Abstract
The importance of pre-existing immune responses to seasonal endemic coronaviruses (HCoVs) for the susceptibility to SARS-CoV-2 infection and the course of COVID-19 is the subject of an ongoing scientific debate. Recent studies postulate that immune responses to previous HCoV infections can either have a slightly protective or no effect on SARS-CoV-2 pathogenesis and, consequently, be neglected for COVID-19 risk stratification. Challenging this notion, we provide evidence that pre-existing, anti-nucleocapsid antibodies against endemic α-coronaviruses and S2 domain-specific anti-spike antibodies against β-coronavirus HCoV-OC43 are elevated in patients with COVID-19 compared to pre-pandemic donors. This finding is particularly pronounced in males and in critically ill patients. Longitudinal evaluation reveals that antibody cross-reactivity or polyclonal stimulation by SARS-CoV-2 infection are unlikely to be confounders. Thus, specific pre-existing immunity to seasonal coronaviruses may increase susceptibility to SARS-CoV-2 and predispose individuals to an adverse COVID-19 outcome, guiding risk management and supporting the development of universal coronavirus vaccines.
Keywords: SARS-CoV-2, COVID-19, seasonal coronaviruses, HCoV, antibodies, humoral immunity, disease severity, susceptibility, common cold, pandemic
Graphical abstract
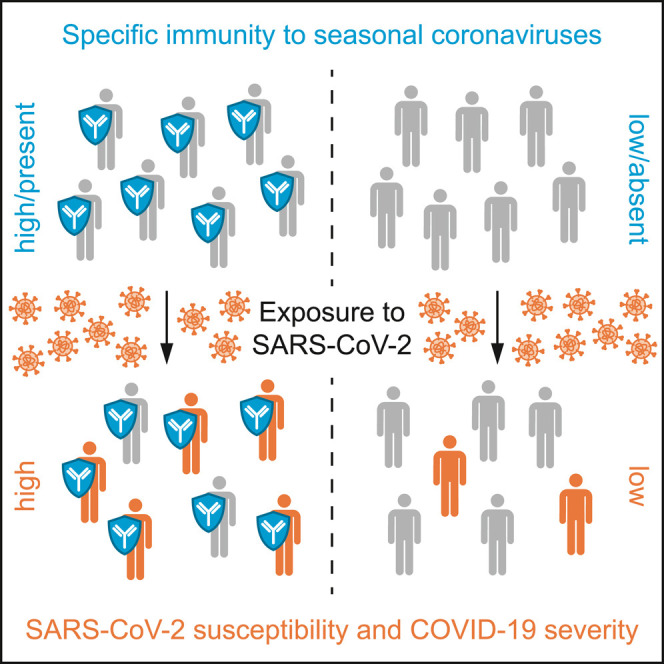
Wratil et al. find specific antibody responses against seasonal human coronaviruses, which cause the common cold, to be elevated in patients with COVID-19 compared to pre-pandemic blood donors. This specific immunity is likely pre-existing in patients and increases their susceptibility to SARS-CoV-2 and severity of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19) caused by the novel human viral pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) developed into a pandemic with more than 242.4 million confirmed cases and 4.93 million deaths thus far (Center for Systems and Science Engineering at John Hopkins University, 2021). Defining parameters that can influence susceptibility to SARS-CoV-2 or that contribute to the high clinical variability of COVID-19 are critical to aid risk stratification, guided application of preventive measures, and COVID-19 management.
There are four species of endemic, seasonal coronaviruses (HCoVs) that typically cause mildly symptomatic respiratory tract infections in humans but are genetically dissimilar and display varying host cell tropism (Pyrc et al., 2006). Two of them, HCoV-229E and HCoV-NL63, belong to the taxonomic genus of α-coronaviruses, while the other two, HCoV-HKU1 and HCoV-OC43, belong to the genus of β-coronaviruses that includes SARS-CoV-2. HCoV infections are frequent (Killerby et al., 2018; Masse et al., 2020; Severance et al., 2008), and a longitudinal survey indicated that protective HCoV immunity may be short-lived (Edridge et al., 2020).
It has been hypothesized that previous encounters with HCoVs provide cross-protective immunity to SARS-CoV-2 (Braun et al., 2020). Corroborating this hypothesis, Sagar et al. (2021) suggested that recent HCoV infections can be associated with reduced COVID-19 severity. Moreover, a protective role of pre-existing T cells reactive to HCoVs in SARS-CoV-2 infection was suggested (Bacher et al., 2020; Loyal et al., 2021).
Anderson et al. (2021) recently reported on the potential influence of humoral HCoV immunity on the susceptibility to SARS-CoV-2 and the course of COVID-19: in pre-pandemic sera collected from individuals who became subsequently infected by SARS-CoV-2, no differences in IgG-type antibody responses to the spike protein of β-coronavirus HCoV-OC43 were observed compared to sera from individuals not infected by SARS-CoV-2. Furthermore, there was no relationship between pre-pandemic anti-HCoV-OC43 spike antibody levels and COVID-19 severity. In patients with COVID-19, IgG antibodies reactive to the spike protein of HCoV-OC43, primarily targeting the S2 domain, were boosted in the first 7 days of hospitalization, but the magnitude of this increase was not correlated to disease severity. The authors concluded that humoral immune responses to HCoVs are not associated with protection against SARS-CoV-2 infection and do not impact the severity of COVID-19. Contradicting this notion, our findings indicate that a genus- and antigen-specific, pre-existing immunity to HCoVs can, in fact, increase SARS-CoV-2 susceptibility and COVID-19 severity.
Results
Levels of specific antibodies reactive to the nucleocapsid or spike antigens of seasonal coronaviruses are elevated in patients with COVID-19 compared to pre-pandemic donors
In a broader methodological approach, we monitored IgG-type antibody levels against the nucleocapsid and the spike S1 domain proteins of SARS-CoV-2 and all four seasonal coronaviruses as well as against full-length spike protein of SARS-CoV-2, HCoV-NL63, and HCoV-OC43 in pre-pandemic sera from 888 healthy adults as well as in 314 sera longitudinally collected from 96 patients with COVID-19 (see STAR Methods and Figure S1). We utilized a newly launched commercial line immunoassay (recomLine) and a recently developed bead-based multiplex immunoassay (MultiCoV-Ab) (STAR Methods and Becker et al., 2021). Specificities and sensitivities of these assays for anti-SARS-CoV-2 antibodies and correlative analyses for anti-HCoV antibodies in pre-pandemic and sera of patients with COVID-19 are provided in STAR Methods, Table S1, and Figure S2.
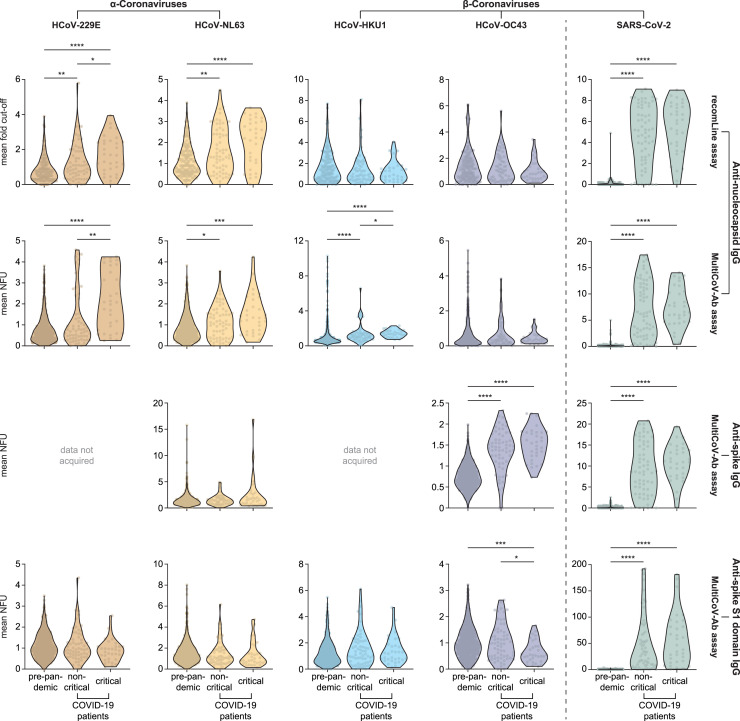
Analyzing the mean of all sampling time points for each donor, we observed drastically increased levels of disease-specific antibodies against the nucleocapsid, full-length spike protein, and spike S1 domain antigen of SARS-CoV-2 in patients with COVID-19 compared to pre-pandemic donors, as expected (Figure 1 , green; Figure S3, green). Surprisingly, in both assays, mean antibody levels against the nucleocapsid of α-coronaviruses, HCoV-229E and HCoV-NL63, were significantly elevated in the COVID-19 cohort compared to the group of pre-pandemic donors (Figure 1, brown and yellow). Anti-nucleocapsid responses to β-coronavirus HCoV-HKU1 were also elevated in patients with COVID-19 compared to pre-pandemic donors, albeit less pronounced and only in the MultiCoV-Ab assay (Figure 1, blue). Anti-nucleocapsid responses to the β-coronavirus HCoV-OC43 were similar between the study groups (Figure 1, purple). Furthermore, critically ill patients compared to less severely affected cases i.e., “non-critical” (defined according to WHO guidelines 2020), had increased antibody titers against the nucleocapsid of the two α-coronaviruses and HCoV-HKU1 (Figure 1, brown, yellow, blue), but not of SARS-CoV-2 or HCoV-OC43 (Figure 1, green and purple). In contrast, full-length spike-specific antibodies targeting HCoV-OC43, but not those targeting HCoV-NL63, were significantly increased in patients with COVID-19 compared to pre-pandemic donors (Figure 1, third row, yellow and purple). Of note, antibody responses to the spike S1 domain of β-coronavirus HCoV-OC43 were, in turn, reduced in critically ill patients compared to pre-pandemic donors (Figure 1, lower row, purple) and indifferent between the study groups for the other HCoVs tested (Figure 1, lower row, brown, orange, and blue). Qualitative evaluation of the recomLine assay showed both significantly increased numbers of patients with COVID-19 positive for anti-nucleocapsid antibodies recognizing α-coronaviruses compared to pre-pandemic donors, and more critically ill patients being positive for antibodies targeting HCoV-229E than non-critically ill (Table S2).
Figure 1.
Anti-nucleocapsid and anti-spike S1 domain antibody levels in sera from pre-pandemic donors and patients with COVID-19
Eight hundred and eighty-eight pre-pandemic sera from healthy adult blood donors (184 in case of the line immunoassay), 153 samples from 32 critically ill patients with COVID-19 (161 in case of the recomLine assay, critical), and 142 samples from 64 less severely affected patients with COVID-19 (143 in case of the recomLine assay, non-critical) were analyzed for their antibody levels against HCoV-229E, -NL63, -HKU1, and -OC43, as well as SARS-CoV-2. Mean antibody levels per donor/patient (dots) are depicted as violin plots for every group (pre-pandemic, as well as critical and non-critical COVID-19). Differences in the assays' antibody responses comparing the groups were tested for their statistical significance via Kruskal-Wallis test and pairwise comparisons using Wilcoxon rank-sum test with continuity correction. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. NFU, normalized fluorescence units.
Anti-HCoV antibody concentrations remain largely unaltered in patients with COVID-19 during the disease course, indicating that high antibody responses against seasonal coronaviruses were pre-existing in these patients
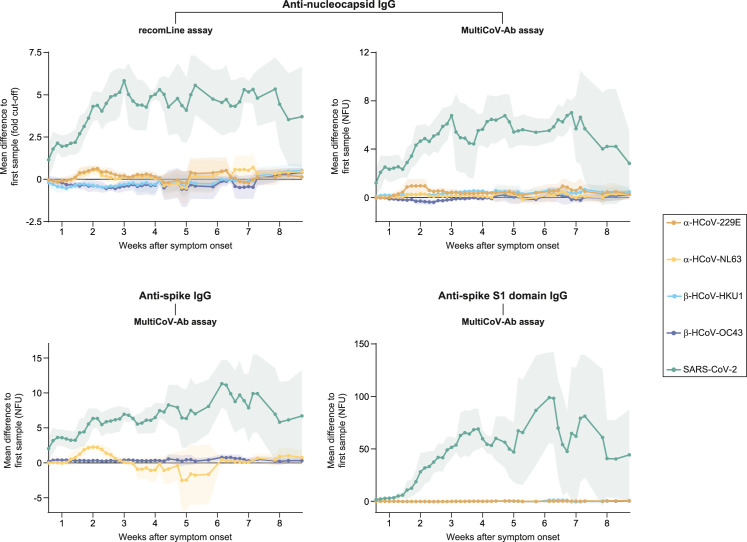
Two confounders could potentially contribute to the increased concentrations of antibodies against seasonal coronaviruses observed in patients with COVID-19: (1) cross-reactivity of anti-SARS-CoV-2 antibodies with the assays’ HCoV antigen components and (2) polyclonal stimulation of pre-existing, HCoV-specific plasma cells by SARS-CoV-2 infection. To address both scenarios, we first explored longitudinal changes in antibody levels of those 28 patients with COVID-19 in our cohort who had donated sera both in the first 2 weeks after symptom onset and at later time points in the disease course. Expectedly, specific antibody responses to the nucleocapsid, full-length spike protein, and spike S1 domain of SARS-CoV-2 drastically increased during the disease course in this cohort (Figure 2 , green). In contrast, titers of antibodies against all four HCoVs remained largely unaltered (Figure 2, brown, yellow, blue, and purple). Only, anti-full-length spike antibody levels against HCoV-NL63 increased in the first 2 weeks after the onset of symptoms and decreased thereafter (Figure 2, lower left, yellow).
Figure 2.
Longitudinal antibody level changes in 28 patients with COVID-19
One-hundred and seventy-four sera from 28 patients with COVID-19 who donated specimens both in the first 2 weeks after symptom onset and at later time points were analyzed. The five time point rolling averages for differences in antibody levels compared to the first sample donated by each individual patient are shown. Shaded areas depict standard deviations. NFU, normalized fluorescence units.
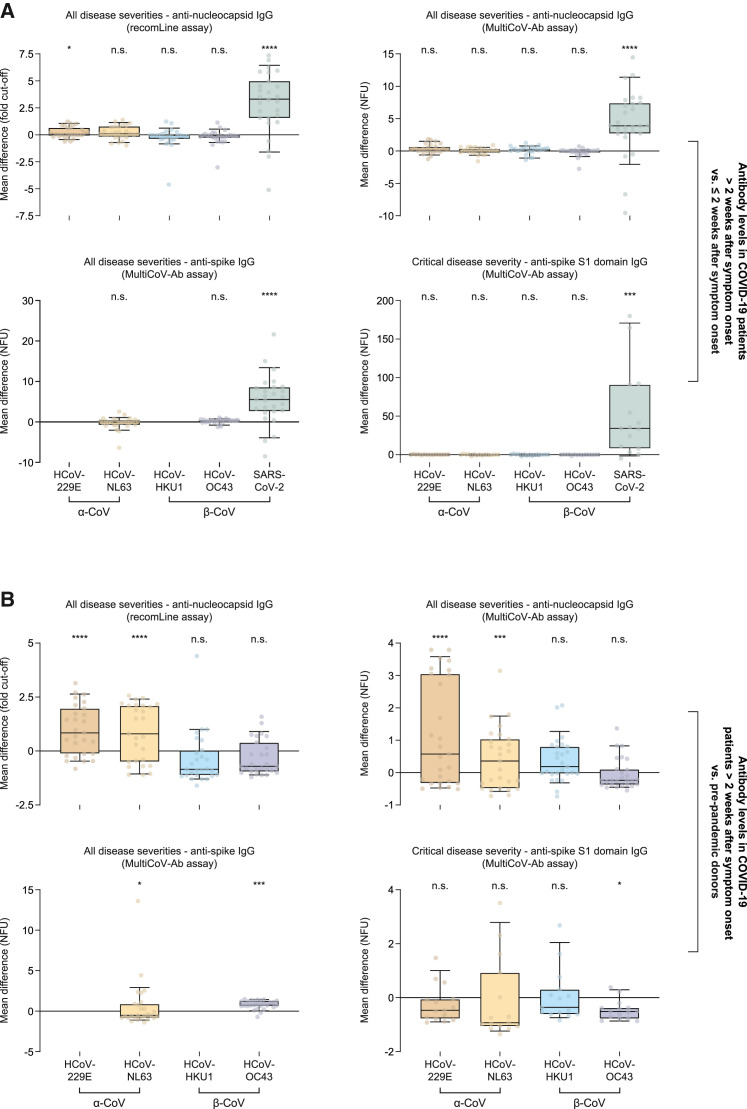
Second, we compared mean antibody levels in these 28 patients with COVID-19 in sera collected from the third week after the onset of symptoms onward relative to those obtained in the first 2 weeks. Inherent to the newly mounting immune response, we observed markedly increased antibody titers against the pandemic SARS-CoV-2 in this longitudinal comparison (Figure 3A, green). In the same comparison of specimens, however, most anti-HCoV antibody responses showed only insignificant changes (Figure 3A, brown, yellow, blue, and purple, respectively). Anti-nucleocapsid antibody levels against HCoV-229E were slightly, but significantly elevated in the recomLine assay, possibly hinting at weak cross-reactivity (Figure 3A, top left, brown). Conversely, anti-nucleocapsid antibody responses to HCoV-229E and HCoV-NL63 (Figure 3B, upper row, brown and yellow) as well as anti-full-length spike antibody levels against HCoV-OC43 (Figure 3B, lower left, purple) were markedly elevated in these 28 individuals with COVID-19 compared to pre-pandemic donors. Anti-spike S1 domain antibody levels against HCoV-OC43 in critically ill patients, on the other hand, were significantly decreased in this comparison (Figure 3B, lower right, purple). Collectively, these results largely exclude a relevant cross-reactivity of anti-SARS-CoV-2 antibodies in HCoV serology or a polyclonal stimulation of HCoV-specific plasma cells after SARS-CoV-2 infection. We conclude that high antibody titers to the nucleocapsid of HCoV-229E and HCoV-NL63, as well as full-length spike antigen of HCoV-OC43, were most likely pre-existing in these patients with COVID-19.
Figure 3.
Comparison of antibody levels in 28 patients with COVID-19 at later time points with earlier time points and pre-pandemic specimens
Sixty-nine sera collected in the first 2 weeks after symptom onset from 28 patients with COVID-19, 105 sera collected after the first 2 weeks after symptom onset from the same patients, and 888 pre-pandemic sera from healthy adult blood donors (184 in case of the recomLine assay) were analyzed. Differences in mean antibody levels for each patient comparing samples obtained more than 2 weeks after symptom onset with those from the first 2 weeks after symptom onset and mean antibody levels from pre-pandemic adults (dots) are depicted as boxplots with whiskers between the 10th and 90th percentiles for the following groups: patients with COVID-19 more than 2 weeks after symptom onset versus less than 2 weeks after symptom onset (A), patients with COVID-19 more than 2 weeks after symptom onset versus pre-pandemic donors (B). Differences in antibody levels in each group were analyzed for their statistical significance using two-tailed, paired t tests in (A) and two-tailed, unpaired t tests in (B). ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; n.s., not significant; NFU, normalized fluorescence units; α-CoV, α-coronaviruses; β-CoV, β-coronaviruses.
In patients with COVID-19, antibody responses to SARS-CoV-2 and seasonal coronaviruses often show sex-specific differences, and interleukin-6 levels at admission correlate significantly with disease severity in multivariate analyses
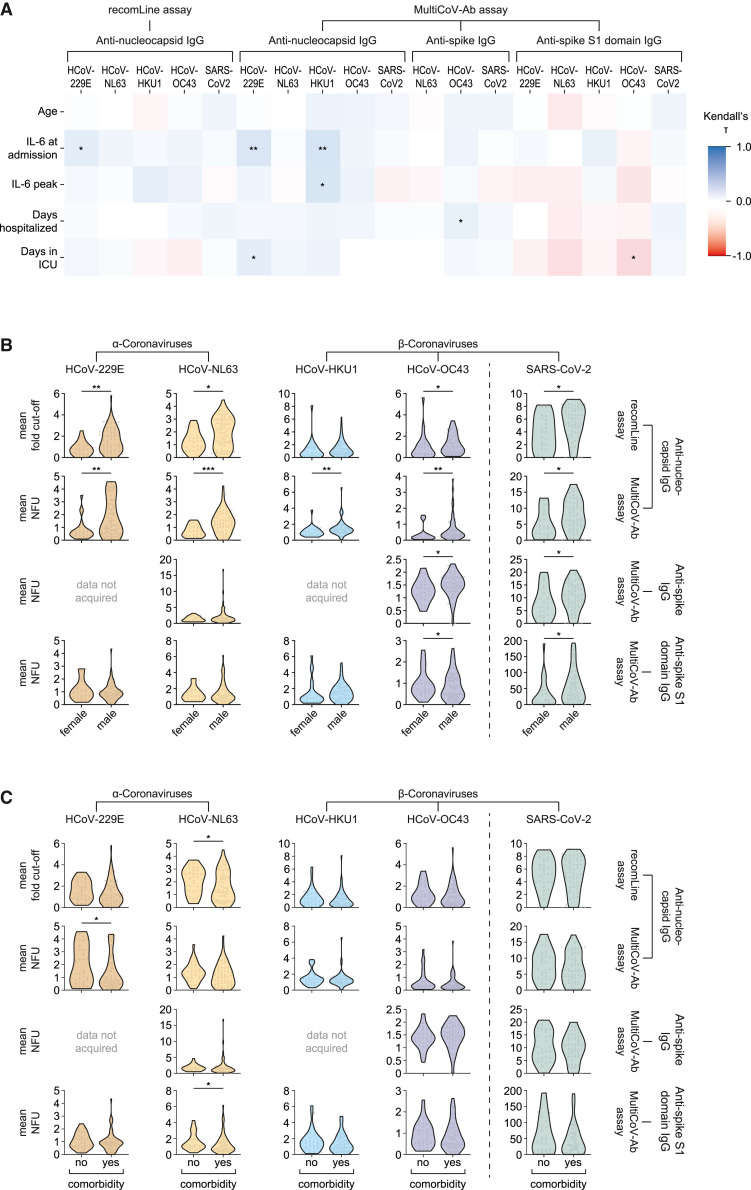
We compared the mean antibody levels for each patient with COVID-19 enrolled in this study to additional health record data. Our analysis revealed that patients’ age, their interleukin-6 (IL-6) levels both upon admission and at their peak, and the duration of their hospitalization or the time they spent on intensive care units (ICUs) showed only weak correlations with their anti-SARS-CoV-2 and anti-HCoV antibody responses, respectively (Figure 4A).
Figure 4.
Comparison of mean antibody levels in patients with COVID-19 with additional health record data
Mean antibody levels in 96 patients with COVID-19 were compared to additional health record data. Kendall’s correlation coefficients (τ) between quantitative assay results and age, interleukin-6 levels at admission as well as at their individual peak, days patients spend hospitalized or admitted to intensive care units (ICUs) are depicted in (A). (B and C) Mean antibody levels in the same patients (dots) compared to sex (B) and presence of comorbidities (C) as violin plots. In (B) and (C), differences between the groups were analyzed for their statistical significance using the Wilcoxon rank-sum test with continuity correction. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. NFU, normalized fluorescence units.
Male patients compared to females had significantly higher antibody levels against SARS-CoV-2 (Figure 4B, green) and against the nucleocapsid of both α-coronaviruses as well as HCoV-OC43, in both assays utilized (Figure 4B, brown, yellow, purple). Anti-nucleocapsid responses to HCoV-HKU1 were significantly elevated in males in the MultiCoV-Ab assay Figure 4B, blue). Anti-OC43 antibodies against full-length spike protein were increased in males compared to females (Figure 4B, third row, purple). Conversely, anti-spike S1 domain antibodies were significantly decreased in the same comparison (Figure 4B, bottom row, purple). Patients’ comorbidities had, in most cases, no effect on anti-SARS-CoV-2 anti-HCoV antibody titers (Figure 4C).
In multivariate analyses, we investigated whether the disease severity of individuals with COVID-19 correlated with antibody levels against SARS-CoV-2 and seasonal coronaviruses while also considering the aforementioned data from patients’ health records. Data on hospitalization and admission to ICU were excluded from this multivariate analysis because they were, among others, used to define the patients' disease severity (WHO guidelines, 2020) and were thus considered dependent variables. In all assays, COVID-19 severity correlated significantly with patients' IL-6 levels at admission (p ≤ 0.0140; supplemental information). Peak IL-6 responses or patients' mean antibody levels correlated with disease severity only in specific assays (supplemental information). However, patients' age, sex, or presence of comorbidities did not correlate with disease severity in this multivariate analysis (supplemental information).
Discussion
In summary, our study provides evidence that specific pre-existing adaptive immunity to seasonal coronaviruses is associated with increased susceptibility to SARS-CoV-2 infection and adverse disease outcome. The mode of action underlying these findings is unclear. We hypothesize a direct or indirect enhancement of early stages of SARS-CoV-2 infection on the nasal or oral mucosa or in the respiratory tract, or an antibody-dependent cellular cytotoxicity influencing immunopathology in lung tissue mediated by specific pre-existing antibodies against seasonal coronaviruses. Regarding anti-nucleocapsid antibody responses, a recent study suggested that lectin pathway recognition molecules of the complement system, including the effector enzyme MASP-2, can directly bind to SARS-CoV-2 nucleocapsid protein, with subsequent activation of lectin pathway-mediated C3b and C4b deposition (Ali et al., 2021). Conceivably, pre-existing anti-nucleocapsid antibodies against seasonal coronaviruses may cross-react with SARS-CoV-2 nucleocapsid released from infected, dying cells in the respiratory tract negatively modulating the development of thromboembolism and aggravating disease outcome.
During the validation of the MultiCoV-Ab assay (Becker et al., 2021), similar, albeit less pronounced, trends for elevated anti-nucleocapsid IgG titers against HCoV-229E and HCoV-NL63 were observed in relation to individuals’ SARS-CoV-2 serostatus. Another study conducted in healthcare workers found decreased levels of nucleocapsid-specific antibodies against seasonal coronaviruses in symptomatic individuals with COVID-19 compared to those with asymptomatic disease (Ortega et al., 2021). Of particular note, the COVID-19 cohorts in the former studies consisted mainly of non-hospitalized patients with asymptomatic or mild disease severity (79.1%; Becker et al., 2021) (99.2%; Ortega et al., 2021), whereas our current study had a substantially lower proportion of mildly affected patients with COVID-19 (26.0%, STAR Methods).
Our data support the notion of a SARS-CoV-2 susceptibility- and COVID-19 severity-enhancing effect related to high abundance of nucleocapsid-specific antibodies against α-coronaviruses and possibly β-coronavirus HCoV-HKU1. Two other studies monitored anti-nucleocapsid responses to seasonal coronaviruses in COVID-19 cases via the recomLine assay and observed decreased anti-HCoV-OC43 antibody titers in critically ill patients compared to less severely affected (Dugas et al., 2020, 2021). Utilizing the same assay, we observed a similar albeit statistically insignificant trend toward low anti-HCoV-OC43 nucleocapsid antibody levels in critically ill patients. This result, however, could not be confirmed in the MultiCoV-Ab assay.
Furthermore, our findings indicate that SARS-CoV-2 susceptibility is enhanced by pre-existing antibodies targeting the spike antigen of HCoV-OC43. Regarding humoral responses to seasonal coronavirus spike protein, several studies observed elevated antibody levels against HCoV-OC43 in patients with COVID-19 (Prévost et al., 2020; Anderson et al., 2021) and vaccinees (Tauzin et al., 2021) compared to uninfected, non-vaccinated individuals, corroborating our results. However, longitudinal and cross-sectional analyses suggested that these increased anti-HCoV-OC43 spike antibody titers were likely not pre-existing, but dependent on either the COVID-19 disease course (Prévost et al., 2020; Anderson et al., 2021) or vaccination (Tauzin et al., 2021), and mainly mediated by antibodies targeting the S2 domain of the viral spike (Anderson et al., 2021). In line with these findings, our data suggest that high anti-HCoV-OC43 spike antibody levels in COVID-19 are likely due to increased concentrations of antibodies targeting the S2 domain. Furthermore, decreased anti-spike S1 domain responses were observed in critically ill patients compared to pre-pandemic donors. Our longitudinal assessment, on the other hand, revealed high, yet stable and COVID-19 disease course-independent antibody levels against the full-length spike antigen of HCoV-OC43 and against the nucleocapsid of seasonal α-coronaviruses, indicating that these elevated antibody concentrations were, indeed, pre-existing. These discrepant results could be due to differences in the COVID-19 patient cohorts: the former studies included lower rates of severely and critically ill patients with COVID-19 (8.9%; Prévost et al., 2020) (14.0%; Anderson et al., 2021), whereas the percentage of such cases was more than 3-fold higher in our patient cohort (44.8%). Thus, our findings could potentially be more applicable to severe COVID-19. Of note, Prévost et al. (2020) did not perform longitudinal antibody analyses in the same patients, but cross-sectional analyses in dissimilar patient groups. The alterations in anti-spike antibodies against HCoV-OC43 in individuals with COVID-19 observed by Prévost et al. (2020), therefore, could underlie inter-individual rather than longitudinal changes in serological responses. Moreover, differences in the type and specificities of the assays utilized to detect anti-full-length spike antibodies against HCoV-OC43 and their cross-reactivity to anti-SARS-CoV-2 antibodies could contribute to the different results obtained by Prévost et al. (2020) and Anderson et al. (2021) compared to ours. Unfortunately, a well-validated, broadly available anti-HCoV spike antibody assay is lacking. All studies on serological responses against these antigens currently relied on self-developed methods and the analytical performances of these are difficult to compare.
Adding to the discussion, Sokal et al. (2021) found fractions of SARS-CoV-2 spike protein-specific memory B cells that were cross-reactive for HCoV-HKU1 and HCoV-OC43 as well as B cells specific for HCoV-HKU1 or HCoV-OC3 spike protein among PBMCs from four patients with COVID-19 3 months after infection with SARS-CoV-2. The abundance of these HCoV antigen-specific cells declined over time. However, the authors were unable to investigate the influence of SARS-CoV-2-specific, cross-reactive memory B cells on the overall serological responses against the novel coronavirus, in particular at earlier time points after infection. Furthermore, it was not addressed in this study whether the declining numbers of HCoV-specific memory B cells were associated with SARS-CoV-2 infection itself or due to COVID-19-independent, rapid fluctuations of HCoV antibody responses as observed by Edridge et al. (2020).
Comparing antibody responses against seasonal coronavirus in patients with COVID-19 with additional health record data we found that these responses are largely independent from age, having comorbidities, the time patients spent hospitalized or on ICU, and IL-6 levels. Interestingly, the group of male patients showed, in most instances, significantly increased anti-nucleocapsid antibody titers against seasonal coronaviruses. In multivariate analyses, we found IL-6 levels, especially those measured at admission, to correlate with disease severity, in line with recent studies (Leisman et al., 2020).
A study conducted by Sagar et al. (2021) proposed that acute HCoV infections can be associated with reduced COVID-19 severity. Data from medical records on PCR testing for acute HCoV infections were analyzed retrospectively in this investigation and not adaptive immune responses to individual HCoVs that we unveil as relevant in our study.
Anderson et al. (2021) suggested that pre-existing IgG-type antibody responses to the spike antigen of β-coronaviruses HCoV-OC43 in patient sera collected up to 7 years before SARS-CoV-2 infection do not influence susceptibility to the novel coronavirus and COVID-19 severity. However, Anderson et al. (2021) did not investigate the role of nucleocapsid-specific antibody responses to α-coronaviruses as a critical and predisposing factor for COVID-19. Moreover, HCoV antibody titers have been reported to decay or fluctuate considerably within months after infection or re-infection (Edridge et al., 2020), questioning the validity of the interpretation of pre-existing HCoV immunity at the time of SARS-CoV-2 exposure in patient-matched reference sera, which sometimes date back many years (Anderson et al., 2021). Based on the limited dataset that only assessed anti-HCoV-antibodies targeting the spike protein, Anderson et al. (2021) concluded that humoral adaptive immunity to seasonal coronaviruses is not associated with protection from infection or an altered disease course. Contradicting this notion, we provide evidence that pre-existing, humoral immunity reflected by specific antibodies recognizing either the nucleocapsid of seasonal α-coronaviruses or the spike antigen of HCoV-OC43 increases SARS-CoV-2 susceptibility. We propose that seasonal coronavirus serology can serve as a marker to guide clinical risk stratification and that individuals with recently resolved seasonal coronavirus infections may benefit from advanced preventive measures against COVID-19. Our findings fuel efforts to develop a universal vaccine that mitigates the immunological crosstalk between coronaviruses of different species and its potentially negative effects on the outcome of subsequent, possibly lethal coronavirus infections.
Limitations of the study
The results of our longitudinal data analysis cannot completely exclude the possibility of cross-reactive anti-SARS-CoV-2 antibodies that bind seasonal coronavirus antigens, thus contributing to the elevated anti-HCoV antibody titers observed in our assays. Furthermore, we cannot fully rule out polyclonal stimulation of HCoV-specific plasma cells after SARS-CoV-2 infection. However, for these scenarios to potentially contribute to our findings they would have to be (1) specific for certain antigens of individual HCoV species, (2) increase quickly already in the earliest days after SARS-CoV-2 infection, and (3) be stable over several weeks and independent from the COVID-19 disease course. Taken together, this seems unlikely. To corroborate our results, studies on matched sera from individuals with COVID-19 collected shortly before infection and during the disease course should be conducted. Also, the role of low antibody responses against the spike S1 domain found in critically ill patients should be investigated further. Furthermore, future work should seek to identify factors that drive humoral immunity toward strong, specific anti-HCoV responses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| R-phycoerythrin labelled goat-anti-human IgG | Dianova | Cat#JIM-109-116-098; RRID: AB_2337678 |
| Biological samples | ||
| Pre-pandemic serum samples from healthy adult blood donors (anonymized) | Blutspendedienst des Bayerischen Roten Kreuzes | N/A |
| Serum specimens from patients with COVID-19 (pseudonymized) | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| HCoV-OC43 spike protein | Sino Biological | Cat#40607-V08B |
| Critical commercial assays | ||
| recomLine SARS-CoV-2 IgG line immunoassay | Mikrogen | Cat#7374 |
| Deposited data | ||
| Pseudonymized patient record data and raw data from serum antibody measurements in patients and pre-pandemic donors | This study | Mendeley: https://doi.org/10.17632/stfw4w4vjh.1 |
| Oligonucleotides | ||
| Primer: CAG promoter forward CTT CTG GCG TGT GAC CGG | This study | N/A |
| Primer: CAG promoter reverse CAT GGT GGC CTT TGC CAA | This study | N/A |
| Primer: T4 foldon forward AAG TGG CCT AGC GGG CGC TTG GTC CCA CGT G | This study | N/A |
| Primer: T4 foldon reverse AAG ATC TGC TAG CTC GAG TCG C | This study | N/A |
| Primer: NL63-S1 forward CAT TTT GGC AAA GGC CAC CAT GAA GCT GTT CCT GAT CCT GC | This study | N/A |
| Primer: NL63-S1 reverse GGA GGA ATT TGC AGG AAT CAG GGA ACC GTC AG | This study | N/A |
| Primer: NL63-S2 forward CCC TGA TTC CTG CAA ATT CCT CCG ACA ACG GTA TCT | This study | N/A |
| Primer: NL63-S2 reverse CCA AGC GCC CGC TAG GCC ACT TGA TGT AGT TCT CGA A | This study | N/A |
| Recombinant DNA | ||
| Plasmid: pCAGGS | NovoPro | Cat#V008798 |
| Plasmid: pCAGGS encoding SARS-CoV-2 trimeric spike | Amanat et al. (2020) | N/A |
| Plasmids: pCAGGS encoding spike S1 domains of SARS-CoV-2, HCoV-229E, HCoV-NL63, HCoV-HKU1 or HCoV-OC43 | Becker et al. (2021) | N/A |
| Plasmids: pRSET2b encoding nucleocapsid proteins of SARS-CoV-2, HCoV-229E, HCoV-NL63, HCoV-HKU1 or HCoV-OC43 | Becker et al. (2021) | N/A |
| Plasmid: pCMV3-C-FLAG encoding HCoV-NL63 spike gene ORF cDNA | Sino Biological | Cat#VG40604-CF |
| Software and algorithms | ||
| recomScan 3.4 | Mikrogen | Cat#31006 |
| xPOTENT 4.3 | Luminex | Cat# XPON-UPGRD-FM3D |
| Prism 9.3.0 | GraphPad | www.graphpad.com |
| R version 4.1.1 | R Foundation | www.r-project.org |
| R package tidyverse 1.3.1 | Wickham et al. (2019) | cran.r-project.org/package=tidyverse |
| R package caret 6.0-90 | RStudio | cran.r-project.org/package=caret |
| R package MASS 7.3-54 | Venables and Ripley (2002) | cran.r-project.org/package=MASS |
| Other | ||
| Dynablot Plus strip processor | Dynex Technologies | Cat#D7144-P6-E |
| Flexmap 3D | Luminex | Cat#FLEXMAP-3D |
| Biomek i7 | Beckman | Cat#B87587 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oliver T. Keppler (keppler@mvp.lmu.de).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Patients and specimens
We established a collection of pre-pandemic serum samples from 888 healthy adult blood donors (collected prior to December 2019 in Germany) whose health record data were anonymized. Furthermore, we included a set of 314 serum specimens collected between March 8, 2020, and July 7, 2020, from 96 patients infected with SARS-CoV-2 at the LMU Klinikum, Munich, Germany. Patients are part of the COVID-19 Registry of the LMU Klinikum (CORKUM, WHO trial id DRKS00021225) and the study was approved by the local ethics committee (No: 20-245). All patients were tested positive for SARS-CoV-2 by rRT-PCR in nasopharyngeal or oropharyngeal swabs. The median age of the 96 patients with COVID-19 examined in this study was 61 years (interquartile range 50 to 71 years), and 26.0% (25/96) of these individuals were female. Clinical data, including symptoms and symptom onsets, were obtained from health records. Immunocompromised individuals were excluded from this study. If the time of symptom onset was not stated e.g., in asymptomatic patients, we substituted this information with the time of the first SARS-CoV-2-PCR-positive result. This was the case for 26.0% (25/96) of patients. We categorized the disease severity of patients with COVID-19 following the WHO guidelines "Clinical Management of COVID-19": asymptomatic (no clinical signs of infection), mild (symptomatic patients without evidence of viral pneumonia or hypoxia), moderate (clinical signs of pneumonia, including fever, cough, dyspnoea), severe (clinical signs of pneumonia, plus one of the following: respiratory rate > 30 /min, severe respiratory distress, SpO2 < 90% on room air), critical (one of the following: acute respiratory distress syndrome, sepsis, septic shock). Five patients contributing a total of 15 samples were categorized as asymptomatic, 19 patients contributing 35 samples as mild, 29 patients contributing 57 samples as moderate, 11 patients contributing 41 samples as severe, and 32 patients contributing 166 samples as critical (Figure S1). Due to the anonymization of pre-pandemic blood donors, these individuals could not be age-matched to the patients with COVID-19 examined in this study.
Method details
Coronavirus antigens
For the expression and purification of SARS-CoV-2 full-length spike protein as well as the nucleocapsid and spike S1 domain antigens of SARS-CoV-2 and HCoVs used in the MultiCoV-Ab assay, well-described plasmids were utilized (Becker et al., 2021; Amanat et al., 2020). To generate the expression construct of HCoV-NL63 full-length spike protein, four DNA fragments were amplified by PCR. Fragment 1 comprising a part of the CAG promotor sequence was amplified from pCAGGS expression plasmid encoding the SARS-CoV-2 trimeric spike (Amanat et al., 2020) using CAG promoter forward and reverse primers. Fragment 2 comprises the DNA encoding the T4 foldon and was amplified from the same plasmid using T4-foldon forward and reverse primers. Fragment 3 corresponds to the S1 fragment of HCoV-NL63 and was amplified from pCMV3-C-FLAG encoding HCoV-NL63 spike gene ORF cDNA using NL63-S1 forward and reverse primers. Fragment 4 corresponding to S2 fragment of HCoV-NL63 was amplified from the same cDNA template using NL63-S2 forward and reverse primers. Individual amplified DNA fragments were fused by overlap extension and cloned into XbaI and NotI sites of a pCAGGS expression vector. The newly generated expression construct encoding the full-length spike protein of HCoV-NL63 was verified by DNA sequencing. Protein expression and purification of antigens used in the MultiCoV-Ab assay was performed as previously described (Becker et al., 2021). HCoV-OC3 spike protein was purchased.
Antibody detection assays
The commercial recomLine SARS-CoV-2 IgG line immunoassay was used to determine the presence of IgG-type anti-nucleocapsid-specific antibodies against SARS-CoV-2 and HCoVs in serum specimens. The assay was performed in accordance with the manufacturers’ instructions. Briefly, test strips were incubated with 20μL serum diluted in 2 mL wash buffer on a Dynablot Plus strip processor. Following washing with wash buffer, conjugation solution and, after additional washing, substrate solution were added. After incubating the test strips for 8 min in substrate solution, strips were rinsed with deionized water and subsequently dried between 2 layers of absorbent paper. Quantitative results for the recomLine assay were obtained by analyzing test strips with the recomScan software. According to the manufacturer’s guidelines, the “fold cut-off” value was determined by subtracting the signal of interest with that of the internal cut-off band.
Further, the previously described MultiCoV-Ab multiplex immunoassay was employed that detects the presence of IgG antibodies against several SARS-CoV-2 and HCoVs antigens, including the S1 domain of viral spike protein and the nucleocapsid antigen (SARS-CoV-2 and all HCoVs), as well as the full-length spike antigen (SARS-CoV-2, HCoV-NL63 and HCoV-OC43) (Becker et al., 2021). Briefly, serum samples diluted 1:400 were incubated with antigens immobilized on magnetic beads in 384-well plates. Following washing with phosphate buffered saline containing 0.05% (v/v) Tween-20 and incubation with R-phycoerythrin labelled goat-anti-human IgG, antibody binding was measured on a FLEXMAP 3D running the xPONENT software v4.3. Normalization values were calculated by dividing the mean fluorescence intensity for each sample by those of plate-by-plate quality controls. Liquid handling was, in part, carried out on a Biomek i7.
Determination of assay specificities
The specificity of the two assays used in this study for SARS-CoV-2-specific IgG-type antibodies was measured in 888 (184 in case of the recomLine assay) pre-pandemic sera from healthy adult blood donors (see STAR Methods section ‘patients and specimens’). With 1/184 false positive results, the recomLine assay had a specificity for anti-nucleocapsid antibodies against SARS-CoV-1 of 99.5% (95% CI – 97.0% to 99.9%, Table S1). While MultiCoV-Ab normally uses a dual full-length spike and RBD cut-off system to determine positivity (Becker et al., 2021), specificities and sensitivities for the detection of antibodies against nucleocapsid, full-length spike and spike S1 domain antigens were analyzed separately for the purposes of this study. The MultiCoV-Ab assay had false positive rates of 4/888 for nucleocapsid-specific, 18/888 for full-length spike-specific and 21/888 spike S1 domain-specific anti-SARS-CoV-2 antibodies translating into specificities of 99.6% (95% CI – 98.9% to 99.8%), 98.0% (95% CI – 96.8 to 98.7) and 97.6% (95% CI – 96.4 to 98.5%), respectively (Table S1). Out of the four false positive samples in the anti-SARS-CoV-2 nucleocapsid antibody component of the Multi-CoV-Ab assay, none was positive in the anti-SARS-CoV-2 full-length spike antibody component, and one was positive in the anti-SARS-CoV-2 spike S1 domain antibody component of the same assay.
Infections with seasonal coronaviruses are frequent (Killerby et al., 2018), especially in children (Masse et al., 2020). We were unable to establish a cohort of individuals that were verifiably never infected by one or more HCoVs and, thus, can be assumed to be negative for long-lasting IgG-type antibodies against these viruses. As a consequence, the specificity for the two assays used in this study to detect anti-HCoV antibodies could not be analyzed.
In sera from pre-pandemic adults analyzed for nucleocapsid-specific antibodies by the recomLine assay, 28.8% (53/184) were positive for antibodies targeting HCoV-229E, 45.1% (83/184) for antibodies targeting HCoV-NL63, 57.6% (106/184) for antibodies targeting HCoV-HKU1, and 53.8% (99/184) for antibodies against HCoV-OC43, respectively (Table S2). 59.2% (109/184) of tested pre-pandemic samples were positive for more than one anti-HCoV-antibody analyzed via the recomLine assay, and 15.8% (29/184) were positive for antibodies against all HCoVs. However, since the specificity of the two assays for detecting HCoV-specific antibodies could not be measured, we mainly focused on comparing rather quantitative antibody levels than qualitative assay results in this study.
Determination of assay sensitivities
Sensitivities of the recomLine and the MultiCoV-Ab assays in detecting anti-SARS-CoV-2 antibodies were calculated from mean antibody levels of every patient in the study cohort (see STAR Methods section ‘patients and specimens’ and Figure S1). With mean IgG-type anti-SARS-CoV-2 nucleocapsid antibody levels of 84/95 patients with COVID-19 being positive, the overall sensitivity of the recomLine assay was 88.4% (95% CI – 80.5% to 93.4%, Table S1). Similarly, the MultiCoV-Ab assay was positive in 83/95 patients for nucleocapsid-specific and in 86/95 patients for full-length spike as well as spike S1 domain-specific anti-SARS-CoV-2 antibodies translating into sensitivities of 87.4% (95% CI – 79.2% to 92.6%) and 90.5% (95% CI – 83.0% to 94.9%), respectively (Table S1).
We were unable to establish a cohort of patients with acute, primary HCoV infection since infections with HCoVs are frequent (Killerby et al., 2018; Masse et al., 2020), the prevalence of long-lasting IgG-type antibodies is high (Severance et al., 2008), and re-infections are likely to occur (Edridge et al., 2020). Accordingly, we were unable to formally determine the sensitivity of the recomLine and MultiCoV-Ab assays for IgG-type anti-HCoV antibodies.
Assay correlation
We correlated quantitative results of all samples from pre-pandemic donors and patients with COVID-19 in both assays for HCoV-specific antibodies. Pearson correlations for different assays and assay components were similar comparing data from pre-pandemic donors and patients with COVID-19 (Figure S2). Results for anti-HCoV antibodies targeting the same antigen from HCoVs of the same taxonomic genus (α- or β-coronaviruses) correlated stronger than those targeting different antigens or HCoVs from different genera (Figure S2). This indicates that the specificity of the assays for similar antigens from HCoVs of the same genus is possibly decreased or that cross-reacting antibodies within the same genus are frequent, in line with data from other studies (Becker et al., 2021; Edridge et al., 2020).
Quantification and statistical analysis
Multivariate analysis was performed using logistic regression, with disease severity (critical or non-critical) as dependent and quantitative antibody levels, age, sex, comorbidities and IL-6 levels as independent variables (Data S1). As no additional health record data was available for pre-pandemic donors, pre-pandemic samples had to be excluded from the multivariate analysis.
Acknowledgments
The CORKUM biobank is funded, in part, by the Federal Ministry of Education and Research (BMBF) initiative “NaFoUniMedCovid19” (01KX2021), LMUexcellent, the Free State of Bavaria under the Excellence Strategy of the Federal Government and the States, and the Faculty of Medicine of the LMU München. MultiCoV-Ab is funded by the Ministry for Economic Affairs, Labor and Housing Construction (Baden-Württemberg, grant no. FKZ 3-4332.62-NMI-68) and the EU Horizon 2020 research and innovation program (grant agreement no. 101003480—CORESMA). We thank all the staff working with sample collection and sample preparation. We are grateful to all CORKUM investigators and staff. The authors thank the patients and their families for their participation in the CORKUM registry.
Author contributions
P.R.W. and O.T.K. conceived the study. P.R.W. and O.T.K. drafted the first version of the manuscript. N.A.S., A.D., N.S.-M., L.K., and V.H. contributed to drafting sections of the manuscript. P.R.W., N.A.S., B.K., A.D., and L.K. performed data analysis. P.R.W., N.A.S., B.K., A.O., P.M.S., A.R., M.G., and S.S. conducted laboratory experiments. A.D., D.J., M.B., U.R., and N.S.-M. performed the MultiCoV-Ab experiments and data normalization. M.M., J.C.H., C.S., J.M., M.R., J.B., S.K., B.Z., M.v.B.-B., J.E., and N.S.-M. participated in the study design. All authors contributed to the interpretation of data and approved the final manuscript.
Declaration of interests
N.S.-M. was a speaker at Luminex user meetings in the past. The Natural and Medical Sciences Institute at the University of Tübingen is involved in applied research projects as a fee for services with Luminex. The remaining authors declare no competing interests.
Published: December 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110169.
Supplemental information
Data and code availability
All raw data corresponding to pseudonymized patient record data and serum antibody measurements in patients with COVID-19 and pre-pandemic donors have been deposited to Mendeley Data (https://doi.org/10.17632/stfw4w4vjh.1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ali Y.M., Ferrari M., Lynch N.J., Yaseen S., Dudler T., Gragerov S., Demopulos G., Heeney J.L., Schwaeble W.J. Lectin pathway mediates complement activation by SARS-CoV-2 proteins. Front. Immunol. 2021;12:714511. doi: 10.3389/fimmu.2021.714511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schröder I., Wieters I., Khodamoradi Y., et al. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271.e5. doi: 10.1016/j.immuni.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Strengert M., Junker D., Kaiser P.D., Kerrinnes T., Traenkle B., Dinter H., Häring J., Ghozzi S., Zeck A., et al. Exploring beyond clinical routine SARS-CoV-2 serology using multiCoV-Ab to evaluate endemic coronavirus cross-reactivity. Nat. Commun. 2021;12:1152. doi: 10.1038/s41467-021-20973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Center for Systems and Science Engineering at John Hopkins University COVID-19 Dashboard. 2020. https://coronavirus.jhu.edu/map.html
- Dugas M., Grote-Westrick T., Vollenberg R., Lorentzen E., Brix T., Schmidt H., Tepasse P.R., Kühn J. Less severe course of COVID-19 is associated with elevated levels of antibodies against seasonal human coronaviruses OC43 and HKU1 (HCoV OC43, HCoV HKU1) Int. J. Infect. Dis. 2020;105:304–306. doi: 10.1016/j.ijid.2021.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas M., Grote-Westrick T., Merle U., Fontenay M., Kremer A.E., Hanses F., Vollenberg R., Lorentzen E., Tiwari-Heckler S., Duchemin J., et al. Lack of antibodies against seasonal coronavirus OC43 nucleocapsid protein identifies patients at risk of critical COVID-19. J. Clin. Virol. 2021;139:104847. doi: 10.1016/j.jcv.2021.104847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014–2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisman D.E., Ronner L., Pinotti R., Taylor M.D., Sinha P., Calfee C.S., Hirayama A.V., Mastroiani F., Turtle C.J., Harhay M.O., et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020;28:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse S., Capai L., Villechenaud N., Blanchon T., Charrel R., Falchi A. Epidemiology and clinical symptoms related to seasonal coronavirus identified in patients with acute respiratory infections consulting in primary care over six influenza seasons (2014-2020) in France. Viruses. 2020;12:630. doi: 10.3390/v12060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega N., Ribes M., Vidal M., Rubio R., Aguilar R., Williams S., Barrios D., Alonso S., Hernández-Luis P., Mitchell R.A., et al. Seven-month kinetics of SARS-CoV-2 antibodies and role of pre-existing antibodies to human coronaviruses. Nat. Commun. 2021;12:4740. doi: 10.1038/s41467-021-24979-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrc K., Dijkman R., Deng L., Jebbink M.F., Ross H.A., Berkhout B., van der Hoek L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:e143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severance E.G., Bossis I., Dickerson F.B., Stallings C.R., Origoni A.E., Sullens A., Yolken R.H., Viscidi R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccin. Immunol.: CVI. 2008;15:1805–1810. doi: 10.1128/CVI.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. Fourth edition. Springer; 2002. Modern Applied Statistics with S. [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- World Health Organization . 2020. Clinical management of COVID-19: interim guidance, 27 May 2020. https://apps.who.int/iris/handle/10665/332196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data corresponding to pseudonymized patient record data and serum antibody measurements in patients with COVID-19 and pre-pandemic donors have been deposited to Mendeley Data (https://doi.org/10.17632/stfw4w4vjh.1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.