Highlights
-
•
Obesity-related SNPs are associated with regional brain volumes.
-
•
FTO SNPs are negatively correlated with brain volumes in visual object recognition.
-
•
Metabolically unhealthy variant is associated with reduced brain volumes in DA motive system.
-
•
5-HT4 pathway might be a potential therapeutic target for human obesity.
Keywords: Obesity, Brain image-derived phenotypes, MLM-based association analysis, Genetic correlation
Abstract
Objective
To determine whether there is a correlation between obesity-related variants and regional brain volumes.
Methods
Based on a mixed linear model (MLM), we analyzed the association between 1,498 obesity-related SNPs in the GWAS Catalog and 164 regional brain volumes from 29,420 participants (discovery cohort N = 19,997, validation cohort N = 9,423) in UK Biobank. The statistically significant brain regions in association analysis were classified into 6 major neural networks (dopamine (DA) motive system, central autonomic network (CAN), cognitive emotion regulation, visual object recognition network, auditory object recognition network, and sensorimotor system). We summarized the association between obesity-related variants (metabolically healthy obesity variants, metabolically unhealthy obesity variants, and unclassified obesity-related variants) and neural networks.
Results
From association analysis, we determined that 17 obesity-related SNPs were associated with 51 regional brain volumes. Several single SNPs (e.g., rs13107325-T (SLC39A8), rs1876829-C (CRHR1), and rs1538170-T (CENPW)) were associated with multiple regional brain volumes. In addition, several single brain regions (e.g., the white matter, the grey matter in the putamen, subcallosal cortex, and insular cortex) were associated with multiple obesity-related variants. The metabolically healthy obesity variants were mainly associated with the regional brain volumes in the DA motive system, sensorimotor system and cognitive emotion regulation neural networks, while metabolically unhealthy obesity variants were mainly associated with regional brain volumes in the CAN and total tissue volumes. In addition, unclassified obesity-related variants were mainly associated with auditory object recognition network and total tissue volumes. The results of MeSH (medical subject headings) enrichment analysis showed that obesity genes associated with brain structure pointed to the functional relatedness with 5-Hydroxytryptamine receptor 4 (5-HT4), growth differentiation factor 5 (GDF5), and high mobility group protein AT-hook 2 (HMGA2 protein).
Conclusion
In summary, we found that obesity-related variants were associated with different brain volume measures. On the basis of the multiple SNPs, we found that metabolically healthy and unhealthy obesity-related SNPs were associated with different brain neural networks. Based on our enrichment analysis, modifications of the 5-HT4 pathway might be a promising therapeutic strategy for obesity.
1. Introduction
With globalization and modernization driving global obesity trends, the prevalence of obesity has increased in every country. In the past 40 years, the number of obese men and women has increased from 100 million to 671 million worldwide (Collaboration, 2017), and at least 2.8 million people die every year from obesity or because they were overweight (World Health Organization, 2020). Considering its distinct pathophysiology, which prevents weight loss but promotes weight gain, obesity is classified as a disease that requires urgent prevention and management (Wolfenden et al., 2019).
Obesity is deemed a brain disease because the brain regulates food intake and energy metabolism (Banks, 2003, Shefer et al., 2013, Small et al., 1997). Increasing evidence suggests a link between obesity and the brain (Banks, 2003, Kuhne and Stengel, 2019, McDougle et al., 2021, Raka et al., 2019, Shefer et al., 2013, Small et al., 1997). The gut satiety hormones produced by the gastrointestinal tract send satiety signals to the brain via the gut-brain axis (Kuhne and Stengel, 2019), and abnormal brain response to satiety signals or high-fat feeding may play an important role in obesity pathogenesis (Smith et al., 2018, Covasa et al., 2000). Moreover, neuroimaging studies have shown that obesity is accompanied by focal structural alternations in many brain regions (Pannacciulli et al., 2006, Dekkers et al., 2019, Taki et al., 2008, Kennedy et al., 2019, Raji, 2010, Hamer and Batty, 2019), including the thalamus (Dekkers et al., 2019, Taki et al., 2008, Kennedy et al., 2019, Raji, 2010), caudate (Dekkers et al., 2019, Taki et al., 2008, Hamer and Batty, 2019), putamen (Pannacciulli et al., 2006, Dekkers et al., 2019, Hamer and Batty, 2019), pallidum (Dekkers et al., 2019, Hamer and Batty, 2019), hippocampus (Dekkers et al., 2019, Raji, 2010, Horstmann et al., 2013), nucleus accumbens (Dekkers et al., 2019, Hamer and Batty, 2019), medial prefrontal cortex (Kennedy et al., 2019), temporal lobe (Taki et al., 2008, Gustafson et al., 2004), cerebral spinal fluid (Dekkers et al., 2019) and brainstem (Dekkers et al., 2019, Kennedy et al., 2019). These altered brain regions are involved in functions including decision-making (Hare et al., 2009), emotion processing (Locke et al., 2015), reward processing (Pannacciulli et al., 2006, Kennedy et al., 2019), and appetite regulation (Pannacciulli et al., 2006, Kennedy et al., 2019, Papageorgiou et al., 2017). Therefore, it is meaningful to identify the factors that contribute to brain volume alterations in obese patients. As a variety of obesity-related genetic variations have been associated with the brain physiology and neurobehavior of obese individuals, genetic factors may contribute to brain structural alterations in obese people. Several articles have reported the high expression of obesity-related genes in the brain (Kennedy et al., 2019, Horstmann et al., 2013, Locke et al., 2015, Vainik et al., 2018). About one-third of body mass index (BMI) and brain structure genes are shared (Kennedy et al., 2019), and genetic factors influence the relationship between BMI and brain regions related to eating behavior (Locke et al., 2015). However, most studies on the association between the obesity gene and brain volumes have focused on one single obesity gene. A GWAS study for BMI showed that the fat mass and the obesity-associated gene (FTO) can influence obesity (Frayling et al., 2007). Several groups reported that the genetic variant rs9939609-A in FTO was associated with the increased volume of the accumbens in children (Rapuano et al., 2017), while it was associated with the decreased volume of the accumbens in the elderly (de Groot, 2015). Other SNPs of the FTO gene, such as rs1421085, rs9930333, rs3751812, have also been reported. FTO rs1421085-C was associated with the increased grey matter volumes of the right middle temporal gyrus, parahippocampal gyrus, lateral occipital cortex (Lugo‐Candelas et al., 2020), while FTO rs3751812-T was associated with the reduced volume of occipital lobe (Ho et al., 2010). FTO rs9930333-G was not associated with ventricular cerebrospinal fluid volume (Melka et al., 2013). However, multiple lines of evidence also indicates that obesity-related genetic polymorphisms are not associated with structural connectivity or structural abnormalities in the brain (Beyer et al., 2021, Weise et al., 2019, Cole et al., 2013). These inconsistent results might be due to differences in participant age, genetic variations, and small sample size. Although obesity-related genetic variants have been found to be associated with brain volume alterations, such research is still in its infancy. There is currently a lack of systematic investigation of obesity-related genetic susceptibility factors in brain volume. At the same time, obesity-related variants can be classified into metabolically healthy obesity-related variants and metabolically unhealthy obesity-related variants (Mulugeta et al., 2021) which have different effects on human health (Mulugeta et al., 2021, Martin, 2021, Winkler et al., 2018). Mendelian randomization studies provide evidence for the protective effects of metabolically healthy obesity variants and the detrimental effects of metabolically unhealthy obesity variants on type 2 diabetes, heart disease, stroke, as well as the differential associations with cranial grey matter volume (Mulugeta et al., 2021, Martin, 2021).
Therefore, we used a large cohort of 29,420 participants with SNP genotypes and brain MRI to explore the correlation between obesity variants and 164 regional brain volumes. We also performed a confirmatory analysis of the association of previously reported FTO SNPs with different brain structures. Further, we explored the association of metabolically healthy versus unhealthy SNPs to provide a more comprehensive and systematic understanding of the changes in brain structure from the perspective of genetics and obesity mechanisms.
2. Methods
2.1. Obesity-related SNPs collection
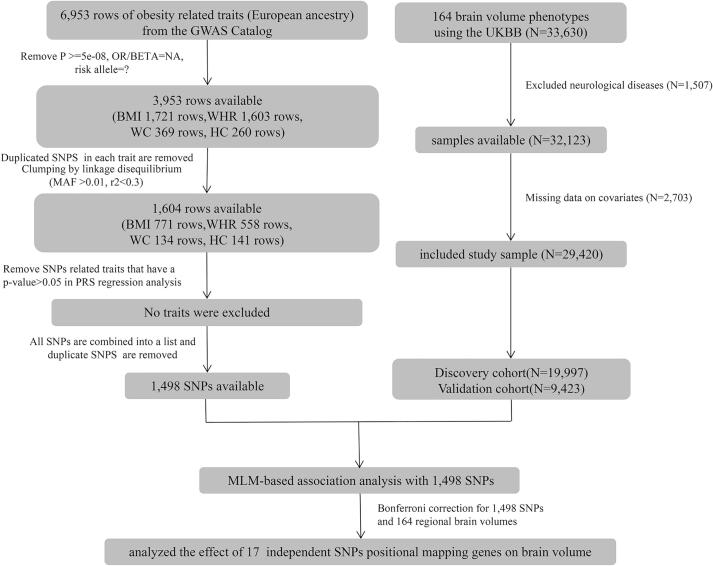
We included 4 obesity-related traits (body mass index (BMI), waist to hip ratio (WHR), waist circumference (WC), hip circumference (HC)) in this study. Obesity-related SNPs in the European ancestry cohort (p < 5 × 10−8, minor allele frequency > 0.01) were obtained from GWAS Catalog database (accessed on March 9, 2020). Duplicate SNPs in each trait were removed. The remaining SNPs of each trait were clumped based on linkage disequilibrium (LD, r2 < 0.3). We performed a polygenic risk score (PRS) on these independent SNPs, and used a linear model to further verify the correlation between the PRS and its related obesity trait. Obesity traits with p > 0.05 in the linear regression analysis were excluded. A flowchart of the process of SNPs and participant selection is shown in Fig. 1.
Fig. 1.
Flowchart describing sample selection.
2.2. Brain volume participants and regional brain volumes acquisition
UK Biobank is a prospective cohort study that includes >500,000 individuals aged 40 and 69 from across the UK (Bycroft et al., 2018). The genotypes of genome-wide simple nucleotide polymorphisms were assayed using Affymetrix Axiom SNP arrays. Data access permission was granted under UKB application 71668.
Since 2014, around 42,928 participants were invited back to the UKB imaging centers (the first imaging visit). Nuclear Magnetic Resonance (NMR) scans were made using Siemens Skyra 3T adopting the 3D Magnetization Prepared Rapid Acquisition Gradient-echo (MPRAGE) protocol. The key parameters included the resolution of 1x1x1 mm, field-of-view of 208x256x256 matrix, and in-plane acceleration of 2. The obtained images were nonlinearly warped into the MNI152 space for spatial standardization using FNIRT (FMRIB’s Nonlinear Image Registration Tool) (Andersson et al., 2007, Anderson et al., 2007). The brain image was then divided into 10 total tissue volumes (both normalised for overall head size as well as not normalised: cerebrospinal fluid, white matter, grey matter, grey + white matter, peripheral cortical grey matter) and 139 regional volumes of grey matter segmentation by FAST (FMRIB’s Automated Segmentation Tool), and 15 subcortical structures by FIRST (FMRIB’s Integrated Registration and Segmentation Tool) (Patenaude et al., 2011, Zhang et al., 2001).
In total, there are 164 regional brain volume traits in the UKB (Supplementary Table 1). Our study included 33,630 participants of European ancestry. Among them, 1,507 participants were excluded due to neurological diseases that may affect brain volume (Supplementary Table 2). We further excluded 2,703 participants who responded with missing information of education attainment, BMI or head motion. Finally, 29,420 participants remained in our analysis of the genetic association between regional brain volumes and obesity. According to the genetic relationship, we took 19,997 genetically unrelated cases as the discovery cohort and 9,423 genetically related cases as the validation cohort.
2.3. Genetic association analysis of regional brain volumes
We tested the association between regional brain volumes and selected obesity-associated SNPs using a mixed linear model adjusted for age, gender, BMI, education attainment, total intracranial volume, and the first 5 genetic principle components. According to the recommendations of the literature, since the head motion data was reasonably similar across all modalities, the head motion data of the resting fMRI scanned in the same period of time was used as the covariables in our study (Alexander-Bloch et al., 2016, Elliott et al., 2018). In association analysis, SNPs with bonferroni adjusted p-value were considered to be significantly associated with the regional brain volume (Bonferroni correction for the numbers of unique SNPs and 164 regional brain volumes). We replicated these significant results in a validation cohort of 9,423 genetically related participants (uncorrected p-value < 0.05). Considering the pleiotropy of traits, we used LDtrait to identify the pleiotropic relationship within the 4 obesity traits in each independent SNP (p-value < 5 × 10−8, r2 > 0.3, beta ≠ NA, https://ldlink.nci.nih.gov/?tab=help#LDtrait). These brain regions verified by the validation group were classified into 6 major neural networks (dopamine (DA) motive system, central autonomic network (CAN), cognitive emotion regulation, visual object recognition network, auditory object recognition network, and sensorimotor system). Previous publications were cited for each brain region and function (Supplementary Table 3). We use this network definition to explain our findings in the results and discussion.
2.3.1. Gene set enrichment analysis of obesity genes relevant in regional brain volumes
Those SNPs that were significantly associated with brain volumes were mapped to genes by their genomic positions. Then, we input statistically significant genes into the gene2function of Functional Mapping and Annotation of GWAS (FUMA GWAS) tool, and obtained tissue expression profile data. We used R software to draw the tissue expression profile of 13 brain tissues through the expression profile data. In addition, we performed enrichment analysis to explore the function of independent genes shared between obesity and brain volumes through the R package (“Meshes” version 4.0.2) (Yu, 2018). The Meshes package is a biological data mining software package based on MeSH terms, which contains comprehensive biomedical vocabulary such as Chemicals and Drugs, Anatomy, and Phenomena. Genes mapped to MeSH terms are manually generated by NCBI (gene2pubmed). Then, the enrichMeSH function investigates MeSH term associations of genes by hypergeometric model. The enrichment analysis in the Meshes package enables to extract a wider range of gene meanings, and enhance and enrich biological significance and unknown functional associations.
2.4. Statistical analysis
SNP clumping was conducted using PLINK version 1.9 (Purcell et al., 2007). Polygenic risk score analyses were performed using PRSice-2 (https://www.prsice.info/). The mixed linear model (MLM) was performed using GCTA software with a pre-compiled kinship matrix (Jiang et al., 2019).
3. Results
3.1. Obesity-related SNPs collection
We extracted 3,953 records with a frequency higher than 0.01 and p < 5 × 10−8 from the GWAS Catalog database. After removing duplicate SNPs and linkage disequilibrium test, 1,604 records were retained. No traits were excluded after linear regression analysis (Supplementary Table 4). Finally, we included 4 obesity-related traits (BMI, WHR, WC, HC) in this study (Supplementary Table 5). The genetic association analysis between obesity and brain volume based on MLM was performed using 1,498 unique SNPs.
3.2. Sample characteristics
Among the 29,420 participants with brain MRI images, the average age was 55.608 ± 7.391 (Table 1). Males made up 47.8% of the participants. Almost half of the participants finished college or university study (47.3%) (Supplementary Table 1). The mean and standard deviation (SD) of each regional brain volume can be found in Supplementary Table 1. The distribution of 4 obesity-related traits in the sample was shown in Table 1. The average BMI was 26.473 ± 4.357 kg/m2. The average WC and HC were 88.223 ± 12.588 cm and 100.912 ± 8.677 cm, separately. The average WHR was 0.873 ± 0.088.
Table 1.
Sample characteristics.
| Variables | Entire cohort | Discovery cohort | Validation cohort | |
|---|---|---|---|---|
| No. of patients | 29,420 | 19,777 | 9,423 | |
| Age | 55.608 ± 7.391 | 55.604 ± 7.368 | 55.615 ± 7.441 | |
| Sex |
Female | 15,357(52.2%) | 10,280(51.6%) | 5,077(53.9%) |
| Male | 14,063(47.8%) | 9,717(48.4%) | 4,346(46.1%) | |
| BMI | 26.473 ± 4.357 | 26.398 ± 4.313 | 26.631 ± 4.444 | |
| WHR | 0.873 ± 0.088 | 0.873 ± 0.088 | 0.874 ± 0.087 | |
| WC | 88.223 ± 12.588 | 88.076 ± 12.547 | 88.536 ± 12.67 | |
| HC | 100.912 ± 8.677 | 100.782 ± 8.621 | 101.187 ± 8.788 | |
| Head motion | 0.122 ± 0.058 | 0.121 ± 0.058 | 0.124 ± 0.058 | |
| Total intracranial volume (mm3) | 1.201,153 ± 114,562.735 | 1,203,410 ± 114,480.1 | 1,196,363 ± 114,596.8 | |
3.3. Obesity-related SNPs are associated with alterations in regional brain volumes
We identified 21 obesity-related SNPs associated with 59 brain regions in the discovery cohort. Eighty-one percent (n = 17) of SNPs were verified (p < 0.05). The 17 obesity-related SNPs, mapped genes, and their related brain regions both in the discovery cohort and validation cohort were shown in Table 2 and Fig. 2. These brain regions were involved in the modulation of multiple brain functions, including reward, cognitive emotion, auditory, visual, sensorimotor, and central autonomic. SNPs that cannot be replicated in the association analysis were listed in Supplementary Table 6. Some of these variants were respectively associated with different sides of the same brain region in the discovery and validation groups (Supplementary Table 6 and Supplementary Table 7). For instance, rs4889606-A was associated with the lower grey matter volume of the right Crus II cerebellum in the discovery cohort (rs4889606-A, right: p = 5.55 × 10−8, beta = -60.749), while it was associated with grey matter of the vermis Crus II cerebellum in the validation group (rs4889606-A, vermis: p = 0.015, beta = -2.781). Another variant, rs10886017-A, was in relation with the reduced grey matter volume of right precentral in discovery cohort (rs10886017-A, right: p = 5.86 × 10−8, beta = -90.805), but it had same effect on the left precentral in the validation group (rs10886017-A, left: p = 0.024, beta = -55.191).
Table 2.
The association of obesity-related variants and regional brain volumes.
| S.N | SNP-EA | Gene | Class | N | EAF | Beta | SE | P.value | Brain |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs1038088-G | SSH2 | BMI+ | 19,997 | 0.524 | 17.659 | 3.041 | 6.39 × 10−9 | Volume of grey matter in Subcallosal Cortex (left) |
| 2 | rs1038088-G | SSH2 | BMI+ | 19,997 | 0.524 | 15.501 | 2.754 | 1.81 × 10−8 | Volume of grey matter in Subcallosal Cortex (right) |
| 3 | rs11727676-C | HHIP | WHR + BMI- | 19,997 | 0.094 | 68.706 | 9.471 | 4.03 × 10−13 | Volume of grey matter in Intracalcarine Cortex (left) |
| 4 | rs11727676-C | HHIP | WHR + BMI- | 19,997 | 0.094 | 73.953 | 8.98 | 1.79 × 10−16 | Volume of grey matter in Intracalcarine Cortex (right) |
| 5 | rs11727676-C | HHIP | WHR + BMI- | 19,997 | 0.094 | 129.419 | 19.425 | 2.69 × 10−11 | Volume of grey matter in Occipital Pole (right) |
| 6 | rs12321904-T | MMAB | BMI+ | 19,997 | 0.492 | −13.264 | 2.32 | 1.08 × 10−8 | Volume of grey matter in VI Cerebellum (vermis) |
| 7 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | −199.145 | 28.303 | 1.97 × 10−12 | Volume of grey matter in Precentral Gyrus (left) |
| 8 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | −163.696 | 25.392 | 1.14 × 10−10 | Volume of grey matter in Postcentral Gyrus (left) |
| 9 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | −136.248 | 24.761 | 3.74 × 10−8 | Volume of grey matter in Postcentral Gyrus (right) |
| 10 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | −89.495 | 14.23 | 3.19 × 10−10 | Volume of grey matter in Angular Gyrus (left) |
| 11 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 71.89 | 12.789 | 1.90 × 10−8 | Volume of grey matter in Caudate (left) |
| 12 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 83.676 | 13.419 | 4.49 × 10−10 | Volume of grey matter in Caudate (right) |
| 13 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 153.62 | 7.931 | 1.37 × 10−83 | Volume of grey matter in Putamen (left) |
| 14 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 171.967 | 8.599 | 5.68 × 10−89 | Volume of grey matter in Putamen (right) |
| 15 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 40.955 | 1.901 | 6.22 × 10−103 | Volume of grey matter in Ventral Striatum (left) |
| 16 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 39.955 | 1.891 | 3.92 × 10−99 | Volume of grey matter in Ventral Striatum (right) |
| 17 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 116.801 | 14.497 | 7.82 × 10−16 | Volume of grey matter in Brain-Stem |
| 18 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 50.719 | 5.269 | 6.17 × 10−22 | Volume of grey matter in I-IV Cerebellum (left) |
| 19 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 40.14 | 5.656 | 1.28 × 10−12 | Volume of grey matter in I-IV Cerebellum (right) |
| 20 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 60.023 | 6.953 | 6.02 × 10−18 | Volume of grey matter in V Cerebellum (left) |
| 21 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 41.691 | 6.751 | 6.59 × 10−10 | Volume of grey matter in V Cerebellum (right) |
| 22 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 127.266 | 17.742 | 7.33 × 10−13 | Volume of grey matter in VI Cerebellum (left) |
| 23 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 131.29 | 17.427 | 4.93 × 10−14 | Volume of grey matter in VI Cerebellum (right) |
| 24 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 86.683 | 12.276 | 1.65 × 10−12 | Volume of grey matter in VIIIa Cerebellum (left) |
| 25 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 25.969 | 3.17 | 2.56 × 10−16 | Volume of grey matter in VIIIa Cerebellum (vermis) |
| 26 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 86.785 | 12.98 | 2.30 × 10−11 | Volume of grey matter in VIIIa Cerebellum (right) |
| 27 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 104.841 | 9.205 | 4.70 × 10−30 | Volume of grey matter in VIIIb Cerebellum (left) |
| 28 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 15.109 | 1.585 | 1.57 × 10−21 | Volume of grey matter in VIIIb Cerebellum (vermis) |
| 29 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 108.063 | 9.964 | 2.11 × 10−27 | Volume of grey matter in VIIIb Cerebellum (right) |
| 30 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 95.695 | 7.196 | 2.37 × 10−40 | Volume of grey matter in IX Cerebellum (left) |
| 31 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 30.419 | 1.667 | 2.29 × 10−74 | Volume of grey matter in IX Cerebellum (vermis) |
| 32 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 104.296 | 7.925 | 1.50 × 10−39 | Volume of grey matter in IX Cerebellum (right) |
| 33 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | 10.036 | 0.813 | 4.90 × 10−35 | Volume of grey matter in X Cerebellum (vermis) |
| 34 | rs13107325-T | SLC39A8 | BMI + WHR- | 19,997 | 0.071 | −2,366.75 | 379.517 | 4.48 × 10−10 | Volume of peripheral cortical grey matter |
| 35 | rs7970350-C | HMGA2 | WC + HC+ | 19,997 | 0.512 | −2,883.9 | 484.584 | 2.66 × 10−9 | Volume of white matter (normalised for head size) |
| 36 | rs7970350-C | HMGA2 | WC + HC+ | 19,997 | 0.512 | −6,084.3 | 1,065.68 | 1.13 × 10−8 | Volume of grey + white matter (normalised for head size) |
| 37 | rs1481012-A | ABCG2 | BMI+ | 19,997 | 0.887 | 18.18 | 3.479 | 1.73 × 10−7 | Volume of pallidum (left) |
| 38 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | 20.454 | 2.758 | 1.20 × 10−13 | Volume of grey matter in Subcallosal Cortex (right) |
| 39 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −10.73 | 1.797 | 2.34 × 10−9 | Volume of grey matter in Planum Polare (left) |
| 40 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −10.793 | 1.819 | 2.94 × 10−9 | Volume of grey matter in Planum Polare (right) |
| 41 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −68.284 | 11.407 | 2.15 × 10−9 | Volume of grey matter in Occipital Pole (right) |
| 42 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −3,882.67 | 532.891 | 3.19 × 10−13 | Volume of peripheral cortical grey matter (normalised for head size) |
| 43 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −4,892.74 | 667.287 | 2.26 × 10−13 | Volume of grey matter (normalised for head size) |
| 44 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −3,319.8 | 480.06 | 4.67 × 10−12 | Volume of white matter (normalised for head size) |
| 45 | rs1538170-T | CENPW | HC+ | 19,997 | 0.454 | −8,212.59 | 1,055.32 | 7.13 × 10−15 | Volume of grey + white matter (normalised for head size) |
| 46 | rs17019336-T | HHIP-AS1 | BMI+ | 19,997 | 0.762 | −32.702 | 6.253 | 1.69 × 10−7 | Volume of grey matter in Intracalcarine Cortex (right) |
| 47 | rs1727294-A | PITPNM2 | HC+ | 19,997 | 0.212 | −3,245.01 | 584.4 | 2.81 × 10−8 | Volume of white matter (normalised for head size) |
| 48 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 126.725 | 23.764 | 9.67 × 10−8 | Volume of grey matter in Frontal Pole (right) |
| 49 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 31.713 | 5.943 | 9.49 × 10−8 | Volume of grey matter in Insular Cortex (left) |
| 50 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 13.833 | 2.419 | 1.08 × 10−8 | Volume of grey matter in Parahippocampal Gyrus (left) |
| 51 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 4,471.49 | 637.428 | 2.30 × 10−12 | Volume of peripheral cortical grey matter (normalised for head size) |
| 52 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 1,401.02 | 231.601 | 1.46 × 10−9 | Volume of peripheral cortical grey matter |
| 53 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 5,289.25 | 798.307 | 3.46 × 10−11 | Volume of grey matter (normalised for head size) |
| 54 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | 1,458.67 | 246.928 | 3.48 × 10−9 | Volume of grey matter |
| 55 | rs1876829-C | CRHR1 | WHR+ | 19,997 | 0.223 | −1,455.13 | 224.861 | 9.72 × 10−11 | Volume of white matter |
| 56 | rs329124-A | JADE2 | BMI+ | 19,997 | 0.577 | −5.597 | 0.995 | 1.83 × 10−8 | Volume of grey matter in Ventral Striatum (left) |
| 57 | rs329124-A | JADE2 | BMI+ | 19,997 | 0.577 | −6.135 | 0.988 | 5.42 × 10−10 | Volume of grey matter in Ventral Striatum (right) |
| 58 | rs3744017-A | TRIM47 | BMI+ | 19,997 | 0.188 | 49.258 | 8.359 | 3.79 × 10−9 | Volume of grey matter in Caudate (left) |
| 59 | rs3744017-A | TRIM47 | BMI+ | 19,997 | 0.188 | 53.697 | 8.771 | 9.24 × 10−10 | Volume of grey matter in Caudate (right) |
| 60 | rs591939-G | NAGLU | WHR+ | 19,997 | 0.248 | −26.011 | 4.724 | 3.66 × 10−8 | Volume of grey matter in Putamen (left) |
| 61 | rs591939-G | NAGLU | WHR+ | 19,997 | 0.248 | −28.603 | 5.125 | 2.39 × 10−8 | Volume of grey matter in Putamen (right) |
| 62 | rs6857-C | NECTIN2 | BMI + WHR + WC+ | 19,997 | 0.832 | −39.875 | 7.068 | 1.68 × 10−8 | Volume of grey matter in Intracalcarine Cortex (right) |
| 63 | rs1243188-C | MLLT10 | WHR + BMI+ | 19,997 | 0.3 | −1,428.21 | 191.636 | 9.14 × 10−14 | Volume of ventricular cerebrospinal fluid (normalised for head size) |
| 64 | rs1243188-C | MLLT10 | WHR + BMI+ | 19,997 | 0.3 | −1,186.24 | 162.27 | 2.67 × 10−13 | Volume of ventricular cerebrospinal fluid |
| 65 | rs1243188-C | MLLT10 | WHR + BMI+ | 19,997 | 0.3 | 1,186.21 | 162.27 | 2.67 × 10−13 | Volume of grey + white matter |
| 66 | rs798502-A | GNA12 | WC + HC+ | 19,997 | 0.699 | 32.16 | 5.455 | 3.74 × 10−9 | Volume of grey matter in Insular Cortex (left) |
| 67 | rs798502-A | GNA12 | WC + HC+ | 19,997 | 0.699 | 36.184 | 5.515 | 5.34 × 10−11 | Volume of grey matter in Insular Cortex (right) |
| 68 | rs806794-A | H2BC7 | HC + WC+ | 19,997 | 0.729 | −26.161 | 5.019 | 1.87 × 10−7 | Volume of grey matter in Putamen (right) |
| 69 | rs806794-A | H2BC7 | HC + WC+ | 19,997 | 0.729 | −33.395 | 6.158 | 5.85 × 10−8 | Volume of thalamus (left) |
SNP-EA, SNP and effect allele; Gene, genes mapping using genomic position; Class, The SNP and its LD variants-related obesity traits; N, sample number; EAF, frequency of effect allele; Beta, effect size of effect allele; P.value, the significance level of each variant.
BMI is the body mass index, WHR is the waist to hip ratio, WC is the waist ratio, HC is the hip raito.
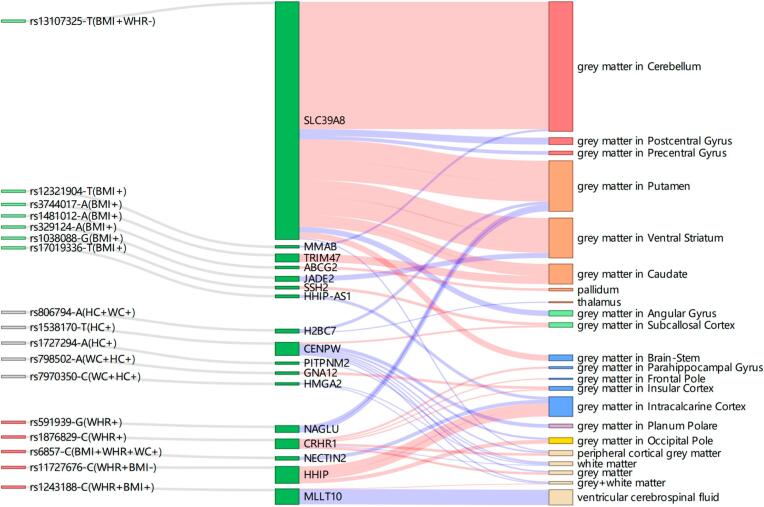
Fig. 2.
Sankey diagram of obesity-related genetics and their related brain regions. The first column is obesity-related SNPs, the green is metabolically healthy variants, the red is metabolically unhealthy variants, the grey is unclassified obesity-related variants; the second column is the mapped genes of obesity-related SNPs by positional mapping; the third column shows the regions of the brain associated with the obesity gene. The red and blue lines represent positive and negative correlations, respectively, the line width is determined by dividing the BETA by the mean of the regional brain volume.
3.4. Single SNP has an association with volumes of multiple brain regions
Our study showed that the missense variant, rs13107325-T, located in SLC39A8 was associated with 28 brain regions (Table 2). These brain regions mainly belong to the DA motive and sensorimotor systems (Fig. 2), as evidenced by increased volumes of 3 DA motive system-related brain regions (red, third column) and increased volumes of the cerebellum grey matter in the sensorimotor system (orange, third column). An intron variant rs1876829-C mapped to CRHR1 was associated with volume alterations in 8 brain regions. These brain regions included frontal pole, parahippocampal gyrus, and insular cortex in CAN, and total tissue volumes (grey matter, white matter, peripheral cortical grey matter). rs1538170-T in CENPW gene was associated with the reduction of brain volume in multiple neural networks, including the grey matter of the occipital pole in visual object recognition network, planum polare in auditory object recognition network, and total tissue volumes (grey matter, white matter, grey + white matter, peripheral cortical grey matter). However, only one increased regional brain volume was associated with the CENPW gene (rs1538170-T, right: p = 1.20 × 10−13, beta = 20.454).
3.5. Single brain region is associated with multiple SNPs
Total tissue volumes (grey matter, white matter, grey + white matter, peripheral cortical grey matter) and the volume of grey matter in intracalcarine cortex were associated with multiple SNPs (Fig. 2, pale yellow and blue, third column). For example, decreased volume of the white matter region was associated with CRHR1, HMGA2, CENPW, PITPNM2 genes. The grey + white matter volume was associated with CENPW, HMGA2 and MLLT10 genes. In addition, the grey matter volume of intracalcarine cortex was associated with HHIP-AS1, NECTIN2, HHIP genes.
3.6. DA motive system
6 genetic variants were associated with the brain volumes of the DA motive system (Fig. 2). We found 2 genes (SLC39A8, JADE2) were associated with alterations in the grey matter volume of ventral striatum. SLC39A8 was associated with higher grey matter volume in bilateral ventral striatum (rs13107325-T, left: p = 6.22 × 10−103, beta = 40.955; right: p = 3.92 × 10−99, beta = 39.955), while rs329124-A in JADE2 was associated with lower grey matter volume in bilateral ventral striatum (rs329124-A, left: p = 1.83 × 10−8, beta = -5.597; right: p = 5.42 × 10−10, beta = -6.135). SLC39A8 and rs3744017-A in TRIM47 were associated with higher grey matter volume in bilateral caudate (rs13107325-T, left: p = 1.90 × 10−8, beta = 71.89; right: p = 4.49 × 10−10, beta = 83.676. rs3744017-T, left: p = 3.79 × 10−9, beta = 49.258; right: p = 9.24 × 10−10, beta = 53.697). However, the reduced volumes of bilateral caudate were associated with rs2650492-A in SBK1 in discovery cohort (Supplementary Table 6) (rs2650492-A, left: p = 2.73 × 10−9, beta = -23.029; right: p = 3.26 × 10−11, beta = -27.358), but it did not show statistical significance in the validation group (Supplementary Table 7) (rs2650492-A, left: p = 0.169, beta = -7.665; right: p = 0.08, beta = -10.377).
3.7. Central autonomic network (CAN)
6 genetic variants were associated with the brain volumes of the CAN (Fig. 2, blue, third column). Most of these obesity-related genes were associated with the increased regional brain volumes of CAN, except for HHIP-AS1 and NECTIN2. CRHR1 was associated with the multiple regional brain volumes in CAN, such as higher grey matter volumes of the right frontal pole, left insular, and left parahippocampal gyrus. Although the reduced volume of left supracalcarine grey matter was associated with rs798502-A in GNA12 in discovery cohort (Supplementary Table 6) (rs798502-A, p = 1.45 × 10−7, beta = -6.02), but it did not show statistical significance in the validation group (Supplementary Table 7) (rs798502-A, p = 0.091, beta = -2.794).
3.8. Metabolically healthy and unhealthy obesity-related SNPs were associated with brain volumes in different brain neural networks, respectively
Given that the subgroups of obesity, we classified the obesity-related variants into “metabolically healthy” variants (7 variants), “metabolically unhealthy” variants (5 variants) and unclassified obesity-related variants (5 variants). The details of classification were given in Supplementary Table 8. As shown in Fig. 2, metabolically healthy obesity-related SNPs were associated with multiple regional brain volumes of sensorimotor system, DA motive system, and cognitive emotion regulation. The volumes of CAN-related brain regions and total tissue volumes were generally associated with metabolically unhealthy obesity-related SNPs. And unclassified obesity-related variants were mainly associated with auditory object recognition network and total tissue volumes.
From discovery and validation groups, these results jointly suggested that increased volumes of the DA motive system were generally associated with metabolically healthy obesity-related genes, such as SLC39A8, TRIM47, ABCG2. Furthermore, increased regional brain volumes in the CAN were generally associated with metabolically unhealthy obesity-related genes, such as CRHR1 and HHIP. Unclassified obesity-related genes were associated with decreased total tissue volumes (grey matter, grey + white matter, peripheral cortical grey matter), such as CENPW, HMGA2, PITPNM2.
3.9. A special report of FTO-related results
Considering the importance of FTO in obesity, we performed a confirmatory analysis of 4 FTO candidate variants that have been reported in the literature. Our rs9939609-A result was not inconsistent with previous report. rs9939609-A was not associated with accumbens (Table 3, Line 1–2). Agreeing with the previous reports, rs9930333-G was not associated with ventricular cerebrospinal fluid volume and rs3751812-T was negatively associated with the left frontal orbital, occipital fusiform and the bilateral grey matter volumes in occipital pole (Table 3, Line 3–7). Contrary to the literature, rs1421085-C was negatively associated with the grey matter volume in right middle temporal gyrus, parahippocampal gyrus, and lateral occipital cortex (Table 3, Line 8–10).
Table 3.
The association of FTO-related variants and regional brain volumes.
| S.N | SNP-EA | N | EAF | Beta | SE | P.value | Brain |
|---|---|---|---|---|---|---|---|
| 1 | rs9939609-A | 19,997 | 0.391 | −0.610 | 1.029 | 0.553 | Volume of accumbens (right) |
| 2 | rs9939609-A | 19,997 | 0.391 | −0110 | 1.118 | 0.922 | Volume of accumbens (left) |
| 3 | rs9930333-G | 19,997 | 0.421 | 89.411 | 151.893 | 0.556 | Volume of ventricular cerebrospinal fluid |
| 4 | rs3751812-T | 19,997 | 0.390 | −15.821 | 6.609 | 0.017 | Volume of grey matter in Frontal Orbital Cortex (left) |
| 5 | rs3751812-T | 19,997 | 0.390 | −11.317 | 5.438 | 0.037 | Volume of grey matter in Occipital Fusiform Gyrus (left) |
| 6 | rs3751812-T | 19,997 | 0.390 | −26.919 | 12.193 | 0.027 | Volume of grey matter in Occipital Pole (left) |
| 7 | rs3751812-T | 19,997 | 0.390 | −23.758 | 11.760 | 0.043 | Volume of grey matter in Occipital Pole (right) |
| 8 | rs1421085-C | 19,997 | 0.4 | −17.181 | 7.250 | 0.018 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 9 | rs1421085-C | 19,997 | 0.4 | −24.859 | 11.191 | 0.026 | Volume of grey matter in Lateral Occipital Cortex, inferior division (right) |
| 10 | rs1421085-C | 19,997 | 0.4 | −9.546 | 3.994 | 0.017 | Volume of grey matter in Parahippocampal Gyrus, anterior division (right) |
| 11 | rs7193144-C | 19,997 | 0.391 | −16.636 | 7.285 | 0.022 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 12 | rs7193144-C | 19,997 | 0.391 | −26.429 | 12.197 | 0.03 | Volume of grey matter in Occipital Pole (left) |
| 13 | rs7193144-C | 19,997 | 0.391 | −23.248 | 11.764 | 0.048 | Volume of grey matter in Occipital Pole (right) |
| 14 | rs9939609-A | 19,997 | 0.391 | −16.633 | 7.279 | 0.022 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 15 | rs9939609-A | 19,997 | 0.391 | −27.664 | 12.187 | 0.023 | Volume of grey matter in Occipital Pole (left) |
| 16 | rs9939609-A | 19,997 | 0.391 | −24.499 | 11.755 | 0.037 | Volume of grey matter in Occipital Pole (right) |
| 17 | rs9930333-G | 19,997 | 0.421 | −18.322 | 7.194 | 0.011 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 18 | rs9930333-G | 19,997 | 0.421 | −28.794 | 12.045 | 0.017 | Volume of grey matter in Occipital Pole (left) |
| 19 | rs9930333-G | 19,997 | 0.421 | −28.980 | 11.618 | 0.013 | Volume of grey matter in Occipital Pole (right) |
| 20 | rs3751812-T | 19,997 | 0.390 | −16.557 | 7.282 | 0.023 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 21 | rs1421085-C | 19,997 | 0.4 | −17.181 | 7.250 | 0.018 | Volume of grey matter in Middle Temporal Gyrus, posterior division (right) |
| 22 | rs1421085-C | 19,997 | 0.4 | −27.379 | 12.139 | 0.024 | Volume of grey matter in Occipital Pole (left) |
| 23 | rs1421085-C | 19,997 | 0.4 | −25.703 | 11.708 | 0.028 | Volume of grey matter in Occipital Pole (right) |
SNP-EA, SNP and effect allele; N, sample number; EAF, frequency of effect allele; Beta, effect size of effect allele; P.value, the significance level of each variant.
In our exploratory analysis, 3 of 1,498 SNPs were independent FTO variants. But these FTO variants were not statistically significant after Bonferroni correction (p < 2.04 × 10−7 (0.05/164/1,498)). We further defined p < 0.05 as a potential association and found that only one SNP, rs7193144, showed potential significance in our cohorts (Table 3, Line 11–13). This FTO gene was negatively correlated with the grey matter volumes of the occipital pole and middle temporal gyrus belonging to the visual object recognition network.
3.10. Pathways shared between obesity and brain volumes
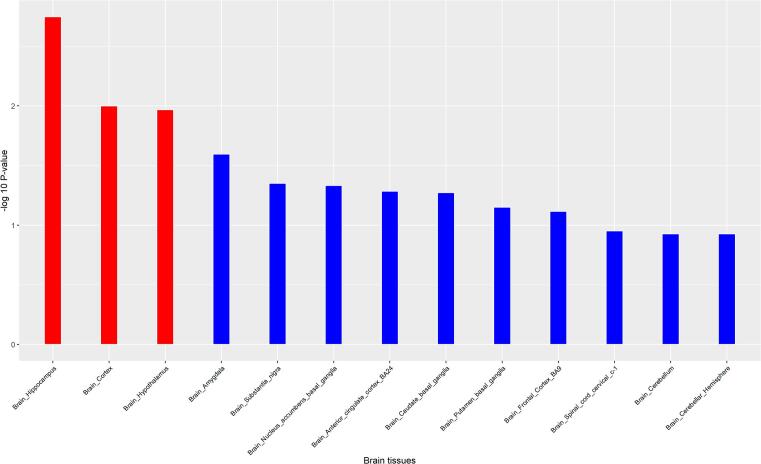
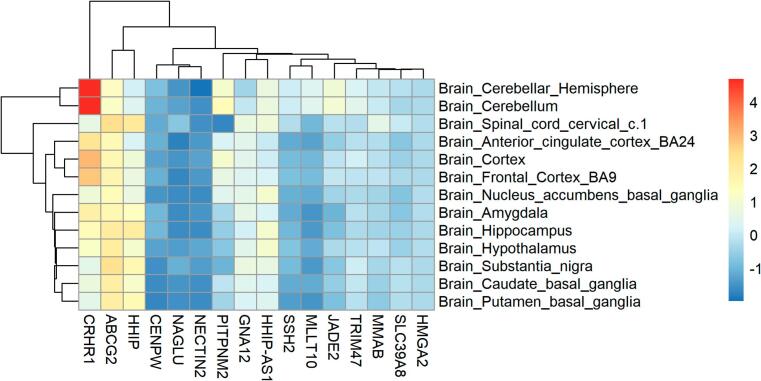
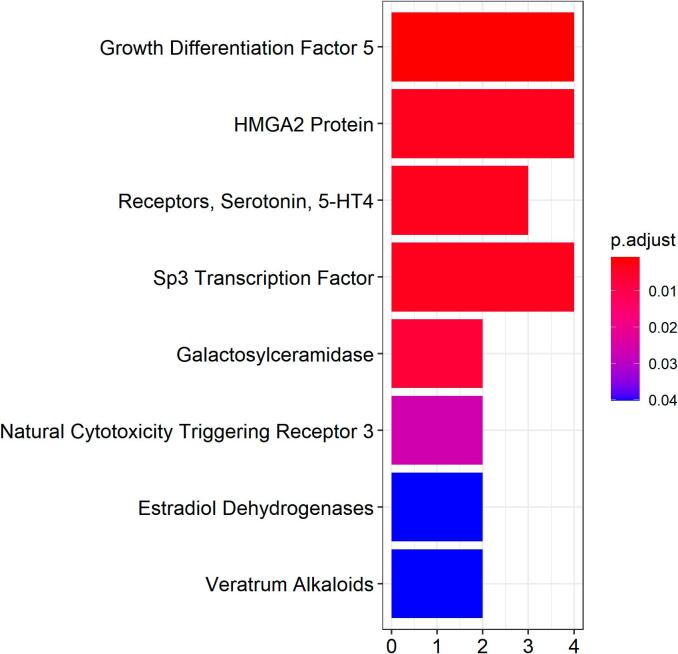
Among the 17 independent genes, the H2BC7 gene was excluded from the gene expression analysis, because FUMA does not have its gene expression information. Then FUMA and R packages were used to analyze the gene expressions of these 16 genes in 13 normal brain tissues (Fig. 3). The figure showed the significant enrichment in the hippocampus, brain cortex, hypothalamus. Gene expression heatmap (Fig. 4) showed that several genes, such as CRHR1, ABCG2, HHIP were highly expressed in multiple brain tissues. The clustering relationships were shown to the heat map, CRHR1 was highly expressed mainly in the cerebellum and cerebellar hemisphere, ABCG2 was highly expressed mainly in grey matter nuclei (substantia nigra, putamen, caudate, amygdala). Some genes showed low expression in multiple brain tissues, such as HMGA2, CENPW and SLC39A8. MeSH enrichment analysis showed that our 17 genes were enriched in Chemicals and Drugs (Fig. 5), such as GDF5 (HMGA2, CENPW, GNA12, HHIP), HMGA2 Protein (HMGA2, CENPW, GNA12, HHIP), and 5-HT4 (CRHR1, HHIP, HHIP-AS1).
Fig. 3.
Tissue expression profile of obesity genes in 13 regional brain tissues.
Fig. 4.
Gene expression of regional brain. x-axis: independent genes in our result; y-axis: 13 regional brain tissues.
Fig. 5.
MeSH enrichment analysis of 17 obesity genes in Chemicals and Drugs.
4. Discussion
In this study, we identified 17 obesity-related SNPs associated with brain structural alterations from a large 29,420 participants of European ancestry. The present study showed that specific SNPs may have an association with volumes of multiple brain regions. For example, SLC39A8 was associated with alterations in the volumes of 28 brain regions. Meanwhile, one single brain region volume may have an association with multiple SNPs. For instance, the reduced white matter volume was observed to be associated with 4 SNPs.
As seen from Fig. 2, the strongest association was found for SLC39A8 (rs13107325-T). SLC39A8 was associated with the DA motive system and sensorimotor system. According to GWAS results, the SLC39A8 gene was reported as a candidate gene for the development of subcortical brain structures, and the effect of rs13107325 was found in the caudate, putamen, ventral striatum, and cerebellar contribution (lobules VI–X) (Elliott et al., 2018, Satizabal et al., 2019). This is consistent with what we observed. SLC39A8 is a shared gene between the obesity and brain, and the relationship between obesity variants and brain volumes was strongly validated in our research.
FTO gene is a well-characterized obesity gene and has also been reported to be associated with brain region volume (Melka et al., 2013). Firstly, we tried to verify the previously reported variants in our discovery cohort. We noticed that the inconsistent associations of FTO SNPs with regional brain volumes were reported in adolescents or children (Rapuano et al., 2017, Lugo‐Candelas et al., 2020), while the consistent association was reported in elder people (Ho et al., 2010). We speculate that the effect of FTO gene on regional brain volumes may interact with age. In our exploratory analysis, the FTO SNPs results did not pass the Bonferroni correction. The reason may lie in that our research has included a large number of obesity variants, increasing the number of multiple testing. Only one SNP was in the potential associations (p < 0.05) in our results. This rs7193144 and 4 variants reported in the literature are in linkage disequilibrium (rs9939609, r2 = 0.98; rs9930333, r2 = 0.87; rs3751812, r2 = 0.93; rs1421085, r2 = 0.96) and have relationships with similar regional brain volumes. For instance, all 5 SNPs were associated with the volumes of multiple brain regions in visual object recognition (middle temporal gyrus, occipital pole) (Table 3, Line 6,7,11–23). Findings of previous literature report support this viewpoint (de Groot, 2015, Ho et al., 2010, Gilbert-Diamond et al., 2017). For example, a study concludes that children with FTO rs9939609-A are more likely to respond to food cues (viewing food advertisement) and may cue overeating (Gilbert-Diamond et al., 2017).
Previous studies have shown that dysregulation of the DA motive system contributes to addiction and obesity (Volkow et al., 2017, Wang et al., 2001, Kroemer and Small, 2016, Stice and Burger, 2019). Some scholars attempted to integrate the existing obesity theories of dysfunctional DA motive system into a complete etiological model, known as the dynamic vulnerability model (Stice and Burger, 2019). Individuals with greater innate reward region responsiveness to high-calorie food receipt were predisposed towards overeating (reward surfeit theory). And increased responsiveness of the reward area to food cues can predict future weight gain (incentive sensitization theory) (Stice and Burger, 2019). Individual differences in striatum (nucleus accumbens) of DA motive system responsiveness to food cues may be one of the main reasons for obese individuals to show addiction-like behavior resulting in weight gain (Demos et al., 2012). Overweight and obese women (BMI:26–38 kg/m2) had larger nucleus accumbens volume and connectivity (Coveleskie et al., 2015). Moreover, children at risk of obesity had larger nucleus accumbens volume and were more responsive to food cues (Rapuano et al., 2017). In our results, metabolically healthy obesity-related variants, rs13107325-T, rs3744017-A and rs1481012-A were associated with the increased grey matter volumes of putamen, ventral striatum and caudate in the DA motive system. These results were in line with the dynamic vulnerability model. The results, however, are heterogeneous. Severe obese (BMI: 42–60 kg/m2) showed reduced striatal dopamine D2 receptor availability, and their BMI was negatively associated with striatal dopamine D2 receptor availability (Wang et al., 2001). Although this is an observational study, the authors speculate that dopamine deficiency (reward deficit theory of obesity) in obese individuals may lead to pathological eating as a means of compensating for those circuits with reduced activation (Wang et al., 2001). However, prospective fMRI studies or experimental studies seem to not support the theory that people whose reward circuits are weakened in response to food stimuli tend to eat more to compensate for the reward deficit (Stice and Burger, 2019). In addition, some authors hold the hypothesis that decreased responses to energy-dense food reflect reduced reward-related learning rather than reward deficiency (Kroemer and Small, 2016).
Given the lack of support for the reward deficit theory, we gathered some hints through the following animal experiments and human studies. Animal experiments showed that the expression of dopamine receptor changes after long-term exposure to palatable food and obesity (Geiger et al., 2009, Alsiö et al., 2010). 5 weeks of access to a high-fat high-sugar diet versus chow-fed controls resulted in down-regulation of striatal dopamine receptor in rats and this effect extends well into high-fat high-sugar diet withdrawal phase (Alsiö et al., 2010). In addition, obese mice induced by 15 weeks cafeteria-diet fed exhibited lower extracellular accumbens dopamine levels and an attenuated dopamine response than normal weight rats (Geiger et al., 2009). However, dopamine receptor availability is reversed after weight loss induced by bariatric gastric bypass surgery in five female subjects (Steele et al., 2010). And the increased dopamine receptor availability was almost proportional to the amount of weight lost (Steele et al., 2010). All of the evidence supports the hypothesis that DA availability is influenced by a period of exposure to eating habits and obesity status. Moreover, increased nucleus accumbens volume at young age and decreased volume at older age also reflect the effect of persistent obesity on brain structure (García‐García et al., 2020). Metabolically unhealthy obesity is an outcome of long-term maintenance of obesity (Stice and Burger, 2019). In our results, metabolically unhealthy obesity-related variant, rs591939-G, was associated with decreased grey matter volume of putamen in the DA system. Thus, we speculate that reward deficit may be secondary damage caused by a period of exposure to habitually high energy intake or the persistent state of obesity. However, this is only speculation and needs to be further confirmed by prospective longitudinal studies.
Notably, we found that volume alterations in many brain regions such as the grey matter in the frontal pole, insular cortex, and parahippocampal gyrus, were associated with metabolically unhealthy obesity-related SNPs, but were rarely significantly associated with metabolically healthy obesity-related SNPs. These brain regions are part of the CAN, which regulates blood pressure (insular) (Kitamura et al., 2018), blood glucose or insulin (frontal pole, parahippocampal) (Rosenthal et al., 2001, Byun, 2019), and heart function (insular, parahippocampal) (Chouchou et al., 2019, Meguro et al., 2017). Thus, the associations between metabolically unhealthy obesity-related SNPs and alterations in the volumes of CAN regions could explain a higher risk of cardiovascular diseases or metabolic burden observed in metabolically unhealthy obesity.
Several mechanisms may be involved in these obesity-related brain morphological alternations, although the nature of this exploratory analysis does not allow us to draw any conclusions in this respect. But the enrichment analysis still gives us some hints. We found many drugs and chemicals related to obesity genes through MeSH enrichment analysis and we analyzed the first 3 important findings. GDF5 promotes brown adipogenesis in vitro and in adipose GDF5 overexpressing mice (Hinoi et al., 2014). It can contribute to protection against high fat diet-induced obesity (Lee, 2018). In addition, GDF5 was found to be expressed in the developing and adult rat brain and was related to neuronal development and differentiation, especially the DA neuron (O'Keeffe et al., 2004). It plays a role in the development and maintenance of DA neurones and protects DA neurones from degeneration (O'Keeffe et al., 2004, Hanson et al., 2012). In conclusion, we speculate that GDF5 can protect against high fat diet-induced obesity and has a beneficial effect on the DA system. The second MeSHes result, HMGA2 is a DNA-binding protein and is involved in adipogenesis (Su et al., 2020). The expression of HMGA2 is up-regulated in obese people (Markowski et al., 2013). The fat cells were severely depleted when HMGA2 was knocked out in mice (Zhou et al., 1995). HMGA2 (rs10784502-T) also has known roles in total brain volume through genetic association (Stein et al., 2012). The rs10784502 and HMGA2 SNP in our results (Fig. 2) are in the same linkage disequilibrium block (rs7970350, r2 = 0.96). They showed similar negative associations with total brain volume or grey + white matter (Stein et al., 2012). Although HMGA2 is low expressed in the brain, it may affect brain structure by a role in peripheral tissues. The underlying mechanism of HMGA2 warrants further study. The third MeSHes result, 5-HT is a monoamine neurotransmitter and is considered as a satiety-generating signal which regulates food intake in both experimental models and human subjects (Ratner et al., 2012, Haahr et al., 2012). A study has found a positive association between BMI and the 5-HT4 receptor density in many brain regions, such as nucleus accumbens, pallidum, hippocampal, and frontal orbital cortex (Haahr et al., 2012). Gene expression heatmap (Fig. 5) showed that the 5-HT4 related genes (HHIP, CRHR1) in our MeSH enrichment analysis results were highly expressed in the brain regions of the DA motive system. Consistent with the previous literature reports, we found that 5-HT4 is critically involved in DA motive (Haahr et al., 2012, Jean et al., 2007). Currently, 5-HT4 receptor agonist has been found to reduce motivation for eating in obese mice (Ratner et al., 2012). In addition, CRHR1 is the receptor of corticotropin-releasing hormone (CRH), which plays an important role in the HPA-axis resulting in obesity. In previous studies, it has been found that patients with obesity and metabolic syndrome often have subtle, chronic HPA-axis hyperactivity, and this central regulatory disorder of CRH can be treated with a CRHR1 antagonist (Contoreggi et al., 2004).
A strength of our study was the extensive number of candidate obesity-related variants and the large sample size, and the results of this study are likely to be representative. In addition, this analysis was adjusted for many important confounders, the results in discovery cohort were nearly sustained in validation cohort. At last, we got some further biological information.
Several limitations should be addressed. Our study only focused on the association between brain structure and obesity-related variants, and did not observe the association between obesity-related traits (e.g., BMI, WHR, WC, HC) and brain volumes, and did not draw any causal inferences. In addition, some brain regions that may be related to obesity are missing in our study, such as hypothalamus or the prefrontal lobe.
5. Conclusion
In summary, we found that 17 obesity-related SNPs were associated with different brain volume measures. We found that metabolically healthy and unhealthy obesity-related SNPs were associated with regions from different brain neural networks. Previously reported associations of FTO and brain volumes were confirmed for rs3751812-T and rs9930333-G. Based on our enrichment analysis, modifications of the 5-HT4 pathway might be a promising therapeutic strategy for obesity.
Funding support
This work was supported by the “Changbai Mountain Scholar” Distinguished Professor Awarding Program of the Department of Education of Jilin Province, China.
Author contributions
Xingchen Pan: Formal analysis, Investigation, Writing – original draft. Miaoran Zhang: Software, Data curation. Aowen Tian: Visualization. Lanlan Chen: Writing – review & editing. Zewen Sun: Writing – review & editing. Liying Wang: Supervision. Peng Chen: Conceptualization, Methodology, Software, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2021.102870.
Contributor Information
Liying Wang, Email: wlying@jlu.edu.cn.
Peng Chen, Email: pchen@jlu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Collaboration N.C.D.R.F. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390(10113):2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden L., Ezzati M., Larijani B., Dietz W. The challenge for global health systems in preventing and managing obesity. Obes. Rev. 2019;20(S2):185–193. doi: 10.1111/obr.12872. [DOI] [PubMed] [Google Scholar]
- Banks W.A. Is obesity a disease of the blood-brain barrier? Physiological, pathological, and evolutionary considerations. Curr. Pharm. Des. 2003;9(10):801–809. doi: 10.2174/1381612033455350. [DOI] [PubMed] [Google Scholar]
- Shefer G., Marcus Y., Stern N. Is obesity a brain disease? Neurosci. Biobehav. Rev. 2013;37(10):2489–2503. doi: 10.1016/j.neubiorev.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Small D.M., Jones-Gotman M., Zatorre R.J., Petrides M., Evans A.C. Flavor processing: more than the sum of its parts. NeuroReport. 1997;8(18):3913–3917. doi: 10.1097/00001756-199712220-00014. [DOI] [PubMed] [Google Scholar]
- Kuhne S.G., Stengel A. Alteration of peptidergic gut-brain signaling under conditions of obesity. J. Physiol. Pharmacol. 2019;70(5) doi: 10.26402/jpp.2019.5.01. [DOI] [PubMed] [Google Scholar]
- McDougle M., Quinn D., Diepenbroek C., Singh A., de la Serre C., de Lartigue G. Intact vagal gut-brain signalling prevents hyperphagia and excessive weight gain in response to high-fat high-sugar diet. Acta Physiol. (Oxf) 2021;231(3) doi: 10.1111/apha.2021.231.issue-310.1111/apha.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raka F., Farr S., Kelly J., Stoianov A., Adeli K. Metabolic control via nutrient-sensing mechanisms: role of taste receptors and the gut-brain neuroendocrine axis. Am. J. Physiol. Endocrinol. Metab. 2019;317(4):E559–E572. doi: 10.1152/ajpendo.00036.2019. [DOI] [PubMed] [Google Scholar]
- Smith R., Alkozei A., Killgore W.D.S. Conflict-related dorsomedial frontal cortex activation during healthy food decisions is associated with increased cravings for high-fat foods. Brain Imaging Behav. 2018;12(3):685–696. doi: 10.1007/s11682-017-9726-7. [DOI] [PubMed] [Google Scholar]
- Covasa M., Grahn J., Ritter R.C. Reduced hindbrain and enteric neuronal response to intestinal oleate in rats maintained on high-fat diet. Auton Neurosci. 2000;84(1-2):8–18. doi: 10.1016/S1566-0702(00)00176-4. [DOI] [PubMed] [Google Scholar]
- Pannacciulli N., Del Parigi A., Chen K., Le D.S.N.T., Reiman E.M., Tataranni P.A. Brain abnormalities in human obesity: a voxel-based morphometric study. Neuroimage. 2006;31(4):1419–1425. doi: 10.1016/j.neuroimage.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Dekkers I.A., Jansen P.R., Lamb H.J. Obesity, brain volume, and white matter microstructure at MRI: a cross-sectional UK biobank study. Radiology. 2019;291(3):763–771. doi: 10.1148/radiol.2019181012. [DOI] [PubMed] [Google Scholar]
- Taki, Y., et al., Relationship between body mass index and gray matter volume in 1,428 healthy individuals. Obesity (Silver Spring), 2008. 16(1): p. 119-124. [DOI] [PubMed]
- Kennedy J.T., Astafiev S.V., Golosheykin S., Korucuoglu O., Anokhin A.P. Shared genetic influences on adolescent body mass index and brain structure: a voxel-based morphometry study in twins. Neuroimage. 2019;199:261–272. doi: 10.1016/j.neuroimage.2019.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji C.A., et al. Brain structure and obesity. Hum. Brain Mapp. 2010;31(3):353–364. doi: 10.1002/hbm.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer M., Batty G.D. Association of body mass index and waist-to-hip ratio with brain structure: UK Biobank study. Neurology. 2019;92(6):e594–e600. doi: 10.1212/WNL.0000000000006879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstmann A., Kovacs P., Kabisch S., Boettcher Y., Schloegl H., Tönjes A., Stumvoll M., Pleger B., Villringer A., Esteban F.J. Common genetic variation near MC4R has a sex-specific impact on human brain structure and eating behavior. PLoS ONE. 2013;8(9):e74362. doi: 10.1371/journal.pone.0074362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson D., Lissner L., Bengtsson C., Bjorkelund C., Skoog I. A 24-year follow-up of body mass index and cerebral atrophy. Neurology. 2004;63(10):1876–1881. doi: 10.1212/01.wnl.0000141850.47773.5f. [DOI] [PubMed] [Google Scholar]
- Hare T.A., Camerer C.F., Rangel A. Self-control in decision-making involves modulation of the vmPFC valuation system. Science. 2009;324(5927):646–648. doi: 10.1126/science.1168450. [DOI] [PubMed] [Google Scholar]
- Locke A.E., Kahali B., Berndt S.I., Justice A.E., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou I., Astrakas L.G., Xydis V., Alexiou G.A., Bargiotas P., Tzarouchi L., Zikou A.K., Kiortsis D.N., Argyropoulou M.I. Abnormalities of brain neural circuits related to obesity: a diffusion tensor imaging study. Magn. Reson. Imaging. 2017;37:116–121. doi: 10.1016/j.mri.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Vainik U., Baker T.E., Dadar M., Zeighami Y., Michaud A., Zhang Y.u., García Alanis J.C., Misic B., Collins D.L., Dagher A. Neurobehavioral correlates of obesity are largely heritable. Proc. Natl. Acad. Sci. U.S.A. 2018;115(37):9312–9317. doi: 10.1073/pnas.1718206115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling T.M., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapuano K.M., Zieselman A.L., Kelley W.M., Sargent J.D., Heatherton T.F., Gilbert-Diamond D. Genetic risk for obesity predicts nucleus accumbens size and responsivity to real-world food cues. Proc. Natl. Acad. Sci. U.S.A. 2017;114(1):160–165. doi: 10.1073/pnas.1605548113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot C., et al. Association of the fat mass and obesity-associated gene risk allele, rs9939609A, and reward-related brain structures. Obesity (Silver Spring) 2015;23(10):2118–2122. doi: 10.1002/oby.21191. [DOI] [PubMed] [Google Scholar]
- Lugo‐Candelas C., Pang Y., Lee S., Cha J., Hong S., Ranzenhofer L., Korn R., Davis H., McInerny H., Schebendach J., Chung W.K., Leibel R.L., Walsh B.T., Posner J., Rosenbaum M., Mayer L. Differences in brain structure and function in children with the FTO obesity-risk allele. Obes Sci Pract. 2020;6(4):409–424. doi: 10.1002/osp4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.J., Stein J.L., Hua X., Lee S., Hibar D.P., et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci U S A. 2010;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melka M.G., et al. FTO, obesity and the adolescent brain. Hum. Mol. Genet. 2013;22(5):1050–1058. doi: 10.1093/hmg/dds504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer F., Zhang R., Scholz M., Wirkner K., Loeffler M., Stumvoll M., Villringer A., Witte A.V. Higher BMI, but not obesity-related genetic polymorphisms, correlates with lower structural connectivity of the reward network in a population-based study. Int. J. Obes. (Lond) 2021;45(3):491–501. doi: 10.1038/s41366-020-00702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise C.M., Bachmann T., Pleger B. Brain structural differences in monozygotic twins discordant for body mass index. Neuroimage. 2019;201 doi: 10.1016/j.neuroimage.2019.07.019. [DOI] [PubMed] [Google Scholar]
- Cole J.H., Boyle C.P., Simmons A., Cohen-Woods S., Rivera M., McGuffin P., Thompson P.M., Fu C.H.Y. Body mass index, but not FTO genotype or major depressive disorder, influences brain structure. Neuroscience. 2013;252:109–117. doi: 10.1016/j.neuroscience.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Mulugeta A., Lumsden A., Hyppönen E. Unlocking the causal link of metabolically different adiposity subtypes with brain volumes and the risks of dementia and stroke: a Mendelian randomization study. Neurobiol. Aging. 2021;102:161–169. doi: 10.1016/j.neurobiolaging.2021.02.010. [DOI] [PubMed] [Google Scholar]
- Martin S., et al. Genetic evidence for different adiposity phenotypes and their opposing influence on ectopic fat and risk of cardiometabolic disease. Diabetes. 2021 doi: 10.2337/db21-0129. [DOI] [PubMed] [Google Scholar]
- Winkler T.W., Günther F., Höllerer S., Zimmermann M., Loos R.JF., Kutalik Z., Heid I.M. A joint view on genetic variants for adiposity differentiates subtypes with distinct metabolic implications. Nat. Commun. 2018;9(1) doi: 10.1038/s41467-018-04124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G., Leslie S., Allen N., Donnelly P., Marchini J. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J.L., M. Jenkinson, and S. Smith, Non-linear registration, aka spatial normalisation. FMRIB technial report TR07JA2, 2007. 22.
- Anderson, J., M. Jenkinson, and S. Smith, Non-Linear Optimisation: FMRIB Technical Report TR07JA1. 2007.
- Patenaude B., Smith S.M., Kennedy D.N., Jenkinson M. A bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56(3):907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Alexander-Bloch A., Clasen L., Stockman M., Ronan L., Lalonde F., Giedd J., Raznahan A. Subtle in-scanner motion biases automated measurement of brain anatomy from in vivo MRI. Hum. Brain Mapp. 2016;37(7):2385–2397. doi: 10.1002/hbm.23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott L.T., Sharp K., Alfaro-Almagro F., Shi S., Miller K.L., Douaud G., Marchini J., Smith S.M. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562(7726):210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics, 2018. 34(21): p. 3766-3767. [DOI] [PubMed]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Zheng Z., Qi T., Kemper K.E., Wray N.R., Visscher P.M., Yang J. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019;51(12):1749–1755. doi: 10.1038/s41588-019-0530-8. [DOI] [PubMed] [Google Scholar]
- Satizabal C.L., Adams H.H.H., Hibar D.P., White C.C., Knol M.J., et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat. Genet. 2019;51(11):1624–1636. doi: 10.1038/s41588-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert-Diamond D., Emond J.A., Lansigan R.K., Rapuano K.M., Kelley W.M., Heatherton T.F., Sargent J.D. Television food advertisement exposure and FTO rs9939609 genotype in relation to excess consumption in children. Int J Obes (Lond) 2017;41(1):23–29. doi: 10.1038/ijo.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Wise R.A., Baler R. The dopamine motive system: implications for drug and food addiction. Nat. Rev. Neurosci. 2017;18(12):741–752. doi: 10.1038/nrn.2017.130. [DOI] [PubMed] [Google Scholar]
- Wang G.-J., Volkow N.D., Logan J., Pappas N.R., Wong C.T., Zhu W., Netusll N., Fowler J.S. Brain dopamine and obesity. Lancet. 2001;357(9253):354–357. doi: 10.1016/s0140-6736(00)03643-6. [DOI] [PubMed] [Google Scholar]
- Kroemer N.B., Small D.M. Fuel not fun: Reinterpreting attenuated brain responses to reward in obesity. Physiol. Behav. 2016;162:37–45. doi: 10.1016/j.physbeh.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice E., Burger K. Neural vulnerability factors for obesity. Clin Psychol Rev. 2019;68:38–53. doi: 10.1016/j.cpr.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demos K.E., Heatherton T.F., Kelley W.M. Individual differences in nucleus accumbens activity to food and sexual images predict weight gain and sexual behavior. J. Neurosci. 2012;32(16):5549–5552. doi: 10.1523/JNEUROSCI.5958-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K. Coveleskie A. Gupta L.A. Kilpatrick E.D. Mayer C. Ashe-McNalley J. Stains J.S. Labus E.A. Mayer Altered functional connectivity within the central reward network in overweight and obese women Nutr Diabetes 5 1 2015 e148 e148. [DOI] [PMC free article] [PubMed]
- Geiger B.M., Haburcak M., Avena N.M., Moyer M.C., Hoebel B.G., Pothos E.N. Deficits of mesolimbic dopamine neurotransmission in rat dietary obesity. Neuroscience. 2009;159(4):1193–1199. doi: 10.1016/j.neuroscience.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsiö J., Olszewski P.K., Norbäck A.H., Gunnarsson Z.E.A., Levine A.S., Pickering C., Schiöth H.B. Dopamine D1 receptor gene expression decreases in the nucleus accumbens upon long-term exposure to palatable food and differs depending on diet-induced obesity phenotype in rats. Neuroscience. 2010;171(3):779–787. doi: 10.1016/j.neuroscience.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Steele K.E., Prokopowicz G.P., Schweitzer M.A., Magunsuon T.H., Lidor A.O., Kuwabawa H., Kumar A., Brasic J., Wong D.F. Alterations of central dopamine receptors before and after gastric bypass surgery. Obes. Surg. 2010;20(3):369–374. doi: 10.1007/s11695-009-0015-4. [DOI] [PubMed] [Google Scholar]
- García‐García I., Morys F., Dagher A. Nucleus accumbens volume is related to obesity measures in an age-dependent fashion. J. Neuroendocrinol. 2020;32(12) doi: 10.1111/jne.v32.1210.1111/jne.12812. [DOI] [PubMed] [Google Scholar]
- Kitamura J., Ueno H., Nagai M., Hosomi N., Honjo K., Nakamori M., Mukai T., Imamura E., Nezu T., Aoki S., Ohshita T., Nomura E., Wakabayashi S., Maruyama H., Matsumoto M. Blood pressure variability in acute ischemic stroke: influence of infarct location in the insular cortex. Eur. Neurol. 2018;79(1-2):90–99. doi: 10.1159/000486306. [DOI] [PubMed] [Google Scholar]
- Rosenthal J.M., Amiel S.A., Yaguez L., Bullmore E., Hopkins D., Evans M., Pernet A., Reid H., Giampietro V., Andrew C.M., Suckling J., Simmons A., Williams S.C.R. The effect of acute hypoglycemia on brain function and activation: a functional magnetic resonance imaging study. Diabetes. 2001;50(7):1618–1626. doi: 10.2337/diabetes.50.7.1618. [DOI] [PubMed] [Google Scholar]
- Byun M.S., et al. Region-specific association between basal blood insulin and cerebral glucose metabolism in older adults. Neuroimage Clin. 2019;22 doi: 10.1016/j.nicl.2019.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchou F., Mauguière F., Vallayer O., Catenoix H., Isnard J., Montavont A., Jung J., Pichot V., Rheims S., Mazzola L. How the insula speaks to the heart: Cardiac responses to insular stimulation in humans. Hum. Brain Mapp. 2019;40(9):2611–2622. doi: 10.1002/hbm.24548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro T., Meguro Y., Kunieda T. Atrophy of the parahippocampal gyrus is prominent in heart failure patients without dementia. ESC Heart Fail. 2017;4(4):632–640. doi: 10.1002/ehf2.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E., Nakamura Y., Takada S., Fujita H., Iezaki T., Hashizume S., Takahashi S., Odaka Y., Watanabe T., Yoneda Y. Growth differentiation factor-5 promotes brown adipogenesis in systemic energy expenditure. Diabetes. 2014;63(1):162–175. doi: 10.2337/db13-0808. [DOI] [PubMed] [Google Scholar]
- Lee, M.J., Transforming growth factor beta superfamily regulation of adipose tissue biology in obesity. Biochim Biophys Acta Mol Basis Dis, 2018. 1864(4 Pt A): p. 1160-1171. [DOI] [PubMed]
- O'Keeffe G.W., Hanke M., Pohl J., Sullivan A.M. Expression of growth differentiation factor-5 in the developing and adult rat brain. Brain Res. Dev. Brain Res. 2004;151(1-2):199–202. doi: 10.1016/j.devbrainres.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Hanson L.R., Fine J.M., Hoekman J.D., Nguyen T.M., Burns R.B., Martinez P.M., Pohl J., Frey W.H. Intranasal delivery of growth differentiation factor 5 to the central nervous system. Drug Deliv. 2012;19(3):149–154. doi: 10.3109/10717544.2012.657720. [DOI] [PubMed] [Google Scholar]
- Su L., Bryan N., Battista S., Freitas J., Garabedian A., D’Alessio F., Romano M., Falanga F., Fusco A., Kos L., Chambers J., Fernandez-Lima F., Chapagain P.P., Vasile S., Smith L., Leng F. Identification of HMGA2 inhibitors by AlphaScreen-based ultra-high-throughput screening assays. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-75890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowski D.N., Thies H.W., Gottlieb A., Wenk H., Wischnewsky M., Bullerdiek J. HMGA2 expression in white adipose tissue linking cellular senescence with diabetes. Genes Nutr. 2013;8(5):449–456. doi: 10.1007/s12263-013-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Benson K.F., Ashar H.R., Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376(6543):771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- Stein J.L., Medland S.E., Vasquez A.A., Hibar D.P., Senstad R.E., et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 2012;44(5):552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2020; Available from: https://www.who.int/news-room/facts-in-pictures/detail/6-facts-on-obesity.
- Ratner, C., et al., Cerebral markers of the serotonergic system in rat models of obesity and after Roux-en-Y gastric bypass. Obesity (Silver Spring), 2012. 20(10): p. 2133-41. [DOI] [PMC free article] [PubMed]
- Haahr M.E., Rasmussen P.M., Madsen K., Marner L., Ratner C., Gillings N., Baaré W.F.C., Knudsen G.M. Obesity is associated with high serotonin 4 receptor availability in the brain reward circuitry. Neuroimage. 2012;61(4):884–888. doi: 10.1016/j.neuroimage.2012.03.050. [DOI] [PubMed] [Google Scholar]
- Jean A., Conductier G., Manrique C., Bouras C., Berta P., Hen R., Charnay Y., Bockaert J., Compan V. Anorexia induced by activation of serotonin 5-HT4 receptors is mediated by increases in CART in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 2007;104(41):16335–16340. doi: 10.1073/pnas.0701471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contoreggi C., Rice K.C., Chrousos G. Nonpeptide corticotropin-releasing hormone receptor type 1 antagonists and their applications in psychosomatic disorders. Neuroendocrinology. 2004;80(2):111–123. doi: 10.1159/000081785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.