Abstract
Diaporthe (Diaporthaceae, Diaporthales) is a common fungal genus inhabiting plant tissues as endophytes, pathogens and saprobes. Some species are reported from tree branches associated with canker diseases. In the present study, Diaporthe samples were collected from Alnusglutinosa, Fraxinusexcelsior and Quercusrobur in Utrecht, the Netherlands. They were identified to species based on a polyphasic approach including morphology, pure culture characters, and phylogenetic analyses of a combined matrix of partial ITS, cal, his3, tef1 and tub2 gene regions. As a result, four species (viz. Diaporthepseudoalnea sp. nov. from Alnusglutinosa, Diaporthesilvicola sp. nov. from Fraxinusexcelsior, D.foeniculacea and D.rudis from Quercusrobur) were revealed from tree branches in the Netherlands. Diaporthepseudoalnea differs from D.eres (syn. D.alnea) by its longer conidiophores. Diaporthesilvicola is distinguished from D.fraxinicola and D.fraxini-angustifoliae by larger alpha conidia.
Keywords: Two new taxa, Diaporthepseudoalnea , Diaporthesilvicola , taxonomy, two new taxa
Introduction
Diaporthe (syn. Phomopsis) is the type genus of Diaporthaceae in Diaporthales, commonly occurring as plant endophytes, pathogens and saprobes (Udayanga et al. 2014, 2015; Guarnaccia et al. 2017, 2018a, 2018b; Tibpromma et al. 2018; Yang et al. 2020; Dissanayake et al. 2020; Jiang et al. 2021). The sexual morph is characterized by immersed perithecial ascomata and an erumpent pseudostroma with more or less elongated perithecial necks, unitunicate clavate to cylindrical asci, and fusoid, ellipsoid to cylindrical, hyaline uni- to bicellular ascospores (Udayanga et al. 2011; Senanayake et al. 2017). The asexual morph is characterized by ostiolate conidiomata, with cylindrical phialides producing up to three types of hyaline, aseptate conidia (Udayanga et al. 2011; Gomes et al. 2013; Yang et al. 2018), and was previously classified as Phomopsis. Following the “one fungus one name” nomenclature, Rossman et al. (2015) recommended to use Diaporthe based on priority, necessitating the transfer of numerous Phomopsis species to Diaporthe.
Species of Diaporthe are known to cause plant diseases including dieback, canker, leaf spot, fruit rot, pod blights and seed decay. For example, D.citri, D.cytosporella and D.foeniculina caused melanose and stem end rot diseases of Citrus spp. (Udayanga et al. 2014), while Daporthelithocarpi caused leaf spot disease of Castaneahenryi in China (Jiang et al. 2021). Up to 19 Diaporthe species were confirmed to be associated with pear cankers in China (Guo et al. 2020), and eight species of Diaporthe were found to be the casual agents of Chinese grapevine dieback (Manawasinghe et al. 2019). Seven Diaporthe species were reported from blueberry twig blight and dieback diseases in Portugal (Hilário et al. 2020). Diaporthebiconispora and an additional six species were identified as endophytes from healthy Citrus tissues in China (Huang et al. 2015). Diaportheconstrictospora and an additional 11 species were isolated as saprobes from dead wood in karst formations in China (Dissanayake et al. 2020).
Diaporthe species were previously classified mainly based on host association and morphology (Rehner and Uecker 1994; Santos and Phillips 2009; Udayanga et al. 2011, 2014). However, several taxonomic studies of Diaporthales proved that phylogeny based on multiple genes is suitable to separate species (Voglmayr et al. 2012, 2017; Fan et al. 2018; Jiang et al. 2019, 2020; Jaklitsch and Voglmayr 2019, 2020). Species of Diaporthe are now characterised and circumscribed both by morphology and phylogeny of multi-locus DNA data, which revealed many cryptic species in recent years (Diogo et al. 2010; Lombard et al. 2014; Gao et al. 2016, 2017; Long et al. 2019; Yang et al. 2020, 2021; Zapata et al. 2020; Huang et al. 2021). To clarify the species boundaries of the Diaportheeres complex, the Genealogical Phylogenetic Species Recognition principle (GCPSR) and the coalescent-based model Poisson Tree Processes (PTPs) were employed, which suggested that the Diaportheeres species complex actually represents only a single species, D.eres (Hilário et al. 2021).
In the present study, Diaporthe samples from cankered branches of several tree species were collected in the Netherlands, and identified based on modern taxonomic approaches. As a result, two new species and two known species were identified, and the new species are described and illustrated herein.
Materials and Methods
Collection, examination and isolation
The fresh specimens of cankered branches were sampled from Alnusglutinosa, Fraxinusexcelsior and Quercusrobur in Utrecht, the Netherlands. Morphological characteristics of the conidiomata were determined under a Nikon AZ100 dissecting stereomicroscope. More than 20 conidiomata were sectioned, and 50 conidia were randomly selected for measurement using a Leica compound microscope (LM, DM 2500). Isolates were obtained by removing a mucoid conidial mass from conidiomata, spreading the suspension onto the surface of 1.8 % potato dextrose agar (PDA), and incubated at 25 °C for up to 24 h. Single germinating conidia were removed and plated onto fresh PDA plates. Cultural characteristics of isolates incubated on PDA in the dark at 25 °C were recorded, including the colony color and conidiomata structures. The cultures were deposited in the China Forestry Culture Collection Center (CFCC; http://www.cfcc-caf.org.cn/), and the specimens in the herbarium of the Chinese Academy of Forestry (CAF; http://museum.caf.ac.cn/).
DNA extraction, PCR amplification and phylogenetic analyses
Genomic DNA was extracted from colonies grown on cellophane-covered PDA using a cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1990). DNA was checked by electrophoresis in 1 % agarose gel, and the quality and quantity were measured using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). Five partial loci, including the 5.8S nuclear ribosomal DNA gene with the two flanking internally transcribed spacer (ITS) regions, the calmodulin (cal), the histone H3 (his3), the translation elongation factor 1-alpha (tef1) and the beta-tubulin (tub2) genes were amplified by the primer pairs and polymerase chain reaction (PCR) process listed in Table 1. The PCR products were assayed via electrophoresis in 2 % agarose gels. DNA sequencing was performed using an ABI PRISM 3730XL DNA Analyser with a BigDye Terminator Kit v.3.1 (Invitrogen, USA) at the Shanghai Invitrogen Biological Technology Company Limited (Beijing, China).
Table 1.
Genes used in this study with PCR primers and process.
| Locus | PCR primers | PCR: thermal cycles: (Annealing temp. in bold) | Reference |
| ITS | ITS1/ITS4 | (95 °C: 30 s, 48 °C: 30 s, 72 °C: 1 min) × 35 cycles | White et al. 1990 |
| cal | CAL228F/CAL737R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| his3 | CYLH3F/H3-1b | (95 °C: 30 s, 57 °C: 30 s, 72 °C: 1 min) × 35 cycles |
Crous et al. 2004
Glass and Donaldson 1995 |
| tef1 | EF1-728F/EF1-986R | (95 °C: 15 s, 54 °C: 20 s, 72 °C: 1 min) × 35 cycles | Carbone and Kohn 1999 |
| tub2 | T1(Bt2a)/Bt2b | (95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass & Donaldson 1995; O’Donnell and Cigelnik 1997 |
The quality of the amplified nucleotide sequences was checked and the sequences assembled using SeqMan v.7.1.0. Reference sequences were retrieved from the National Center for Biotechnology Information (NCBI), based on recent publications on the genus Diaporthe (Dissanayake et al. 2021; Gao et al. 2021; Huang et al. 2021; Sun et al. 2021, Wang et al. 2021; Yang et al. 2021). Sequences were aligned using MAFFT v. 6 (Katoh and Toh 2010) and corrected manually using MEGA 7.0.21. The best-fit nucleotide substitution models for each gene were selected using jModelTest v. 2.1.7 (Darriba et al. 2012) under the Akaike Information Criterion.
The phylogenetic analyses of the combined gene regions were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was implemented on the CIPRES Science Gateway portal (https://www.phylo.org) using RAxML-HPC BlackBox 8.2.10 (Stamatakis 2014), employing a GTRGAMMA substitution model with 1000 bootstrap replicates. While BI was performed using a Markov Chain Monte Carlo (MCMC) algorithm in MrBayes v. 3.0 (Ronquist et al. 2003). Two MCMC chains, started from random trees for 1000000 generations and trees, were sampled every 100th generation, resulting in a total of 10000 trees. The first 25 % of trees were discarded as burn-in of each analysis. Branches with significant Bayesian Posterior Probabilities (BPP) were estimated in the remaining 7500 trees. Phylogenetic trees were viewed with FigTree v.1.3.1 and processed by Adobe Illustrator CS5. The nucleotide sequence data of the new taxa were deposited in GenBank and are listed in Table 2.
Table 2.
Isolates and GenBank accession numbers used in the phylogenetic analyses of Diaporthe.
| Species | Strain | Host | Origin | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | cal | his3 | tef1 | tub2 | ||||
| Diaportheacaciigena | CBS 129521 | Acaciaretinodes | Australia | KC343005 | KC343247 | KC343489 | KC343731 | KC343973 |
| D.acericola | MFLUCC 17-0956 | Acernegundo | Italy | KY964224 | KY964137 | NA | KY964180 | KY964074 |
| D.acerigena | CFCC 52554 | Acertataricum | China | MH121489 | MH121413 | MH121449 | MH121531 | NA |
| D.acerigena | CFCC 52555 | Acertataricum | China | MH121490 | MH121414 | MH121450 | MH121532 | NA |
| D.acuta | PSCG 047 | Pyruspyrifolia | China | MK626957 | MK691125 | MK726161 | MK654802 | MK691225 |
| D.acutispora | LC6161 | Coffea | China | KX986764 | KX999274 | KX999235 | KX999155 | KX999195 |
| D.alangii | CFCC 52556 | Alangiumkurzii | China | MH121491 | MH121415 | MH121451 | MH121533 | MH121573 |
| D.alangii | CFCC 52557 | Alangiumkurzii | China | MH121492 | MH121416 | MH121452 | MH121534 | MH121574 |
| D.albosinensis | CFCC 53066 | Betulaalbosinensis | China | MK432659 | MK442979 | MK443004 | MK578133 | MK578059 |
| D.albosinensis | CFCC 53067 | Betulaalbosinensis | China | MK432660 | MK442980 | MK443005 | MK578134 | MK578060 |
| D.alleghaniensis | CBS 495.72 | Betulaalleghaniensis | Canada | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| D.ambigua | CBS 114015 | Pyruscommunis | South Africa | KC343010 | KC343252 | KC343494 | KC343736 | KC343978 |
| D.ampelina | STE-U 2660 | Vitisvinifera | France | NA | AY745026 | NA | AY745056 | NA |
| D.amygdali | CBS 126679 | Prunusdulcis | Portugal | MH864208 | KC343264 | KC343506 | KC343748 | KC343990 |
| D.anacardii | CBS 720.97 | Anacardiumoccidentale | East Africa | KC343024 | KC343266 | KC343508 | KC343750 | KC343992 |
| D.angelicae | CBS 111592 | Heracleumsphondylium | Austria | KC343027 | KC343269 | KC343511 | KC343753 | KC343995 |
| D.apiculatum | CFCC 53068 | Rhuschinensis | China | MK432651 | MK442973 | MK442998 | MK578127 | MK578054 |
| D.apiculatum | CFCC 53069 | Rhuschinensis | China | MK432652 | MK44297 | MK442999 | MK578128 | MK578055 |
| D.aquatica | IFRDCC 3051 | Aquatic habitat | China | JQ797437 | NA | NA | NA | NA |
| D.arctii | DP0482 | Arctiumlappa | Austria | KJ590736 | KJ612133 | KJ659218 | KJ590776 | KJ610891 |
| D.arecae | CBS 161.64 | Arecacatechu | India | KC343032 | KC343274 | KC343516 | KC343758 | KC344000 |
| D.arengae | CBS 114979 | Arengaengleri | Hong Kong | MF773664 | KC343276 | KC343518 | KC343760 | KC344002 |
| D.aseana | MFLUCC 12-0299a | Unknown | Thailand | KT459414 | KT459464 | NA | KT459448 | KT459432 |
| D.asheicola | CBS 136967 | Vacciniumashei | Chile | KJ160562 | KJ160542 | NA | KJ160594 | KJ160518 |
| D.aspalathi | CBS 117169 | Aspalathuslinearis | South Africa | KC343036 | KC343278 | KC343520 | KC343762 | KC344004 |
| D.australafricana | CBS 111886 | Vitisvinifera | Australia | KC343038 | KC343280 | KC343522 | KC343764 | KC344006 |
| D.australiana | CBS 146457 | Macadamia | Australia | MN708222 | NA | NA | MN696522 | MN696530 |
| D.baccae | CBS 136972 | Vacciniumcorymbosum | Italy | MK370623 | MG281695 | MF418264 | KJ160597 | MF418509 |
| D.batatas | CBS 122.21 | Ipomoeabatatas | USA | KC343040 | KC343282 | KC343524 | KC343766 | KC344008 |
| D.bauhiniae | CFCC 53071 | Bauhiniapurpurea | China | MK432648 | MK442970 | MK442995 | MK578124 | MK578051 |
| D.bauhiniae | CFCC 53072 | Bauhiniapurpurea | China | MK432649 | MK442971 | MK442996 | MK578125 | MK578052 |
| D.bauhiniae | CFCC 53073 | Bauhiniapurpurea | China | MK432650 | MK442972 | MK442997 | MK578126 | MK578053 |
| D.beilharziae | BRIP 54792 | Indigoferaaustralis | Australia | JX862529 | NA | NA | JX862535 | KF170921 |
| D.benedicti | SBen914 | Diaporthebenedicti | USA | KM669929 | KM669862 | NA | KM669785 | NA |
| D.betulae | CFCC 50469 | Betulaplatyphylla | China | KT732950 | KT732997 | KT732999 | KT733016 | KT733020 |
| D.betulae | CFCC 50470 | Betulaplatyphylla | China | KT732951 | KT732998 | KT733000 | KT733017 | KT733021 |
| D.betulicola | CFCC 51128 | Betulaalbo-sinensis | China | KX024653 | KX024659 | KX024661 | KX024655 | KX024657 |
| D.betulicola | CFCC 51129 | Betulaalbo-sinensis | China | KX0246554 | KX024660 | KX024662 | KX0246556 | KX024658 |
| D.betulina | CFCC 52560 | Betulaalbo-sinensis | China | MH121495 | MH121419 | MH121455 | MH121537 | MH121577 |
| D.betulina | CFCC 52561 | Betulaalbo-sinensis | China | MH121496 | MH121420 | MH121456 | MH121538 | MH121578 |
| D.biconispora | ZJUD62 | Citrusmaxima | China | KJ490597 | NA | KJ490539 | KJ490476 | KJ490418 |
| D.biguttulata | ZJUD47 | Citruslimon | China | KJ490582 | NA | KJ490524 | KJ490461 | KJ490403 |
| D.bohemiae | CBS 143347 | Vitisvinifera | Czech Republic | MK300012 | MG281710 | MG281361 | MG281536 | MG281188 |
| D.brasiliensis | CBS 133183 | Aspidospermatomentosum | Brazil | KC343042 | KC343284 | KC343526 | KC343768 | KC344010 |
| D.caatingaensis | URM7485 | Tacingainamoena | Brazil | KY085927 | KY115598 | NA | KY115604 | KY115601 |
| D.camelliae-sinensis | SAUCC194.92 | Camelliasinensis | China | MT822620 | MT855699 | MT855588 | MT855932 | MT855817 |
| D.canthii | CPC 19740 | Canthiuminerme | South Africa | JX069864 | NA | NA | NA | NA |
| D.caryae | CFCC 52563 | Caryaillinoinensis | China | MH121498 | MH121422 | MH121458 | MH121540 | MH121580 |
| D.caryae | CFCC 52564 | Caryaillinoinensis | China | MH121499 | MH121423 | MH121459 | MH121541 | MH121581 |
| D.cassines | CPC 21916 | Cassineperagua | South Africa | KF777155 | NA | NA | KF777244 | NA |
| D.caulivora | CBS 127268 | Glycinemax | Croatia | MH864501 | KC343287 | KC343529 | KC343771 | KC344013 |
| D.cercidis | CFCC 52565 | Cercischinensis | China | MH121500 | MH121424 | MH121460 | NA | MH121582 |
| D.cercidis | CFCC 52566 | Cercischinensis | China | MH121501 | MH121425 | MH121461 | NA | MH121583 |
| D.chamaeropis | CBS 454.81 | Chamaeropshumilis | Greece | KC343048 | KC343290 | KC343532 | KC343774 | KC344016 |
| D.charlesworthii | BRIP 54884m | Rapistrumrugostrum | Australia | KJ197288 | NA | NA | KJ197250 | KJ197268 |
| D.chensiensis | CFCC 52567 | Abieschensiensis | China | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| D.chensiensis | CFCC 52568 | Abieschensiensis | China | MH121503 | MH121427 | MH121463 | MH121545 | MH121585 |
| D.chongqingensis | PSCG 435 | Pyruspyrifolia | China | MK626916 | MK691209 | MK726257 | MK654866 | MK691321 |
| D.chrysalidocarpi | SAUCC194.35 | Chrysalidocarpuslutescens | China | MT822563 | MT855646 | MT855532 | MT855760 | MT855876 |
| D.cichorii | MFLUCC 17-1023 | Cichoriumintybus | Italy | KY964220 | KY964133 | NA | KY964176 | KY964104 |
| D.cinnamomi | CFCC 52569 | Cinnamomum | China | MH121504 | NA | MH121464 | MH121546 | MH121586 |
| D.cinnamomi | CFCC 52570 | Cinnamomum | China | MH121505 | NA | MH121465 | MH121547 | MH121587 |
| D.cissampeli | CPC 27302 | Cissampeloscapensis | South Africa | KX228273 | NA | KX228366 | NA | KX228384 |
| D.citri | AR3405 | Citrus | USA | KC843311 | KC843157 | KJ420881 | KC843071 | KC843187 |
| D.citri | CFCC 53079 | Citrussinensis | China | MK573940 | MK574579 | MK574595 | MK574615 | MK574635 |
| D.citriasiana | CGMCC 3.15224 | Citrusunshiu | China | JQ954645 | KC357491 | KC490515 | JQ954663 | KC357459 |
| D.citrichinensis | CGMCC 3.15225 | Citrus | China | JQ954648 | KC357494 | NA | JQ954666 | NA |
| D.collariana | MFLU 17-2770 | Magnoliachampaca | Thailand | MG806115 | MG783042 | NA | MG783040 | MG783041 |
| D.compactum | LC3083 | Camelliasinensis | China | KP267854 | NA | KP293508 | KP267928 | NA |
| D.conica | CFCC 52571 | Alangiumchinense | China | MH121506 | MH121428 | MH121466 | MH121548 | MH121588 |
| D.conica | CFCC 52572 | Alangiumchinense | China | MH121507 | MH121429 | MH121467 | MH121549 | MH121589 |
| D.constrictospora | CGMCC 3.20096 | Unknown | China | MT385947 | MT424718 | MW022487 | MT424682 | MT424702 |
| D.convolvuli | CBS 124654 | Convolvulusarvensis | Turkey | KC343054 | KC343296 | KC343538 | KC343780 | KC344022 |
| D.coryli | CFCC 53083 | Corylusmandshurica | China | MK432661 | MK442981 | MK443006 | MK578135 | MK578061 |
| D.coryli | CFCC 53084 | Corylusmandshurica | China | MK432662 | MK442982 | MK443007 | MK538176 | MK578062 |
| D.corylicola | CFCC 53986 | Corylusheterophylla | China | MW839880 | MW836684 | MW836717 | MW815894 | MW883977 |
| D.corylicola | CFCC 53987 | Corylusheterophylla | China | MW839867 | MW836685 | MW836718 | MW815895 | MW883978 |
| D.crotalariae | CBS 162.33 | Crotalariaspectabilis | USA | MH855395 | JX197439 | KC343540 | GQ250307 | KC344024 |
| D.crousii | CAA 823 | Vacciniumcorymbosum | Portugal | MK792311 | MK883835 | MK871450 | MK828081 | MK837932 |
| D.cucurbitae | DAOM 42078 | Cucumis | Canada | KM453210 | NA | KM453212 | KM453211 | KP118848 |
| D.cuppatea | CBS 117499 | Aspalathuslinearis | South Africa | MH863021 | KC343299 | KC343541 | KC343783 | KC344025 |
| D.cynaroidis | CBS 122676 | Proteacynaroides | South Africa | KC343058 | KC343300 | KC343542 | KC343784 | KC344026 |
| D.cytosporella | FAU461 | Citruslimon | Italy | KC843307 | KC843141 | NA | KC843116 | KC843221 |
| D.diospyricola | CPC 21169 | Diospyroswhyteana | South Africa | KF777209 | NA | NA | NA | NA |
| D.discoidispora | ZJUD89 | Citrusunshiu | China | KJ490624 | NA | KJ490566 | KJ490503 | KJ490445 |
| D.dorycnii | MFLUCC 17-1015 | Dorycniumhirsutum | Italy | KY964215 | NA | NA | KY964171 | KY964099 |
| D.drenthii | CBS 146453 | Macadamia | Australia | MN708229 | NA | NA | MN696526 | MN696537 |
| D.elaeagni-glabrae | LC4802 | Elaeagnusglabra | China | KX986779 | KX999281 | KX999251 | KX999171 | KX999212 |
| D.ellipicola | CGMCC 3.17084 | Lithocarpusglaber | China | KF576270 | NA | NA | KF576245 | KF576294 |
| D.endophytica | CBS 133811 | Schinusterebinthifolius | Brazil | KC343065 | KC343307 | KC343549 | KC343791 | KC344033 |
| D.eres | CBS 146.46 | Alnus | Netherlands | KC343008 | KC343250 | KC343492 | KC343734 | KC343976 |
| D.eres | CBS 121004 | Juglans | USA | KC343134 | KC343376 | KC343618 | KC343860 | KC344102 |
| D.eres | CGMCC 3.17081 | Lithocarpusglabra | China | KF576282 | NA | NA | KF576257 | KF576306 |
| D.eres | CFCC 51632 | Camptothecaacuminata | China | KY203726 | KY228877 | KY228881 | KY228887 | KY228893 |
| D.eres | CBS 139.27 | Celastrus | USA | KC343047 | KC343289 | KC343531 | KC343773 | KC344015 |
| D.eres | CBS 143349 | Vitisvinifera | United Kingdom | MG281017 | MG281712 | MG281363 | MG281538 | MG281190 |
| D.eres | AR5193 | Ulmus | Germany | KJ210529 | KJ434999 | KJ420850 | KJ210550 | KJ420799 |
| D.eres | CFCC 52575 | Castaneamollissima | China | MH121510 | NA | MH121470 | MH121552 | MH121592 |
| D.eres | CFCC 52576 | Castaneamollissima | China | MH121511 | MH121432 | MH121471 | MH121553 | MH121593 |
| D.eres | CFCC 52577 | Acanthopanaxsenticosus | China | MH121512 | MH121433 | MH121472 | MH121554 | MH121594 |
| D.eres | CFCC 52578 | Sorbus | China | MH121513 | MH121433 | MH121473 | MH121555 | MH121595 |
| D.eres | CFCC 52579 | Juglansregia | China | MH121514 | NA | MH121474 | MH121556 | NA |
| D.eres | CFCC 52580 | Meliaazedarace | China | MH121515 | NA | MH121475 | MH121557 | MH121596 |
| D.eres | CFCC 52581 | Rhododendronsimsii | China | MH121516 | NA | MH121476 | MH121558 | MH121597 |
| D.eres | MAFF 625034 | Pyruspyrifolia | Japan | NA | KJ435023 | KJ420868 | NA | KJ420819 |
| D.eres | AR5211 | Hederahelix | France | KJ210538 | KJ435043 | KJ420875 | KJ210559 | KJ420828 |
| D.eres | CGMCC 3.17089 | Lithocarpusglabra | China | KF576267 | NA | NA | KF576242 | KF576291 |
| D.eres | MFLUCC 17-0963 | Lonicera | Italy | KY964190 | KY964116 | NA | KY964146 | KY964073 |
| D.eres | DAOM 695742 | Picearuben | Canada | KU552025 | NA | NA | KU552023 | KU574615 |
| D.eres | MFLUCC 16-0113 | Prunuspersica | China | KU557563 | NA | KU557611 | KU557631 | KU55758 |
| D.eres | CBS 144.27 | Spiraea | USA | KC343144 | KC343386 | KC343628 | KC343870 | KC344112 |
| D.eres | CBS 587.79 | Pinusparvifloravar | Japan | KC343153 | KC343395 | KC343637 | KC343879 | KC344121 |
| D.eres | CBS 338.89 | Hederahelix | Yugoslavia | KC343152 | KC343394 | KC343636 | KC343878 | KC344120 |
| D.eres | MFLU 17-0646 | Rosa | United Kingdom | MG828895 | MG829274 | NA | MG829270 | MG843877 |
| D.eucalyptorum | CBS 132525 | Eucalyptus | China | MH305525 | NA | NA | NA | NA |
| D.foeniculacea | CBS 111553 | Foeniculumvulgare | Spain | MH854926 | KC343343 | KC343585 | KC343827 | KC344069 |
| D.foeniculacea | CFCC 54192 | Quercusrobur | Netherlands | MZ727033 | NA | MZ753474 | MZ816339 | MZ753483 |
| D.foeniculacea | M35 | Quercusrobur | Netherlands | MZ727034 | NA | MZ753475 | MZ816340 | MZ753484 |
| D.foeniculacea | M40-1 | Quercusrobur | Netherlands | MZ727035 | NA | MZ753476 | MZ816341 | MZ753485 |
| D.foeniculacea | M84 | Quercusrobur | Netherlands | MZ727036 | NA | MZ753477 | MZ816342 | MZ753486 |
| D.fraxini-angustifoliae | BRIP 54781 | Fraxinusangustifolia | Australia | JX862528 | KT459462 | NA | JX862534 | NA |
| D.fraxinicola | CFCC 52582 | Fraxinuschinensis | China | MH121517 | MH121435 | NA | MH121560 | NA |
| D.fraxinicola | CFCC 52583 | Fraxinuschinensis | China | MH121518 | MH121436 | NA | MH121559 | NA |
| D.fulvicolor | PSCG 051 | Pyruspyrifolia | China | MK626859 | MK691132 | MK726163 | MK654806 | MK691236 |
| D.fusicola | CGMCC 3.17087 | Lithocarpusglabra | China | KF576281 | KF576233 | NA | KF576256 | KF576305 |
| D.ganjae | CBS 180.91 | Cannabissativa | USA | KC343112 | KC343354 | KC343596 | KC343838 | KC344080 |
| D.ganzhouensis | CFCC 53087 | Unknown | China | MK432665 | MK442985 | MK443010 | MK578139 | MK578065 |
| D.ganzhouensis | CFCC 53088 | Unknown | China | MK432666 | MK442986 | MK443011 | MK578140 | MK578066 |
| D.garethjonesii | MFLUCC 12-0542a | Unknown | Thailand | KT459423 | KT459470 | NA | KT459457 | KT459441 |
| D.goulteri | BRIP 55657a | Helianthusannuus | Australia | KJ197290 | NA | NA | KJ197252 | KJ197270 |
| D.grandiflori | SAUCC194.84 | Heterostemmagrandiflorum | China | MT822612 | MT855691 | MT855580 | MT855809 | MT855924 |
| D.guangxiensis | JZB320087 | Vitisvinifera | China | MK335765 | MK736720 | NA | MK500161 | MK523560 |
| D.gulyae | BRIP 54025 | Helianthusannuus | Australia | NA | NA | NA | JN645803 | KJ197271 |
| D.guttulata | CGMCC 3.20100 | Unknown | China | MT385950 | MW022470 | MW022491 | MT424685 | MT424705 |
| D.helianthi | CBS 592.81 | Helianthusannuus | Serbia | KC343115 | KC343357 | KC343599 | KC343841 | KC344083 |
| D.heliconiae | SAUCC194.77 | Heliconiametallica | China | MT822605 | MT855684 | MT855573 | MT855802 | MT855917 |
| D.heterophyllae | CPC 26215 | Acaciaheterophylla | France | MG600222 | MG600218 | MG600220 | MG600224 | MG600226 |
| D.heterostemmatis | SAUCC194.85 | Heterostemmagrandiflorum | China | MT822613 | MT855692 | MT855581 | MT855810 | MT855925 |
| D.hickoriae | CBS 145.26 | Caryaglabra | USA | KC343118 | KC343360 | NA | KC343844 | KC344086 |
| D.hispaniae | CBS 143351 | Vitisvinifera | Spain | MG281123 | MG281820 | MG281471 | MG281644 | MG281296 |
| D.hongkongensis | CBS 115448 | Dichroafebrifuga | China | MK304388 | KC343361 | KC343603 | KC343845 | KC344087 |
| D.hubeiensis | JZB320123 | Vitisvinifera | China | MK335809 | MK500235 | NA | MK523570 | MK500148 |
| D.incompleta | LC6754 | Camelliasinensis | China | KX986794 | KX999289 | KX999265 | KX999186 | KX999226 |
| D.inconspicua | CBS 133813 | Maytenusilicifolia | Brazil | NA | KC343365 | KC343607 | KC343849 | KC344091 |
| D.infecunda | CBS 133812 | Schinusterebinthifolius | Brazil | KC343126 | KC343368 | KC343610 | KC343852 | KC344094 |
| D.irregularis | CGMCC 3.20092 | Unknown | China | MT385951 | MT424721 | NA | MT424686 | MT424706 |
| D.isoberliniae | CPC 22549 | Isoberliniaangolensis | Zambia | KJ869190 | NA | NA | NA | KJ869245 |
| D.juglandicola | CFCC 51134 | Juglansmandshurica | China | KU985101 | KX024616 | KX024622 | KX024628 | KX024634 |
| D.kadsurae | CFCC 52586 | Kadsuralongipedunculata | China | MH121521 | MH121439 | MH121479 | MH121563 | MH121600 |
| D.kadsurae | CFCC 52587 | Kadsuralongipedunculata | China | MH121522 | MH121440 | MH121480 | MH121564 | MH121601 |
| D.kochmanii | BRIP 54033 | Helianthusannuus | Australia | NA | NA | NA | JN645809 | NA |
| D.kongii | BRIP 54031 | Helianthusannuus | Australia | NA | NA | NA | NA | KJ197272 |
| D.lenispora | CGMCC 3.20101 | Unknown | China | MT385952 | MW022472 | MW022493 | MT424687 | MT424707 |
| D.litchicola | BRIP 54900 | Litchichinensis | Australia | LC041036 | NA | NA | JX862539 | NA |
| D.litchii | SAUCC194.22 | Litchichinensis | China | MT822550 | MT855635 | MT855519 | MT855747 | MT855863 |
| D.lithocarpus | CGMCC 3.15175 | Lithocarpusglabra | China | KC135104 | KF576235 | NA | KC153095 | KF576311 |
| D.longicolla | FAU599 | Glycinemax | USA | KJ590728 | KJ612124 | KJ659188 | KJ590767 | KJ610883 |
| D.longispora | CBS 194.36 | Ribes | Canada | MH855769 | KC343377 | KC343619 | KC343861 | KC344103 |
| D.lusitanicae | CBS 123212 | Foeniculumvulgare | Portugal | MH863279 | KC343378 | KC343620 | KC343862 | KC344104 |
| D.lutescens | SAUCC194.36 | Chrysalidocarpuslutescens | China | MT822564 | MT855647 | MT855533 | MT855761 | MT855877 |
| D.macadamiae | CBS 146455 | Macadamia | Australia | MN708230 | NA | NA | MN696528 | MN696539 |
| D.macintoshii | BRIP 55064a | Rapistrumrugosum | Australia | KJ197289 | NA | NA | KJ197251 | KJ197269 |
| D.mahothocarpus | CGMCC 3.15181 | Lithocarpusglabra | China | KC153096 | NA | NA | KC153087 | KF576312 |
| D.malorum | CAA 734 | Malusdomestica | Portugal | KY435638 | KY435658 | KY435648 | KY435627 | KY435668 |
| D.masirevicii | BRIP 54256 | Glycinemax | Australia | KJ197277 | NA | NA | KJ197238 | KJ197256 |
| D.mayteni | CBS 133185 | Maytenusilicifolia | Brazil | KC343139 | KC343381 | KC343623 | KC343865 | KC344107 |
| D.maytenicola | CPC 21896 | Maytenusacuminata | South Africa | KF777157 | NA | NA | NA | KF777250 |
| D.mediterranea | SAUCC194.111 | Machiluspingii | China | MT822639 | MT855718 | MT855606 | MT855836 | MT855951 |
| D.melastomatis | SAUCC194.55 | Melastomamalabathricum | China | MT822583 | MT855664 | MT855551 | MT855780 | MT855896 |
| D.melonis | CBS 435.87 | Glycinesoja | Indonesia | KC343141 | KC343383 | KC343625 | KC343867 | KC344109 |
| D.middletonii | BRIP 54884e | Rapistrumrugosum | Australia | KJ197286 | NA | NA | KJ197248 | KJ197266 |
| D.minima | CGMCC 3.20097 | Unknown | China | MT385953 | MT424722 | MW022496 | MT424688 | MT424708 |
| D.minusculata | CGMCC 3.20098 | Unknown | China | MT385957 | MW022475 | MW022499 | MT424692 | MT424712 |
| D.miriciae | BRIP 54736j | Helianthusannuus | Australia | KJ197282 | NA | NA | KJ197244 | KJ197262 |
| D.multigutullata | CFCC 53095 | Citrusmaxima | China | MK432645 | MK442967 | MK442992 | MK578121 | MK578048 |
| D.multigutullata | CFCC 53096 | Citrusmaxima | China | MK432646 | MK442968 | MK442993 | MK578122 | MK578049 |
| D.musigena | CBS 129519 | Musa | Australia | KC343143 | KC343385 | KC343267 | KC343869 | KC344111 |
| D.neoarctii | CBS 109490 | Ambrosiatrifida | USA | KC343145 | KC343387 | KC343629 | KC343871 | KC344113 |
| D.neoraonikayaporum | MFLUCC 14-1136 | Tectonagrandis | Thailand | KU712449 | KU749356 | NA | KU749369 | KU743988 |
| D.nothofagi | BRIP 54801 | Nothofaguscunninghamii | Australia | JX862530 | NA | NA | JX862536 | KF170922 |
| D.novem | CBS 127269 | Glycinemax | Croatia | KC343155 | KC343397 | KC343639 | KC343881 | KC344123 |
| D.ocoteae | CPC 26217 | Ocoteabullata | France | KX228293 | NA | NA | NA | KX228388 |
| D.oraccinii | LC3166 | Camelliasinensis | China | KP267863 | NA | KP293517 | KP267937 | KP293443 |
| D.ovalispora | ZJUD93 | Citruslimon | China | KJ490628 | NA | KJ490570 | KJ490507 | KJ490449 |
| D.ovoicicola | CGMCC 3.17093 | Lithocarpusglabra | China | KF576265 | KF576223 | NA | KF576240 | KF576289 |
| D.oxe | CBS 133186 | Maytenusilicifolia | Brazil | KC343164 | KC343406 | KC343648 | KC343890 | KC344132 |
| D.padina | CFCC 52590 | Padusracemosa | China | MH121525 | MH121443 | MH121483 | MH121567 | MH121604 |
| D.padina | CFCC 52591 | Padusracemosa | China | MH121526 | MH121444 | MH121484 | MH121568 | MH121605 |
| D.pandanicola | MFLUCC 17-0607 | Pandanaceae | Thailand | MG646974 | NA | NA | NA | MG646930 |
| D.paranensis | CBS 133184 | Maytenusilicifolia | Brazil | KC343171 | KC343413 | KC343655 | KC343897 | KC344139 |
| D.parapterocarpi | CPC 22729 | Pterocarpusbrenanii | Zambia | KJ869138 | NA | NA | NA | KJ869248 |
| D.parvae | PSCG 035 | Pyrusbretschneideri | China | MK626920 | MK691169 | MK726211 | MK654859 | MK691249 |
| D.pascoei | BRIP 54847 | Perseaamericana | Australia | MK111097 | NA | NA | JX862538 | KF170924 |
| D.passiflorae | CPC 19183 | Passifloraedulis | Netherlands | JX069860 | NA | NA | NA | NA |
| D.passifloricola | CPC 27480 | Passiflorafoetida | Malaysia | KX228292 | NA | KX228367 | NA | KX228387 |
| D.penetriteum | LC3215 | Camelliasinensis | China | KP267879 | NA | NA | KP293532 | KP267953 |
| D.perjuncta | CBS 109745 | Ulmusglabra | Austria | KC343172 | KC343414 | KC343656 | KC343898 | KC344140 |
| D.perseae | CBS 151.73 | Perseagratissima | Netherlands | KC343173 | KC343415 | NA | NA | NA |
| D.pescicola | MFLUCC 16-0105 | Prunuspersica | China | KU557555 | KU557603 | NA | KY400831 | KU557579 |
| D.phaseolorum | AR4203 | Phaseolusvulgaris | USA | KJ590738 | KJ612135 | KJ659220 | KJ590739 | KJ610893 |
| D.phillipsii | CAA 817 | Vacciniumcorymbosum | Portugal | MK792305 | MK883831 | MK871445 | MK828076 | MN000351 |
| D.podocarpi-macrophylli | LC6155 | Podocarpusmacrophyllus | Japan | KX986774 | KX999278 | KX999246 | KX999167 | KX999207 |
| D.pometiae | SAUCC194.72 | Pometiapinnata | China | MT822600 | MT855679 | MT855568 | MT855797 | MT855912 |
| D.pseudoalnea | CFCC 54190 | Alnusglutinosa | Netherlands | MZ727037 | MZ753468 | MZ781302 | MZ816343 | MZ753487 |
| D.pseudoalnea | M2A | Alnusglutinosa | Netherlands | MZ727038 | MZ753469 | MZ753478 | MZ816344 | MZ753488 |
| D.pseudomangiferae | CBS 101339 | Mangiferaindica | Dominican Republic | KC343181 | KC343423 | KC343665 | KC343907 | KC344149 |
| D.pseudophoenicicola | CBS 176.77 | Mangiferaindica | Iraq | KC343183 | KC343425 | KC343667 | KC343909 | KC344151 |
| D.pseudotsugae | MFLU 15-3228 | Pseudotsugamenziesii | Italy | KY964225 | KY964138 | NA | KY964181 | KY964108 |
| D.psoraleae | CPC 21634 | Psoraleapinnata | South Africa | KF777158 | NA | NA | KF777245 | KF777251 |
| D.psoraleae-pinnatae | CPC 21638 | Psoraleapinnata | South Africa | KF777159 | NA | NA | NA | KF777252 |
| D.pterocarpicola | MFLUCC 10-0580a | Pterocarpusindicus | Thailand | JQ619887 | JX197433 | NA | JX275403 | JX275441 |
| D.pungensis | SAUCC194.112 | Elaeagnuspungens | China | MT822640 | MT855719 | MT855607 | MT855837 | MT855952 |
| D.pyracanthae | CAA483 | Pyracanthacoccinea | Portugal | KY435635 | KY435645 | KY435656 | KY435625 | KY435666 |
| D.racemosae | CPC 26646 | Euclearacemosa | South Africa | MG600223 | MG600219 | MG600221 | MG600225 | MG600227 |
| D.raonikayaporum | CBS 133182 | Spondiasmombin | Brazil | KC343188 | KC343430 | KC343672 | KC343914 | KC344156 |
| D.ravennica | MFLUCC 16-0997 | Clematisvitalba | Italy | NA | NA | NA | MT394670 | NA |
| D.rhusicola | CPC 18191 | Rhuspendulina | South Africa | JF951146 | NA | NA | NA | NA |
| D.rosae | MFLUCC 17-2658 | Rosa | United Kingdom | MG828894 | MG829273 | NA | NA | MG843878 |
| D.rosiphthora | COAD 2914 | Rosa | Brazil | MT311197 | MT313691 | NA | MT313693 | NA |
| D.rossmaniae | CAA 762 | Vacciniumcorymbosum | Portugal | MK792290 | MK883822 | MK871432 | MK828063 | MK837914 |
| D.rostrata | CFCC 50062 | Juglansmandshurica | China | KP208847 | KP208849 | KP208851 | KP208853 | KP208855 |
| D.rostrata | CFCC 50063 | Juglansmandshurica | China | KP208848 | KP208850 | KP208852 | KP208854 | KP208856 |
| D.rudis | AR3422 | Laburnumanagyroides | Austria | KC843331 | KC843146 | NA | KC843090 | KC843177 |
| D.rudis | CFCC 54193 | Quercusrobur | Netherlands | MZ727039 | MZ753470 | MZ753479 | MZ816345 | MZ753489 |
| D.rudis | M86 | Quercusrobur | Netherlands | MZ727040 | MZ753471 | MZ753480 | MZ816346 | MZ753490 |
| D.saccarata | CBS 116311 | Protearepens | South Africa | KC343190 | KC343432 | KC343674 | KC343916 | KC344158 |
| D.sackstonii | BRIP 54669b | Helianthusannuus | Australia | KJ197287 | NA | NA | KJ197249 | KJ197267 |
| D.salicicola | BRIP 54825 | Salixpurpurea | Australia | JX862531 | NA | NA | JX862537 | KF170923 |
| D.sambucusii | CFCC 51986 | Sambucuswilliamsii | China | KY852495 | KY852499 | KY852503 | KY852507 | KY852511 |
| D.sambucusii | CFCC 51987 | Sambucuswilliamsii | China | KY852496 | KY852500 | KY852504 | KY852508 | KY852512 |
| D.schimae | CFCC 53103 | Schimasuperba | China | MK442640 | MK442962 | MK442987 | MK578116 | MK578043 |
| D.schimae | CFCC 53104 | Schimasuperba | China | MK442641 | MK442963 | MK442988 | MK578117 | MK578044 |
| D.schimae | CFCC 53105 | Schimasuperba | China | MK442642 | MK442964 | MK442989 | MK578118 | MK578045 |
| D.schini | CBS 133181 | Schinusterebinthifolius | Brazil | KC343191 | KC343433 | KC343675 | KC343917 | KC344159 |
| D.schisandrae | CFCC 51988 | Schisandrachinensis | China | KY852497 | KY852501 | KY852505 | KY852509 | KY852513 |
| D.schisandrae | CFCC 51989 | Schisandrachinensis | China | KY852498 | KY852502 | KY852506 | KY852510 | KY852514 |
| D.schoeni | MFLU 15-1279 | Schoenusnigricans | Italy | KY964226 | KY964139 | NA | KY964182 | KY964109 |
| D.sclerotioides | CBS 296.67 | Cucumissativus | Netherlands | MH858974 | KC343435 | KC343677 | KC343919 | KC344161 |
| D.searlei | CBS 146456 | Macadamia | Australia | MN708231 | NA | NA | NA | MN696540 |
| D.sennae | CFCC 51636 | Sennabicapsularis | China | KY203724 | KY228875 | NA | KY228885 | KY228891 |
| D.sennae | CFCC 51637 | Sennabicapsularis | China | KY203725 | KY228876 | NA | KY228886 | KY228892 |
| D.sennicola | CFCC 51634 | Sennabicapsularis | China | KY203722 | KY228873 | KY228879 | KY228883 | KY228889 |
| D.sennicola | CFCC 51635 | Sennabicapsularis | China | KY203723 | KY228874 | KY228880 | KY228884 | KY228890 |
| D.serafiniae | BRIP 55665a | Helianthusannuus | Australia | KJ197274 | NA | NA | KJ197236 | KJ197254 |
| D.shaanxiensis | CFCC 53106 | on branches of liana | China | MK432654 | MK442976 | MK443001 | MK578130 | NA |
| D.shaanxiensis | CFCC 53107 | on branches of liana | China | MK432655 | MK432977 | MK432002 | MK578131 | NA |
| D.siamensis | MFLUCC 10-0573a | Dasymaschalon | Thailand | NA | JQ619897 | NA | JX275393 | JX275429 |
| D.silvicola | CFCC 54191 | Fraxinusexcelsior | Netherlands | MZ727041 | MZ753472 | MZ753481 | MZ816347 | MZ753491 |
| D.silvicola | M79 | Fraxinusexcelsior | Netherlands | MZ727042 | MZ753473 | MZ753482 | MZ816348 | MZ753492 |
| D.sojae | FAU635 | Glycinemax | USA | KJ590719 | KJ612116 | KJ659208 | KJ590762 | KJ610875 |
| D.spartinicola | CPC 24951 | Spartium junceμm | Spain | KR611879 | NA | KR857696 | NA | KR857695 |
| D.spinosa | PSCG 383 | Pyruspyrifolia | China | MK626849 | MK691129 | MK726156 | MK654811 | MK691234 |
| D.sterilis | CBS 136969 | Vacciniumcorymbosum | Italy | KJ160579 | KJ160548 | MF418350 | KJ160611 | KJ160528 |
| D.stictica | CBS 370.54 | Buxussampervirens | Italy | KC343212 | KC343454 | KC343696 | KC343938 | KC344180 |
| D.subclavata | ZJUD95 | Citrusunshiu | China | KJ490630 | NA | KJ490572 | KJ490509 | KJ490451 |
| D.subcylindrospora | KUMCC 17-0151 | Unknown | China | MG746629 | NA | NA | MG746630 | MG746631 |
| D.subellipicola | KUMCC 17-0153 | Unknown | China | MG746632 | NA | NA | MG746633 | MG746634 |
| D.subordinaria | CBS 464.90 | Plantagolanceolata | South Africa | KC343214 | KC343456 | KC343698 | KC343940 | KC344182 |
| D.taoicola | MFLUCC 16-0117 | Prunuspersica | China | KU557567 | NA | NA | KU557636 | KU557591 |
| D.tectonae | MFLUCC 12-0777 | Tectonagrandis | Thailand | KU712430 | KU749345 | NA | KU749359 | KU743977 |
| D.tectonendophytica | MFLUCC 13-0471 | Tectonagrandis | Thailand | KU712439 | KU749354 | NA | KU749367 | KU743986 |
| D.tectonigena | MFLUCC 12-0767 | Camelliasinensis | China | KX986782 | KX999284 | KX999254 | KX999174 | KX999214 |
| D.terebinthifolii | CBS 133180 | Schinusterebinthifolius | Brazil | KC343216 | KC343458 | KC343700 | KC343942 | KC344184 |
| D.ternstroemia | CGMCC 3.15183 | Ternstroemiagymnanthera | China | KC153098 | NA | NA | KC153089 | NA |
| D.thunbergii | MFLUCC 10-0576a | Thunbergialaurifolia | Thailand | JQ619893 | JX197440 | NA | JX275409 | NA |
| D.thunbergiicola | MFLUCC 12-0033 | Thunbergialaurifolia | Thailand | KP715097 | NA | NA | KP715098 | NA |
| D.tibetensis | CFCC 51999 | Juglandisregia | China | MF279843 | MF279888 | MF279828 | MF279858 | MF279873 |
| D.tibetensis | CFCC 52000 | Juglandisregia | China | MF279844 | MF279889 | MF279829 | MF279859 | MF279874 |
| D.torilicola | MFLUCC 17-1051 | Torilisarvensis | Italy | KY964212 | KY964127 | NA | KY964168 | KY964096 |
| D.toxica | CBS 534.93 | Lupinusangustifolius | Australia | KC343220 | KC343462 | KC343704 | KC343946 | KC344188 |
| D.tulliensis | BRIP 62248a | Theobromacacao | Australia | KR936130 | NA | NA | KR936133 | KR936132 |
| D.ueckerae | FAU656T | Cucumismelo | USA | KJ590726 | KJ612122 | KJ659215 | KJ590747 | KJ610881 |
| D.ukurunduensis | CFCC 52592 | Acerukurunduense | China | MH121527 | MH121445 | MH121485 | MH121569 | NA |
| D.ukurunduensis | CFCC 52593 | Acerukurunduense | China | MH121528 | MH121446 | MH121486 | MH121570 | NA |
| D.undulata | LC6624 | Unknown | China | KX986798 | NA | KX999269 | KX999190 | KX999230 |
| D.unshiuensis | ZJUD52 | Citrusunshiu | China | KJ490587 | NA | KJ490529 | KJ490466 | KJ490408 |
| D.unshiuensis | CFCC 52594 | Caryaillinoensis | China | MH121529 | MH121447 | MH121487 | MH121571 | MH121606 |
| D.unshiuensis | CFCC 52595 | Caryaillinoensis | China | MH121530 | MH121448 | MH121488 | MH121572 | MH121607 |
| D.vaccinii | CBS 160.32 | Oxycoccusmacrocarpos | USA | MH121502 | MH121426 | MH121462 | MH121544 | MH121584 |
| D.vangueriae | CBS 137985 | Vangueriainfausta | Zambia | KJ869137 | NA | NA | NA | KJ869247 |
| D.vawdreyi | BRIP 57887a | Psidiumguajava | Australia | KR936126 | NA | NA | KR936129 | KR936128 |
| D.velutina | LC4421 | Neolitsea | China | KX986790 | NA | KX999261 | KX999182 | KX999223 |
| D.verniciicola | CFCC 53109 | Verniciamontana | China | MK573944 | MK574583 | MK574599 | MK574619 | MK574639 |
| D.verniciicola | CFCC 53110 | Verniciamontana | China | MK573945 | MK574584 | MK574600 | MK574620 | MK574640 |
| D.viniferae | JZB320071 | Vitisvinifera | China | MK341551 | MK500119 | NA | MK500107 | MK500112 |
| D.virgiliae | CMW 40748 | Virgiliaoroboides | South Africa | KP247556 | NA | NA | NA | KP247575 |
| D.xishuangbanica | LC6707 | Camelliasinensis | China | KX986783 | NA | KX999255 | KX999175 | KX999216 |
| D.xunwuensis | CFCC 53085 | Unknown | China | MK432663 | MK442983 | MK443008 | MK578137 | MK578063 |
| D.xunwuensis | CFCC 53086 | Unknown | China | MK432664 | MK442984 | MK443009 | MK578138 | MK578064 |
| D.yunnanensis | LC6168 | Unknown | China | KX986796 | KX999290 | KX999267 | KX999188 | KX999228 |
| D.zaobaisu | PSCG 031 | Pyrusbretschneideri | China | MK626922 | NA | MK726207 | MK654855 | MK691245 |
| Diaporthellacorylina | CBS 121124 | Corylus | NA | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
Note: NA, not applicable. Strains in this study are marked in bold.
Results
Phylogenetic analyses
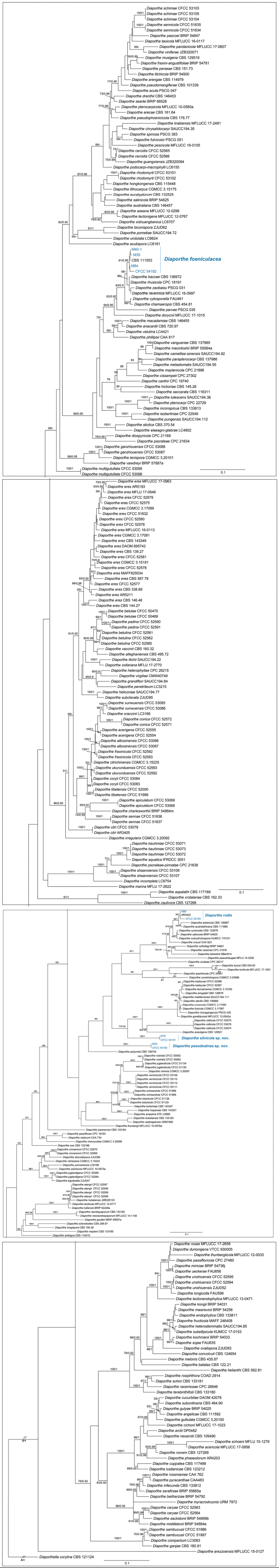
The five-gene sequence dataset (ITS, cal, his3, tef1 and tub2) was analysed to infer the interspecific relationships within Diaporthe. The dataset consisted of 307 sequences including one outgroup taxon, Diaporthellacorylina (CBS 121124). A total of 2649 characters including gaps (516 for ITS, 576 for cal, 526 for his3, 507 for tef1 and 524 for tub2) were included in the phylogenetic analysis. Of these characters, 844 were constant, 318 were variable and parsimony-uninformative, and 1487 were parsimony-informative. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent (Fig. 1). Isolates from the present study formed four individual clades representing four species of Diaporthe, of which isolates CFCC 54192, M35, M40-1 and M84 from Quercusrobur represent D.foeniculacea, while CFCC 54193 and M86 from Q.robur represent D.rudis. CFCC 54191 and M79 from Fraxinusexcelsior and CFCC 54190 and M2A from Alnusglutinosa represent two new species which are here described as D.silvicola and D.pseudoalnea, respectively.
Figure 1.
Phylogram of Diaporthe resulting from a maximum likelihood analysis based on a combined matrix of ITS, cal, his3, tef1 and tub2. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50 %) and Bayesian Posterior Probabilities (right, BPP ≥ 0.75). The tree is rooted with Diaporthellacorylina. Isolates from present study are marked in blue.
Taxonomy
. Diaporthe pseudoalnea
N. Jiang sp. nov.
ABC17B34-3B5B-5E2F-A337-152C8D79C8D9
840714
Figure 2.
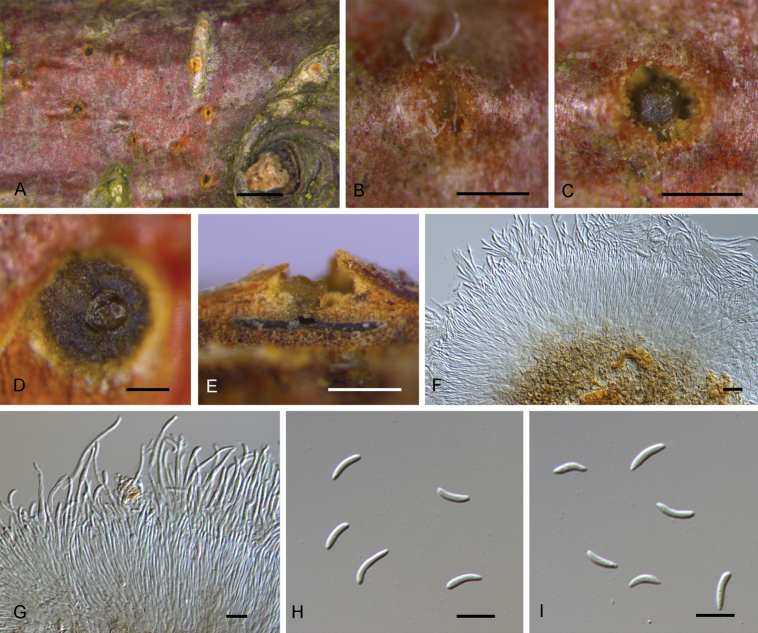
Diaporthepseudoalnea from AlnusglutinosaA–C habit of conidiomata on branches D transverse section of conidiomata E longitudinal section through conidiomata F, G conidiophores and conidia H, I conidia. Scale bars: 2 mm (A), 500 μm (B, C, E), 200 μm (D), 10 μm (F–I).
Etymology.
With reference to D.alnea, which was described from the same host genus, Alnus.
Description.
Conidiomata pycnidial, discoid, immersed in bark, scattered, erumpent through the bark surface, with a solitary locule. Locule 800–1250 μm diam., undivided. Conidiophores 22–68.5 × 1.5–3 μm (av. = 39.8 × 2.2 μm, n = 50), cylindrical, attenuate towards the apex, hyaline, slightly brown at base, phialidic, unbranched, straight or slightly curved. Alpha conidia (5.8–)7.1–8.9(–11.2) × (1.5–)1.8–2.2(–2.7) μm (av. = 7.9 × 2.0 μm, n = 50), L/W = 3.2–4.7 (av. = 3.8, n = 50), hyaline, aseptate, subcylindrical with a nearly rounded apex, multi-guttulate, sometimes acute at both ends. Beta conidia not observed.
Culture characters.
Colonies are initially white with fluffy aerial mycelium, becoming dirty white after 2 weeks, and conidiomata are randomly distributed with orange conidial drops oozing out of the ostioles.
Specimens examined.
NETHERLANDS. Utrecht City, on branches of Alnusglutinosa, 5°11’32” E, 52°05’22” N, 8 Apr. 2019, N. Jiang (holotype CAF800005 = JNH0001; ex-type living culture: CFCC 54190; other living culture: M2A).
Notes.
Diaporthenivosa and D.alnea were recorded from the host genus Alnus. Udayanga et al. (2014) investigated the lectotype of Diaporthenivosa and revealed it as a Melanconis species based on a well-developed ectostromata and the ascospores characteristics, and Jaklitsch and Voglmayr (2020) treated it as a synonym of Melanconismarginalisssp.marginalis. D.alnea has been reported from the Czech Republic, Germany, the Netherlands and the USA, and both sexual and asexual morphs have been described (Udayanga et al. 2014). However, applying the GCPSR principle, D.alnea has recently been considered to be a synonym of Diaportheeres (Hilário et al. 2021), which has also been confirmed in our analyses where the ex-epitype isolate CBS 146.46 of D.alnea is placed within the D.eres clade (Fig. 1). Diaporthepseudoalnea morphologically differs from D.alnea (now D.eres) by its longer conidiophores (22–68.5 × 1.5–3 μm in D.pseudoalnea vs. 9–16 × 1–2 μm in D.alnea; Udayanga et al. 2014). In our multi-gene analyses, D.pseudoalnea forms a distinct phylogenetic lineage which is placed remotely from the isolate CBS 146.46 of D.alnea (Fig. 1).
Diaporthe silvicola
N. Jiang sp. nov.
A563ADB6-35CD-5A06-9D98-8A27FCC9D26A
840715
Figure 3.
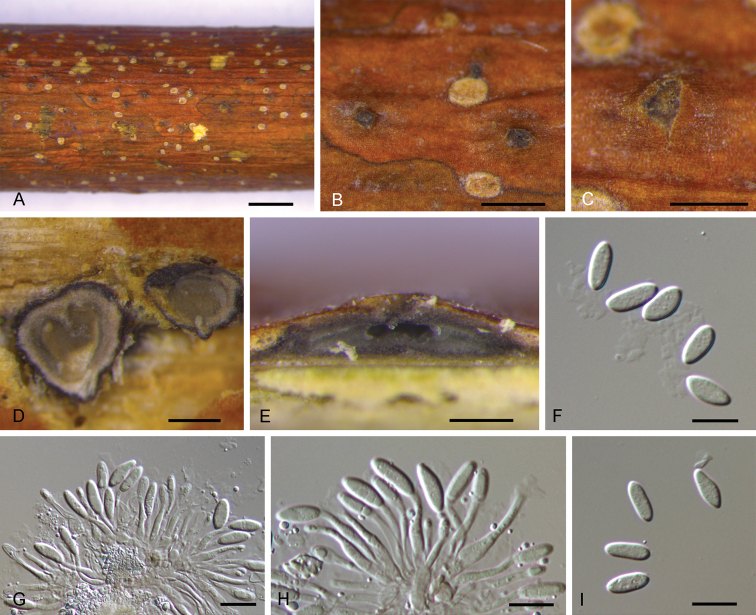
Diaporthesilvicola from FraxinusexcelsiorA–C habit of conidiomata on branches D transverse section of conidiomata E longitudinal section through conidiomata F, I conidia G, H conidiophores and conidia. Scale bars: 2 mm (A), 1 mm (B), 500 μm (C), 200 μm (D, E), 10 μm (F–I).
Etymology.
Name from “silva” = forest and “-cola” = inhabiting; with reference to its woody host.
Description.
Conidiomata pycnidial, conical, immersed in bark, scattered, erumpent through the bark surface, with a solitary locule. Locule 450–700 μm diam., undivided. Conidiophores 6.5–25 × 1.5–4 μm (av. = 15.4 × 2.4 μm, n = 50), cylindrical, attenuate towards the apex, hyaline, slightly brown, phialidic, unbranched, slightly curved. Alpha conidia (9.2–)10.1–12.3(–13.5) × (3.8–)4.2–4.9(–5.2) μm (av. = 11.5 × 4.5 μm, n = 50), L/W = 2.0–3.2 (av. = 2.5, n = 50), hyaline, aseptate, fusiform to oval, multi-guttulate, acute at both ends. Beta conidia not observed.
Culture characters.
Colonies are initially white, aerial mycelium turning grey at edges of plate, yellowish pigmentation developing in centre, conidiomata not produced until 2 weeks.
Specimens examined.
NETHERLANDS. Utrecht City, on branches of Fraxinusexcelsior in the forest ecosystem, 5°10’36” E, 52°05’32” N, 6 Jun. 2019, N. Jiang (holotype CAF800006 = JNH0002; ex-type living culture: CFCC 54191; other living culture: M79).
Notes.
Diaporthefraxini-angustifoliae was reported from Fraxinusangustifoliasubsp.oxycarpa cv. Claret Ash in Australia (Tan et al. 2013). D.fraxinicola was described from Fraxinuschinensis in China (Yang et al. 2018). However, D.silvicola from Fraxinusexcelsior in Netherlands differs from D.fraxini-angustifoliae and D.fraxinicola by obviously larger alpha conidia (9.2–13.5 × 3.8–5.2 μm in D.silvicola vs. 4–10 × 2–3 μm in D.fraxini-angustifoliae vs. 7–10 × 2.9–3.2 μm in D.fraxinicola; Tan et al. 2013; Yang et al. 2018).
Discussion
In this study, branch-inhabiting Diaporthe species were sampled from Alnusglutinosa, Fraxinusexcelsior and Quercusrobur in Utrecht, the Netherlands. Ten Diaporthe isolates were obtained and identified based on five combined loci (ITS, cal, his3, tef1 and tub2), as well as morphological characters from the natural substrates. The phylogenetic and morphological analyses revealed Diaporthepseudoalnea sp. nov. from Alnusglutinosa, Diaporthesilvicola sp. nov. from Fraxinusexcelsior, and D.foeniculacea and D.rudis from Quercusrobur.
Phylogenetic analyses were conducted based on a combined DNA sequence matrix of five loci (ITS, cal, his3, tef1 and tub2) reported as useful markers to distinguish species of Diaporthe (Udayanga et al. 2014, 2015; Guarnaccia et al. 2017, 2018a, 2018b; Tibpromma et al. 2018; Yang et al. 2020; Dissanayake et al. 2020; Huang et al. 2021; Sun et al. 2021, Wang et al. 2021). The two novel species in this study can be distinguished from the other known species by all genes studied, but most effectively by cal, his3, tef1 and tub2. The multi-locus phylogenetic analysis grouped the isolates in two new clades, which support the introduction of the new species.
The utility of host association for Diaporthe species identification is limited because several species have wide host ranges (e.g., D.ere inhabits 282 different hosts; D.rudis inhabits 44 different hosts), and multiple Diaporthe species can infect a single host (e.g., nineteen Diaporthe species are associated with pear cankers in China) (Guo et al. 2020; Farr and Rossman 2021). Thus, a polyphasic approach of morphological, cultural, ecological and molecular data to identify Diaporthe samples or to introduce new species is essential.
Supplementary Material
Acknowledgements
This research was funded by the National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2021-7).
Citation
Jiang N, Voglmayr H, Piao C-G, Li Y (2021) Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31–56. https://doi.org/10.3897/mycokeys.85.73107
Funding Statement
National microbial resource center from Ministry of Science and Technology of the People's Republic of China
References
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 3: 553–556. 10.2307/3761358 [DOI] [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004) Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- Diogo E, Santos JM, Phillips AJ. (2010) Phylogeny, morphology and pathogenicity of Diaporthe and Phomopsis species on almond in Portugal. Fungal Diversity 44: 107–115. 10.1007/s13225-010-0057-x [DOI] [Google Scholar]
- Dissanayake AJ, Chen YY, Liu JK. (2020) Unravelling Diaporthe species associated with woody hosts from karst formations (Guizhou) in China. Journal of Fungi 6: 251. 10.3390/jof6040251 [DOI] [PMC free article] [PubMed]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Fan XL, Bezerra JDP, Tian CM, Crous PW. (2018) Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40: 119–134. 10.3767/persoonia.2018.40.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr DF, Rossman AY. (2021) Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Retrieved August 3, 2021, from https://nt.ars-grin.gov/fungaldatabases/
- Gao H, Pan M, Tian CM, Fan XL. (2021) Cytospora and Diaporthe species associated with hazelnut canker and dieback in Beijing, China. Frontiers in Cellular and Infection Microbiology 11: 664366. 10.3389/fcimb.2021.664366 [DOI] [PMC free article] [PubMed]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14: 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA Fungus 8: 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/AEM.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8: 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Crous PW. (2018a) Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathologia Mediterranea 57: 307–319. 10.14601/Phytopathol_Mediterr-23254 [DOI] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A, Kaliterna J, Kiss L, Larignon P, Luque J, Mugnai L, Naor V, Raposo R, Sándor E, Váczy KZ, Crous PW. (2018b) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40: 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YS, Crous PW, Bai Q, Fu M, Yang MM, Wang XH, Du YM, Hong N, Xu WX, Wang GP. (2020) High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45: 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilário S, Amaral IA, Gonçalves MF, Lopes A, Santos L, Alves A. (2020) Diaporthe species associated with twig blight and dieback of Vacciniumcorymbosum in Portugal, with description of four new species. Mycologia 112: 293–308. 10.1080/00275514.2019.1698926 [DOI] [PubMed] [Google Scholar]
- Hilário S, Gonçalves MF, Alves A. (2021) Using genealogical concordance and coalescent-based species delimitation to asess species boundaries in the Diaportheeres complex. Journal of Fungi 7: 507. 10.3390/jof7070507 [DOI] [PMC free article] [PubMed]
- Huang F, Udayanga D, Wang X, Hou X, Mei X, Fu Y, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119: 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Huang ST, Xia JW, Zhang XG, Sun WX. (2021) Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. 10.3897/mycokeys.78.60878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2019) European species of Dendrostoma (Diaporthales). MycoKeys 59: 1–26. 10.3897/mycokeys.59.37966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. (2020) The genus Melanconis (Diaporthales). MycoKeys 63: 69–117. 10.3897/mycokeys.63.49054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Fan XL, Crous PW, Tian CM. (2019) Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. MycoKeys 48: 67–96. 10.3897/mycokeys.48.31715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Fan XL, Tian CM, Crous PW. (2020) Reevaluating Cryphonectriaceae and allied families in Diaporthales. Mycologia 112: 267–292. 10.1080/00275514.2019.1698925 [DOI] [PubMed] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi 7: 64. 10.3390/jof7010064 [DOI] [PMC free article] [PubMed]
- Katoh K, Toh H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed]
- Lombard L, Van Leeuwen GCM, Guarnaccia V, Polizzi G, Van Rijswick PC, Karin Rosendahl KC, Gabler J, Crous PW. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53: 287–299. 10.14601/Phytopathol_Mediterr-14034 [DOI] [Google Scholar]
- Long H, Zhang Q, Hao YY, Shao XQ, Wei XX, Hyde KD, Wang Y, Zhao DG. (2019) Diaporthe species in south-western China. MycoKeys 57: 113–127. 10.3897/mycokeys.57.35448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manawasinghe IS, Dissanayake AJ, Li X, Liu M, Wanasinghe DN, Xu J, Zhao W, Zhang W, Zhou Y, Hyde KD, Brooks S, Yan J. (2019) High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Frontiers in Microbiology. 10: 1936. 10.3389/fmicb.2019.01936 [DOI] [PMC free article] [PubMed]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Canadian Journal of Botany 72: 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6(1): 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculumvulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat DJ, Perera RH, Li QR, Li WJ. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Huang S, Xia J, Zhang X, Li Z. (2021) Morphological and molecular identification of Diaporthe species in south-western China, with description of eight new species. MycoKeys 77: 65–95. 10.3897/mycokeys.77.59852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Edwards J, Grice KRE, Shivas RG. (2013) Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Diversity 61: 251–260. 10.1007/s13225-013-0242-9 [DOI] [Google Scholar]
- Tibpromma S, Hyde KD, Bhat JD, Mortimer PE, Xu J, Promputtha I, Doilom M, Yang JB, Tang AMC, Karunarathna SC. (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33: 25–67. 10.3897/mycokeys.33.23670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthesojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119: 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Hyde KD. (2014) Species limits in Diaporthe: molecular re-assessment of D.citri, D.cytosporella, D.foeniculina and D.rudis. Persoonia 32: 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50: 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Voglmayr H, Castlebury LA, Jaklitsch WM. (2017) Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales). Persoonia 38: 136–155. 10.3767/003158517X694768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Rossman AY, Castlebury LA, Jaklitsch WM. (2012) Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales). Fungal Diversity 57: 1–44. 10.1007/s13225-012-0175-8 [DOI] [Google Scholar]
- Wang X, Guo Y, Du Y, Yang Z, Huang X, Hong N, Wang G. (2021) Characterization of Diaporthe species associated with peach constriction canker, with two novel species from China. MycoKeys 80: 77–90. 10.3897/mycokeys.80.63816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39: 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2021) New species and records of Diaporthe from Jiangxi Province, China. MycoKeys 77: 41–64. 10.3897/mycokeys.77.59999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Palma MA, Aninat MJ, Piontelli E. (2020) Polyphasic studies of new species of Diaporthe from native forest in Chile, with descriptions of Diaporthearaucanorum sp. nov., Diaporthefoikelawen sp. nov. and Diaporthepatagonica sp. nov. International Journal of Systematic and Evolutionary Microbiology 70(5): 3379–3390. 10.1099/ijsem.0.004183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.