Abstract
SARS-CoV-2 infection causes diverse outcomes ranging from asymptomatic infection to respiratory distress and death. A major unresolved question is whether prior immunity to endemic, human common cold coronaviruses (hCCCoVs) impacts susceptibility to SARS-CoV-2 infection or immunity following infection and vaccination. Therefore, we analyzed samples from the same individuals before and after SARS-CoV-2 infection or vaccination. We found hCCCoV antibody levels increase after SARS-CoV-2 exposure, demonstrating cross-reactivity. However, a case-control study indicates that baseline hCCCoV antibody levels are not associated with protection against SARS-CoV-2 infection. Rather, higher magnitudes of pre-existing betacoronavirus antibodies correlate with more SARS-CoV-2 antibodies following infection, an indicator of greater disease severity. Additionally, immunization with hCCCoV spike proteins before SARS-CoV-2 immunization impedes the generation of SARS-CoV-2-neutralizing antibodies in mice. Together, these data suggest that pre-existing hCCCoV antibodies hinder SARS-CoV-2 antibody-based immunity following infection and provide insight on how pre-existing coronavirus immunity impacts SARS-CoV-2 infection, which is critical considering emerging variants.
Keywords: SARS-CoV-2, COVID-19, HKU1, OC43, 229E, NL63, antibody, pre-existing immunity
Graphical abstract
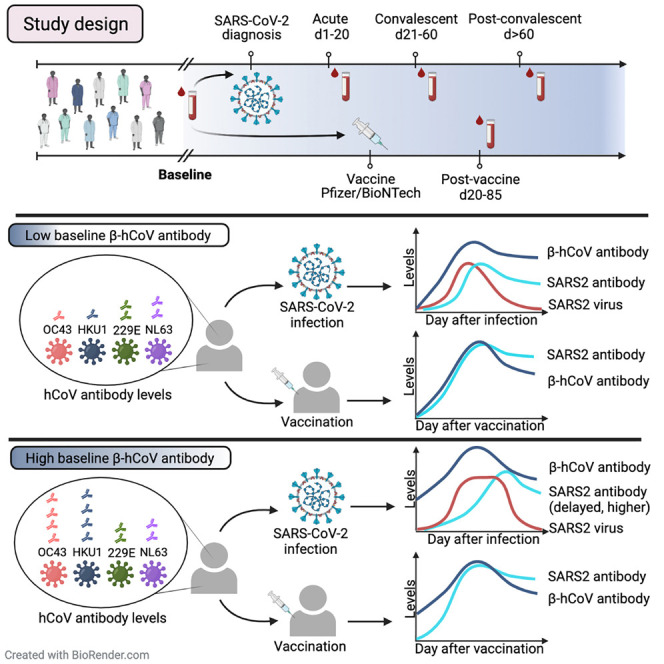
A major unresolved question is whether prior immunity to endemic, human common cold coronaviruses (hCCCoVs) impacts susceptibility to SARS-CoV-2 infection. Lin et al. analyze hCCCoV antibodies in the same individuals before and after SARS-CoV-2 infection, finding pre-existing betacoronavirus antibodies may hinder SARS-CoV-2-effective immunity following infection.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces highly variable disease ranging from very mild or no symptoms to severe respiratory distress and death. Certain comorbidities contribute to the diverse outcomes; however, these factors do not account for all the heterogeneity observed between infected individuals. A major unresolved question is whether susceptibility to SARS-CoV-2 infection and disease severity after infection are impacted by immunity to human common cold coronaviruses (hCCCoVs) that were circulating prior to the SARS-CoV-2 pandemic. Four hCCCoVs that are prevalent worldwide have been endemic in humans for decades and typically induce mild upper respiratory disease and account for ∼30% of “common colds” (Forni et al., 2017). HKU1 and OC43 are betacoronaviruses, as is SARS-CoV-2, which are evolutionarily distinct from the alphacoronaviruses, 229E and NL63. Despite dramatic difference in disease severity induced by the viruses, SARS-CoV-2 and the endemic hCCCoVs share ∼30% homology within the spike proteins (Hicks et al., 2021). Studies identified cross-reactive antibodies that bind both SARS-CoV-2 and hCCCoVs (Ladner et al., 2020; Ng et al., 2020; Wec et al., 2020). However, it is unclear how pre-existing hCCCoV antibodies impact the immune response against SARS-CoV-2 infection (Sealy and Hurwitz, 2021). Prior hCCCoV infections could augment SARS-CoV-2 immunity if hCCCoV antibodies are sufficiently cross-reactive with SARS-CoV-2 to be induced through immunological recall or “back-boosting” and support viral clearance (Fonville et al., 2014). Conversely, pre-existing hCCCoV humoral immunity could hinder the generation of effective SARS-CoV-2-specific antibodies by expanding cross-reactive antibodies that do not neutralize SARS-CoV-2. Further, existing hCCCoV immunity may exacerbate disease by facilitating viral entry into Fc receptor (FcR)-expressing cells to cause antibody-dependent enhancement of disease (Arvin et al., 2020; Iwasaki and Yang, 2020). Since hCCCoV immunity could influence the outcome of SARS-CoV-2 infection in several ways, it is critical to ascertain the impact of pre-existing hCCCoV antibodies on immunity to SARS-CoV-2 infection.
Reports investigating whether antibodies specific for hCCCoVs are boosted following SARS-CoV-2 infection yielded conflicting results. Some data suggested antibodies specific for hCCCoVs were not boosted following SARS-CoV-2 infection (Dugas et al., 2021a, 2021b; Loos et al., 2020), while others reported a boost only in OC43-specific antibodies (Anderson et al., 2021; Guo et al., 2021; Nguyen-Contant et al., 2020; Prévost et al., 2020). Additional studies found a boost in both HKU1 and OC43 antibodies (Aydillo et al., 2021; Cohen et al., 2021; Gouma et al., 2021; Westerhuis et al., 2021) or in antibodies specific for all four hCCCoVs following SARS-CoV-2 infection (Ng et al., 2020; Shrock et al., 2020). Yet, other reports surprisingly found a boost predominantly in antibodies specific for the alphacoronaviruses (Becker et al., 2021; Ortega et al., 2021). A major factor contributing to these inconsistencies is that prior studies did not examine the level of hCCCoV antibodies in the same individual before and after SARS-CoV-2 infection.
Assessing whether prior hCCCoV immunity impacts SARS-CoV-2 disease susceptibility has also yielded inconsistent results (Sealy and Hurwitz, 2021). While some studies reported that the levels of hCCCoV antibodies did not correlate with disease severity or likelihood of becoming infected (Anderson et al., 2021; Gombar et al., 2021; Loos et al., 2020), others concluded that higher levels of hCCCoV antibodies were associated with milder disease (Becker et al., 2021; Dugas et al., 2021a, 2021b; Henss et al., 2021; Ortega et al., 2021; Sagar et al., 2021; Shrock et al., 2020) or with a shorter duration of symptoms (Gouma et al., 2021). Conversely, others found higher levels of hCCCoV antibodies correlated with increased SARS-CoV-2 disease severity (Aydillo et al., 2021; Guo et al., 2021; Prévost et al., 2020; Westerhuis et al., 2021). The health status varied greatly in the cohorts tested in the previous studies, and most of these studies did not test samples from the same individual before and after SARS-CoV-2 infection, which likely contributes to the discrepancy in conclusions. Thus, the impact of pre-existing hCCCoV immunity on susceptibility to SARS-CoV-2 infection remains unresolved.
Here, we measured hCCCoV immunoglobin (Ig) G, IgM, and IgA antibodies in samples obtained from the same individual before and after PCR-confirmed SARS-CoV-2 infection. We observed significant increases of betacoronaviruses IgG antibodies; however, high levels of hCCCoV antibodies were not associated with protection against SARS-CoV-2 infection. Conversely, a greater increase in hCCCoV antibodies correlated with higher antibody levels to SARS-CoV-2 following infection, which were associated with increased disease severity. Moreover, mice immunized with hCCCoV spike proteins prior to SARS-CoV-2 spike exhibited a profound decrease in SARS-CoV-2-neutralizing antibodies relative to mice only immunized with SARS-CoV-2 spike. Overall, these data suggest that pre-existing hCCCoV IgG antibodies may hinder the immune response to SARS-CoV-2.
Results
Common hCCCoV antibody isotypes associate with age and direct patient contact
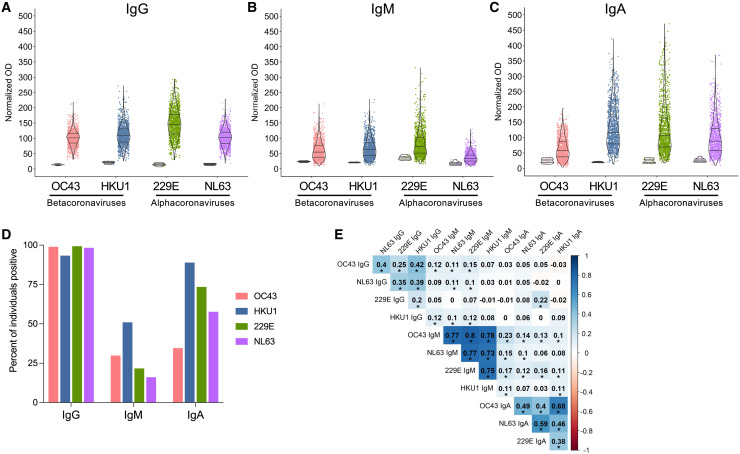
We established a prospective, longitudinal cohort (St. Jude Tracking of Viral and Host Factors Associated with COVID-19 study, SJTRC) of St. Jude employees who provided a baseline blood sample at enrollment and underwent weekly nasal swab screening for SARS-CoV-2 infection by PCR (Table S1). Individuals who tested positive during the study provided samples at two time points following infection. Additionally, participants who did not become infected gave samples after vaccination. This design allowed analysis of samples from the same individuals taken before and after SARS-CoV-2 infection or vaccination. Importantly, weekly nasal swab screening identified asymptomatic infections throughout the study period. To assess hCCCoV immunity prior to SARS-CoV-2 infection, we analyzed 1,202 baseline samples for antibodies specific for the spike proteins of OC43, HKU1, 229E, and NL63 by ELISA. To control for plate-to-plate variability, the same positive control samples were tested on each plate, and the normalized optical density (OD) for each sample was presented. Although antibody levels varied among individuals, IgG antibodies specific for all four of the hCCCoV spike proteins were identified in nearly all participants (Figures 1A and 1D). hCCCoV IgM antibodies were less prevalent than IgG and IgA, with IgA antibodies exhibiting the greatest variability (Figures 1A–1D). Interestingly, there were stronger correlations between antibody isotypes rather than specificity to a particular virus (Figure 1E). For example, individuals with high levels of HKU1 IgM were more likely to have IgM antibodies specific for the other three hCCCoVs rather than HKU1 IgG and IgA. Further, individuals with high levels of HKU1 IgG did not necessarily have high levels of HKU1 IgA and IgM. Together, these data indicate that nearly every individual had antibodies specific for all four hCCCoVs prior to SARS-CoV-2 infection or vaccination. Moreover, the stronger correlations with antibody isotype compared to virus type suggest there is cross-reactivity among hCCCoV-specific antibodies, with a higher degree of promiscuity in the IgM response followed by IgA then IgG, consistent with previous studies (Becker et al., 2021; Poston et al., 2021).
Figure 1.
Wide variation in baseline hCCCoV antibody levels
(A–C) Samples from 1,202 individuals taken prior to SARS-CoV-2 infection were analyzed by ELISA for (A) IgG, (B) IgM, and (C) IgA antibodies specific for spike proteins of OC43, HKU1, 229E, and NL63. Normalized ODs are presented, which is the percent ratio of the sample OD relative to the OD of the positive control of the plate. Negative control samples from young individuals in the FLU09 cohort are shown on the left for each antigen.
(D) The percent of individuals with a positive value for each isotype as determined by a normalized OD greater than three times the average of the negative controls.
(E) Clustered heatmap of Spearman’s correlation coefficients between the hCCCoV antibodies in baseline samples (n = 1,202). Asterisks indicate significant correlations after adjustment for multiple comparisons with the Bonferroni correction (∗p < 0.05).
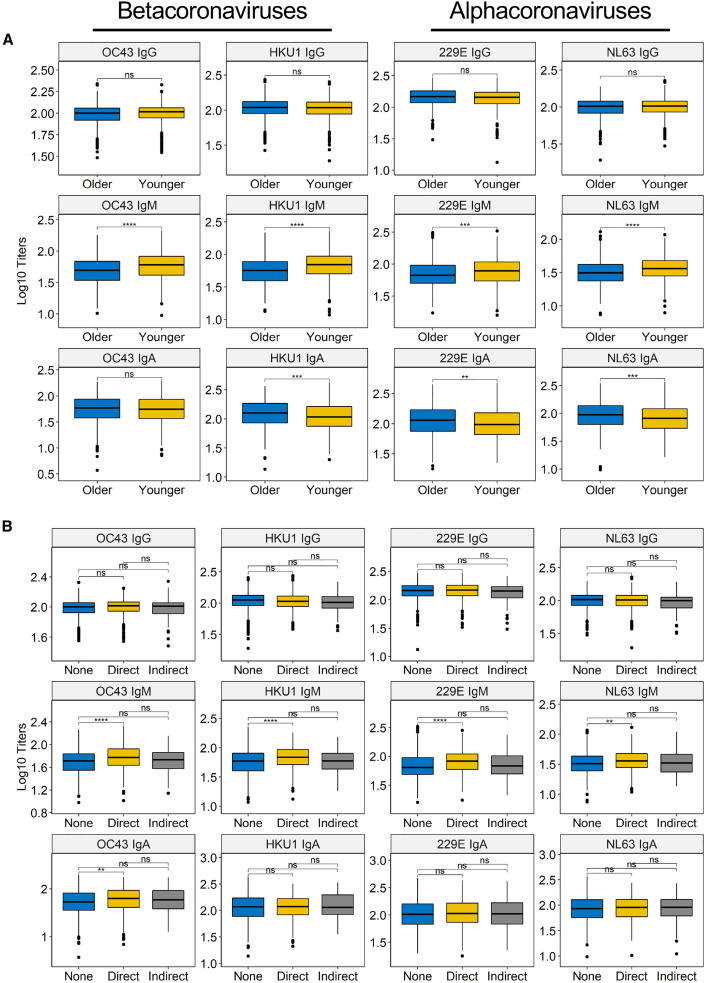
We next examined whether the level of hCCCoV antibodies at baseline correlated with age, sex, race, or direct patient contact. We compared antibody levels in individuals above and below the median age at the time of enrollment, which was 43 years of age (Table S1). We found that older individuals had significantly higher levels of IgA against HKU1, 229E, and NL63 (Figure 2 A). Conversely, younger individuals had significantly higher IgM levels reactive with all four of the hCCCoVs compared to older individuals. We also found that females had higher levels of all four hCCCoV IgM antibodies and higher OC43 IgA antibodies compared to males (Figure S1A). Additionally, IgG and IgA antibody levels differed across race/ethnicity groups in approximately 10% of the analyses (Figure S1B). Since the study participants are employees at a pediatric hospital and interactions with children may increase exposure to hCCCoVs, we assessed the correlation between hCCCoV antibodies and direct patient contact. Individuals with direct patient contact had higher levels of IgM antibodies specific for all four hCCCoVs, as well as OC43 IgA (Figure 2B). Together, these data indicate that in the SJTRC cohort, younger, female participants with direct patient contact were more likely to have elevated levels of hCCCoV IgM.
Figure 2.
hCCCoV IgM levels inversely correlate with age and are higher in individuals with direct patient contact
(A) hCCCoV-normalized ODs were compared between younger (<43 years) versus older (³43 years) individuals based on median age of the cohort.
(B) Participants self-reported whether they had direct, indirect, or no patient contact. Statistical significance was determined by the Wilcoxon–Mann–Whitney test with Bonferroni adjustment (ns, not significant; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
Since the SJTRC cohort did not include individuals younger than 20 years of age, we also analyzed hCCCoV antibody levels in samples collected from a previous study, the FLU09 cohort, that included a wider age range of participants. Similar to previous reports (Selva et al., 2021), we found higher levels of most of the hCCCoV IgG antibodies and all of the IgA antibodies in older individuals compared to younger individuals (Figures S2A–S2I). Unexpectedly, the levels of IgM antibodies for most of the hCCCoVs were low in young individuals, peaked around 20 years of age, and then declined with age (Figures S2B and S2E). Therefore, we examined whether there was a correlation with antibody levels and age in individuals 0–14 (Figures S2J–S2L) or 17–54 years of age (Figures S2M–S2O). We found that most IgG and IgA antibody levels increased with age during the younger years (Figures S2G and S2I) and then remained stable (Figures S2M and S2O). Conversely, there was not a significant association with IgM and age in the younger group (Figure S2K), but a significant decline in IgM was found with age for participants 17–54 years of age (Figure S2N). The decline in IgM in the 17–54 age group is consistent with the SJTRC cohort where we found higher levels of hCCCoV IgM in younger individuals (20–43 years of age) compared to older participants (Figure 2A). These data indicate that IgG and IgA hCCCoV antibodies begin to accumulate very early in life. It is intriguing that IgM levels tend to peak between 10–30 years of age rather than declining linearly with age. As younger individuals are more likely to be recently exposed to hCCCoVs and would have a higher proportion of naive IgM+ B cells relative to older individuals, we expected to see higher IgM levels in younger individuals. Overall, these data show the wide degree of heterogeneity in hCCCoV immunity between individuals and demonstrate that most individuals have antibodies specific for all four hCCCoVs from a very early age.
hCCCoV antibodies are increased after infection with SARS-CoV-2
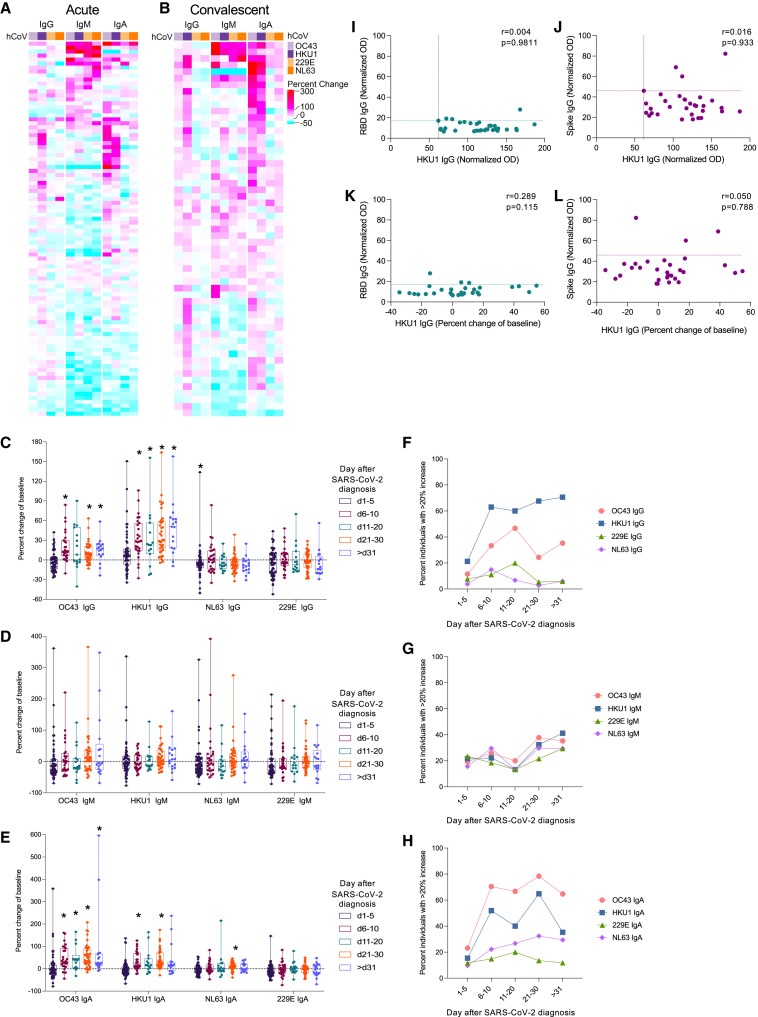
While studies identified cross-reactive antibodies that bind both SARS-CoV-2 and hCCCoVs (Ladner et al., 2020; Ng et al., 2020; Wec et al., 2020), there is significant controversy regarding whether hCCCoV antibodies are boosted after SARS-CoV-2 infection (Anderson et al., 2021; Aydillo et al., 2021; Becker et al., 2021; Dugas et al., 2021a, 2021b; Gouma et al., 2021; Guo et al., 2021; Loos et al., 2020; Ng et al., 2020; Nguyen-Contant et al., 2020; Ortega et al., 2021; Prévost et al., 2020; Shrock et al., 2020; Westerhuis et al., 2021). If pre-existing hCCCoV-specific antibodies cross-react to SARS-CoV-2, the levels of hCCCoV-specific antibodies would increase following SARS-CoV-2 infection. Alternatively, if antibodies specific for hCCCoVs do not cross-react to SARS-CoV-2, the levels of hCCCoV antibodies would not change after infection. We analyzed samples taken before and at two time points after confirmed SARS-CoV-2-infection. The first sample after infection was collected during the acute phase (1–20 days) (Figure 3 A), and a subsequent sample was taken during the convalescent phase (>20 days) (Figure 3B). Interestingly, several individuals exhibited reduced hCCCoV antibody levels shortly after SARS-CoV-2 infection relative to baseline, indicated by a negative percent change of baseline (Figures 3C–3E and S3A–S3C; Table S2). This decrease was most evident in samples taken within the first 20 days after infection. The decrease in hCCCoV antibodies shortly after SARS-CoV-2 infection highlights the caveat of not analyzing hCCCoV antibodies in paired samples collected prior to SARS-CoV-2 infection. Similar to associations prior to infection, individuals exhibiting an increase in IgM antibodies to one subtype of hCCCoVs typically showed increases in IgM reactive to all hCCCoVs (Figures 3A and 3B). In contrast, IgA antibodies specific for both betacoronaviruses typically increased concurrently. Interestingly, HKU1 IgG levels increased the most after SARS-CoV-2 infection compared to the other hCCCoV IgG antibodies, while OC43 IgA showed the greatest increase of the IgA antibodies (Figures 3A and 3B; Table S2). Overall, HKU1 and OC43 IgG and IgA antibodies showed the highest and most consistent increase over baseline levels compared to antibodies specific for the alphacoronaviruses (Figures 3A–3H; Table S2), which is consistent with greater homology among the betacoronaviruses. Importantly, hCCCoV antibody levels did not change in individuals infected with influenza virus (Figures S3D–S3F), demonstrating that the increase in hCCCoV antibodies reflected cross-reactivity with SARS-CoV-2 infection rather than a non-specific consequence of infection.
Figure 3.
Antibodies specific for OC43 and HKU1 increase following SARS-CoV-2 infection
(A and B) Samples taken from individuals during the (A) acute (1–20 days) or the (B) convalescent (>20 days) phase after PCR-confirmed infection were analyzed by ELISA for IgG, IgM, and IgA antibodies specific for spike proteins of OC43, HKU1, 229E, and NL63. The percent change of the normalized OD in the sample after infection relative to the baseline is depicted in the heatmap.
(C–E) The percent change of (C) IgG, (D) IgM, and (E) IgA antibodies relative to the baseline sample was calculated for samples at indicated times following SARS-CoV-2 infection. Asterisks indicate significant difference compared to no fold change determined by Wilcoxon signed-rank test with the Benjamini, Krieger, and Yekutieli method.
(F–H) Proportion of individuals with greater than a 20% increase in (F) IgG, (G) IgM, or (H) IgA. Fold change of hCCCoV antibodies for all acute and convalescent samples compared to baseline samples are reported in Table S2.
(I–L) Normalized OD of (I and K) SARS-CoV-2 RBD IgG and (J and L) spike IgG in samples collected within 5 days of SARS-CoV-2 diagnosis were compared to the (I,J) normalized OD of HKU1 IgG in the same sample or the (K,L) boost in HKU1 IgG in the sample relative to baseline. The r value computed by the Spearman method is shown. Dashed lines indicate cut-offs for positive values.
The increase in hCCCoV antibodies following SARS-CoV-2 infection could be due to activation of pre-existing memory B cells that were generated after prior hCCCoV infection. Alternatively, the elevated levels of hCCCoV antibodies after SARS-CoV2 infection could be due to the generation of new antibodies that cross-react to hCCCoVs in response to SARS-CoV-2. To distinguish these possibilities, we measured antibody levels in samples collected at various times after SARS-CoV-2 diagnosis. We reasoned that an increase in hCCCoV antibodies due to a boost of pre-existing memory B cells would be detected rapidly following diagnosis, while an increase in hCCCoV antibodies resulting from newly generated antibodies would be evident later. Remarkably, the levels of HKU1 IgG rapidly increased in several individuals within the first 5 days after SARS-CoV-2 diagnosis (Figures 3C and 3F), and OC43 and HKU1 IgA levels increased within 10 days in over 50% of individuals (Figures 3E and 3H). The early rise in betacoronavirus hCCCoV IgG and IgA antibodies suggests that infection with SARS-CoV-2 activates pre-existing memory B cells to boost antibodies generated during prior hCCCoV infections. Further, if the increase in hCCCoV antibodies was due to newly generated antibodies in response to SARS-CoV-2 infection, then we would expect these antibodies to also be SARS-CoV-2-specific. Therefore, we examined whether individuals with high levels of HKU1 IgG antibodies within 5 days of diagnosis also had antibodies that recognized SARS-CoV-2 spike or the receptor binding domain (RBD) of the spike. While a few individuals had positive levels of SARS-CoV-2 spike and RBD IgG within 5 days of diagnosis (Figures 3I–3L and S4), there was no correlation between the level of SARS-CoV-2 spike or RBD IgG and HKU1 IgG (Figures 3I and 3J) or a correlation between SARS-CoV-2 spike or RBD IgG and the increase of HKU1 IgG (Figures 3K and 3L). Interestingly, IgM antibodies specific for the SARS-CoV-2 proteins were not typically observed prior to IgG or IgA (Figure S4), which would be expected after exposure to a novel virus or vaccine (Li et al., 2014; Wolf et al., 2011). Thus, the antibody response to SARS-CoV-2 displays a pattern similar to what would be expected after boosting of a memory response. Together, these data are consistent with the notion that SARS-CoV-2 activates pre-existing memory B cells to boost antibodies that were generated after prior hCCCoV infection. The hCCCoV antibodies detected at later time points are likely a combination of boosted, pre-existing antibodies and newly generated antibodies that cross-react to hCCCoVs. If the pre-existing antibodies recognize epitopes on SARS-CoV-2, they could reduce infection severity by promoting viral clearance. Alternatively, if the antibodies do not bind SARS-CoV-2 with sufficient avidity, these antibodies could delay the generation of effective antibodies specific for SARS-CoV-2 by competing with naive B cells for antigen and cytokines. The fact that individuals with an early increase or high levels of hCCCoV antibodies within 5 days of SARS-CoV-2 diagnosis did not have SARS-CoV-2-specific antibodies at this time suggests that the hCCCoV antibodies do not bind SARS-CoV-2 with sufficient avidity to be detected by ELISA.
hCCCoV antibodies do not impact the probability of becoming infected with SARS-CoV-2
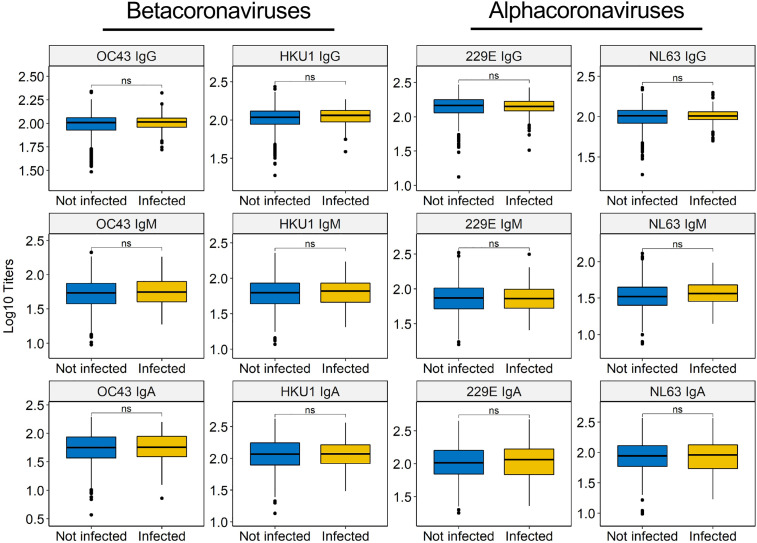
Since hCCCoV-specific antibodies cross-react with SARS-CoV-2 as demonstrated by the early increase after infection and prior studies (Ladner et al., 2020; Ng et al., 2020), we performed a large, case-control study to test whether pre-existing hCCCoV IgG, IgM, and IgA antibodies were different between individuals who became infected during the study compared to individuals that remained negative. It is important to note that all individuals underwent weekly nasal swab screening, which allowed us to identify asymptomatic infections and confirm all SARS-CoV-2 infections by PCR. We assessed baseline hCCCoV antibodies in 121 individuals that subsequently became positive during the study and compared them to baseline samples of 1,081 individuals that remained uninfected. Even though hCCCoV antibodies exhibit sufficient cross-reactivity with SARS-CoV-2 to increase after infection, baseline levels of hCCCoV antibodies were not different between individuals that became infected compared to those that remained SARS-CoV-2 negative during the study period (Figure 4 ; Table S3). These data imply that prior infection with hCCCoVs does not protect against infection with SARS-CoV-2, which is consistent with the inability of hCCCoV-specific antibodies to neutralize SARS-CoV-2 (Aguilar-Bretones et al., 2021; Legros et al., 2021; Poston et al., 2021).
Figure 4.
Baseline hCCCoV antibody levels do not correlate with protection from SARS-CoV-2 infection
Baseline hCCCoV-normalized ODs were compared between individuals that became infected (n = 121) during the study to individuals that remained SAR-CoV-2 negative (n = 1,081) using the Wilcoxon-Mann-Whitney test and adjusted with Bonferroni method (ns, not significant).
Baseline hCCCoV antibodies do not provide protective immunity against SARS-CoV-2 infection
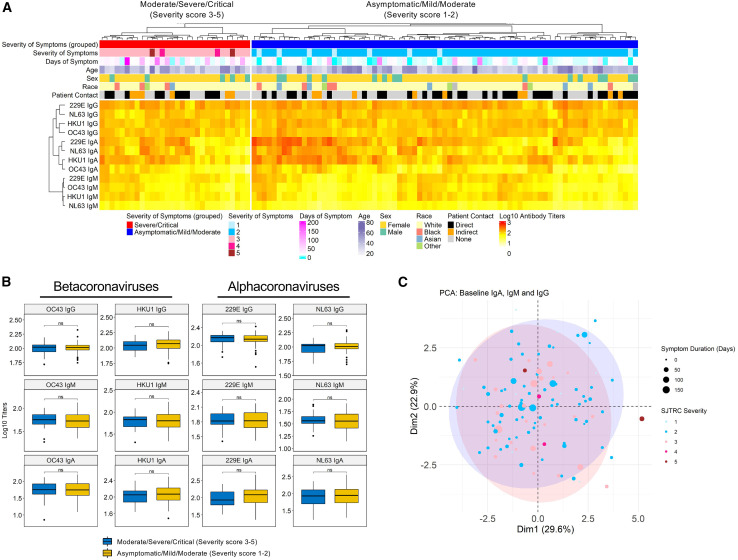
While baseline levels of hCCCoV antibodies were not different between participants that became infected compared to those who remained SARS-CoV-2 negative, hCCCoV antibodies could influence the severity or duration of symptoms. Therefore, we assessed whether there was a correlation with disease severity and baseline levels of hCCCoV antibodies. Infected individuals were given a score of 1–5 based on an a priori ordinal scale as follows: (1) asymptomatic, (2) mild-moderate, (3) moderate-severe illness, (4) severe illness, and (5) critical illness. This scale allowed us to distinguish truly asymptomatic, minimally symptomatic, and more severely symptomatic individuals. Most participants in this cohort had mild-moderate and moderate-severe severity scores (severity scores 2 to 3). Since only a few individuals were asymptomatic, severe, or critical, we compared baseline hCCCoV antibodies between individuals that were either asymptomatic or had mild disease (severity score of 1 to 2) to individuals that experienced moderate, severe, or critical disease (severity score 3–5). We found no significant difference between baseline hCCCoV antibody levels and disease severity when comparing these two groups (Figures 5A and 5B). Moreover, symptom duration did not correlate with baseline hCCCoV antibody levels (Figures 5A and S5). These data suggest that the baseline levels of hCCCoV antibodies do not provide significant protection against SARS-CoV-2 infection. However, as there were few cases of severe COVID-19 requiring hospitalization or critical illness in the included participants, our ability to identify predictors of these states is limited.
Figure 5.
Baseline hCCCoV antibody levels do not correlate with disease severity following SARS-CoV-2 infection
(A) Baseline hCCCoV-normalized ODs are depicted in the heatmap along with demographic information and severity scores. Individuals were given a severity score based on self-reported symptoms: (1) Asymptomatic (n = 8), (2) mild-moderate (n = 69), (3) moderate-severe (n = 26), (4) severe (n = 2), and (5) critical (n = 2).
(B) Comparison of baseline hCCCoV antibody between infected subjects with severity score 3–5 (n = 30) and severity score 1 to 2 (n = 77).
(C) Principal component analysis (PCA) of baseline betacoronavirus IgG-, IgA-, and IgM-normalized ODs. First two components (Dim1 and Dim2) are on the x and y axes, and numbers in parenthesis indicate percent variation explained by each component. The size and color of each bubble represent days and severity of symptoms for 107 SARS-CoV-2-infected subjects. The blue and red shaded areas represent 90% ellipses (Fox and Weisberg, 2019) for severity 3–5 and severity 1 to 2, respectively.
Existing hCCCoV antibodies influence SARS-CoV-2 antibody response
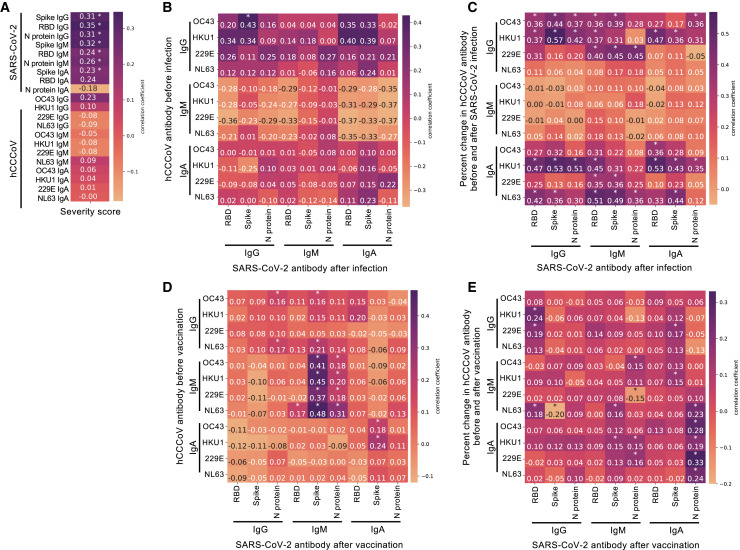
Since the SJTRC cohort consists primarily of individuals with mild-moderate disease severity and only four individuals had severe or critical disease, the impact of hCCCoV antibodies on very severe cases may not be evident in this cohort. Many studies reported that the level of SARS-CoV-2 spike or RBD IgG or IgA following infection correlated with disease severity (Aguilar-Bretones et al., 2021; Becker et al., 2021; Dobaño et al., 2021; Garcia-Beltran et al., 2021; Guthmiller et al., 2021; Legros et al., 2021; Ortega et al., 2021; Shrock et al., 2020). This may be due to the fact that individuals with more severe disease likely have more viral replication and, therefore, greater antigen exposure. Thus, the antibody response after infection may provide a means to further stratify disease severity within the groups, independent of self-reported symptoms. Therefore, we examined whether the antibody response 16–40 days following SARS-CoV-2 infection correlated with disease severity in the SJTRC cohort, in which most participants had mild-moderate disease severity. Importantly, none of the infected individuals had received a vaccine prior to collection of samples used for this comparison or other comparisons reported here. Similar to other studies, the level of IgG specific for SARS-CoV-2 spike, RBD, and N protein significantly correlated with increased disease severity scores (Figures 6 A and S6). Higher spike and RBD IgM and spike IgA levels also correlated with more severe disease. These data indicate that, although most participants had mild-moderate disease, the levels of SARS-CoV-2-specific IgG and IgM correlated with severity. Consequently, we compared baseline hCCCoV antibody levels to SARS-CoV-2 antibody levels following infection to further assess association of baseline hCCCoVs and a distinct correlate of disease severity. Interestingly, higher levels of OC43 IgG prior to infection correlated with increased SARS-CoV-2 IgG after infection (Figure 6B), raising the possibility that high baseline OC43 IgG may be associated with more severe disease.
Figure 6.
Existing hCCCoV antibody levels associate with the magnitude of the SARS-CoV-2 antibody response after infection, but not vaccination
(A) The normalized OD of antibodies in samples taken 16–40 days after SARS-CoV-2 diagnosis (n = 123) was compared to the five severity scores. Kendall rank correlation coefficients are indicated in the heatmap. P values were corrected by false discovery rate. ∗p < 0.05.
(B–E) Pearson’s formulation was utilized to calculate correlation coefficients, with multiple testing correction with the TestCor package between (B) normalized ODs of baseline hCCCoV antibodies compared to normalized ODs of SARS-CoV-2 antibody in samples collected 16–40 days after infection (n = 41), (C) the percent change from baseline of hCCCoV antibodies compared to SARS-CoV-2 antibody in samples collected between 1 and 15 days after infection (n = 43), (D) baseline hCCCoV-normalized ODs compared to SARS-CoV-2 antibody 20–85 days after vaccination with Pfizer/BioNTech BNT162b2 (n = 256), and (E) the increase in hCCCoV antibodies relative to the baseline sample compared to SARS-CoV-2 antibody in samples collected after vaccination (n = 256).
To further examine the impact of hCCCoV immunity on the immune response to SARS-CoV-2, we tested whether the magnitude of the hCCCoV antibody increase or decrease following SARS-CoV-2 infection impacted SARS-CoV-2 antibody levels. The amount that the hCCCoV antibody levels increase in the initial days after SARS-CoV-2 infection is indicative of the extent that memory B cells are activated to produce antibody. Thus, we calculated percent change of hCCCoV antibody in the baseline sample to the sample taken within the first 15 days after diagnosis. Increases in hCCCoV antibody levels in this time frame would reflect the extent of memory B cell activation. We compared this change to the SARS-CoV-2 antibody levels 16–40 days after infection, as these levels correlated with disease severity in our cohort as well as several other studies. Interestingly, a greater increase in betacoronavirus IgG and IgA was associated with higher levels of SARS-CoV-2 IgG and IgM antibodies after infection (Figure 6C). Since increased levels of SARS-CoV-2 IgG and IgM are associated with greater disease severity, these data raise the possibility that the early increase (1–15 days after infection) in hCCCoV antibodies could be associated with higher disease severity. Alternatively, the association between the increase in hCCCoV antibody levels with higher SARS-CoV-2-induced antibodies could be due to newly generated antibodies in response to SARS-CoV-2 infection that cross-react with hCCCoVs. However, analysis of samples taken within the first 5 days of SARS-CoV-2 diagnosis demonstrated that the increase in hCCCoV antibodies preceded detection of SARS-CoV-2 antibodies (Figures 3I–3L), indicating that the early hCCCoV-reactive antibodies do not bind SARS-CoV-2 spike.
If the correlation between the early increase of betacoronavirus antibodies and higher SARS-CoV-2 antibodies after infection was due to newly generated antibodies in response to SARS-CoV-2 infection that cross-react with hCCCoVs rather than an association with disease severity, then we would predict that the baseline hCCCoV levels or boosts would have a similar correlation in response to vaccination in individuals that were not infected with SARS-CoV-2. Therefore, we compared baseline hCCCoV antibody levels in individuals before vaccination to the level of SARS-CoV-2 antibodies after vaccination. For this analysis, none of the vaccinated participants were previously infected with SARS-CoV-2. The fact that all participants were screened weekly by nasal swab and PCR reduced the probability of individuals with asymptomatic infections being included in this group. We first assessed whether hCCCoV antibodies increased following vaccination similar to infection. We observed an increase in HKU1 IgG after Pfizer/BioNTech BNT162b2 vaccination compared to samples taken at baseline (Figure S7). However, there was not a significant increase in OC43 IgG as seen after SARS-CoV-2 infection. Moreover, the increase in HKU1 IgG antibodies after vaccination was not as great as the increase observed in SARS-CoV-2-infected participants. We also noted a significant decrease in all hCCCoV IgA and IgM antibodies following vaccination. Importantly, neither baseline levels of hCCCoV antibodies nor an increase in hCCCoV antibodies after vaccination correlated with increased SARS-CoV-2 antibodies after vaccination (Figures 6D and 6E). In fact, correlations of baseline or boost of hCCCoV antibodies and SARS-CoV-2 antibodies showed strikingly distinct patterns in infected versus vaccinated individuals (Figures 6B–6E). Interestingly, there were significant correlations with baseline hCCCoV IgM and SARS-CoV-2 IgM after vaccination. As IgM antibodies exhibit greater cross-reactivity among the hCCCoVs compared to IgG and IgA, this could reflect existing hCCCoV IgM antibodies that cross-react with SARS-CoV-2. Alternatively, individuals with higher hCCCoV IgM may have a higher proportion of naive B cells capable of responding to a novel antigen. As the vaccine does not induce a robust IgM response in most individuals, it is currently not known whether IgM antibody levels after vaccination impact vaccine efficacy. Together, these data indicate that pre-existing betacoronavirus IgA and IgG correlate with a higher antibody response to SARS-CoV-2 following infection, but not vaccination. As increased SARS-CoV-2 antibodies after infection correlated with greater disease, these findings raise the possibility that pre-existing betacoronavirus IgG and IgA negatively impact the immune response to SARS-CoV-2, which results in greater duration of antigen and therefore more SARS-CoV-2 antibodies.
Prior immunization with hCCCoV spike proteins limits the antibody response to SARS-CoV-2 RBD in mice
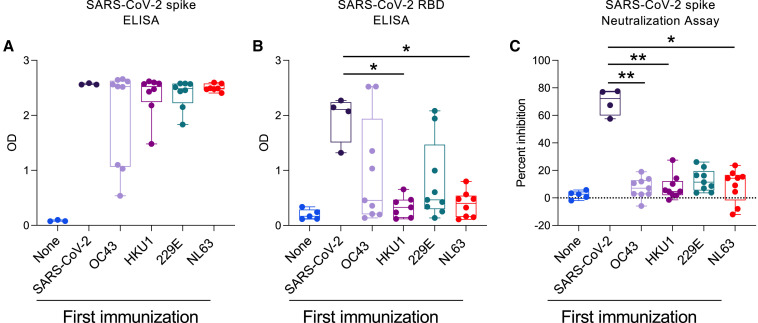
Since most individuals have positive levels of antibodies specific for all four hCCCoVs (Figure 1D), it is not possible to directly examine whether prior exposure to a particular hCCCoV impacts the antibody response to SARS-CoV-2. Therefore, we performed a series of immunizations in mice that had no prior exposure to coronaviruses. C57BL/6 mice were immunized with the spike proteins of SARS-CoV-2, OC43, HKU1, 229E, or NL63. Four weeks later, all mice were immunized with the SARS-CoV-2 spike protein. Thus, mice either received two immunizations of SARS-CoV-2 spike or one immunization of hCCCoV spike followed by one of SARS-CoV-2 spike. Two weeks following immunization with SARS-CoV-2 spike, we measured RBD and spike IgG antibodies to determine if prior exposure to hCCCoV spike proteins impacted the antibody response to SARS-CoV-2 spike and RBD. Prior immunization with hCCCoV spike proteins did not significantly impact antibody levels to SARS-CoV-2 full-length spike (Figure 7 A). However, RBD IgG was significantly decreased in mice that received a prior immunization with HKU1 and NL63 spike proteins compared to mice only immunized with SARS-CoV-2 spike protein (Figure 7B). Importantly, prior immunization with any of the hCCCoV spike proteins inhibited neutralizing antibodies following SARS-CoV-2 spike immunization as detected by a pseudo-neutralization assay (Figure 7C). These data, which are consistent with a prior study utilizing a different strain of mice and adjuvant (Lapp et al., 2021), directly demonstrate that prior exposure to hCCCoV spike proteins has the potential to inhibit generation of neutralizing antibodies specific for the RBD of SARS-CoV-2. Together, these findings illustrate that prior immunity to a virus with a certain degree of homology can impede the immune response to a novel virus.
Figure 7.
Prior immunization with hCCCoV spike proteins limits the antibody responses to SARS-CoV-2 RBD
(A–C) C57BL/6 mice were immunized with spike proteins in CFA, interperitoneally. Four weeks later, all mice were immunized with SARS-CoV-2 spike protein. Control mice (none) only received CFA at both time points. Serum taken 2 weeks after the second immunization was analyzed by ELISA for reactivity to (A) SARS-CoV-2 spike protein or (B) SARS-CoV-2 RBD. (C) Serum was tested in a SARS-CoV-2 surrogate neutralization assay. p values calculated using Kruskal-Wallis test and Dunn’s multiple comparisons test (∗p < 0.05; ∗∗p < 0.01).
Discussion
Immune imprinting refers to preferential activation of memory B cells that were generated during a prior infection with an antigenically related virus rather than naive B cells specific for the novel virus (Guthmiller and Wilson, 2018; Henry et al., 2018; Monto et al., 2017). This concept is well documented for influenza infections whereby humans are repeatedly exposed to antigenically distinct viruses containing regions of homology. Imprinting can hinder immunity to a novel virus if pre-existing antibodies against conserved epitopes dominate the immune response but do not neutralize the novel virus. Since pre-existing memory B cells are present at higher precursor frequencies relative to naive B cells and are primed to be activated, they can outcompete B cells specific for novel epitopes and hinder immunity to the novel virus (Cobey and Hensley, 2017). In addition, antibodies generated to a related virus could block antibodies specific for the novel virus via steric hinderance by binding conserved epitopes near the novel epitopes.
Humans are repeatedly infected with endemic hCCCoVs (Edridge et al., 2020; Kiyuka et al., 2018), and our data indicate that nearly every individual possesses antibodies specific for all four of the endemic hCCCoVs. A recent study demonstrated that memory B cells specific for hCCCoVs dominated the early immune response following SARS-CoV-2 infection; however, these antibodies did not neutralize SARS-CoV-2 (Dugan et al., 2021). This study illustrates how hCCCoV immunity can hinder protective immunity to SARS-CoV-2 by usurping resources to amplify non-neutralizing antibodies. Our data are consistent with these findings as we show correlations with the baseline level or boost of hCCCoV antibodies and levels of SARS-CoV-2 antibodies after infection, which correlated with greater severity following SARS-CoV-2 infection. Mouse experiments further verified that existing hCCCoV immunity reduced neutralizing antibodies specific for the RBD. It is intriguing that hCCCoV antibodies are boosted following SARS-CoV-2 infection and show clear correlations with the SARS-CoV-2 antibody response yet do not significantly affect the incidence of becoming infected or symptom duration. As several factors contribute to susceptibility to SARS-CoV-2 infection, including age, gender, and underlying disease conditions (Fang et al., 2020), it may be difficult to detect the impact of hCCCoV immunity on self-reported symptoms among other confounding factors. It is important to note that the participants in this cohort were primarily Caucasian females with mild to moderate symptoms. Therefore, we were not able to thoroughly assess associations with more severe disease. Regardless, our data suggest that hCCCoV immunity may be an additional factor that can impede effective immunity to SARS-CoV-2 infection. Considering the continued circulation of SARS-CoV-2 variants, it will be important to further investigate mechanisms in which pre-existing immunity impacts the immune response to a novel, but related, virus.
Prior studies investigating whether hCCCoV antibodies contributed to disease severity yielded particularly contradictory results. One main reason for these divergent conclusions is that most of the previous studies lacked baseline samples from the same individual before and after infection. Due to wide variation in hCCCoV antibody levels, it is not possible to accurately assess baseline hCCCoV immunity without analyzing samples from each individual prior to SARS-CoV-2 infection. Importantly, our data demonstrate that hCCCoV antibody levels can increase or decrease as early as 5 days after SARS-CoV-2 infection. Thus, samples taken after SARS-CoV-2 infection are not indicative of pre-existing hCCCoV immunity. Another factor contributing to the divergent conclusions is the composition and range of severity in the different cohorts. While most participants in the SJTRC cohort exhibited mild to moderate symptoms, other studies only included hospitalized individuals. Additionally, the antigens, antibody isotypes, and type of assays varied widely among the previous studies, which may also have influenced the inconsistency in results.
There is extensive cross-reactivity among antibodies specific for hCCCoVs (Ladner et al., 2020; Poston et al., 2021; Wec et al., 2020), and our data illustrate how serology may not be a reliable indicator of the hCCCoV to which an individual was most recently exposed. This is evident in the greater correlation between antibody isotypes specific for different hCCCoVs rather than an association with high levels of IgA, IgM, and IgG specific for a particular hCCCoV. Consistent with previous studies, we found that in older individuals, hCCCoV immunity is more biased toward IgA and IgG compared to IgM in younger individuals (Selva et al., 2021). Each time an individual is exposed to a hCCCoV, the memory B cells are further fine-tuned through affinity maturation and clonal selection to generate higher affinity hCCCoV-specific antibodies. Accordingly, as individuals age, repeated exposure to hCCCoVs creates a more specific and less adaptable repertoire of hCCCoV-specific memory B cells. Since SARS-CoV-2 is a novel virus that individuals had not encountered, it was unexpected that IgM antibodies did not precede IgG antibodies (Figure S3). These data are consistent with a previous report and suggest that the early immune response to SARS-CoV-2 is dominated by reactivation of memory B cells generated during prior hCCCoV infection (Dugan et al., 2021). We hypothesize that betacoronavirus IgG and IgA antibody levels are more indicative of an individual’s cumulative response to hCCCoVs rather than the timing of a recent infection. Accordingly, higher levels of betacoronavirus IgG and IgA antibodies imply a more narrow and less adaptable antibody repertoire, which would be advantageous for immunity to the hCCCoV but detrimental to the immune response to a novel coronavirus. Thus, although younger individuals may be exposed to hCCCoVs more often than older individuals, the hCCCoV IgM bias in younger participants is consistent with a more adaptable repertoire, which may explain why younger individuals exhibit less disease severity than older individuals.
Although baseline hCCCoV antibody levels correlated with SARS-CoV-2 antibody levels following infection, we did not observe an association between baseline hCCCoV immunity and SARS-CoV-2 antibodies after vaccination. Many factors differ between the immune response to vaccination compared to infection. One possibility is that pre-existing hCCCoV antibodies may impede the generation of SARS-CoV-2 neutralizing antibodies, thereby extending viral exposure and enhancing the antibody response after infection. However, inhibition of neutralizing antibodies would not impact antigen load in the context of a vaccination, and therefore hCCCoV immunity would not have a similar impact on infection and vaccination. Alternatively, it is also possible that there is no correlation between baseline hCCCoV antibody levels and antibody levels following vaccination because the mRNA vaccines induce such a robust immune response to the SARS-CoV-2 spike protein that the efficacy of these vaccines may override the effect of imprinting. Interestingly, a recent report showed that imprinting also led to divergent outcomes following influenza virus infection versus vaccination (Dugan et al., 2020).
In summary, our data demonstrate that SARS-CoV-2 infection and vaccination activate existing memory B cells specific for hCCCoVs. Baseline levels of hCCCoV antibodies and the magnitude that these antibodies increased after infection or vaccination varied dramatically among individuals. Higher baseline levels or an increase of betacoronavirus IgG and IgA after infection were associated with increased SARS-CoV-2 antibody levels, which correlated with greater disease severity. These findings suggest that, similar to influenza virus, prior exposure to coronaviruses with sufficient homology can hinder the immune response to a novel coronavirus.
Limitations of study
Limitations of our study include the low number of participants that experienced severe disease. Thus, we performed comparisons of baseline hCCCoV antibody levels to disease severity by grouping individuals with severity scores of 1 to 2 versus 3–5, which may not have revealed factors that specifically correlate with greater disease severity. Moreover, as our cohort consisted of employees, it did not include any individuals below 18 years of age and not many older than 65 years of age. Additionally, in the mouse immunization studies, comparisons were made between mice immunized twice with SARS-CoV-2 spike and mice immunized with an hCCCoV spike followed by SARS-CoV-2 spike. It is possible that the decreased neutralizing antibodies observed in mice receiving hCCCoV spike prior to SARS-CoV-2 are due to the fact that two immunizations with SARS-CoV-2 is required to generate neutralizing antibodies. However, it is important to note that antibodies to the full-length SARS-CoV-2 spike were not decreased in mice immunized with hCCCoV prior to SARS-CoV-2 compared to mice only immunized with SARS-CoV-2 spike, indicating that the overall antibody levels are similar.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human-IgG | Invitrogen | Cat# A18805; RRID: AB_2535582 |
| Goat anti-human-IgM | Invitrogen | Cat# A18835; RRID: AB_253612 |
| Goat anti-human-IgA | Southern Biotech | Cat# 2050-05; RRID: AB_2687526 |
| Goat anti-mouse IgG | Southern Biotech | Cat# 1033-05; RRID: AB_2737432 |
| Biological samples | ||
| Plasma from SJTRC participants | St. Jude Tracking of Viral and Host Factors Associated with COVID-19 study (SJTRC) | NCT04362995 |
| Plasma from FLU09 participants | St. Jude Children’s Research Hospital and the University of Tennessee Health Science Center/Le Bonheur Children’s Hospital | (Allen et al., 2017; Oshansky et al., 2014) |
| Chemicals, peptides, and recombinant proteins | ||
| hCoV-OC43 spike protein | Sino Biological | 40607-V08B |
| hCoV-HKU-1 spike protein | Sino Biological | 40606-V08B |
| hCoV-NL63 spike protein | Sino Biological | 40604-V08B |
| hCoV-229E spike protein | Sino Biological | 40605-V08B |
| SARS-CoV-2 spike protein | Sino Biological | 40589-V08B1 |
| His-tag blocking peptide | BioVision | 3998BP |
| Omniblok™ non-fat milk | AmericanBio | AB10109-01000 |
| OPD (o-phenylenediamine dihydrochloride) | Sigma-Aldrich | P8287 |
| SIGMAFAST OPD | Sigma-Aldrich | P9187 |
| Complete Freund’s Adjuvant (including 5mg/mL Mycobacterium tuberculosis) | Chondrex | 7023 |
| Critical commercial assays | ||
| SARS-CoV-2 Surrogate Virus Neutralization Test Kit | GenScript | L00847-A |
| Expi293™ Expression Medium | Thermo Fisher Scientific | A1435101 |
| ExpiFectamine 293 transfection kit | Thermo Fisher Scientific | A14524 |
| Deposited data | ||
| R script used to run statistical analyses | This paper | https://github.com/SYL16/SJTRC-CCoV |
| R script used to run statistical analyses | This paper | https://github.com/MacauleyLockeml/St-Jude-Trace-study-SARS-CoV-2 |
| Experimental models: Cell lines | ||
| Expi293F cells | Life Technologies | Cat# A14527; RRID:CVCL_D615 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6: wildtype | Jackson Laboratories | Cat# 000664; RRID: IMSR_JAX:000664 |
| Recombinant DNA | ||
| SARS-CoV-2 nucleocapsid (N) protein plasmid (from Wuhan-Hu-1 isolate) | Florian Krammer | N/A |
| SARS-CoV-2 spike (S) protein plasmid (from Wuhan-Hu-1 isolate) | Florian Krammer | N/A |
| SARS-CoV-2 spike protein receptor binding domain (RBD) plasmid (from Wuhan-Hu-1 isolate) | Florian Krammer | N/A |
| Software and algorithms | ||
| R version 4.0.3 and version 3.6.2 | The R Core Team | https://www.R-project.org/ |
| R package “TestCor” | (Irene, 2020) | https://cran.r-project.org/web/packages/TestCor/index.html |
| R package “Psych” | (Revelle, 2021) | https://cran.r-project.org/web/packages/psych/index.html |
| R package “tidyverse” | (Wickham et al., 2019) | https://cran.r-project.org/web/packages/tidyverse/index.html |
| R package “vegan” | (Oksanen et al., 2008) | https://cran.r-project.org/web/packages/vegan/index.html |
| R package “ComplexHeatmap” | (Gu et al., 2016) | http://www.bioconductor.org/packages/devel/bioc/html/ComplexHeatmap.html |
| R package “Factoextra” | (Kassambara and Mundt, 2017) | https://cran.r-project.org/web/packages/factoextra/index.html |
| R package “cluster” | (Maechler et al., 2021) | https://cran.r-project.org/web/packages/cluster/index.html |
| R package “rstatix” | (Kassambara, 2021) | https://cran.r-project.org/web/packages/rstatix/index.html |
| R package “corrplot” | (Wei et al., 2017) | https://github.com/taiyun/corrplot |
| R package “circlize” | (Gu et al., 2014) | https://cran.r-project.org/web/packages/circlize/index.html |
| R package “digest” | (Eddelbuettel et al., 2021) | https://cran.r-project.org/web/packages/digest/index.html |
| R package “survival” | (Therneau and Lumley, 2015) | https://github.com/therneau/survival |
| Python version 3.9.8 | Python Software Foundation | https://www.python.org/ |
| Seaborn package | (Waskom, 2021) | https://seaborn.pydata.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Maureen McGargill (Maureen.mcgargill@stjude.org).
Materials availability
This study did not generate unique reagents.
Experimental model and subject details
Mice
C57BL/6 female mice, 7-8 weeks of age were purchased from Jackson Laboratories and randomly assigned to experimental groups. All mice were maintained under specific pathogen-free conditions at St. Jude Children’s Research Hospital, and all animal studies were approved by the Institutional Animal Care and Use Committee.
SJTRC cohort
The St. Jude Tracking of Viral and Host Factors Associated with COVID-19 study (SJTRC, NCT04362995) is a prospective, Institutional Review Board-approved, longitudinal cohort study of St. Jude Children’s Research Hospital (St. Jude) adult employees. Participants provide written informed consent prior to enrollment and then complete regular questionnaires about demographics (at baseline only), medical history and treatment (at baseline and every 8 weeks), and symptoms (at baseline and every 2 weeks). Study data are collected and managed using REDCap electronic data capture tools hosted at St. Jude. Participants provided a baseline blood sample at enrollment, then underwent nasal swab screening for SARS-CoV-2 infection by PCR approximately weekly when on campus. Study participants who were diagnosed with SARS-CoV-2 provided additional research blood samples within two weeks (acute sample) and then three to eight weeks (convalescent) after diagnosis. Participants who received SARS-CoV-2 vaccination provided an additional blood sample three to eight weeks after completion of the vaccine series. For analyses examining antibody levels after vaccine or infection, the data were limited to individuals that were either infected, but not vaccinated yet, or vaccinated, but not infected. Blood samples were collected in CPT tubes and separated within 24 h of collection into cellular and plasma components, and aliquoted and frozen for future analysis. Vaccinations were administered as standard-of-care.
FLU09 cohort
The FLU09 cohort was previously described (Allen et al., 2017; Oshansky et al., 2014). Briefly, the inclusion criteria required that participants meet the clinical case definition of influenza virus infection at the time of enrollment or be asymptomatic household contacts of a participant with confirmed influenza infection. This study was conducted in compliance with 45 CFR46 and the Declaration of Helsinki. The institutional review boards of St. Jude Children’s Research Hospital and the University of Tennessee Health Science Center/Le Bonheur Children’s Hospital approved the study. Written, informed consent was acquired from participants’ parents or guardians and written assent from age-appropriate subjects was acquired at the time of enrollment. Index cases provided nasal swabs, nasal lavages, and blood on the day of enrollment (day 0) and days 3, 7, 10, and 28, whereas household contacts provided nasal swabs on days 0, 3, 7, and 14 and blood and nasal lavages on days 0 and 28. The population used for these analyses was predominantly African American (81.4%) with 18.6% Caucasian participants (n = 86). Metadata collected from this study included information on several symptoms that were ranked daily (self-reported) according to a visual analog scale. Samples included in the analysis of Figure S2 included individuals that were negative for influenza infection upon enrollment and became infected during the study.
Cell lines
Expi293F cells were cultured in suspension using PETG Erlenmeyer flasks within a 37°C incubator with 380% relative humidity and 8% CO2 on an orbital shaker platform rotating at 135rpm. They were cultured in Expi293 Expression Medium (Thermo Fisher Scientific; A1435101) according to the manufacturer’s instructions until transfection (described below). These cells were derived from the HEK293 human embryonic kidney cell line, which was karyotyped as female. Cells were purchased from a commercial vendor and not further authenticated.
Method details
Severity assessment
Participants who were diagnosed with SARS-CoV-2 during the study provided information four weeks after diagnosis about the symptoms and interventions they had during the period of infection. Severity was classified on an a priori ordinal scale based on data provided by the participants as: 1. Asymptomatic (no attributable symptoms); 2. Mild-moderate (any attributable symptoms, other than shortness of breath, that did not require hospitalization or supplemental oxygen); 3. Moderate-severe illness with shortness of breath not requiring hospitalization or supplemental oxygen; 4. Severe illness requiring hospitalization or supplemental oxygen for ³1 h; and 5. Critical illness (requiring admission to ICU, vasopressors or hemodialysis). Weekly surveillance by nasal swab and PCR enabled us to identify asymptomatic infections. Therefore, this scale was chosen to distinguish truly asymptomatic, minimally symptomatic, and more severely symptomatic individuals.
Recombinant proteins
Expression plasmids for the nucleocapsid (N) protein, spike protein, and the spike protein receptor binding domain (RBD) from the Wuhan-Hu-1 isolate were obtained from Florian Krammer. Proteins were transfected into Expi293F cells using a ExpiFectamine 293 transfection kit (Thermo Fisher Scientific) as previously described (Amanat et al., 2020). Supernatants from transfected cells were harvested and purified with a Ni-NTA column. Full length spike proteins from the endemic hCCCoV (OC43, HKU1, NL63, 229E) and SARS-CoV-2 spike protein used in murine experiments were purchased from Sino Biological.
ELISA
For hCCCoV and SARS-CoV-2 antibody detection in human serum samples, 384-well microtiter plates were coated overnight at 4°C, with recombinant proteins diluted in PBS. Optimal concentrations for each protein and isotype were empirically determined to optimize sensitivity and specificity. SARS-CoV-2 spike RBD was coated at 2 μg/mL in PBS for each isotype detection. Full-length spike was coated at 2 μg/mL for IgG and 4 μg/mL for either IgM or IgA detection. N protein was coated at 1 μg/mL for IgG detection and 2 μg/mL for either IgM or IgA detection. The spike proteins of 229E (Sino Biological, 40605-V08B), NL63(Sino Biological, 40604-V08B), HKU1(Sino Biological, 40606-V08B), or OC43 (Sino Biological, 40607-V08B) were coated at 1 μg/mL for IgG detection and 1.5 μg/mL for IgM and IgA detection. For all ELISAs, plates were washed the next day three times with 0.1% PBS-T (0.1% Tween-20) and blocked with 3% Omniblok™ non-fat milk (AmericanBio; AB10109-01000) in PBS-T for one h. Plates were washed, then incubated with plasma samples diluted 1:50 in 1% milk in PBS-T for 90 min at room temperature. Prior to dilution, plasma samples were incubated at 56°C for 15 min. ELISA plates were washed and incubated for 30 min at room temperature with anti-human secondary antibodies diluted in 1% milk in PBS-T: anti-IgG (1:10,000; Invitrogen, A18805), anti-IgM (1:6000; Invitrogen, A18835), or anti-IgA (1:2,000; Southern Biotech, 2050-05). The plates were washed and incubated at room temperature with OPD (Sigma-Aldrich; P8287) for ten minutes (for hCCCoV ELISAs) or SIGMAFAST OPD (Sigma-Aldrich; P9187) for eight minutes (for SARS-CoV-2 ELISAs). The chemiluminescence reaction was stopped by addition of 3N HCl and absorbances were measured at 490 nm on a microplate reader. To ensure the specificity of this assay, we first screened samples from a prior study that included young children to identify samples to serve as negative controls. In addition, as a control for plate-to-plate variability, we selected two positive samples from the SJTRC cohort that were tested on each plate and used to calculate the percent ratio, which is the OD of each sample relative to the OD of the control samples. Samples with a percent ratio greater than three times the average of the negative controls were considered positive for the hCCCoV. The negative control samples were identified by screening samples from the FLU09 cohort that included young participants. For the SARS-CoV-2 antigens, samples were considered positive if they were greater than two times the average of the mean for all the uninfected samples.
Mouse serum sample analysis was conducted as described above with the secondary antibody anti-mouse IgG (Southern Biotech; 1033-05) diluted 1:6000 in 1% milk in PBS-T. Potential His-tag-specific antibodies were neutralized before addition to the coated ELISA plate using a His-tag blocking peptide (BioVision; 3998BP) by mixing equal volume of serum samples with the peptide and incubating at 37°C for one h. All mouse sera ELISAs utilized the SIGMAFAST OPD substrate.
Murine immunization studies
C57BL/6 female mice were purchased from Jackson Laboratories. An emulsification of Complete Freund’s Adjuvant containing 100μg M. tuberculosis with 50μg of the indicated protein was delivered interperitoneally to each mouse. Twenty-six days after the initial immunization, mice were given a boost with SARS-CoV-2 spike protein as above. Blood samples were obtained through the submandibular vein. Serum was isolated from the samples and stored at −80°C until analysis by ELISA.
SARS-CoV-2 surrogate virus neutralization
Detection of potential neutralizing antibodies against SARS-CoV-2 was performed using a surrogate neutralizing test according to the manufacturer’s directions (GenScript; L00847-A). Briefly, 96-well plates were coated with ACE2 protein. Positive and negative control antibody samples as well as mouse sera were incubated separately with HRP-tagged recombinant RBD. The resulting mixtures were added to the wells of the ACE2-coated plate and incubated at 37°C for 15 min. Plates were washed and then TMB solution was added to each well. After a 15-min incubation in the dark, “Stop” solution was added and the OD (450 nm) measurements for each well were recorded immediately via plate reader. Inhibition was calculated as (1 - (ODsample /ODnegative control)) x 100%.
Quantification and statistical analysis
Descriptive analyses were conducted on log10 transformed hCCCoV antibody concentrations, and Wilcoxon-Mann-Whitney tests were used for pairwise comparisons with the Bonferroni correction for multiple comparisons. Wilcoxon signed rank tests were used for paired data. Cox proportional hazards models were applied to examine the association between baseline hCCCoV antibody concentrations and cumulative incidence of SARS-CoV-2 infection. Two-sided p values less than 0.05 were considered statistically significant. Changes in hCCoV antibody levels after SARS-CoV-2 infection relative to baseline levels were compared with Wilcoxon signed-rank test adjusted using the Benjamini, Krieger, and Yekutieli method. The Spearman method was used to compare hCCCoV and SARS-CoV-2 antibody levels early after infection (Figures 3I–3L).
The correlation coefficients between the pre-existing hCCCoV antibodies and the post-infection or vaccination antibody response (Figures 6B–6E) were calculated by Pearson’s formulation with multiple correlation testing correction assessed by utilization of TestCor package (Irene, 2020). Kendall’s coefficient was applied to understand correlation between SARS-CoV-2 antibody levels and disease severity (Figure 6A) where p- values were adjusted using false discovery rate method in Psych package (Revelle, 2021). Statistical analysis performed using R version 4.0.3, with heatmaps generated using Python programming language Seaborn package (Waskom, 2021). R packages Tidyverse (Wickham et al., 2019), vegan (Oksanen et al., 2008), ComplexHeatmap (Gu et al., 2016), Factoextra (Kassambara and Mundt, 2017), cluster (Maechler et al., 2021), rstatix (Kassambara, 2021), corrplot (Wei et al., 2017), circlize (Gu et al., 2014), digest (Eddelbuettel et al., 2021), and survival (Therneau and Lumley, 2015) were utilized for data analysis.
Acknowledgments
We thank the donors who volunteered for the SJTRC study; Florian Krammer for suppling critical reagents; Rachael Keating for critical reading of the manuscript; Benjamin A. Wilander, Michael Meagher, Timothy Lockey, and the St. Jude GMP for technical assistance; and Tamanna Shamrin and Rishi Kodela for creation and management of the clinical database. This work was funded by ALSAC, NIAID for the St. Jude Center of Excellence for Influenza Research and Surveillance (CEIRS) (HHSN272201400006C to S.S.-C., M.A.M., and P.G.T. and 3U01AI144616-02S1 to P.G.T., M.A.M., S.S.-C., and Richard J. Webby), Centers of Excellence for Influenza Research and Response (CEIRR) (75N93021C00016 to P.G.T., M.A.M., S.S.-C., and Richard J. Webby), Collaborative Influenza Vaccine Innovation Centers (CIVIC-HRP) (75N93019C00052 to S.S.-C. and P.G.T.), and NCI (5P30CA021765-42 to L.T.).
Author contributions
Conceptualization, C.-Y.L., J.W., D.C.B., M.W., S.S.-C., P.G.T., and M.A.M.; methodology, C.-Y.L., S.C., S.A.B., and M.A.M.; investigation, C.-Y.L., D.C.B., S.C., A.H.C., M.W., K.J.A., E.K.A., S.A.B., J.H.E., and the SJTRC study team; formal analysis, Y.S., M.L., J.C.C., V.I.Z., D.D., C.T., C.M.-P., and L.T.; writing, C.-Y.L., J.W., D.C.B., M.W., J.C.C., A.H.M., P.G.T., and M.A.M; visualization, C.-Y.L., Y.S., M.L., and A.H.M.
Declaration of interests
P.G.T. has consulted for Illumina and 10X and serves on the advisory board of Immunoscape and Cyotagents. P.G.T. and J.C.C. filed patents related to treatment of severe respiratory infections, including SARS-CoV-2 (not based on research in this paper).
Published: December 6, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.12.005.
Contributor Information
The SJTRC Study Team:
Aditya H. Gaur, James M. Hoffman, Tomi Mori, Elaine I. Tuomanen, Richard J. Webby, Hana Hakim, Randall T. Hayden, Diego R. Hijano, Walid Awad, Resha Bajracharya, Brandi L. Clark, Valerie Cortez, Ronald H. Dallas, Thomas Fabrizio, Pamela Freiden, Ashleigh Gowen, Jason Hodges, Allison M. Kirk, Ericka Kirkpatrick Roubidoux, Robert C. Mettelman, Jamie Russell-Bell, Aisha Souquette, James Sparks, Lee-Ann Van de Velde, Ana Vazquez-Pagan, Kendall Whitt, Taylor L. Wilson, David E. Wittman, Nicholas Wohlgemuth, and Gang Wu
Supplemental information
Data and code availability
-
•
Data reported in the paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at https://github.com/SYL16/SJTRC-CCoV and https://github.com/MacauleyLockeml/St-Jude-Trace-study-SARS-CoV-2 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.
References
- Aguilar-Bretones M., Westerhuis B.M., Raadsen M.P., de Bruin E., Chandler F.D., Okba N.M., Haagmans B.L., Langerak T., Endeman H., van den Akker J.P., et al. Seasonal coronavirus-specific B cells with limited SARS-CoV-2 cross-reactivity dominate the IgG response in severe COVID-19. J. Clin. Invest. 2021;131:e150613. doi: 10.1172/JCI150613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E.K., Randolph A.G., Bhangale T., Dogra P., Ohlson M., Oshansky C.M., Zamora A.E., Shannon J.P., Finkelstein D., Dressen A., et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. UPenn COVID Processing Unit Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., García-Sastre A. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12:3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Strengert M., Junker D., Kaiser P.D., Kerrinnes T., Traenkle B., Dinter H., Häring J., Ghozzi S., Zeck A., et al. Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity. Nat. Commun. 2021;12:1152. doi: 10.1038/s41467-021-20973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobey S., Hensley S.E. Immune history and influenza virus susceptibility. Curr. Opin. Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2021;2:100354. doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobaño C., Santano R., Jiménez A., Vidal M., Chi J., Rodrigo Melero N., Popovic M., López-Aladid R., Fernández-Barat L., Tortajada M., et al. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: utility and limitations in seroprevalence and immunity studies. Transl. Res. 2021;232:60–74. doi: 10.1016/j.trsl.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan H.L., Guthmiller J.J., Arevalo P., Huang M., Chen Y.-Q., Neu K.E., Henry C., Zheng N.-Y., Lan L.Y.-L., Tepora M.E., et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med. 2020;12:eabd3601. doi: 10.1126/scitranslmed.abd3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan H.L., Stamper C.T., Li L., Changrob S., Asby N.W., Halfmann P.J., Zheng N.-Y., Huang M., Shaw D.G., Cobb M.S., et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity. 2021;54:1290–1303.e7. doi: 10.1016/j.immuni.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas M., Grote-Westrick T., Vollenberg R., Lorentzen E., Brix T., Schmidt H., Tepasse P.-R., Kühn J. Less severe course of COVID-19 is associated with elevated levels of antibodies against seasonal human coronaviruses OC43 and HKU1 (HCoV OC43, HCoV HKU1) Int J Infect Dis. 2021;105:304–306. doi: 10.1016/j.ijid.2021.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas M., Grote-Westrick T., Merle U., Fontenay M., Kremer A.E., Hanses F., Vollenberg R., Lorentzen E., Tiwari-Heckler S., Duchemin J., et al. Lack of antibodies against seasonal coronavirus OC43 nucleocapsid protein identifies patients at risk of critical COVID-19. J Clin Virol. 2021;139:104847. doi: 10.1016/j.jcv.2021.104847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddelbuettel D., Lucas A., Tuszynski J., Bengtsson H., Urbanek S., Frasca M., Lewis B., Stokely M., Muehleisen H., Murdoch D., et al. Comprehensive R Archive Network; 2021. Create Compact Hash Digests of R Objects. [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Fang X., Li S., Yu H., Wang P., Zhang Y., Chen Z., Li Y., Cheng L., Li W., Jia H., Ma X. Epidemiological, comorbidity factors with severity and prognosis of COVID-19: a systematic review and meta-analysis. Aging (Albany N.Y.) 2020;12:12493–12503. doi: 10.18632/aging.103579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville J.M., Wilks S.H., James S.L., Fox A., Ventresca M., Aban M., Xue L., Jones T.C., Le N.M.H., Pham Q.T., et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J., Weisberg S. SAGE Publications, Inc.; 2019. An R Companion to Applied Regression. [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombar S., Bergquist T., Pejaver V., Hammarlund N.E., Murugesan K., Mooney S., Shah N., Pinsky B.A., Banaei N. SARS-CoV-2 infection and COVID-19 severity in individuals with prior seasonal coronavirus infection. Diagn Microbiol Infect Dis. 2021;100:115338. doi: 10.1016/j.diagmicrobio.2021.115338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouma S., Weirick M.E., Bolton M.J., Arevalo C.P., Goodwin E.C., Anderson E.M., McAllister C.M., Christensen S.R., Dunbar D., Fiore D., et al. Health care worker seromonitoring reveals complex relationships between common coronavirus antibodies and COVID-19 symptom duration. JCI Insight. 2021;6:150449. doi: 10.1172/jci.insight.150449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guo L., Wang Y., Kang L., Hu Y., Wang L., Zhong J., Chen H., Ren L., Gu X., Wang G., et al. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study. Emerg. Microbes Infect. 2021;10:664–676. doi: 10.1080/22221751.2021.1905488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller J.J., Wilson P.C. Harnessing immune history to combat influenza viruses. Curr. Opin. Immunol. 2018;53:187–195. doi: 10.1016/j.coi.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., Zheng N.-Y., Utset H., Stamper C.T., Dugan H.L., et al. SARS-CoV-2 Infection Severity Is Linked to Superior Humoral Immunity against the Spike. MBio. 2021;12:e02940-20. doi: 10.1128/mBio.02940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C., Palm A.E., Krammer F., Wilson P.C. From Original Antigenic Sin to the Universal Influenza Virus Vaccine. Trends Immunol. 2018;39:70–79. doi: 10.1016/j.it.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henss L., Scholz T., von Rhein C., Wieters I., Borgans F., Eberhardt F.J., Zacharowski K., Ciesek S., Rohde G., Vehreschild M., et al. Analysis of Humoral Immune Responses in Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2021;223:56–61. doi: 10.1093/infdis/jiaa680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.-P., Snead K.R., Drew M., Corbett K.S., Graham B.S., et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J. Clin. Immunol. 2021;41:906–913. doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irene G. Comprehensive R Archive Network; 2020. TestCor: FWER and FDR Controlling Procedures for Multiple Correlation Tests. [Google Scholar]
- Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. Comprehensive R Archive Network; 2021. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. [Google Scholar]
- Kassambara A., Mundt F. Comprehensive R Archive Network; 2017. Package ‘factoextra’. Extract and visualize the results of multivariate data analyses. [Google Scholar]
- Kiyuka P.K., Agoti C.N., Munywoki P.K., Njeru R., Bett A., Otieno J.R., Otieno G.P., Kamau E., Clark T.G., van der Hoek L., et al. Human Coronavirus NL63 Molecular Epidemiology and Evolutionary Patterns in Rural Coastal Kenya. J. Infect. Dis. 2018;217:1728–1739. doi: 10.1093/infdis/jiy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W., et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with an endemic human CoV. bioRxiv. 2020 doi: 10.1101/2020.07.27.222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapp S.A., Edara V.V., Lu A., Lai L., Hussaini L., Chahroudi A., Anderson L.J., Suthar M.S., Anderson E.J., Rostad C.A. Original antigenic sin responses to Betacoronavirus spike proteins are observed in a mouse model, but are not apparent in children following SARS-CoV-2 infection. PLoS ONE. 2021;16:e0256482. doi: 10.1371/journal.pone.0256482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros V., Denolly S., Vogrig M., Boson B., Siret E., Rigaill J., Pillet S., Grattard F., Gonzalo S., Verhoeven P., et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell. Mol. Immunol. 2021;18:318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Rouphael N., Duraisingham S., Romero-Steiner S., Presnell S., Davis C., Schmidt D.S., Johnson S.E., Milton A., Rajam G., et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos C., Atyeo C., Fischinger S., Burke J., Slein M.D., Streeck H., Lauffenburger D., Ryan E.T., Charles R.C., Alter G. Evolution of Early SARS-CoV-2 and Cross-Coronavirus Immunity. MSphere. 2020;5:e00622-20. doi: 10.1128/mSphere.00622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K., Studer M., Roudier P., Gonzalez J., Kozlowski K., Schubert E., Murphy K. Comprehensive R Archive Network; 2021. cluster: “Finding Groups in Data”: Cluster Analysis Extended Rousseeuw et al. [Google Scholar]
- Monto A.S., Malosh R.E., Petrie J.G., Martin E.T. The Doctrine of Original Antigenic Sin: Separating Good From Evil. J. Infect. Dis. 2017;215:1782–1788. doi: 10.1093/infdis/jix173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11:e01991-20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Kindt R., Legendre P., O’hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H. Comprehensive R Archive Network; 2008. vegan: Community Ecology Package. R package version 1.17-11. [Google Scholar]
- Ortega N., Ribes M., Vidal M., Rubio R., Aguilar R., Williams S., Barrios D., Alonso S., Hernández-Luis P., Mitchell R.A., et al. Seven-month kinetics of SARS-CoV-2 antibodies and role of pre-existing antibodies to human coronaviruses. Nat. Commun. 2021;12:4740. doi: 10.1038/s41467-021-24979-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshansky C.M., Gartland A.J., Wong S.-S., Jeevan T., Wang D., Roddam P.L., Caniza M.A., Hertz T., Devincenzo J.P., Webby R.J., Thomas P.G. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am. J. Respir. Crit. Care Med. 2014;189:449–462. doi: 10.1164/rccm.201309-1616OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poston D., Weisblum Y., Wise H., Templeton K., Jenks S., Hatziioannou T., Bieniasz P. Absence of Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Activity in Prepandemic Sera From Individuals With Recent Seasonal Coronavirus Infection. Clin Infect Dis. 2021;73:e1208–e1211. doi: 10.1093/cid/ciaa1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-Sectional Evaluation of Humoral Responses against SARS-CoV-2 Spike. Cell Rep Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W. Comprehensive R Archive Network; 2021. psych: Procedures for Psychological, Psychometric, and Personality Research. [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealy R.E., Hurwitz J.L. Cross-Reactive Immune Responses toward the Common Cold Human Coronaviruses and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Mini-Review and a Murine Study. Microorganisms. 2021;9:1643. doi: 10.3390/microorganisms9081643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva K.J., van de Sandt C.E., Lemke M.M., Lee C.Y., Shoffner S.K., Chua B.Y., Davis S.K., Nguyen T.H.O., Rowntree L.C., Hensen L., et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021;12:2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T.M., Lumley T. Package ‘survival.’. R Top Doc. 2015;128:28–33. [Google Scholar]
- Waskom M.L. seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021. [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Simko V., Levy M., Xie Y., Jin Y., Zemla J. Package ‘corrplot’. Statistician. 2017;56:e24. [Google Scholar]
- Westerhuis B.M., Aguilar-Bretones M., Raadsen M.P., de Bruin E., Okba N.M.A., Haagmans B.L., Langerak T., Endeman H., van den Akker J.P.C., Gommers D.A.M.P.J., et al. Seasonal coronavirus-specific B cells with limited SARS-CoV-2 cross-reactivity dominate the IgG response in severe COVID-19. J Clin Invest. 2021;131:e150613. doi: 10.1172/JCI150613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- Wolf A.I., Mozdzanowska K., Quinn W.J., 3rd, Metzgar M., Williams K.L., Caton A.J., Meffre E., Bram R.J., Erickson L.D., Allman D., et al. Protective antiviral antibody responses in a mouse model of influenza virus infection require TACI. J. Clin. Invest. 2011;121:3954–3964. doi: 10.1172/JCI57362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in the paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at https://github.com/SYL16/SJTRC-CCoV and https://github.com/MacauleyLockeml/St-Jude-Trace-study-SARS-CoV-2 and is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.