function of miR171 in PpGRAS12 expression. We observed a noticeable expression of the PpGRAS12 gene in the archegonia and egg cells of the mPpGRAS12::GUS protein fusion reporter lines compared with the PpGRAS12::GUS lines and WT suggesting that miR171 controls the expression of PpGRAS12 in P. patens archegonia and egg cells. Loss of function ∆PpGRAS12 lines displayed a fewer number of sporophytes compared to the WT. Based on the elevated expression of PpGRAS12 in the egg cells of the mPpGRAS12::GUS protein fusion reporter lines and reduced sporophyte production in the ∆PpGRAS12 lines, we suggest that PpGRAS12 plays a role in egg cell regulation and sporophyte production.
A callus-like mass at the gametophore base and strong cell fate and proliferation defects were observed in the PpGRAS12-iOE lines at the protonema stage upon the induction of PpGRAS12 expression. Furthermore, we observed the formation of multiple apical simplex meristems at the tips of gametophores upon PpGRAS12 induction in the PpGRAS12-iOE lines. The shoot apical meristem (SAM) is responsible for the post-embryonic growth and generates plant aerial structures. Widespread variations in shoot meristem structure have evolved among living plant lineages such as the single-celled apices of the non-vascular bryophytes, the multicellular meristems with prominent apical cells in the seedless lycophytes, and the multiple cell-layered shoot meristems of angiosperms. An appropriate continuous growth in plants depends on the SAM ability to maintain the balance between self-renewal of stem cells and cell recruitment for lateral organ formation (Lee et al. 2019). The WUS and CLV signaling pathways are key factors of meristematic activity in the SAM (Laux et al. 1996). The A. thaliana clv1 mutant develops enlarged shoot apical meristems (Clark et al. 1995). Furthermore, mutation of the CLV1 gene has resulted in an increased number of all floral organ types (Leyser and Furner 1992). On the other hand, disruption of any gene (PpCLV1a-b, PpCLE1-7 and PpRPK2) involved in the MAPK signaling cascade which is responsible for 2D to 3D transition in P. patens resulted in the formation of supernumerary buds and defective gametophores. We observed the same phenomenon, the formation of enlarged and multiple apical simplex meristems in PpGRAS12-iOE lines in response to an elevated level of PpGRAS12. The qRT-PCR analyses revealed the upregulation of PpCLE genes and concomitant downregulation of PpCLV1a/b, PpAPB, PpNOG1, PpDEK1 and PpRPK2 genes upon elevated levels of PpGRAS12 indicating that PpGRAS12 acts upstream of these genes and induces the formation of multiple and enlarged apical simplex meristems. A detailed analysis at earlier time points revealed that both PpCLV1a and PpCLV1b were initially upregulated and then downregulated. Despite the upregulation of PpCLE genes, the downregulation of genes encoding membrane-localized receptor kinases (PpCLV1 and PpRPK2) led to downregulation of the MAPK signaling pathway and resulted in downregulation of PpAPB genes. Surprisingly, we could not observe an antagonistic interaction between PpDEK1 and PpNOG1 in controlling PpAPB genes, like in the previously proposed model for 3D growth regulation in P. patens. This difference might be due to the fact that the proposed model explains underlying molecular changes during the formation of gametophore initial cells from caulonemal filaments and the subsequent acquisition of a tetrahedral apical cell (Moody 2019). Hence, the antagonistic interaction between PpDEK1 and PpNOG1 in controlling PpAPB genes might only occur during the formation of gametophore initial cells, but not in multiple apical simplex meristem formation. Taken together, our results indicate the involvement and key role of PpGRAS12 in simplex meristem regulation, maintenance and identity control and provides new insights into the MAPK signaling pathway. Further investigations are needed to elucidate how these genes and their feedback loops together with other players in the MAPK signaling pathway as well as cytokinin signaling function synergistically in the formation and development of gametophores. Consequently, this will help us to understand innovations related to an increased body plan complexity during the water-to-land transition of the earliest land plants.
Supplementary Information
Below is the link to the electronic supplementary material.
Electronic supplementary material 1 (DOC 766 kb)
Author contributions
WF designed and supervised the research; HB, CS, MAA, AK and OT performed experiments; HB, CS, MAA, AK, OT and WF analyzed data; HB, MAA, OT and WF wrote the paper.
Funding
Open Access funding enabled and organized by Projekt DEAL. We gratefully acknowledge financial support from the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) to WF (FR 1677/3-1 and FR 1677/3-2).
Compliance with ethical standards
Conflict of interest
All authors did not have any conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Footnotes
Association numbers
For P. patens genes, accession numbers from V1.6 and V3.3 are indicated.
PpGRAS12: Pp1s205_1V6.1 / Pp3c12_10V3.1. PpGRAS7: Pp1s130_63V6.1 / Pp3c7_1494V3.1.
AtSCL6-II: At2g45160. AtSCL6-III: At3g60630. AtSCL6-IV: At4g00150.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arif MA, Alseekh S, Harb J, Fernie A, Frank W. Abscisic acid, cold and salt stimulate conserved metabolic regulation in the moss Physcomitrella patens. Plant Biol. 2018;20:1014–1022. doi: 10.1111/plb.12871. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana an analysis of development in the wild-type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- Bolle C, Koncz C, Chua NH. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu P, Wu S, Lu Y, Sun J, Cao Q, Li Z, Xu T (2019) Identification and expression analysis of GRAS transcription factors in the wild relative of sweet potato Ipomoea trifida. BMC Genomics 20(1):911 [DOI] [PMC free article] [PubMed]
- Clark SE, Running MP, Meyerowitz EM. Clavata1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as Clavata1. Development. 1995;121:2057–2067. [Google Scholar]
- Cove DJ, Perroud PF, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS (2009) The moss Physcomitrella patens: A novel model system for plant development and genomic Studies. Cold Spring Harbor Protocols 2009(2):pdb.emo115 [DOI] [PubMed]
- Demko V, Perroud PF, Johansen W, Delwiche CF, Cooper ED, Remme P, Ako AE, Kugler KG, Mayer KFX, Quatrano R, Olsen OA. Genetic analysis of Defective Kernel1 loop function in three-dimensional body patterning in Physcomitrella patens. Plant Physiol. 2014;166:903–919. doi: 10.1104/pp.114.243758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, WysockaDiller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doerner P. Plant meristems: a merry-go-round of signals. Curr Biol. 2003;13:R368–R374. doi: 10.1016/s0960-9822(03)00280-x. [DOI] [PubMed] [Google Scholar]
- Engstrom EM. HAM proteins promote organ indeterminacy: but how? Plant Signal Behav. 2012;7:227–234. doi: 10.4161/psb.18958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom EM, Andersen CM, Gumulak-Smith J, Hu J, Orlova E, Sozzani R, Bowman JL. Arabidopsis homologs of the Petunia HAIRY MERISTEM gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011;155:735–750. doi: 10.1104/pp.110.168757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Decker EL, Reski R. Molecular tools to study Physcomitrella patens. Plant Biol (Stuttg) 2005;7:220–227. doi: 10.1055/s-2005-865645. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 2009;57:785–797. doi: 10.1111/j.1365-313X.2008.03735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Hendelman A, Kravchik M, Stav R, Frank W, Arazi T. Tomato HAIRY MERISTEM genes are involved in meristem maintenance and compound leaf morphogenesis. J Exp Bot. 2016;67:6187–6200. doi: 10.1093/jxb/erw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, Lee MM, Lim J. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108:2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Oldroyd GE. GRAS-domain transcription factors that regulate plant development. Plant Signal Behav. 2009;4:698–700. doi: 10.4161/psb.4.8.9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiss M, Meyberg R, Westermann J, Haas FB, Schneider L, Schallenberg-Rüdinger M, Ullrich KK, Rensing SA. Sexual reproduction, sporophyte development and molecular variation in the model moss Physcomitrella patens: introducing the ecotype Reute. Plant J. 2017;90(3):606–620. doi: 10.1111/tpj.13501. [DOI] [PubMed] [Google Scholar]
- Huang W, Peng S, Xian Z, Lin D, Hu G, Yang L, Ren M, Li Z. Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol J. 2017;15:472–488. doi: 10.1111/pbi.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen W, Ako AE, Demko V, Perroud PF, Rensing SA, Mekhlif AK, Olsen OA. The DEK1 calpain Linker functions in three-dimensional body patterning in Physcomitrella patens. Plant Physiol. 2016;172:00925. doi: 10.1104/pp.16.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Ossowski S, Weigel D, Reski R, Frank W. Specific gene silencing by artificial microRNAs in Physcomitrella patens: An alternative to targeted gene knockouts. Plant Physiol. 2008;148:684–693. doi: 10.1104/pp.108.128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Hasebe M. Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol. 2014;17:13–21. doi: 10.1016/j.pbi.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Kubo M, Imai A, Nishiyama T, Ishikawa M, Sato Y, Kurata T, Hiwatashi Y, Reski R, Hasebe M. System for stable beta-estradiol-inducible gene expression in the moss Physcomitrella patens. PLoS ONE. 2013;8:e77356. doi: 10.1371/journal.pone.0077356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lee ZH, Hirakawa T, Yamaguchi N, Ito T. The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int J Mol Sci. 2019;20:4065. doi: 10.3390/ijms20164065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Furner IJ. Characterization of three shootapical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mandel T, Moreau F, Kutsher Y, Fletcher JC, Carles CC, Eshed Williams L. The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development. 2014;141:830–841. doi: 10.1242/dev.104687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Lang D, Heckmann J, Zimmer AD, Vervliet-Scheebaum M, Reski R. A uniquely high number of ftsZ genes in the moss Physcomitrella patens. Plant Biol. 2009;11:744–750. doi: 10.1111/j.1438-8677.2008.00174.x. [DOI] [PubMed] [Google Scholar]
- Mayrose M, Ekengren SK, Melech-Bonfil S, Martin GB, Sessa G. A novel link between tomato GRAS genes, plant disease resistance and mechanical stress response. Mol Plant Pathol. 2006;7:593–604. doi: 10.1111/j.1364-3703.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- Moody LA. The 2D to 3D growth transition in the moss Physcomitrella patens. Curr Opin Plant Biol. 2019;47:88–95. doi: 10.1016/j.pbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Moody LA, Kelly S, Rabbinowitsch E, Langdale JA. Genetic regulation of the 2D to 3D growth transition in the moss Physcomitrella patens. Curr Biol. 2018;28:473–478.e5. doi: 10.1016/j.cub.2017.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Minami M, Takase H, Hotta Y, Hiratsuka K. Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J Biol Chem. 2003;278:20865–20873. doi: 10.1074/jbc.M301712200. [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijó JA, Becker JD. A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant. 2016;9(2):205–220. doi: 10.1016/j.molp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Park J, Nguyen KT, Park E, Jeon JS, Choi G. DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell. 2013;25:927–943. doi: 10.1105/tpc.112.108951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud PF, Demko V, Johansen W, Wilson RC, Olsen OA, Quatrano RS. Defective Kernel 1 (DEK1) is required for threedimensional growth in Physcomitrella patens. New Phytol. 2014;203:794–804. doi: 10.1111/nph.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties—lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Reski R, Abel WO. Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta. 1985;165:354–358. doi: 10.1007/BF00392232. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schulze S, Schafer BN, Parizotto EA, Voinnet O, Theres K. LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 2010;64:668–678. doi: 10.1111/j.1365-313X.2010.04359.x. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci USA. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 2015;82:328–336. doi: 10.1111/tpj.12817. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM. Regeneration of the potato shoot apex. Nature. 1952;170:755–757. [PubMed] [Google Scholar]
- Tian CG, Wan P, Sun SH, Li JY, Chen MS. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol. 2004;54:519–532. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- Torres-Galea P, Hirtreiter B, Bolle C. Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013;161:291–304. doi: 10.1104/pp.112.206607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- Whitewoods CD, Cammarata J, Venza ZN, Sang S, Crook AD, Aoyama T, Wang XY, Waller M, Kamisugi Y, Cuming AC, Szövényi P, Nimchuk ZL, Roeder AHK, Scanlon MJ, Harrison CJ. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr Biol. 2018;28:2365–2376. doi: 10.1016/j.cub.2018.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Ohno C, Heisler M, Girke T, Jonsson H, Reddy GV. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol Syst Biol. 2013;9:654. doi: 10.1038/msb.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, Tada Y, Ohme-Takagi M, Matsuoka M, Ueguchi-Tanaka M. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc Natl Acad Sci USA. 2014;111:7861–7866. doi: 10.1073/pnas.1321669111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Mi LM, Xu L, Yu CX, Li C, Chen CL. Genome-wide identification, characterization, interaction network and expression profile of GRAS gene family in sweet orange (Citrus sinensis) Sci Rep. 2019;9:2156. doi: 10.1038/s41598-018-38185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Roy B, Dunlap JR, Enganti R, von Arnim AG. Translational control of Arabidopsis meristem stability and organogenesis by the eukaryotic translation factor eIF3h. PLoS ONE. 2014;9:e95396. doi: 10.1371/journal.pone.0095396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517:377–380. doi: 10.1038/nature13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yan A, Han H, Li T, Geng Y, Liu X, Meyerowitz EM. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science. 2018;361:502–506. doi: 10.1126/science.aar8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplementary material 1 (DOC 766 kb)