Abstract
Not all individuals exposed to a pathogen develop illness: some are naturally resistant whereas others develop an asymptomatic infection. Epidemiological studies suggest that there is similar variability in susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. We propose that natural resistance is part of the disease history in some individuals exposed to this new coronavirus. Epidemiological arguments for natural resistance to SARS-CoV-2 are the lower seropositivity of children compared to adults, studies on closed environments of ships with outbreaks, and prevalence studies in some developing countries. Potential mechanisms of natural resistance include host genetic variants, viral interference, cross-protective natural antibodies, T cell immunity, and highly effective innate immune responses. Better understanding of natural resistance can help to advance preventive and therapeutic measures against infections for improved preparedness against potential future pandemics.
Keywords: innate immunity, natural resistance, trained immunity
Resistance to infection is variable between individuals
Not all individuals encountering a pathogen develop an ensuing disease. Most pathogenic microorganisms in nature do not infect humans and are instead specific for other hosts. By contrast, some individuals are fully resistant to infection with particular pathogens. Should an infection occur, the severity of the disease greatly varies between patients, and this is believed to be due to genetic, non-genetic (age, diet, lifestyle), and environmental factors. The capacity of a pathogen to cause an epidemic, or even a pandemic, greatly depends on the susceptibility of the host to infection, the mode of transmission of the infectious pathogen, and severity of the disease. In the current article we focus on the immunological mechanisms of natural resistance to pathogens. A strong argument for the relevance of immunological mechanisms to natural resistance to pathogens is given by the type of immune response elicited against an infection in humans versus other species: for example, bats, unlike humans, respond to coronavirus infection with high production of interferons (IFNs) and low inflammation, which is a likely mechanism that induces a high level of tolerance against such infections [1] (Box 1 ). By contrast, humans are known to respond with a strong inflammatory reaction during many viral infections such as those caused by influenza virus or coronaviruses [2,3].
Box 1. The immune system of bats.
There are more than 1400 species of bats, representing more than 20% of the world's mammalian species. Bats act as reservoirs of multiple human pathogens including fearsome viruses such as rabies, Ebola, Marburg, Nipah, MERS-CoV, SARS-CoV, and SARS-CoV-2. Surprisingly, these viruses are well-tolerated by bats and do not cause pathology.
Why are bats able to tolerate infections which are deadly for other mammals? Many details of bat immunology remain unknown, but the key seems to lie in their ability to mount a mild immune response and avoid the excessive inflammatory response that could lead to tissue damage and organ failure. First, as flying mammals, bats can increase their metabolic activity rates to levels twice as high as those found in rodents when running [59]. Second, the increased metabolic rate and high body temperature support an immune response that controls infections in a manner comparable to the rise in metabolic activity and the development of fever in humans affected by infection [60]. Third, several bat species express IFN-α constitutively, even in the absence of viral infections [61], which helps to limit the spread of viruses in the bat without developing an exacerbated systemic immune response. Combined with a reduced ability to detect viral DNA and bat-specific inflammasome adaptations (lack of inflammatory genes, low NLRP3 inflammasome function) [48,49], constitutive IFN-α expression helps to maintain low production of inflammatory cytokines, thus limiting the development of inflammatory responses that could cause tissue damage.
Alt-text: Box 1
The variability in the capacity of pathogens to cause infection is not only restricted to interspecies differences but also to immunological variation between individuals within a population. Why do some people not develop illness? Are there interindividual mechanisms similar to those that explain why some animal species are naturally resistant to a particular infection? To understand this we must conceptually demarcate two situations: natural resistance versus asymptomatic infection (see Glossary). These processes are mediated by different mechanisms: namely genetically and innate immune-mediated for natural resistance, versus potentially induced adaptive immunity for asymptomatic infections (Box 2 ).
Box 2. Host-dependent mechanisms associated with resistance to pathogens.
Natural resistance
The infectious agent is unable to infect the host; this may be due to genetic variants in genes important for entry mechanisms (e.g., the absence of a host cell receptor that is crucial for viral entry) or by rapid elimination of the pathogen by host resistance mechanisms. Such rapid clearance may be accomplished by potent components of our rapid-acting innate immune system, by effective heterologous crossreactive immunity by natural antibodies, by crossreactive T cells, or by the synergistic actions of these mechanisms [62]. Under such circumstances the disease process stops a few days after infection without engaging adaptive immune mechanisms.
Asymptomatic infection
The host goes through the entire cascade of interactions between the infectious agent and host defense mechanisms. In this situation, host defense mechanisms can mount a response strong enough to control the pathogen which will be ultimately eliminated or tolerated (e.g., HIV-1-infected long-term non-progressors). Both situations avoid an immunopathological inflammatory response. In these individuals, adaptive immunity will ensue and long-term immunological memory may be induced.
Alt-text: Box 2
Over the past 1.5 years, thousands of articles have been published on the development of immunity against SARS-CoV-2 in patients with mild or severe disease. Many of these studies describe antibody responses occurring during the natural course of infection and their role in recovery and protection [4]. Nevertheless, at least one-third of SARS-CoV-2-infected subjects, and perhaps even the majority, do not develop any clinical symptoms [5]. We briefly review the extensive knowledge on natural resistance in humans against infections and outline the epidemiological and immunological arguments that support the presence of natural resistance to SARS-CoV-2 infection in humans. We also propose that, among various types of infections, SARS-CoV-2 is no exception in that some individuals are naturally resistant to infection.
Natural resistance to infections in humans
Natural resistance against pathogens has been described for many infections, including some of the most severe and widespread infections in human populations. Exposure to Mycobacterium tuberculosis can lead to latent (asymptomatic) infection which can progress to active tuberculosis disease. However, some individuals, even when heavily exposed to a contagious tuberculosis patient, do not develop latent infection. This phenotype has been termed 'early clearance' [6]. Malaria is another well-known example of a human Plasmodium sp. infection in which natural resistance has been described [7]. Although malaria exerts strong evolutionary pressure on humans [8], a multitude of studies published more than 50 years ago have shown that some segments of the population are resistant to developing malaria. This resistance is associated with the presence of sickle-cell disease mutations in hemoglobin B or C genes [9], thalassemias [10], or the presence of the Duffy antigen receptor gene (DARC) in erythrocytes [11]. Genetic polymorphisms inducing natural resistance have also been described in HIV-1 infection, which in evolutionary terms represents a relatively recent infection. Some 25 years ago, ~10% of sex workers in Kenya who were highly exposed to HIV-1 were reported to remain uninfected after 3 years of follow-up [12]. It was later found that this was caused by genetic defects in the CCR5 entry receptor on CD4+ T cells: indeed, the CCR5-Δ32 deletion mutation eliminates this HIV-1 coreceptor on lymphocytes, and has provided robust protection against HIV-1 infection and AIDS [13].
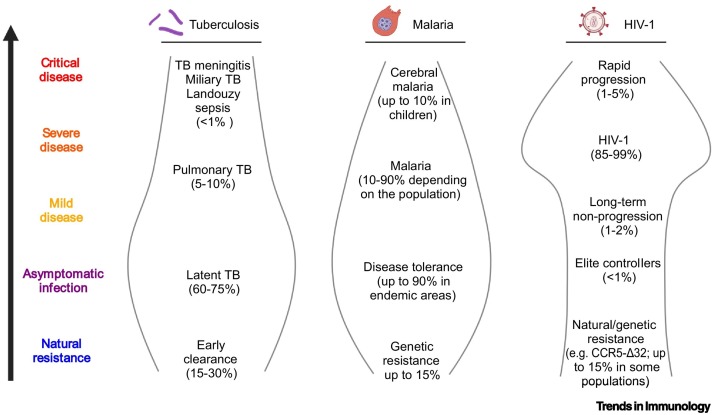
These studies present a picture in which the disease history of an individual can greatly vary depending of the variability of his or her susceptibility to disease: one person can be naturally resistant to an infection, another can be infected but remain asymptomatic or exhibit mild symptoms, whereas another person can become severely ill (Figure 1 ). We thus propose that natural resistance is an important component of disease history in most human infections. There is no reason to believe that this would be different for SARS-CoV-2 infection.
Figure 1.
Examples of natural resistance and disease phenotypes in human infections: tuberculosis (TB), malaria, and HIV-1.
Even though TB, malaria, and HIV-1 infection present high mortality rates, there is a large spectrum of responses ranging from natural resistance to severe forms. The prevalence of the various clinical endotypes varies per disease and depends on intrinsic factors such as the genetic makeup of individuals, as well as on extrinsic factors such as exposure to other environmental agents and pathogens; these can lead to disease tolerance or favor heterologous protective immunity, thereby decreasing morbidity and increasing the likelihood of host survival [30,50,63,64]. This figure was created using BioRender (https://biorender.com/).
Epidemiological arguments for natural resistance against SARS-CoV-2
Similarly to other viral infections such as HIV-1, SARS-CoV-1, and MERS, human infection with SARS-CoV-2 is evidently recent: it has caused a pandemic since the end of 2019. The burden of morbidity and mortality, where tens of millions have become infected and several million individuals have perished from the disease, is mainly due to the absence of protective immunological memory to the disease; indeed, because of the short time-period in which the disease has been present in the global population, evolutionary pressure to presumably increase resistance to infection and/or disease has yet to occur. However, mortality due to coronavirus disease 2019 (COVID-19) is highly prevalent in individuals who have passed reproductive age, and for this reason is likely to result in diminished evolutionary pressure, if at all. Nevertheless, evidence is starting to accumulate that not everyone is equally susceptible to SARS-CoV-2 infection [14], and some of us may even be naturally resistant (Figure 2 ).
Figure 2.
Demographic subgroups of individuals within a population, susceptibility to SARS-CoV-2 infection, and immunological defense mechanisms during COVID-19.
Individuals can be classified according to their degree of susceptibility, from higher to lower, and resistant individuals. These profiles are defined by the inflammatory responses developed in response to infection, interferon (IFN) release, and antibody production [3]. Potent IFN responses and lower inflammatory responses are generally linked to better prognosis during viral infections (i.e., SARS-CoV-2) because they are associated with enhanced early responses that prevent replication and limit viral invasiveness [65]. The specific mechanisms leading to the development of natural resistance to COVID-19 remain to be elucidated but most likely include genetic resistance, the development of heterologous B and T cell responses, and trained immunity. Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2. This figure was created using BioRender (https://biorender.com/).
The first likely mechanism that might mediate natural resistance against SARS-CoV-2 can be considered to be genetically mediated: low expression of the viral receptor ACE2 might be associated with resistance to disease, and such a potential mechanism has been suggested for children [15], although this possibility remains to be rigorously tested [16]. Moreover, although genetic variation is important and might explain up to 30–50% of interindividual immune variations (as revealed by functional genomics studies) [17], environmental factors are also important and might have more impact later in life [18].
A strong argument for the presence of naturally resistant individuals to SARS-CoV-2 infections in the population comes from the strong age-dependency in the epidemiology of COVID-19 [19]. Specifically, the low severity of COVID-19 in children is a fact [20], but a meta-analysis of epidemiological surveys of specific anti-SARS-CoV-2 antibodies in serum also show lower anti-SARS-CoV-2 antibody titers in children and adolescents compared to adults, even if the former cohabit with adults in the same environment and hence presumably receive equal exposure to the virus [21]. In a large survey in southwest Germany, the seroprevalence of SARS-CoV-2 antibodies in children aged 1–10 years was threefold lower compared to their parents [22], and the SARS-CoV-2 seroprevalence of children (from 5 months to 4.4 years) versus adults in a day-care environment in France was 4.3% versus 7.7%, respectively [23]. Thus, at least some children seem to be able to repel the infection without the need to strongly engage adaptive immunity, either through resistance to infection (perhaps low expression of SARS-CoV-2 receptors), or presumably because of effective innate immune responses, although this remains conjectural. Thus, the characteristics that influence a state of natural resistance at a young age seem to be lost – at least partly – with aging, but it is also likely that they might still prevail in some individuals for reasons that remain unknown.
Arguments supporting natural resistance to developing COVID-19 have also been provided by several studies assessing SARS-CoV-2-specific seroprevalence in different populations; results show that there are discrepancies between mathematical models of infection and the measured seroprevalence of antibodies in humans. Specifically, Sweden reported a seroprevalence of 7.3% in the Stockholm area in April 2020 despite mathematical models predicting an infection rate of 20–25% [24]. An interesting case study is that of the Diamond Princess cruise ship that in February 2020 was confronted with a COVID-19 outbreak while in the western Pacific ocean, and reported half of the global COVID-19 cases at that time outside China [25,26]. The infection rates on the cruise ship were mathematically projected to be higher than those occurring in open spaces because complete isolation was not possible, and members of the crew were also exposed to the virus. Different studies have estimated the basic reproduction number (R 0 ) during the outbreak period in the ship to be between 5.71 and 14.8 [25,26], which suggests that the number of exposed individuals was higher than the number of those who actually became infected. Subsequently, in contrast to the officially reported 712 individuals with PCR-positive results for COVID-19, one study reported a calculation in which this number should have been at least 1000 [26]. Although sensitivity issues with the diagnostic tests may explain some of the missing cases, the large difference is unlikely to be due to this sole cause. The discrepancy likely includes a significant number of individuals who encountered the virus but, presumably, were naturally resistant.
Notwithstanding the likely existence of natural resistance in some populations, there are large differences in COVID-19-related morbidity and mortality rates between countries [27]. After the first wave of the pandemic, studies from India reported 11 million COVID-19 cases and 155 000 deaths as of February 26, 2021i. Later, during the months of March to May 2021, a second wave of the pandemic was recorded caused by the SARS-CoV-2 delta variant, despite high COVID-19 seropositivity in major urban areas. For example, New Delhi showed 56% seropositivity in an Indian government surveyii; the delta variant infected up to 400 000 individuals per day in April–May 2021, attesting to the infectivity of this variant. However, even considering this second wave of the pandemic and potential deficiencies in case reporting, the death rate per capita in India (29 per 100 000 population) was substantially lower than the mortality rates in countries such as the USA (184), Brazil (243), Italy (211), and the UK (192 per 100 000)iii. Similarly, a lower case-fatality rate has been observed in India compared to other countries [28]. Thus, India has demonstrated relatively low mortality rates despite its high population density, lack of restrictive measurements, and suboptimal health infrastructure. Notably, in the state of Karnataka in India, 91% of the reported COVID-19 infections were asymptomatic [29].
On the one hand, the contrast in morbidity and mortality is remarkable between India, developing countries in sub-Saharan Africa, and developing countries in Latin America (such as Peru, Brazil, and Mexico). The demographic structure of the populations of these developing countries, where there are high percentages of young individuals in all these geographical locations, is unlikely to explain the differences in the burden of COVID-19 when comparing India, sub-Saharan Africa, and Latin America. On the other hand, it has been hypothesized that high infectious pressure in India and sub-Saharan Africa might contribute, at least in part, to a degree of natural resistance to pathogens [30]. The large pathogen burden in developing countries [31] may constantly engage the innate immune system, thus protecting individuals against new infections, and even leading to evolutionary adaptations in the long term, akin to the hygiene hypothesis [30].
In line with this, an example of possible large-scale natural resistance is reflected in the COVID-19 pandemic in sub-Saharan Africa. Although sub-Saharan countries might be expected to experience rapid expansion of the pandemic and considerable numbers of victims (compounded by the absence of a strong healthcare infrastructure and limited restrictive measures adopted), the incidence and mortality of COVID-19 have been much lower than in other regions of the world where countries have more prepared and advanced healthcare systems and stronger containment measuresi. This difference might be in part due to factors such as the relatively low age of the population in Africa [32], but might also suggest a high degree of natural resistance in the population, although this remains to be tested. Of note, a recent study compared the prevalence of crossreactive antibodies against SARS-CoV-2 in COVID-19 patient samples obtained before the full-blown start of the pandemic in sub-Saharan Africa versus the USA; seroprevalence was six- to eightfold higher in plasma samples from Tanzania and Zambia than in samples from the USA [33]. It is reasonable to speculate that these crossreactive antibodies arose in response to previous contacts with other coronaviruses present in the environment in sub-Saharan countries, and that they induced cross-protection against SARS-CoV-2 infection (and thus natural resistance) in African populations. We argue that, collectively, these studies suggest that natural resistance to SARS-CoV-2 can be present in human populations and may impact on the outcomes of the pandemic for different age groups and countries.
Immunological mechanisms explaining natural resistance
One important aspect to consider is the fact that two types of responses might protect a host from the deleterious effects of an infection: on the one hand, there are resistance mechanisms that aim to eliminate a microorganism, on the other hand, there are tolerance mechanisms that aim to avoid collateral damage caused by inflammation or overactivation of the immune response. Indeed, disease tolerance represents a crucial component for host defense by ensuring that the collateral damage induced by an effective immune system is limited and does not place the host in danger [34]. A proper balance between resistance and tolerance is crucial for the most effective survival strategy during infections [35], including for COVID-19, in which overwhelming inflammation is dangerous in critically ill patients [36].
An important environmental process that can provide natural resistance against viral infections, even if only for a relatively short period, is viral interference. This is the process by which previous exposure of cells to another virus induces inhibition of viral reproduction [37,38]. For instance, rhinovirus infection can protect against influenza A virus (IAV) infection in an experimental model. In seminal experiments published in 1957, Isaacs and Lindemann experimentally demonstrated this phenomenon in vitro; eggs exposed to IAV produced a factor, 'interferon', that when transferred to new eggs could prevent infection with the virus [39]. Recently, interference between rhinoviruses and SARS-CoV-2 was also demonstrated where rhinoviruses were able to block the replication of SARS-CoV-2 in human epithelial cells [40]. In addition to IFN-dependent mechanisms, other mechanisms might also play a role in viral interference: for instance, some studies have suggested that there is competition for the receptors used for viral entry, and furthermore that occupancy or downmodulation of cellular receptors for particular viruses might also be involved [41]. In addition, recent epidemiological data also suggest that there is viral interference between IAV and coronaviruses [42]. We therefore posit that higher circulation/dissemination of different viruses in India or sub-Saharan Africa might contribute to explaining the lower susceptibility to SARS-CoV-2 infections in these populations compared to Europe and the Americas, and where the process of viral interference might also play an important role.
As alluded to above, natural resistance against an infection might also be afforded by pre-existing adaptive immunity. For instance, approximately half of individuals not exposed to SARS-CoV harbor reactive CD4+ T cells against SARS-CoV-2 in peripheral blood [43]. Moreover, pre-existing memory CD4+ T cells reactive to SARS-CoV-2 and with comparable affinity to the common-cold human coronaviruses (i.e., OC43, NL63, HKU1 and 229E) have been reported in human unexposed individuals; this finding might support the idea that exposure to common-cold viruses might result in some degree of protection against SARS-CoV-2, although this remains to be thoroughly tested [44]. In addition, pre-existing polymerase-specific CD4+ T cells have been described in persistently seronegative healthcare workers, and these might presumably lead to abortive infection and protect these individuals from COVID-19 [45]. However, other studies suggest that crossreactive antibodies and CD4+ T cells do not contribute to the clearance of SARS-CoV-2 infections. Specifically, one study reported that infections with SARS-CoV-2 increased the circulating titers of seasonal human coronavirus antibodies but did not neutralize SARS-CoV-2 virion particles [46]. Another group measured the anti-SARS-CoV-2 neutralizing activity in sera obtained from individuals infected with seasonal coronavirus before the pandemic and did not detect any crossreactive neutralizing activity against SARS-CoV-2, suggesting that the crossreactive coronavirus antibodies may have not been neutralizing [47]. However, most of these studies have been conducted in small samples from urban settlements in highly developed cities from the USA or Europe, where individuals presumably maintain high hygiene standards and are exposed to low pathogen burdens, including coronaviruses. Evidently, such studies would benefit from the inclusion of larger and more diverse population samples, and from comparison of the neutralization potential of sera obtained from individuals living under different socioeconomic conditions and environments. Finally, one aspect that deserves attention in future studies is that of potentially protective regulatory T cell (Treg) effects during SARS-CoV-2 infections; these populations, by inhibiting hyperinflammation and ensuring a tolerant phenotype, might be more relevant than was previously anticipated [48].
There is another potential immune mechanism via which natural resistance to SARS-CoV-2 might be induced – the presence of effective innate immune responses which might eliminate the virus before adaptive immunity is engaged. Specifically, an innate immune profile characterized by high cytokine production upon stimulation with pathogens has been associated with early clearance/natural resistance; for example, this has been described in detail for tuberculosis (TB) [49]. Indeed, stronger heterologous cytokine responses upon blood lymphocyte stimulation with bacterial antigens were reported in an Indonesian population in which close contacts exhibited natural resistance to TB [50], compared to a population susceptible to the disease. This suggested that, for various pathogen scenarios, the increased ability of innate immune cells to respond to infection by producing cytokines upon stimulation can contribute to establishing natural resistance in some individuals.
Of note, the quality and nature of innate immune responses is definitely not static and should not be ignored. For instance, Bacillus Calmette–Guérin (BCG) vaccination not only provides some protection against TB but can also enhance innate immunity, as evidenced by higher cytokine production upon stimulation of circulating monocytes with non-mycobacterial stimuli compared to controls [51]. The latter process, wherein vaccines or particular infections can induce long-term strengthening of innate immune responses, also termed trained immunity, is mediated through long-lasting epigenetic and metabolic rewiring of myeloid cells [52,53]. Induction of trained immunity by live attenuated vaccines such as BCG, measles-mumps-rubella (MMR), and oral polio vaccine (OPV) has been suggested as a potential approach for eliciting protection against COVID-19 because it increases innate antiviral responses early during an infection; indeed, several clinical trials are investigating this hypothesis [54,55]. A recently published initial observational study suggested an association between BCG vaccination and reduced COVID-19 symptoms in a population of elderly individuals from Greece [56], and a case–control study investigated the impact of MMR on immunity and reported significant protection against COVID-19 [57], supporting the hypothesis that some live attenuated vaccines may induce nonspecific protection against COVID-19. Moreover, abundant exposure to environmental microorganisms might also play a role in inducing trained immunity, and thereby enhance putative natural resistance to pathogens such as SARS-CoV-2. Collectively, we argue that these studies suggest multiple mechanisms that might be implicated in inducing a state of natural resistance against infections in humans, including with SARS-CoV-2: these may consist of viral interference as well as heterologous innate and adaptive immune processes. Thorough and robust preclinical and clinical assessments are eagerly awaited to test these hypotheses (Figure 3, Key figure).
Figure 3.
Key figure. Mechanisms of human natural resistance to viral infections.
The mechanisms of natural resistance to viral infections include genetic polymorphisms leading to the inability of a virus to infect human cells (e.g., absence of the entry receptor); viral interference [mainly interferon (IFN)-mediated], where a virus-infected cell becomes resistant against a second infection; trained innate immunity, which is induced by epigenetic reprogramming of innate immune cells (e.g., myeloid cells and natural killer cells); and heterologous antibodies and T cells that can recognize and respond to unrelated antigens/pathogens upon exposure to an antigen/pathogen of a different nature. This figure was created using BioRender (https://biorender.com/).
Concluding remarks
Understanding the entire clinical spectrum of an infection is crucial to elucidating its impact at the individual as well as population levels. Knowing the extent to which an infection (especially one that can instigate a pandemic) can cause severe, mild, or asymptomatic disease is important for determining public health measures to limit its burden. Similarly, identifying individuals within the population who are naturally resistant against a particular infection is important for understanding host defense as well as for informing novel putative strategies for prevention and therapy.
Presently, the data regarding natural resistance to COVID-19 are accumulating, albeit scattered. Efforts should be made on several fronts to understand these data more comprehensively. Pandemics are not exceptions in human history. The emergence of new pathogens or new variants of old pathogens can lead to the global spread of infections in the absence of immunological memory. Although the development of classical types of vaccines should and will remain a fundamental tool for the prevention of future pandemics, the design, development, testing, production, and distribution of future vaccines needs at least 12–24 months – even in the most optimistic scenarios. In the initial phase of a pandemic the spread of the infection can have dire consequences for global health and economy, as has been witnessed for COVID-19. It is therefore crucial that we become better prepared for a future epidemic/pandemic, not if, but when. We propose that the development of parallel preventive strategies to boost general natural resistance of a host (until specific vaccines can be developed) will be crucial. One such strategy might be the development of vaccines that can stimulate heterologous/trained immune responses, and thereby induce broad (partial) protection against multiple pathogens (Figure 4 ). One might also envisage testing additional live attenuated vaccines to induce trained immunity, and/or the development of novel adjuvants with such properties. Indeed, in addition to previously studied live attenuated vaccines such as BCG, MMR, and OPV, recent work testing the AS03-adjuvanted IAV vaccine in humans also demonstrated that such vaccination could stimulate epigenetic changes in myeloid cells which persisted for several weeks to months, and conferred heightened protection against unrelated viruses such as dengue and Zika [58]. Understanding natural resistance to COVID-19 in years to come may thus represent a vital step for future pandemic preparedness (see Outstanding questions).
Figure 4.
Proposal for promoting pandemic preparedness: the use of immunological boosters and novel vaccines to trigger natural resistance against infections.
Upon emergence of a new pathogen, there is a period of uncertainty and potentially large morbidity and mortality until the development of a specific novel vaccine is achieved. Until that moment, the use of existing (or novel) vaccines that induce natural resistance and/or trained immunity might be used to increase the immunological responses of the population to such threatening pathogen(s). This may serve to bridge vaccination with reduced transmission, morbidity, and mortality elicited by the new pathogen. This figure was created using BioRender (https://biorender.com/).
Outstanding questions.
What is the extent of natural resistance to SARS-CoV-2 infection in different populations across the world?
Are there environmental factors that substantially influence natural resistance against SARS-CoV-2 infection, such as infectious pressure, seasonality, or even nutrition?
What are the genetic, immunological, and molecular factors that render some individuals naturally resistant to SARS-CoV-2 infection?
Can we take advantage of the mechanisms of natural resistance to design and develop immunomodulators and/or therapeutic vaccines against COVID-19?
Are the qualitative and quantitative immune responses of individuals in different populations substantially similar or different in response to a specific SARS-CoV-2 vaccine?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
The work of M.G.N. was supported by a European Research Council (ERC) Advanced Grant (833247), a Spinoza Grant of the Netherlands Organization for Scientific Research, and a COVID-19 FastGrant. R.v.C was supported by National Institute of Health (R01AI145781). J.D-A. was supported by a Veni grant of The Netherlands Organization for Scientific Research (09150161910024). Work in the laboratory of B.P. was supported by grants from the National Institutes of Health (U19AI090023, R01AI048638, R01 DK057665, U19AI057266), the Bill and Melinda Gates Foundation, and Open Source Philanthropy.
Declaration of interests
The authors declare no conflicts of interest.
Glossary
- Asymptomatic infection
also known as subclinical infection, a condition is considered to be asymptomatic if the patient is a carrier of a disease or infection but does not experience the symptoms that are normally associated with it.
- Bacillus Calmette-Guérin (BCG) vaccination
vaccination against Mycobacterium tuberculosis has been shown to increase the likelihood of eliminating mycobacteria early during infection (so-called 'early clearers').
- Basic reproduction number (R0)
the average number of individuals that one infected person will pass on a virus to.
- Crossreactive antibodies
antibodies that recognize antigens other than the antigens they were originally generated against.
- Hygiene hypothesis
this proposes that childhood exposure to germs and infections can enable the immune system to develop a balanced response to infections and allergens.
- Morbidity
the number of people who become ill in a given place and time-period relative to the total population.
- Mortality
the number of people who die in a given place and time-period relative to the total population.
- Seroprevalence
the proportion of people in a population who harbor antibodies against a specific antigen in their serum – an indication that they have been exposed to a specific infectious organism.
- Trained immunity
long-term functional reprogramming of innate immune cells evoked by exogenous or endogenous stimuli; leads to an altered response to a second challenge after return to a non-activated state.
- Viral interference
the process by which previous cell exposure to one virus can inhibit the replication of a second virus.
Resources
ihttps://coronavirus.jhu.edu/map.htmliihttps://www.mygov.in/covid-19/iiihttps://coronavirus.jhu.edu/data/mortalityReferences
- 1.Banerjee A., et al. Bats and coronaviruses. Viruses. 2019;11:41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Tang H. Aberrant coagulation causes a hyper-inflammatory response in severe influenza pneumonia. Cell. Mol. Immunol. 2016;13:432–442. doi: 10.1038/cmi.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giamarellos-Bourboulis E.J., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poland G.A., et al. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sah P., et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verrall A.J., et al. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology. 2014;141:506–513. doi: 10.1111/imm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariuki S.N., Williams T.N. Human genetics and malaria resistance. Hum. Genet. 2020;139:801–811. doi: 10.1007/s00439-020-02142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization . WHO; 2018. World Malaria Report 2018. [Google Scholar]
- 9.Allison A.C. Protection afforded by sickle-cell trait against subtertian malareal infection. Br. Med. J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams T.N., Weatherall D.J. World distribution, population genetics, and health burden of the hemoglobinopathies. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabeti P.C., et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 12.Fowke K.R., et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 13.Church J.A. Long-term control of HIV by CCR5 delta32/delta32 stem-cell transplantation. Pediatrics. 2009;124:S159. [Google Scholar]
- 14.Pollard C.A., et al. The COVID-19 pandemic: a global health crisis. Physiol. Genomics. 2020;52:549–557. doi: 10.1152/physiolgenomics.00089.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunyavanich S., et al. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann P., Curtis N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch. Dis. Child. 2020;106:429–439. doi: 10.1136/archdischild-2020-320338. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., et al. A functional genomics approach to understand variation in cytokine production in humans. Cell. 2016;167:1099–1110. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Brodin P., et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 20.Bialek S., et al. Coronavirus disease 2019 in children – United States, February 12–April 2, 2020. Morb. Mortal. Wkly Rep. 2020;69:422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viner R.M., et al. Susceptibility to SARS-CoV-2 infection among children and adolescents compared with adults: a systematic review and meta-analysis. JAMA Pediatr. 2021;175:143–156. doi: 10.1001/jamapediatrics.2020.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tönshoff B., et al. Prevalence of SARS-CoV-2 infection in children and their parents in southwest Germany. JAMA Pediatr. 2021;175:586–593. doi: 10.1001/jamapediatrics.2021.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachassinne E., et al. SARS-CoV-2 transmission among children and staff in daycare centres during a nationwide lockdown in France: a cross-sectional, multicentre, seroprevalence study. Lancet Child Adolesc. Health. 2021;5:256–264. doi: 10.1016/S2352-4642(21)00024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlowski E.J.W., Goldsmith D.J.A. Four months into the COVID-19 pandemic, Sweden’s prized herd immunity is nowhere in sight. J. R. Soc. Med. 2020;113:292–298. doi: 10.1177/0141076820945282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocklöv J., et al. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai C.C., et al. The Bayesian susceptible-exposed-infected-recovered model for the outbreak of COVID-19 on the Diamond Princess cruise ship. Stoch. Environ. Res. Risk Assess. 2021;2021:1. doi: 10.1007/s00477-020-01968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atzrodt C.L., et al. A guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020;287:3633–3650. doi: 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen J. Will India’s devastating COVID-19 surge provide data that clear up its death ‘paradox’? Science. 2021 https://www.science.org/content/article/will-india-s-devastating-covid-19-surge-provide-data-clear-its-death-paradox Published online April 30, 2021. [Google Scholar]
- 29.Kumar N., et al. Descriptive epidemiology of SARS-CoV-2 infection in Karnataka state, South India: transmission dynamics of symptomatic vs. asymptomatic infections. EClinicalMedicine. 2021;32 doi: 10.1016/j.eclinm.2020.100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domínguez-Andrés J., Netea M.G. Impact of historic migrations and evolutionary processes on human immunity. Trends Immunol. 2019;40:1105–1119. doi: 10.1016/j.it.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbafati C., et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. doi: 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawal Y. Africa’s low COVID-19 mortality rate: a paradox? Int. J. Infect. Dis. 2021;102:118–122. doi: 10.1016/j.ijid.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tso F.Y., et al. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int. J. Infect. Dis. 2021;102:577–583. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medzhitov R., et al. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider D.S., Ayres J.S. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Q., et al. Targeting inflammation and cytokine storm in COVID-19. Pharmacol. Res. 2020;159 doi: 10.1016/j.phrs.2020.105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz-Cherry S. Viral interference: the case of influenza viruses. J. Infect. Dis. 2015;212:1690–1691. doi: 10.1093/infdis/jiv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., et al. Antibody seroprevalence in the epicenter Wuhan, Hubei, and six selected provinces after containment of the first epidemic wave of COVID-19 in China. Lancet Reg. Health West. Pac. 2021;8 doi: 10.1016/j.lanwpc.2021.100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaacs A., Lindemann J. Virus interference. II. Some properties of interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:268–273. [Google Scholar]
- 40.Dee K., et al. Human rhinovirus infection blocks SARS-CoV-2 replication within the respiratory epithelium: implications for COVID-19 epidemiology. J. Infect. Dis. 2021;224:31–38. doi: 10.1093/infdis/jiab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remion A., et al. Co-infection, super-infection and viral interference in HIV. Retrovirology. 2013;10:P72. [Google Scholar]
- 42.Nickbakhsh S., et al. Virus–virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. U. S. A. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Bert N., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 44.Mateus J., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swadling L., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021 doi: 10.1038/S41586-021-04186-8. Published online November 10, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson E.M., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poston D., et al. Absence of severe acute respiratory syndrome coronavirus 2 neutralizing activity in prepandemic sera from individuals with recent seasonal coronavirus infection. Clin. Infect. Dis. 2020;73:e1208–e1211. doi: 10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galgani M., et al. Role of metabolism in the immunobiology of regulatory T cells. J. Immunol. 2016;197:2567–2575. doi: 10.4049/jimmunol.1600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster M., et al. BCG-induced protection against Mycobacterium tuberculosis infection: evidence, mechanisms, and implications for next generation vaccines. Immunol. Rev. 2021;301:122–144. doi: 10.1111/imr.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verrall A.J., et al. Early clearance of Mycobacterium tuberculosis: the INFECT case contact cohort study in Indonesia. J. Infect. Dis. 2019;221:1351–1360. doi: 10.1093/infdis/jiz168. [DOI] [PubMed] [Google Scholar]
- 51.Verrall A.J., et al. Early clearance of Mycobacterium tuberculosis is associated with increased innate immune responses. J. Infect. Dis. 2020;221:1342–1350. doi: 10.1093/infdis/jiz147. [DOI] [PubMed] [Google Scholar]
- 52.Domínguez-Andrés J., et al. Advances in understanding molecular regulation of innate immune memory. Curr. Opin. Cell Biol. 2020;63:68–75. doi: 10.1016/j.ceb.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Bekkering S., et al. Trained immunity: reprogramming innate immunity in health and disease. Annu. Rev. Immunol. 2021;39:667–669. doi: 10.1146/annurev-immunol-102119-073855. [DOI] [PubMed] [Google Scholar]
- 54.Ten Doesschate T., et al. Two randomized controlled trials of bacillus Calmette–Guérin vaccination to reduce absenteeism among health care workers and hospital admission by elderly persons during the COVID-19 pandemic: a structured summary of the study protocols for two randomised controlled trials. Trials. 2020;21:481. doi: 10.1186/s13063-020-04389-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madsen A.M.R., et al. Using BCG vaccine to enhance non-specific protection of health care workers during the COVID-19 pandemic: a structured summary of a study protocol for a randomised controlled trial in Denmark. Trials. 2020;21:799. doi: 10.1186/s13063-020-04714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsilika M., et al. ACTIVATE-2: a double-blind randomized trial of BCG vaccination against COVID19 in individuals at risk. MedRxiv. 2021 doi: 10.1101/2021.05.20.21257520. Published online May 24, 2021. [DOI] [Google Scholar]
- 57.Gujar N., et al. A case control study to assess effectiveness of measles containing vaccines in preventing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children. Hum. Vaccin. Immunother. 2021;17:3316–3321. doi: 10.1080/21645515.2021.1930471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wimmers F., et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell. 2021;17:3316–3321. doi: 10.1016/j.cell.2021.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Shea T.J., et al. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hock R.J. The metabolic rates and body temperatures of bats. Biol. Bull. 1951;101:289–299. [Google Scholar]
- 61.Zhou P., et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arunachalam P.S., et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat. Med. 2020;26:932–940. doi: 10.1038/s41591-020-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vannberg F.O., et al. Human genetic susceptibility to intracellular pathogens. Immunol. Rev. 2011;240:105–116. doi: 10.1111/j.1600-065X.2010.00996.x. [DOI] [PubMed] [Google Scholar]
- 64.Galvani A.P., Slatkin M. Evaluating plague and smallpox as historical selective pressures for the CCR5-delta32 HIV-resistance allele. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15276–15279. doi: 10.1073/pnas.2435085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye Q., et al. The pathogenesis and treatment of the ‘cytokine storm’ in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]