Abstract
A fluorogenic PCR-based method (TaqMan-PCR) was developed for typing and subtyping of influenza virus genomes in clinical specimens. The TaqMan-PCR employs a probe technology that exploits the endogenous 5′–3′ nuclease activity of the Taq DNA polymerase to allow direct detection of the amplicon by release of a fluorescent reporter during the PCR. Therefore, post-PCR analysis is avoided since hybridization with the fluorogenic probe and quantification of the amplified product is performed simultaneously during PCR cycling. The specificity of the method was evaluated on 86 influenza A (25 H1N1 and 61 H3N2) and 49 influenza B virus reference strains and isolates. The sensitivity of the technique was found to be at the level of 0.1 50% tissue culture infective dose. This TaqMan-PCR was applied prospectively to surveillance work by community-based sampling in Germany during the last two influenza seasons. Seven hundred five throat swabs were analyzed during the winter of 1997–1998. A total of 195 of 705 samples (28%) were positive by PCR. Influenza viruses could be isolated from 125 specimens (18%). During the 1998–1999 season, 1,840 respiratory samples were received. Influenza viruses were isolated from 281 specimens (15%) out of 525 throat swabs (29%) which were positive for influenza A or B virus by TaqMan-PCR. Further differentiation of influenza A virus-positive swabs revealed an intensive circulation of the subtype H3N2 during both seasons, 1997–1998 and 1998–1999. The TaqMan-PCR was much more sensitive than culture and revealed an excellent correlation for typing and subtyping of influenza viruses when samples were positive by both methods.
Influenza A and B virus infections are major causes of morbidity and mortality worldwide. The influenza viruses are divided into three types, A, B, and C, on the basis of antigenic differences in nucleoprotein and matrix (M) proteins. Influenza A viruses are further classified into subtypes based on the antigenic differences of the surface proteins hemagglutinin (HA) and neuraminidase (NA). Currently, there are 15 distinct HA (H1 to H15) and 9 NA (N1 to N9) subtypes. Three subtypes of HA (H1 to H3) and two subtypes of NA (N1 and N2) are found among influenza A viruses that have caused epidemics among humans. Influenza A virus HA and NA glycoproteins undergo mutational changes known as antigenic drift. Antigenic shift occurs when RNA segments reassort between different subtypes in coinfected cells. Influenza B viruses show a higher genetic stability than influenza A viruses; however, antigenic variation is detectable (reviewed in reference 19). For this reason, major epidemics of respiratory disease caused by new variants of influenza virus continue to occur.
A variety of techniques are available for the detection and identification of influenza viruses. Standard laboratory methods are based on virus isolation in tissue culture or embryonated eggs. The final identification of the isolated virus is usually accomplished by immunofluorescence tests or enzyme immunoassays. Hemagglutination inhibition (HI) procedures are used for the subtyping of influenza A viruses as well as for analysis of antigenic variation of both type A and type B viruses. These methods are very sensitive but depend upon the presence of infectious particles in the original sample. More rapid tests, such as antigen detection by enzyme-linked immunosorbent assay or direct immunofluorescence (7, 12, 14), are carried out for the detection of the virus as well. However, these techniques are of limited sensitivity and specificity. PCR is one of the most sensitive and specific techniques which has been described for typing and subtyping of influenza viruses. In some of these studies, agarose gel analysis was used for the detection of the amplified gene region (1, 8, 25, 26). Other groups performed probe hybridization (5, 20), nested PCR (27, 28), or PCR-enzyme immunoassay (EIA) (4, 21) to evaluate the specificity of the amplified fragments.
The application of PCR and subsequent techniques to confirm the specificity of PCR product in diagnostic laboratories require simple and fast, but highly reproducible, procedures with a minimal number of steps. In this regard, fluorogenic PCRs taking advantage of the 5′–3′ nuclease activity of Taq DNA polymerase have been described (13, 16, 18). A fluorogenic probe, labeled with both a fluorescent reporter and a quencher dye, was used in the TaqMan-PCR. After hybridization of the probe to the target sequence, the Taq DNA polymerase cleaved the TaqMan probe with its 5′–3′ nuclease activity. The reporter and quencher dyes were separated upon cleavage, and the target was amplified, resulting in increased fluorescence of the reporter. In this paper, we describe the application of the TaqMan assay for detection of influenza virus genomes. Influenza type A- and B-virus-specific primer/probe sets were designed for the rapid detection of influenza virus in respiratory specimens. Influenza A virus-positive samples were further analyzed by using specific primer pairs and fluorogenic probes for the identification of HA (H1 and H3) and NA (N1 and N2) subtypes. The PCR assay described here for typing and subtyping of influenza viruses is especially attractive because it eliminates post-PCR processing steps since fluorescence was measured online during the amplification reactions.
MATERIALS AND METHODS
Virus strains and clinical specimens.
Influenza A and B virus reference strains (see Table 2) as well as influenza viruses isolated during the 1995–1996 and 1996–1997 seasons were from the collection of the National Reference Centers for Influenza in Berlin and Hannover, respectively. The reference strains were propagated in embryonated eggs at 37°C. Clinical samples (throat swabs) from patients with respiratory disease symptoms were mainly provided by general practitioners and pediatricians working in the influenza surveillance scheme (Arbeitsgemeinschaft Influenza [24]). The specimens were collected using Virocult swabs (Medical Wire & Equipment, Corsham, England) containing transport medium and were sent through the mail without any refrigeration. The time in transit was between 1 and 4 days. The swabs were vortexed with 5 ml of medium and 200 μl was inoculated onto confluent Madin-Darby canine kidney (MDCK) cells. The cells were maintained in rolling tubes containing serum-free minimum essential medium (Gibco BRL, Life Technologies GmbH, Karlsruhe, Germany) supplemented with 1.25 μg of trypsin per ml (Gibco BRL). The cultures were incubated at 33°C and were examined every day for detection of a cytopathic effect. The medium was tested continuously for hemadsorption and hemagglutination of guinea pig red cells (0.5% vol/vol). Every hemagglutination-positive culture was identified by using the classical HI procedure (3). Briefly, specific antisera raised in ferrets or sheep were treated with receptor-destroying enzyme. The HI tests were carried out by using 4 hemagglutination units of virus and 1.0% (vol/vol) guinea red blood cells. The HI tests of influenza viruses cultivated in embryonated eggs were performed by using 0.5% (vol/vol) chicken erythrocytes.
TABLE 2.
Influenza virus reference strains used to evaluate the specificity of the TaqMan-PCRa
| Influenza virus strains |
CT ofb:
|
|||
|---|---|---|---|---|
| Type B-PCR | Type A-PCR | Subtype A H1-PCR | Subtype A H3-PCR | |
| Influenza A (H1N1) | ||||
| A/PR/8/34 | Neg | 22 | 23 | Neg |
| A/Singapore/6/86 | Neg | 24 | 24 | Neg |
| A/Taiwan/1/86 | Neg | 22 | 23 | Neg |
| A/Texas/36/91 | Neg | 23 | 22 | Neg |
| A/Bayern/7/95 | Neg | 21 | 22 | Neg |
| Influenza A (H3N2) | ||||
| A/Leningrad/360/86 | Neg | 19 | Neg | 20 |
| A/Sichuan/2/87 | Neg | 18 | Neg | 20 |
| A/Shanghai/11/87 | Neg | 22 | Neg | 19 |
| A/England/427/88 | Neg | 20 | Neg | 21 |
| A/Guizhou/54/89 | Neg | 19 | Neg | 19 |
| A/Beijing/353/89 | Neg | 21 | Neg | 20 |
| A/Beijing/32/92 | Neg | 18 | Neg | 20 |
| A/Shangdong/9/93 | Neg | 19 | Neg | 22 |
| A/Johannesburg/33/94 | Neg | 19 | Neg | 21 |
| A/Wuhan/353/95 | Neg | 20 | Neg | 19 |
| A/Nanchang/933/95 | Neg | 19 | Neg | 18 |
| A/Sydney/5/97 | Neg | 18 | Neg | 18 |
| Influenza B | ||||
| B/USSR/100/83 | 23 | Neg | Neg | Neg |
| B/Ann Arbor/1/86 | 24 | Neg | Neg | Neg |
| B/Victoria/2/87 | 22 | Neg | Neg | Neg |
| B/Yamagata/16/88 | 22 | Neg | Neg | Neg |
| B/Panama/45/90 | 23 | Neg | Neg | Neg |
| B/Quindao/102/91 | 20 | Neg | Neg | Neg |
| B/Beijing/184/93 | 21 | Neg | Neg | Neg |
| B/Harbin/7/94 | 19 | Neg | Neg | Neg |
| B/Belarus/1/95 | 22 | Neg | Neg | Neg |
PCR amplification was performed by running 45 cycles with each of the type- and subtype-specific primer/probe systems.
Neg, negative (no increase in fluorescence and, therefore, no amplification was detected during the 45 cycles).
RNA extraction and cDNA synthesis.
Viral RNA was extracted by using a commercial kit (QIAamp Viral RNA Kit; Qiagen, Hilden, Germany). Briefly, 150 μl of clinical throat swab specimen, allantoic fluid, or tissue culture supernatant was mixed with an equal volume of lysis buffer, was heated for 15 min at 70°C, and was applied to a spin column. Unbound material was removed by several washing steps, and the RNA was eluted by using 50 μl of RNase-free water. The cDNA synthesis was carried out at 37°C for 1 h by using 10 μl of RNA, 100 U of murine leukemia virus reverse transcriptase (Gibco BRL), 10 mM dithiothreitol, 20 U of RNasin (Promega, Mannheim, Germany), and 0.25 μM random hexamer primers.
PCR and sequence analysis. (i) Nested RT-PCR.
For influenza surveillance during the 1996–1997 season, virus isolation in cell culture as well as detection by PCR was performed. The primers used for detection and differentiation of influenza viruses have been described by Zhang and Evans (28). For differentiation of NA subtypes, a PCR-coupled DNA enzyme immunoassay was used (21).
(ii) TaqMan-PCR.
The TaqMan probes were designed so that the predicted melting temperature was at least 5°C higher than the predicted melting temperature of the PCR primers. The optimum size of the amplicons was between 200 and 300 bp but not exceeding 400 bp in order to ensure a high sensitivity of the PCR. The probes consisted of oligonucleotides with the 5′-reporter dye FAM (6-carboxy-fluorescein), the internal quencher dye TAMRA (6-carboxy-tetramethyl-rhodamine), and a 3′ end-blocking phosphate. Primer and probes used for typing and subtyping of influenza viruses are listed in Table 1 and were obtained from TIB MOLBIOL, Berlin, Germany. The TaqMan-PCR was carried out in a 96-well flat-bottomed microtiter plate format (Perkin-Elmer) by using special MicroAmp vials. The PCR mix was made up to a volume of 25 μl, containing 5 μl of cDNA, 50 mM Tris-hydrochloride (pH 9), 50 mM KCl, 4 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dUTP, 0.5 U of uracil-N-glycosylase (UNG) (Gibco BRL), 1.25 U of Taq DNA polymerase (InViTek, Berlin, Germany), 0.25 μM concentrations (each) the forward and reverse primers, 0.2 μM of a fluorescence-labeled probe, and 1 μM ROX as a passive reference. After UNG treatment at 50°C for 2 min and UNG inactivation at 95°C for 10 min, the cDNA was amplified by 45 two-step cycles (1 min at 92°C, 1 min at 60°C).
TABLE 1.
Oligonucleotide primers and probes designed for typing and subtyping of influenza viruses
| Influenza virus type/subtype | Primer or probea | Sequence | Coordinatesb |
|---|---|---|---|
| A | AM-151 | 5′ CATGGAATGGCTAAAGACAAGACC | 151–174 |
| AM-397 | 5′ AAGTGCACCAGCAGAATAACTGAG | 374–397 | |
| Probe AM-245 | 5′ CTGCAGCGTAGACGCTTTGTCCAAAATG | 245–272 | |
| A/H1 | HA1-583 | 5′ GGTGTTCATCACCCGTCTAACAT | 583–605 |
| HA1-895 | 5′ GTGTTTGACACTTCGCGTCACAT | 873–895 | |
| Probe HA1-783 | 5′ TGCCTCAAATATTATTGTGTCCCCGGGT | 756–783 | |
| A/H3 | HA3-115 | 5′ GCTACTGAGCTGGTTCAGAGTTC | 115–137 |
| HA3-375 | 5′ GAAGTCTTCATTGATAAACTCCAG | 352–375 | |
| Probe HA3-208 | 5′ CTATTGGGAGACCCTCATTGTGATGG | 208–233 | |
| A/N1 | NA1-1078 | 5′ ATGGTAATGGTGTTTGGATAGGAAG | 1078–1102 |
| NA1-1352 | 5′ AATGCTGCTCCCACTAGTCCAG | 1331–1352 | |
| Probe NA1-1138 | 5′ TGATTTGGGATCCTAATGGATGGACAG | 1138–1164 | |
| A/N2 | NA2-560 | 5′ AAGCATGGCTGCATGTTTGTG | 560–580 |
| NA2-858 | 5′ ACCAGGATATCGAGGATAACAGGA | 835–858 | |
| Probe NA2-821 | 5′ TGCTGAGCACTTCCTGACAATGGGCT | 796–821 | |
| B | BHA-188 | 5′ AGACCAGAGGGAAACTATGCCC | 188–209 |
| BHA-347 | 5′ CTGTCGTGCATTATAGGAAAGCAC | 324–347 | |
| Probe BHA-273 | 5′ ACCTTCGGCAAAAGCTTCAATACTCCA | 273–299 |
AM, M gene of influenza A viruses; HA1 and HA3, HA gene of influenza A viruses of subtypes H1 and H3, respectively; NA1 and NA2, NA gene of influenza A viruses of subtypes N1 and N2, respectively; BHA, HA gene of influenza B viruses.
Primer and probe positions for type A-specific primers correspond to the M gene of A/Bangkok/1/79 (GenBank accession no. Z26862), for subtyping of H1N1 viruses to A/USSR/90/77 (GenBank accession no. K01330 [HA gene] and K02018 [NA gene]), for subtyping of H3N2 viruses to A/Beijing/32/92 (GenBank accession no. Z46392 [HA gene] and U42770 [NA gene]) and for detection of influenza B viruses to the HA gene of B/Kobe/1/94 (D38646).
(iii) Post-PCR analysis.
The amplification in the TaqMan-PCR was followed on the ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, Calif.). The plate was scanned at 518 nm (FAM) and 582 nm (TAMRA). Data acquisition analysis was handled by using the Fluorescence Data Manager (Perkin-Elmer) and Excel (Microsoft Corporation, Redmond, Wash.) spreadsheets. ROX was used as a passive reference to which the reporter dye signal was normalized (Rn) during data analysis. The degree of probe hydrolysis was estimated by calculating the deltaRn, which is the difference between the Rn value of a reaction containing all components, including the template, and the Rn value of an unreacted sample. The threshold cycle (CT) represents a detection threshold for the sequence detector. It is the cycle at which a statistically significant increase in deltaRn is first detected.
(iv) Sequence analysis.
A 548-bp region of the HA1 domain of the HA gene (nucleotides 43 to 501) was amplified by using primer P1 (5′ CTGGGACATCATGCAGTGC) and primer P2 (5′ AGTCACGTTCAGCGCTGGAT). The primer coordinates corresponded to A/Wuhan/359/95 (GenBank accession no. AF 008722). The generated amplicons were purified by using a gel extraction spin kit (Jetquick; Genomed, Bad Oeynhausen, Germany) and were directly sequenced with the Big Dye terminator cycle sequencing kit (Applied Biosystems, Warrington, Great Britain) on a 377 DNA automated sequencer (Applied Biosystems).
RESULTS
Design of primers and probes.
Two oligonucleotide primer/probe sets were designed to differentiate influenza A and B viruses (Table 1). For further subtyping of influenza A viruses, four specific primer/probe sets were selected to differentiate the HA subtypes H1 and H3 as well as the NA subtypes N1 and N2 (Table 1).
A 246-bp segment of the M gene, conserved for influenza A viruses but substantially different from that of influenza B viruses, was selected for amplification. The primers were designed after sequence comparison of 12 human strains of influenza A viruses, representing the subtype H1N1 (A/PR/8/34, A/WSN/33, A/USSR/90/77, A/Forth Monmouth/1/47), the subtype H2N2 (A/Ann Arbor/6/60, A/Singapore/1/57, A/Leningrad/134/57, A/NT/60/68), and H3N2 strains (A/Udorn/72, A/Port Chalmers/1/73, A/Bangkok/1/79, A/Guandong/39/89). Moreover, a highly pathogenic avian strain of the subtype H7N7 (A/FPV/Dobson) was included in the analysis. The GenBank database was used for sequence information. The primers AM-151 and AM-397 and the probe AM-245 derived from the M gene showed 100% homology with all available sequences of human influenza A viruses, with the exception of AM-151, which had a sequence mismatch at the seventh base at the 5′ end present in A/Singapore/1/57 (H2N2).
The primers for influenza B virus were targeted to highly conserved regions of the HA gene. For the alignment of type B sequences, 20 strains were selected, including 10 strains isolated between 1990 and 1994, to assess the variability of the recent isolates. Primer BHA-347 and the probe BHA-273 had 100% homology with all strains used in the alignment. With primer BHA-188, three strains showed a mismatch at the second base away from the 5′ end, and two strains showed a mismatch at the fifth base away from the 5′ end.
Subtype-specific primer/probe sets were also designed by multiple alignment, comparing the HA genes of 16 strains each of type A influenza virus subtype H1 and H3, respectively. For selection of NA-specific oligonucleotides, the sequences of nine N1 strains and 16 N2 strains were aligned. The primers and probes designed to subtype influenza A viruses had 100% homology with 90 to 94% of the strains included in the alignments (the exceptions were a few strains having a mismatch in one of the first bases of the 5′ end).
Sequence analysis of circulating strains of influenza virus A (H3N2) viruses.
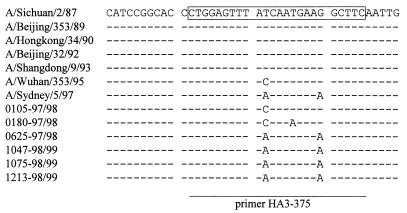
In performing primer and probe design, it was important that the strains included in the alignment should represent influenza viruses isolated over a long period of time as well as during recent years. This could easily be done for N2 subtyping since the sequences of the last three years' reference strains, A/Johannesburg/33/94 and A/Wuhan/359/95, were available from the database. For selection of the H3-specific probe and primers it was possible to use only published sequences of strains up to 1994. To include recent HA sequences, the HA genes of the strains A/Wuhan/359/95 and A/Sydney/5/97 were sequenced along with selected influenza viruses isolated during the past two years in Germany. The H3-specific probe and the primer HA3-115 were 100% homologous with both the reference strains and our own isolates. The results of sequence analysis of the region selected for the reverse primer HA3-375 are shown in Fig. 1. The homology of these strains with the primer HA3-375 was between 92 and 96%. At base 11 from the 3′ end, all Wuhan-like isolates had a cytosine instead of a thymine residue. For Sydney/5/97 and the Sydney-like viruses analyzed here, mismatches at positions 11 and 19 from the 3′ end seemed to be a common feature.
FIG. 1.
Identification of a part of the HA1 genome segment chosen for detection of influenza A (H3N2) viruses. A selection of reference strains, including the latest H3N2 reference strains A/Wuhan/353/95 and A/Sydney/5/97 and influenza A (H3N2) viruses isolated during the last two influenza seasons in Germany, are shown in the alignment. Only regions of sequence immediately flanking the binding site of the reverse primer HA3-375 are shown. The regions targeted by the primer are boxed and underlined.
Amplification of RNA from virus stocks.
The specificity of the designed primer/probe sets was evaluated by using nine reference strains of influenza B virus, twelve reference strains of influenza A virus (H3N2), and five reference strains of influenza A virus (H1N1) (Table 2). Increased fluorescent signals and, therefore, positive results were obtained only with the homologous type of virus as shown in Table 2. Moreover, to estimate the specificity of the PCR assays with currently circulating strains, 53 serologically characterized influenza virus isolates of the 1995–1996 and 1996–1997 seasons were examined. All reference strains and virus isolates investigated gave the expected results, differentiating influenza A and B viruses as well as influenza A virus subtypes.
Sensitivity of the TaqMan-PCR.
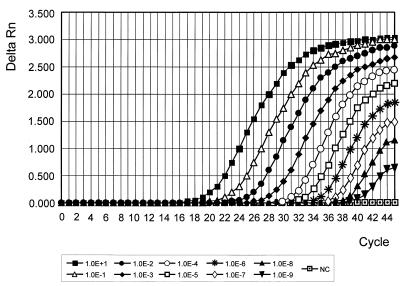
The sensitivity of the TaqMan-PCR was evaluated for each type- and subtype-specific PCR. Virus titration of egg-grown influenza A and B viruses was performed by a serial dilution of the allantoic fluid, followed by infection of MDCK cells in 96-well plates. The 50% tissue culture infective dose (TCID50) values for different strains varied between 107 and 108/ml. Virus RNA was prepared by using 10-fold serial dilutions of the strains A/Nanchang/933/95 (H3N2), A/Bayern/7/95 (H1N1), and B/Belarus/1/95. The sensitivity of the PCR for detection of subtype H3-specific sequences is shown in Fig. 2. Dilutions containing as little as 0.1 TCID50 were positive by all of the type- and subtype-specific influenza TaqMan assays.
FIG. 2.
Sensitivity of the TaqMan-PCR. Tenfold serial dilutions of the influenza virus strain A/Nanchang/933/95 (H3N2) were used to evaluate the sensitivity of the TaqMan-PCR for detection of the subtype H3N2. Cultivation of the strain was performed in embryonated hen eggs. The TCID50 was approximately 8.6 × 108 per ml. RNA was extracted and reverse transcribed as described in Materials and Methods. Amplification of subtype H3-specific sequences of the HA gene was performed by using the primer pair HA3-115/HA3-375 and the probe HA3-208. Uninfected allantoic fluid served as the negative control (NC).
Comparison of nested RT- and TaqMan-PCR.
A retrospective study was performed as a second approach to estimate the sensitivity of the newly developed TaqMan-PCR. More than 100 swabs from the 1996–1997 season which were stored at −70°C were subjected to different PCR assays for typing (influenza A and B viruses) and subtyping (H1, H3, N1, and N2). Seventy-four of these clinical specimens have been shown to be positive by nested reverse transcription (RT)-PCR, and influenza viruses could be cultivated from 57 of these samples. Seventy-three of the 74 samples positive by nested PCR (48 of type A and 26 of type B) were also positive by the TaqMan assay, and none of the samples negative by nested PCR was positive in the TaqMan-PCR. Further subtyping of influenza-A-virus-positive swabs by the TaqMan-PCR revealed the same results as obtained by the nested RT-PCR. Of the 48 samples which were influenza A virus-positive, 11 were of subtype H1N1 and 37 were of subtype H3N2. It could be shown that the sensitivity of the TaqMan-PCR evaluated on clinical samples was comparable to that of the well-established nested RT-PCR.
Prospective study during the 1997–1998 and 1998–1999 seasons.
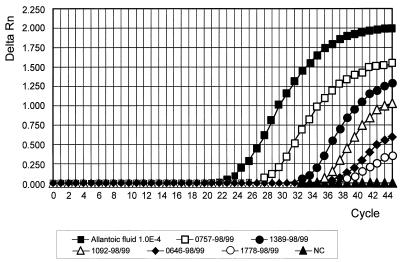
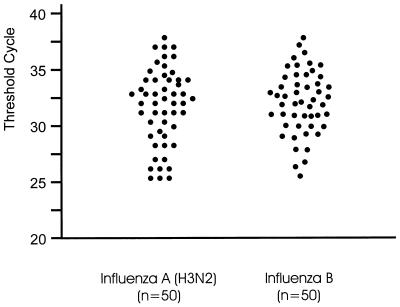
Clinical specimens collected by general practitioners and pediatricians between December 1997 and April 1998 and between November 1998 and April 1999 were analyzed for influenza virus RNA by TaqMan-PCR and virus isolation in cell culture. The results of the first, type-specific PCR were available on the same day the specimen was received, whereas the results of the virus isolation were obtained only after 3 to 14 days. In the case of a positive type A PCR, further subtyping was done by using specific primer/probe sets for currently circulating HA (H1 and H3) and NA (N1 and N2) subtypes. Online measurement during PCR cycling is shown in Fig. 3 for detection of the M gene of influenza A (H3N2) viruses. The intensity of the fluorescent signal (Fig. 3) and the value of the CT varied among the specimens, depending on the amount of virus in the swab. Typical results concerning threshold cycles, the cycle at which an increase in fluorescence was detected, are shown in Fig. 4.
FIG. 3.
Detection of influenza A viruses in clinical samples by TaqMan-PCR. The specimens were collected during the 1998–1999 season. RNA was extracted and reverse transcribed as described in Materials and Methods. Amplification of M gene-specific sequences was performed by using the primer pair AM-151/AM-397 and the probe AM-245. A 10−4-fold dilution of the strain A/Sydney/5/97 cultivated in embryonated hen eggs served as the control. Uninfected allantoic fluid was used as the negative control (NC).
FIG. 4.
Representative clinical samples analyzed by TaqMan-PCR. The specimens were collected during the 1996 through 1999 influenza seasons. Fifty throat swabs positive for influenza virus A and 50 swabs positive for influenza virus B were selected. RNA was extracted and reverse transcribed as described in Materials and Methods. Amplification of subtype-H3-specific sequences of the HA gene was performed by using the primer pair HA3-115/HA3-375 and the probe HA3-208. Detection of influenza-B-specific sequences was performed by using the primer pair BHA-188/BHA-347 and the probe BHA3-273. The threshold cycle is the cycle at which a statistically significant increase of the reporter fluorescence is first detected.
During the 1997–1998 season, a total of 705 clinical samples were investigated. Of the 195 samples identified as type A influenza virus by PCR, 125 were culture positive for influenza A virus. Further differentiation of influenza A virus-positive swabs revealed an extensive circulation of the subtype H3N2. Among the 195 PCR-positive samples, only eight belonged to influenza A virus subtype H1N1. None of the specimens were positive for influenza B virus, either by PCR or by cultivation methods.
During the 1998–1999 season, 1,840 respiratory samples were received. Influenza A virus could be isolated from 250 of 440 throat swabs which were positive by type A-specific TaqMan-PCR. Further investigation of the influenza A virus-positive samples resulted in the detection of only the subtype H3N2. No influenza A (H1N1) viruses were detected during the 1998–1999 season in Germany. Eighty-five samples were found to be PCR positive for influenza B virus, from 31 of which virus could be cultivated.
DISCUSSION
The immunofluorescence test (IF) and EIAs are widely used for rapid identification of influenza viruses. These assays are intended to assist the health care provider in the rapid diagnosis of influenza virus infection since test kits are commercially available and can be performed without special equipment. However, well-evaluated enzyme-linked immunosorbent assays were available only for the detection of influenza A virus antigen (15, 17). The IF requires a sufficient amount of ciliated cells. In addition, most of the swabs reach the laboratory within 2 to 3 days after sampling, especially when collected by community-based sampling. Under these conditions, the ciliated cells are destroyed and become improper for IF. These limitations made it necessary to develop other rapid assays for the identification of influenza viruses, which is especially important for rapid epidemiological and surveillance studies. PCR assays have been designed for the identification of influenza A, B, or C virus (4, 5, 20, 27, 28). Further differentiation between HA and NA subtypes of influenza A virus were performed by using subtype-specific primers (1, 25, 26, 28). Although it was shown that different PCR protocols can be used for typing and subtyping of influenza viruses, there is a lack of prospective clinical evaluations. Few studies exist where RT-PCR for influenza viruses was used to investigate a large number of clinical specimens (6, 25). In a recent report, a multiplex RT-PCR was described for influenza surveillance involving community-based sampling (9). Application of PCR for epidemiological studies and rapid surveillance during the influenza season requires that the procedures are simple and reproducible, permitting the processing of a large number of samples. This necessity is addressed by the TaqMan-PCR for influenza virus described here. Time-consuming post-PCR analyses such as blotting or hybridization procedures are omitted since the amplification of a specific PCR product is measured during PCR cycling.
The specificities of the primer/probe sets designed in this investigation for typing and subtyping of influenza viruses were evaluated by using a variety of reference strains and well-characterized isolates. All 77 reference strains and serotyped isolates were positive in the respective TaqMan-PCR, differentiating the specimens into the correct type or subtype. These results obtained with isolated influenza viruses could be confirmed by a retrospective study by using more than 100 frozen clinical samples collected in the 1996–1997 season. These specimens have been previously characterized by virus isolation in cell culture and by nested RT-PCR.
The comparison of nested RT-PCR and TaqMan-PCR proved to be useful in assessing the sensitivity of the newly developed method. With one exception, all swab samples positive by nested RT-PCR were positive by the TaqMan procedure using type- and subtype-specific assay systems. The sensitivity of the TaqMan-PCR was further evaluated using 10-fold serial dilutions of virus-containing allantoic fluids. We were able to detect the presence of as little as 0.1 TCID50 (corresponding to approximately 10 virus particles), if a particle ratio of 100:1 was assumed. The sensitivities were comparable for all the type- and subtype-specific influenza virus TaqMan assays. The detection limit of other PCR systems varied between 1 and 2 PFU, corresponding to 100 to 200 particles (4, 5, 9, 28) and 6 × 102 to 5 × 103 particles (25) when only gel analysis was used for characterization of the amplicon.
All of the primer/probe sets for typing and subtyping of influenza viruses designed for the TaqMan system are targeted to highly conserved regions of the genes coding for the M protein, the HA, and the NA. The antigenic drift occurring in influenza A and B viruses is mostly evident in influenza A (H3N2) viruses. Thus, it was of interest to check the presently circulating strains for degree of homology with the H3-specific probe and primers. The H3-specific probe and the primer HA3-115 showed a 100% homology with German isolates as well as the reference strains A/Wuhan/353/95 and A/Sydney/5/97.
The homology of the isolates with the reverse primer HA3-375 varied between 92 and 96%. All of the A/Wuhan/353/95-like isolates revealed a mismatch at base 11 from the 3′ end, and all of the A/Sydney/5/97-like viruses showed mismatches at positions 11 and 19 from the 3′ end of the primer. However, it is agreed that a mutational change of an internal base in the primer sequence can be tolerated by PCR. Testing of various primer sequences for their ability to amplify a drosophila hunchback gene suggested that several mismatches can be tolerated by Taq polymerase as long as they are not found at the immediate 3′ end of the primer (23).
The mutations identified in currently circulating strains did not influence the sensitivity and specificity of the H3-specific TaqMan-PCR. However, we plan to replace the thymine at position 11 with an inosine. Inosine substitution was described for the primer pair SK68/SK69 to improve the amplification of a variety of human immunodeficiency virus type 1 strains (2). Preliminary studies using a newly designed reverse primer obtained from a more conserved region of the HA gene gave comparable results, but will have to be largely evaluated during the 1999–2000 season. Nevertheless, the region selected for targeting of the primer HA3-375 is highly stable since a perfect correlation could be shown for H3 strains isolated from 1987 to 1993. But, as our results indicate, it may be necessary to check primer and probe sequences each winter season to ensure an efficient amplification of currently circulating strains.
Since very few studies have used RT-PCR for detection of influenza viruses in surveillance work involving community-based sampling, a prospective study was performed during the 1997–1998 and 1998–1999 seasons to establish the diagnostic value of the new TaqMan-PCR. During the winter of 1997–1998, a total of 705 specimens were received, 125 of which (18%) yielded influenza virus by culture, but 195 of which (28%) gave a positive result by TaqMan-PCR. In Germany, the 1997–1998 season was characterized by an intensive circulation of influenza A (H3N2) viruses and sporadic outbreaks of H1N1 viruses. Of the 700 influenza viruses isolated in Germany in this season, only three were of type B.
However, the sensitivity and specificity of the influenza B virus-specific primer/probe set were shown in the prospective study during the 1998–1999 season. Influenza B virus RNA was found in 85 samples out of 1,840 swabs investigated (5%), but only 31 of them (1.7%) were positive by virus culture. The majority of clinical specimens received came from the northern and eastern parts of the country. These regions were characterized by an intensive circulation of influenza A (H3N2) viruses. Typing and subtyping by the TaqMan-PCR revealed 440 (24%) influenza-A-(H3N2)-positive samples. Viruses could be isolated from 250 of the PCR-positive swabs. Moreover, there was a perfect correlation between types and subtypes obtained by TaqMan-PCR and by virus cultivation when samples were positive by both methods. In general, the high specificity and sensitivity of the TaqMan-PCR evaluated with well-characterized influenza isolates and frozen respiratory specimens could be confirmed by the prospective studies performed during the last two seasons.
The prospective studies performed in 1997–1998 and 1998–1999 revealed an overall increase in detection of influenza viruses of approximately 12%. During the surveillance work, we found an overall detection rate of 15 to 18% by virus culture and 28 to 29% by the TaqMan-PCR for swabs that we received by community-based sampling during that period. These data were obtained for all specimens received between 1 November and 30 April of the following year. During the weeks of peak influenza activity of the last season (weeks 3 to 9 of 1999), a total of 937 samples were investigated, 195 (21%) of which yielded influenza virus by culture. However, influenza RNA was found in 441 samples (47%) of the same set, providing an increase in detection of influenza virus during the peak activity of approximately 26%. The improvement of virus detection by TaqMan-PCR was especially evident during the period of the highest clinical activity registered by sentinel physicians and increased virus isolation, indicating the circulation of influenza viruses in the community.
The increase of 26% influenza virus detection is comparable to the results reported from influenza surveillance programs in England and Wales (9), where 39% of the samples received during the peak period yielded influenza virus by culture but 58% were positive by multiplex-PCR, providing an increase in detection of influenza viruses of 20%. The Royal College of General Practitioners surveillance network set up in the United Kingdom (11) is based on reporting influenza-like illness and obtaining samples from such patients. In Germany, acute respiratory illness is reported to the Arbeitsgemeinschaft Influenza, and swabs are not taken only from patients with influenza-like illness, which might explain the higher PCR detection rate of 58% mentioned above compared to 47% reported in this study.
It has been demonstrated that clinical symptoms and the shedding of viral RNA can persist after a virus infection becomes undetectable by cultivation methods. On days 5 through 9, PCR-EIA could detect influenza A virus RNA in 17 of 45 samples, while only 4 of 45 samples were culture positive (4). In this study, the exact date of the onset of disease was not always reported by the physicians. Therefore, it seems to be realistic that some of the specimens described here were not collected during the first 4 days, and therefore a higher detection rate by PCR is to be assumed for these swabs. Moreover, the isolation procedure is dependent upon the presence of infectious particles in the clinical sample. During the surveillance studies reported here, all of the swabs were sent by mail without any refrigeration. Therefore, the length of time in transit as well as the unchilled transport of the samples may have influenced the sensitivity of tissue culture and contributed to the higher sensitivity of the TaqMan-PCR.
The detection of influenza virus-specific nucleic acids in samples which are negative by the virus culture “gold standard” assay shows the sensitivity of the PCR protocol used, but also clearly shows that care should be taken to avoid false-positive results. Thus, internal as well as external control measures are necessary to monitor the quality of PCR. UNG treatment can be used to prevent contamination by amplicon carryover, but false-positive results have been reported despite the UNG procedure (22). The effectiveness of the UNG inactivation protocol is shown to vary widely, depending on the length and guanine-plus-cytosine content of the product (10).
The most striking advantage of the TaqMan-PCR is its capacity to save time and avoid contamination. Contamination can occur by cross-contamination during sample preparation and setup of PCRs, but most frequently by amplicon carryover which is almost completely avoided by using the TaqMan protocol. Saving time and avoiding false-positive results due to contamination is important for surveillance work during the season when a large number of samples are processed. Strong safety guidelines are observed in our laboratory. These measures include separate locations for swab processing, RNA extraction, cDNA synthesis, mix preparations, and the amplification procedure. In addition, treatment with UNG is generally performed in our protocols, and 20% of the PCR run consists of negative controls. Moreover, there is no amplicon processing at all since post-PCR analysis is eliminated. A further step was taken towards quality control. All of the PCR-positive samples which could not be confirmed by virus culture were investigated in independent second PCR settings. Therefore, false-positive results probably do not cause the higher sensitivity of the TaqMan-PCR than that of virus cultivation shown in this study.
In conclusion, the TaqMan-PCR described here for typing and subtyping of influenza viruses has been shown to be very sensitive and specific. This procedure was evaluated during two influenza seasons for surveillance work by community-based sampling, investigating more than 2,500 specimens. The TaqMan-PCR was much more sensitive than culture and revealed an excellent correlation for typing and subtyping of influenza viruses when samples were positive by both methods. The processing of such a large number of specimens showed the reliability and practicability of the TaqMan-PCR, especially as results can be obtained within a few hours.
REFERENCES
- 1.Bressoud A, Whitcomb J, Pourzand C, Haller O, Cerutti P. Rapid detection of influenza virus H1 by the polymerase chain reaction. Biochem Biophys Res Commun. 1990;167:425–430. doi: 10.1016/0006-291x(90)92040-7. [DOI] [PubMed] [Google Scholar]
- 2.Cassol S, Salas T, Lapointe N, Arella M, Rudnik J, O'Shaughnessy M. Improved detection of HIV-1 envelope sequences using optimized PCR and inosine-substituted primers. Mol Cell Probes. 1991;5:157–160. doi: 10.1016/0890-8508(91)90011-8. [DOI] [PubMed] [Google Scholar]
- 3.Chakraverty P. Antigenic relationship between influenza B viruses. Bull W H O. 1971;45:755–766. [PMC free article] [PubMed] [Google Scholar]
- 4.Cherian T, Bobo L, Steinhoff M C, Karron R A, Yolken R H. Use of PCR-enzyme immunoassay for identification of influenza A virus matrix RNA in clinical samples negative for cultivable virus. J Clin Microbiol. 1994;32:623–628. doi: 10.1128/jcm.32.3.623-628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claas E C J, Sprenger M J W, Kleter G E M, van Beek R, Quint W G V, Masurel N. Type-specific identification of influenza viruses A, B and C by the polymerase chain reaction. J Virol Methods. 1992;39:1–13. doi: 10.1016/0166-0934(92)90120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claas E C J, van Milaan A J, Sprenger M J W, Ruiten-Stuiver M, Arron G I, Rothbarth P H, Masurel N. Prospective application of reverse transcriptase polymerase chain reaction for diagnosing influenza infections in respiratory samples from a children's hospital. J Clin Microbiol. 1993;31:2218–2221. doi: 10.1128/jcm.31.8.2218-2221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coonrod J D, Karathanasis P, Betts R F, Donofrio J C. Enzyme-linked immunosorbent assay of core antigens for clinical diagnosis of influenza. J Med Virol. 1988;25:399–409. doi: 10.1002/jmv.1890250404. [DOI] [PubMed] [Google Scholar]
- 8.Donofrio J C, Coonrod J D, Davidson J N, Betts R F. Detection of influenza A and B in respiratory secretions with the polymerase chain reaction. PCR Methods Appl. 1992;1:263–268. doi: 10.1101/gr.1.4.263. [DOI] [PubMed] [Google Scholar]
- 9.Ellis J S, Fleming D M, Zambon M C. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J Clin Microbiol. 1997;35:2076–2082. doi: 10.1128/jcm.35.8.2076-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espy M J, Smith T F, Persing D H. Dependence of polymerase chain reaction product inactivation protocols on amplicon length and sequence composition. J Clin Microbiol. 1993;31:2361–2365. doi: 10.1128/jcm.31.9.2361-2365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming D M, Chakraverty P, Sadler C, Litton P. Combined clinical and virological surveillance of influenza in winters of 1992 and 1993. Br Med J. 1995;311:290–291. doi: 10.1136/bmj.311.7000.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gleaves C A, Brown J A. Detection of influenza A in clinical specimens and cell culture fluid by a commercial EIA. Clin Diagn Virol. 1993;1:123–127. doi: 10.1016/0928-0197(93)90020-6. [DOI] [PubMed] [Google Scholar]
- 13.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston S L G, Siegel C S. A comparison of direct immunofluorescence, shell vial culture, and conventional cell culture for the rapid detection of influenza A and B. Diagn Microbiol Infect Dis. 1991;14:131–134. doi: 10.1016/0732-8893(91)90047-j. [DOI] [PubMed] [Google Scholar]
- 15.Johnston S L G, Bloy H. Evaluation of a rapid enzyme immunoassay for detection of influenza A virus. J Clin Microbiol. 1993;31:142–143. doi: 10.1128/jcm.31.1.142-143.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonardi G P, Leib H, Birkhead G S, Smith C, Costello P, Conron W. Comparison of rapid detection methods for influenza A virus and their value in health-care management of institutionalized geriatric patients. J Clin Microbiol. 1994;32:70–74. doi: 10.1128/jcm.32.1.70-74.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 19.Murphy B R, Webster R G. Orthomyxoviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 1337–1420. [Google Scholar]
- 20.Pisareva M, Bechtereva T, Plyusnin A, Dobretsova A, Kisselev O. PCR-amplification of influenza A virus specific sequences. Arch Virol. 1992;125:313–318. doi: 10.1007/BF01309648. [DOI] [PubMed] [Google Scholar]
- 21.Schweiger B, Lange I, Heckler R, Willers H, Schreier E. Rapid detection of influenza A neuraminidase subtypes by cDNA amplification coupled to a simple DNA enzyme immunoassay. Arch Virol. 1994;139:439–444. doi: 10.1007/BF01310805. [DOI] [PubMed] [Google Scholar]
- 22.Schweiger B, Pauli G, Zeichhardt H, Kücherer C. A multicentre quality assessment study to monitor the performance of HIV-1 PCR. J Virol Methods. 1997;67:45–55. doi: 10.1016/s0166-0934(97)00075-x. [DOI] [PubMed] [Google Scholar]
- 23.Sommer R, Tautz D. Minimal homology requirements for PCR primers. Nucleic Acids Res. 1989;17:6749. doi: 10.1093/nar/17.16.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szecsenyi J, Uphoff H, Ley S, Brede H D. Influenza surveillance: experiences from establishing a sentinel surveillance system in Germany. J Epidemiol Community Health. 1995;49:9–13. doi: 10.1136/jech.49.suppl_1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright K E, Wilson G A R, Novosad D, Dimock C, Tan D, Weber J M. Typing and subtyping of influenza viruses in clinical samples by PCR. J Clin Microbiol. 1995;33:1180–1184. doi: 10.1128/jcm.33.5.1180-1184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada A, Imanishi J, Nakajima E, Nakajima K, Nakajima S. Detection of influenza viruses in throat swab by using polymerase chain reaction. Microbiol Immunol. 1991;35:259–265. doi: 10.1111/j.1348-0421.1991.tb01555.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamada A, Imanishi J. Detection of influenza B virus in throat swabs using the polymerase chain reaction. Acta Virol. 1992;36:320–325. [PubMed] [Google Scholar]
- 28.Zhang W, Evans D H. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods. 1991;33:165–189. doi: 10.1016/0166-0934(91)90017-t. [DOI] [PubMed] [Google Scholar]