Abstract
The recent torrent of structures of chromatin complexes determined by cryoelectron microscopy provides an opportunity to discern general principles for how chromatin factors and enzymes interact with their nucleosome substrate. We find that many chromatin proteins employ strikingly similar arginine anchor and variant arginine interactions to bind to the nucleosome acidic patch. We also observe that many chromatin proteins target the H3 and H2B histone fold α1-loop1 elbows and the H2B C-terminal helix on the nucleosomal histone face. These interactions with the histones can be complemented with interactions with and distortions of nucleosomal DNA.
Keywords: nucleosome, chromatin, histone, acidic patch, arginine anchor, structural biology
Introduction
When we reviewed how chromatin factors and enzymes recognize the nucleosome in 2016 [1], there were published atomic structures for only 6 proteins or peptides bound to the nucleosome, all determined by X-ray crystallography, and one docking model based on low resolution cryoelectron microscopy (cryo-EM) data [2–8]. Five years later, the resolution revolution in cryo-EM has produced an explosion of new chromatin complex structures. It is therefore an opportune time to examine the more than 40 unique chromatin complex structures now available to identify general principles for how chromatin factors and enzymes recognize the nucleosome.
Our 2016 review highlighted two general principles for nucleosome recognition: multivalent interactions and the arginine anchor paradigm. We noted that chromatin factors and enzymes often interacted with multiple subunits of the nucleosome across multiple nucleosome surfaces. We also observed that 5 of the then 7 available chromatin complex structures employed an arginine residue from the chromatin protein to engage the histone H2A-H2B dimer acidic patch on the nucleosome surface in essentially an identical manner. We termed this the arginine anchor nucleosome binding motif [5]. The availability of many new chromatin complex structures confirms these two key principles of multivalency and the arginine anchor, but also allows us to identify new structural paradigms for interactions with both the histone and DNA components of the nucleosome (Fig. 1).
Fig. 1:
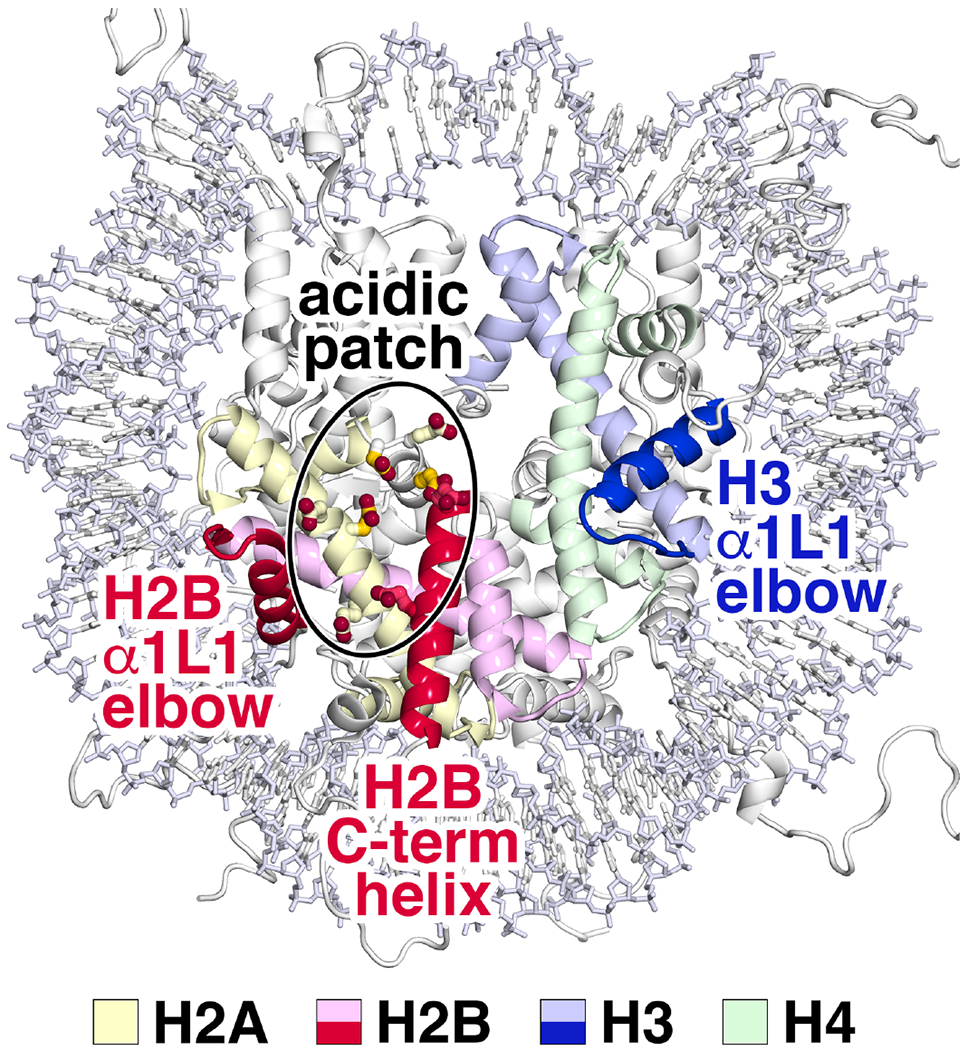
Nucleosome features targeted by chromatin proteins. The nucleosome acidic patch created by histone H2A and H2B acidic residues (encircled) is frequently contacted by chromatin protein arginine residues. In addition, the histone H3 α1L1 elbow region of the histone fold (blue), the corresponding histone H2B α1L1 elbow (red) and the histone H2B C-terminal helix are sites of interaction by multiple chromatin proteins. The H2A, H2B, H3 and H4 core histones are shown in shades of yellow, red, blue and green respectively. The carbon side chain atoms of H2A acidic patch residues E61, D90 and E92 are shown in a more saturated yellow color.
The arginine anchor and variant arginines interact with the acidic patch
Early nucleosome complex structures exposed the H2A-H2B acidic patch as a hot spot for nucleosome recognition based on the shared motif of interaction, the arginine anchor [9,10]. Despite this emerging paradigm of nucleosome recognition, it was unclear how many nucleosome binding proteins required the acidic patch for nucleosome engagement and whether other hot spots existed and remained undiscovered. Recent studies have demonstrated the pervasiveness of acidic patch binding proteome-wide and heightened our understanding of the molecular details underlying acidic patch interactions. The nucleosome acidic patch is formed by an evolutionarily conserved cluster of acidic amino acids in histones H2A (E56, E61, E64, D90, E91, and E92) and H2B (E105, E113) [11]. These acidic residues line a depression that is split into two halves by a low ridge (Fig. 2a). In 2020, one of us (R.K.M.) reported an unbiased proteomic analysis of the nucleosome interactome to establish the fundamental principles governing recognition of the nucleosome disk surface [12]. Consistent with early structural studies, the region centered on the acidic patch, including the surface that interacts with the arginine anchor, emerged as a clear hot spot, contributing to nucleosome binding by the majority of proteins overall and more than 90% of the subset of proteins that interact with the nucleosome disk face in this study.
Fig. 2:
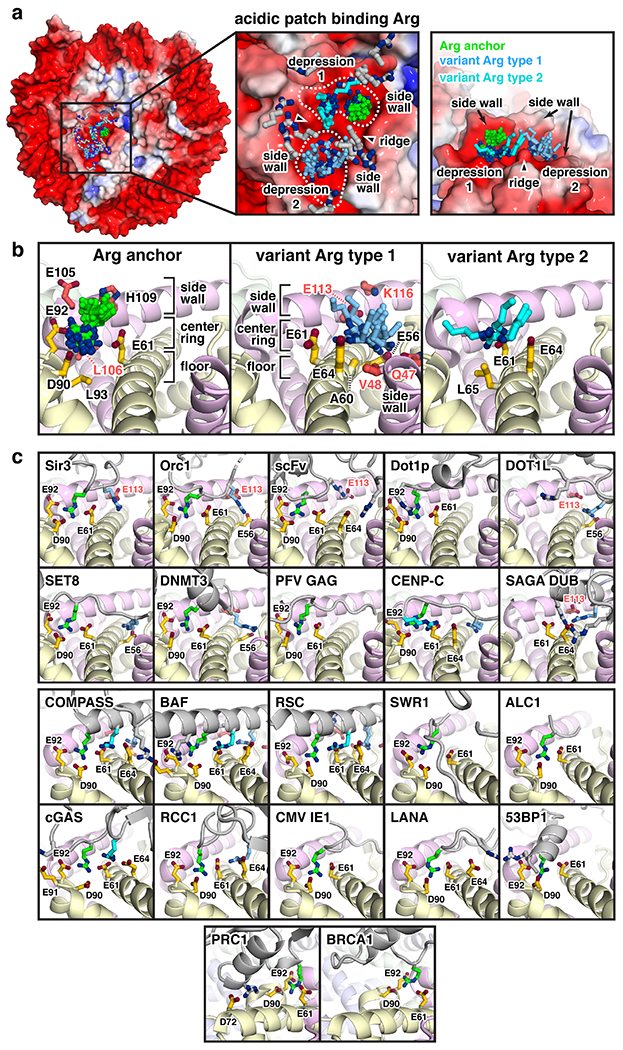
Arginine-acidic patch interactions. (a) Left, electrostatic surface disk view of nucleosome generated with APBS (PDB id 3MVD [3]) overlaid with all chromatin protein acidic patch-binding arginines colored by type – arginine anchors (green), type 1 variant arginines (light blue), type 2 variant arginines (cyan), and atypical variant arginines (gray). Center, zoomed top view with atypical variant arginines from Dot1L (PDB id 6NJ9; R278 [21]), SAGA DUB (PDB id 4ZUX; R91 [44]), and COMPASS (PDB id 6VEN; R936 [27]) omitted for clarity. Right, zoomed side view of all arginine anchors and type 1 and 2 variant arginines. (b) All arginine anchors (left), type 1 variant arginines (center), and type 2 variant arginines (right) with nearby histone residues annotated. (c) Interactions of chromatin proteins (gray), highlighting arginines (colored as in panels a and b) that interact with the nucleosome acidic patch. Structures are clustered in groups with identical views and shown in cartoon representation with key side chains depicted. Acidic patch binding proteins and protein complexes are Sir3 (PDB id 3TU4; R29 (anchor); R28 (type1) [6]), Orc1 (PDB id 6OM3; R31 (anchor); R29 (type1) [31]), scFV (PDB id 6DZT; R124 (anchor); R126/188 (atypical) [65]), Dot1p (PDB id 7K6Q; R571 (anchor); R572 (atypical) [32]), Dot1L (PDB id 6NJ9; R282 (anchor); R278 (atypical) [21]), Set8 (PDB id 7D1Z; R192 (anchor); R188 (type 1) [29]), DNMT3 (PDB id 6PA7; R743 (anchor); R740 (type 1) [66]), PFV GAG (PDB id 5MLU; R540 (anchor) [67]), CENP-C (PDB id 4X23; R717 (anchor); R714 (type 1); R719 (type 2) [4]), SAGA DUB (PDB id 4ZUX; Sgf11 R84 (type 1); R78/91 (atypical) [44]), COMPASS (PDB id 6VEN; Set1 R901 (anchor); R908 (type 1); R904 (type 2); R936 (atypical) [27]), BAF (PDB id 6LTJ; SMARCB1 R370 (anchor); R377 (type 1); R373 (type 2); R366/374 (atypical) [24]), RSC (PDB id 6TDA; Sfh1 R397 (anchor); R404 (type 1); R400 (type 2); R401/408 (atypical) [25]), SWR1 (PDB id 6GEN; Swc6 R44 (anchor) [51]), ALC1 (PDB id 6ZHX; R9 (anchor) [68]), cGAS (PDB id 7JO9; R241 (anchor); R222 (type 2); R244 (atypical) [14]), RCC1 (PDB id 3MVD; R223 (anchor); R216 (type 1) [3]), CMV IE1 (PDB id 5E5A; R486 (anchor) [43]), LANA (PDB id 1ZLA; R9 (anchor); R12 (atypical) [2]), 53BP1 (PDB id 5KGF; R1627 (anchor); R1630 (atypical) [69]), PRC1 (PDB id 4R8P; Ring1B R98 (anchor); R81 (atypical) [5]), and BRCA1 (PDB id 7JZV; R71 (anchor) [30]).
The surge of new nucleosome complex structures solved by cryo-EM not only reinforces the acidic patch as a primary hot spot for nucleosome binding (23 of 41 structures that interact with the histone components of the nucleosome) and provides additional examples of the arginine anchor motif, but also illuminates several new patterns for acidic patch recognition. It is now clear that most proteins use two or more arginines to bind to the acidic patch. Alignment of all acidic patch-interacting arginines from unique structures demonstrates that the canonical arginine anchor is the most common arginine position, but many arginines in variant positions also participate in acidic patch binding (Fig. 2a). With more than twenty examples, the canonical arginine anchors (green) insert with strikingly similar conformations into a narrow cavity bounded by the α2 and α3 helices of H2A and the C-terminal helix of H2B (Fig. 2b). The arginine anchor cavity is formed from three layers: 1) the floor (H2A L93, H2B L106); 2) the middle ring comprising the acidic triad (H2A E61, D90, and E92); and 3) the side wall (H2B E105 and H109) that corrals the arginine anchor and orients it for interaction with the acidic triad. Recently, a de novo R95Q mutation was reported in the arginine anchor of the Polycomb Repressive Complex 1 (PRC1) component RING1A [13]. This mutation is causative in an epigenetic neurodevelopmental disorder due to aberrant developmental gene regulation, underscoring the fundamental importance of arginine anchors in health and disease.
Analysis of variant arginines demonstrates a diversity of side chain orientations and binding positions within the acidic patch in addition to the canonical arginine anchor (Fig. 2a). However, a subset of variant arginines converge on one of two acidic patch surfaces, herein described as type 1 and type 2. Type 1 variant arginines (light blue) bind on the opposite half of the acidic patch and commonly interact with H2A E56 and/or H2B E113 (Fig. 2b). The type 1 variant arginine depression is wider that the arginine anchor cavity but equally deep, and like the arginine anchor cavity contains three layers: 1) the floor (H2A A60 and mainchain atoms of the H2A α2 helix); 2) the middle ring of acidic residues (H2A E56, E61, and E64; H2B E113); and 3) two side walls, one constructed by the H2B C-terminal helix (H2B E113 and K116) and the other by the H2B α1L1 elbow (H2B Q47 and V48, see next section). Similar to the arginine anchor cavity, these side walls corral the type 1 variant arginine side chains and direct them to the bottom of the depression. In contrast, type 2 variant arginines (cyan) project their side chain guanidinium groups into a shallow cleft formed by H2A E61, E64, and L65 that is immediately adjacent to the canonical arginine anchor binding cavity (Fig. 2b). We refer to all other variant arginines as atypical variant arginines due to the absence of clear patterns of acidic patch binding.
Based on existing structures, arginine anchors and variant arginines are found roughly equally in structured protein loops, α-helices, and/or extended protein chains (Fig. 2c). For example, cGAS, a cytosolic double-stranded DNA sensor that functions in the innate immune system, is negatively regulated by binding the nucleosome acidic patch using two adjacent loops, one with an arginine anchor and an atypical variant arginine, and the other, a type 2 variant arginine [14–19]. In another example, the histone H3 K79 methyltransferase Dot1L binds the acidic patch using a loop containing a type 1 variant arginine along with an atypical variant arginine [20–23]. Two orthologs of the SWI/SNF family of remodeling complexes, the human BAF complex [24] and the yeast RSC complex [25,26], bind the acidic patch using a single α-helix with four or five arginines all aligned on the nucleosome-facing surface of the helix. These helices include an arginine anchor, type 1 and type 2 variant arginines, and one or more atypical variant arginines. Interestingly, the Set1 catalytic subunit of the yeast COMPASS H3 K4 methyltransferase complex employs a helix with near identical orientation and arginine composition, suggesting evolutionary convergence on a nucleosome binding mechanism [27,28]. Finally, some proteins use arginines in extended peptide chains to interact with the acidic patch, including the H4 K20 methyltransferase Set8 that interacts with its target sequence in the H4 tail and projects an N-terminal extension with an arginine anchor that interacts with the acidic patch [29].
In some instances, related domains use nearly identical modes of acidic patch recognition. Two examples are the RING domains of E3 ligases PRC1 [5] and BRCA1 [30], which both have an arginine anchor imbedded within a similar loop sequence, and the Bromo-associated homology (BAH) domains of yeast proteins Sir3 [6] and Orc1 [31] that interact with the acidic patch using loops with an arginine anchor and a type 1 variant arginine. Interestingly, while several orthologous proteins have been shown to interact similarly with the acidic patch, this is by no means a requirement, as in the case of human Dot1L and its yeast ortholog Dot1p that utilize a type 1 variant arginine and a canonical arginine anchor, respectively [20–23,32]. Even with an increasing set of examples of acidic patch-interacting proteins, this structural variability presents challenges to prediction of new arginine anchors and variant arginines except in select cases.
Several questions regarding acidic patch interactions remain. The relative contributions of each arginine to nucleosome binding and/or enzymatic activity has only been reported for a few nucleosome complexes [5,15,16,20,27]. These studies suggest a hierarchy of importance for acidic patch-binding: arginine anchor > type 1/type 2 variant arginines >> atypical variant arginines, but further detailed analysis is required to confirm this hypothesis. Moreover, given the convergence of many chromatin proteins on one surface, it is largely unclear how nucleosome acidic patch binding is regulated. Nearby histone post-translational modification can tune acidic patch interactions, but this may only provide crude regulation [20,33]. A recent discovery that methylation of an acidic patch binding arginine in RCC1 enhances its chromatin interaction, presents a more elegant solution [34]. It remains unclear if arginine methylation or other modifications are used broadly to positively or negatively regulate acidic patch binding. However, there is now no doubt that arginine-acidic patch interactions are extraordinarily pervasive in nucleosome recognition and critical to diverse genome-templated processes.
Interactions with the histone elbow
The arginine anchor is the most prominent structural paradigm used by chromatin proteins and enzymes to engage the nucleosome, but it is not the only one. The plethora of new chromatin complex structures now makes it possible to recognize other structural motifs on the nucleosome targeted by chromatin proteins. Two structural motifs we would like to highlight here are what we term the histone elbow and the histone H2B C-terminal helix.
The histone fold, which enables the pairing of histones H3 with H4 and histones H2A with H2B, is composed of three α-helices separated by two loops. The resulting H3/H4 and H2A/H2B histone fold pairs form crescent structures with outer surfaces that bind DNA in the nucleosome [35] (Fig. 1). The junction of the H3 histone fold α1 helix and L1 loop (H3 α1L1) is exposed on the nucleosome surface and we observe that chromatin proteins interact with this region in multiple nucleosome complex structures. The α1 helix and the L1 loop combination resembles an elbow, and we therefore call this the H3 α1L1 elbow. Over 40% of unique structures where chromatin proteins interact with histones (18 of 41) make close contact with the H3 α1L1 elbow (Fig. 3a).
Fig. 3:
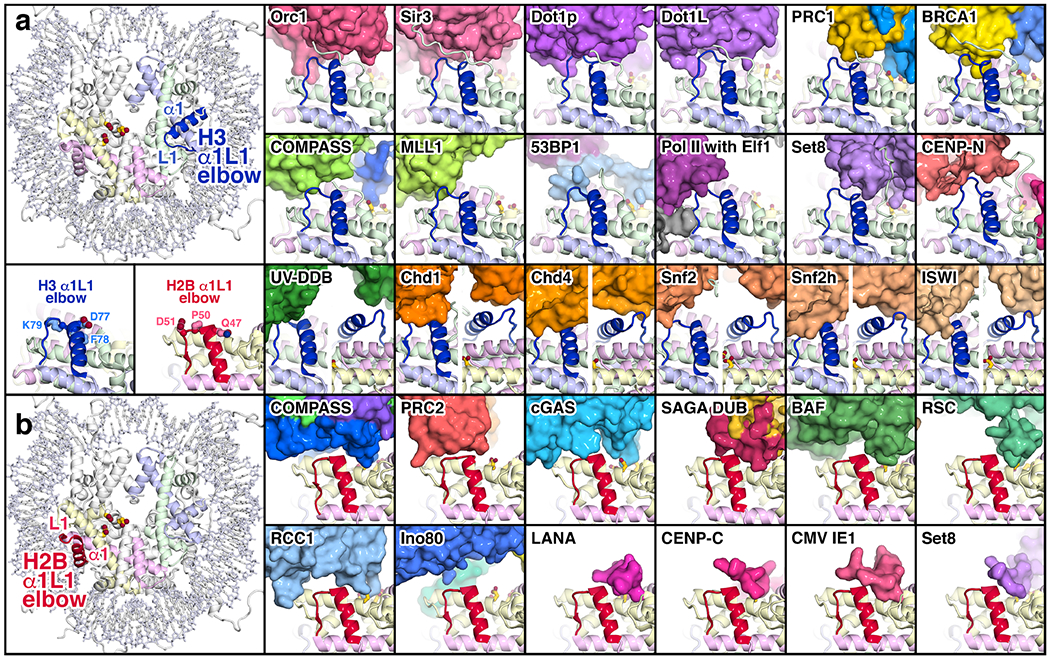
Interactions of chromatin proteins with the histone H3 and H2B α1L1 elbows. (a) Interactions with the H3 α1L1 elbow. The location of the H3 α1L1 elbow is shown on the left in the nucleosome crystal structure (PDB id 1KX5 [70]) together with the H2A acidic patch residues E61, D90 and E92. To the right are shown proteins and protein complexes in surface representation that interact with the H3 α1L1 elbow (blue): Orc1 (PDB id 6OM3 [31]), Sir3 (PDB id 3TU4 [6]), Dot1p (PDB id 7K6Q [32]), Dot1L (PDB id 6NQA [21]), PRC1 (PDB id 4R8P [5]), BRCA1 (PDB id 7JZV [30]), COMPASS (PDB id 6VEN [27]), MLL1 (PDB id 6KIU [36]), 53BP1 (PDB id 5KGF [69]), PolII elongation complex with Elf1 (PDB id 6IR9 [52]), Set8 (PDB id 7D1Z [29]), CENP-N (PDB id 6MUP [71]), UV-DDB (PDB id 6R8Y [62]), Chd1 (PDB id 6FTX [38]), Chd4 (PDB id 6RYR [39]), Snf2 (PDB id 5X0Y [40]), Snf2h(PDB id 6NE3 [41]) and ISWI (PDB id 6K1P [42]). In the CENP-CN structure, the α1L1 elbow of the centromeric histone variant, CENP-A, contains two extra residues compared to the histone H3 it replaces. The histones are shown in cartoon representation, and the nucleosomal DNA omitted for clarity. The H3 α1L1 elbow as it occurs in the nucleosome crystal structure is shown in blue in the same orientation together with elbow residues D77, F78 and K79 on the left below the full nucleosome structure. An additional view from the back is provided for Chd1, Chd4, Snf2, Snf2h and ISWI. (b) Interactions with the H2B α1L1 elbow. The location of the H2B α1L1 elbow is shown on the left in the nucleosome crystal structure (PDB id 1k5x) together with the H2A acidic patch residues E61, D90 and E92. Proteins and protein complexes shown that interact with the H2B α1L1 elbow (red) are COMPASS (PDB id 6VEN [27]), PRC2 (PDB id 6WKR [45]), cGAS (PDB id 7JO9 [14]), SAGA DUB (PDB id 4ZUX [44]), BAF (PDB id 6LTJ [24]), RSC (PDB id 6TDA [25]), RCC1 (PDB id 3MVD [3]), Ino80 (PDB id 6FML [46]), LANA (PDB id 1ZLA [2]), CENP-C (PDB id 4X23 [4]), CMV IE1 (PDB id 5E5A [43]) and Set8 (PDB id 7D1Z [29]). The H2B α1L1 elbow as it occurs in the nucleosome crystal structure is shown in red in the same orientation together with elbow residues Q47, P50 and D51 on the left above the full nucleosome structure.
Although the specific interactions at the H3 α1L1 elbow interface differ among the different chromatin proteins, common themes do emerge. The chromatin proteins that bind the H3 α1L1 elbow usually make close contact through a combination of van der Waal interactions, hydrogen bonding and ionic interactions without significantly altering the structure of the H3 α1L1 elbow. The exceptions to the absence of conformational change are the yeast Dot1p and the human Dot1L proteins where the H3 α1L1 elbow is distorted to position the H3 K79 target of the Dot1 methyltransferase into the enzyme’s active site [21,32]. Many of the interactions are made to the L1 loop at the tip of the H3 α1L1 elbow region corresponding to H3(76-81) with the sequence QDFKTD. H3 D77 and K79 can make charged interactions with the chromatin protein [5,6,21,29,31,36]. In contrast, the side chain of H3 F78 between these two residues does not appear to interact with chromatin proteins and instead packs against the H4 α2 helix.
The catalytic subunit of several chromatin remodeling enzymes, including Chd1, Chd4, Snf2, Snf2h and ISWI also interact with the H3 α1L1 elbow [37–42]. These enzymes, which share the Snf2 family ATPase domain, bind to the nucleosome at DNA positions SHL2 and SHL6. When bound at SHL2, the ATPase domains are positioned similarly through extensive interactions with the DNA and additional contacts with the H4 N-terminal tail and the H3 α1L1 elbow which is adjacent to SHL2.
H2B is the only other core histone with an exposed α1L1 elbow and like H3, the H2B α1L1 elbow interacts with multiple chromatin proteins (Fig. 3b). (The α1L1 regions of the other core histones, H2A and H4, are buried in the interior of the nucleosome and not solvent accessible.) The tip of the H2B α1L1 elbow corresponding to H2B(47-51) with the sequence QVHPD interacts in almost 30% of the unique chromatin complex structures containing histone interactions (12 of 41). Q47 often hydrogen bonds or makes van der Waal contact with interacting chromatin proteins, while H49 can hydrogen bond or stack with residues from the chromatin protein [2,4,14,24,25,27,43–45]. In addition, P50 and D51 residues in the H2B L1 loop interact with several chromatin proteins [14,27,43–46].
Unlike the histone acidic patch with its well defined residues that can be targeted for study by mutating to alanine or basic residues, the variety of interactions made by chromatin proteins to the H3 and H2B α1L1 elbows make it more difficult to recommend specific mutations to disrupt such interactions. Furthermore, given the multivalent interactions made by most chromatin proteins to multiple surfaces on the nucleosome, more study will be needed to determine how critical the observed histone elbow interactions are. One possible strategy to investigate potential interactions of a chromatin protein with the H3 α1L1 elbow might be to swap the charges on H3 D77 and K79. Similarly, the H2B α1L1 elbow could be targeted by mutating Q47, H49 and D51.
Interactions with the H2B C-terminal helix
The other major protein feature on the nucleosome targeted by chromatin proteins is the H2B C-terminal helix (Fig. 4). It is perhaps not surprising that this helix is contacted in about half of the unique structures with histone interactions (20 of 41) given its prominently exposed position on the surface of the nucleosome and its proximity to the acidic patch targeted by the arginine anchor. However it should be noted that most chromatin proteins do not simply tangentially contact the H2B C-terminal helix as a consequence of the arginine anchor binding to the histone acidic patch. Many make multiple contacts along the H2B C-terminal helix, including chromatin proteins that do not employ an arginine anchor to bind to the histone acidic patch. Some like COMPASS nearly engulf the entire length of the H2B C-terminal helix [27,28]. The interactions made by chromatin proteins to the H2B C-terminal helix include van der Waals interactions, hydrogen bonding and ionic interactions and there does not appear to be a predominant mode of binding employed.
Fig. 4:
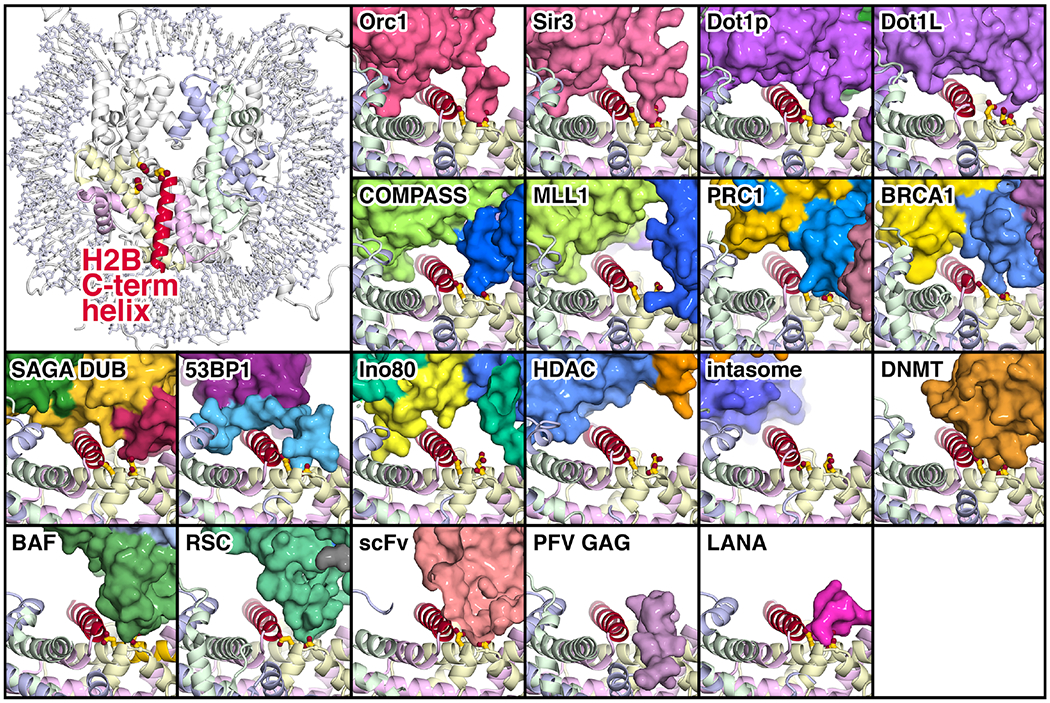
Interactions of chromatin proteins with the histone H2B C-terminal helix. The location of the H2B C-terminal helix is shown in the upper left in the nucleosome crystal structure (PDB id 1KX5 [70]) together with the adjacent H2A acidic patch residues E61, D90 and E92. Proteins and protein complex shown in surface representation that interact with the H2B C-terminal helix (red) are Orc1 (PDB id 6OM3 [31], Sir3 (PDB id 3TU4 [6]), Dot1p (PDB id 7K6Q [32]), Dot1L (PDB id 6NQA [21]), COMPASS (PDB id 6VEN [27]), MLL1 (PDB id 6KIU [36]), PRC1 (PDB id 4R8P [5]), BRCA1 (PDB id 7JZV [30]), SAGA DUB (PDB id 4ZUX [44]), 53BP1 (PDB id 5KGF [69]), Ino80 (PDB id 6FML [46]), HDAC (PDB id 6Z6P [72]), intasome (PDB id 6RNY [73]), DNMT (PDB id 6PA7 [66]), BAF (PDB id 6LTJ [24]), RSC (PDB id 6TDA [25]), scFv (PDB id 6DZT [65]), PFV GAG (PDB id 5MLU [67]) and LANA (PDB id 1ZLA [2]). The histones are shown in cartoon representation, and the nucleosomal DNA omitted for clarity.
The fore mentioned nucleosome interactome study from the McGinty laboratory provides experimental evidence of widespread interactions with the H2B and H3 α1L1 elbows and the H2B C-terminal helix proteome-wide [12]. Besides the acidic patch, the two nucleosome disk regions that are most critical for protein binding include a face of the H2B C-terminal helix and parts of either the H2B or H3 α1L1 elbow.
Conformational changes to nucleosomal DNA
In addition to binding to exposed histone surfaces of the nucleosome, many proteins interact with the contorted DNA wrapping the histone octamer and/or to linker DNA projecting from the nucleosome core particle. While typically only minor conformational rearrangements are observed in histone disk surfaces upon protein binding, in several cases, substantial rearrangements are observed in nucleosomal and linker DNA. The majority of these DNA rearrangements can be classified into three categories: 1) unwrapping of entry/exit DNA, 2) bulging of DNA away from the histone octamer surface, and 3) translocation of the DNA sequence around the histone octamer surface (Fig. 5).
Fig. 5:
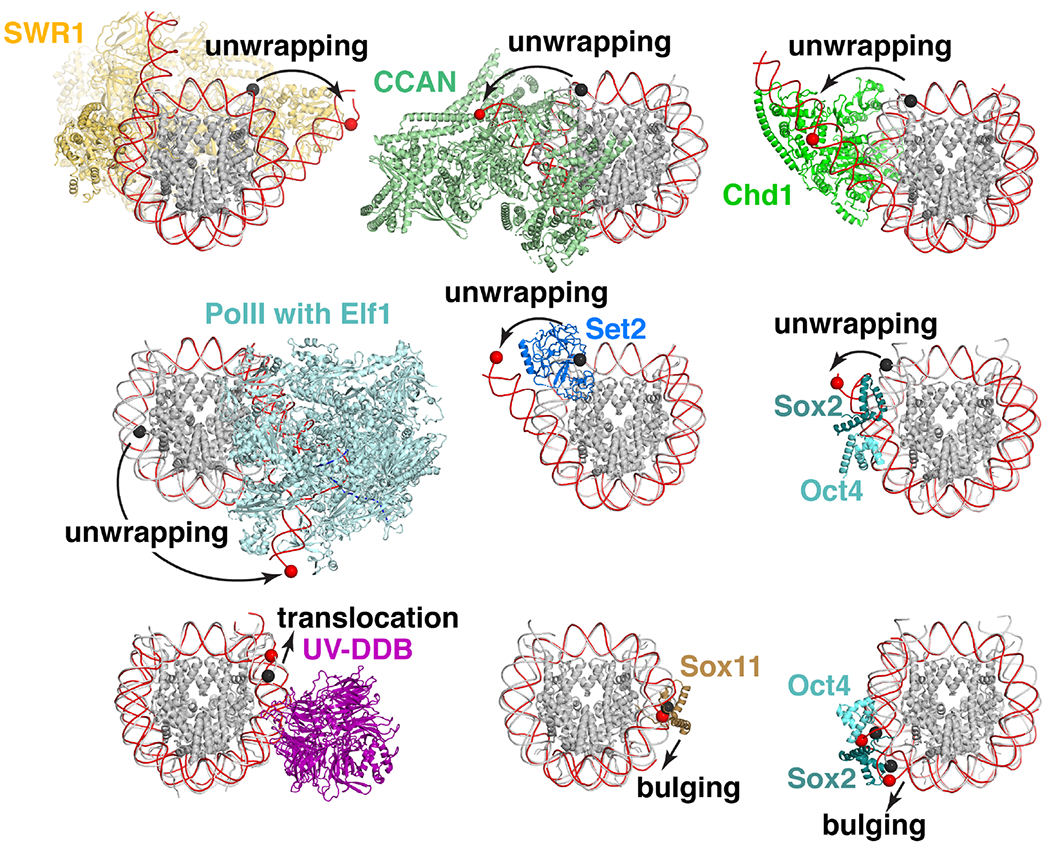
DNA conformational changes upon nucleosome binding. Chromatin proteins and complexes induce conformational changes to DNA. Overlay of nucleosome-bound (red DNA) and nucleosome only (white DNA) structures with identical DNA positions for bound and nucleosome only structures highlighted with red and black spheres, respectively. Nucleosome only structure PDB id 6PX1 used when matched structure is unavailable. Conformational changes include unwrapping as depicted for SWR1 (PDB id 6GEN [51]), CCAN (PDB id 6QLD [53]), Chd1 (PDB id 5O9G [37]), PolII with Elf1 and Spt4/5 stalled at SHL-1 (PDB id 6IR9 [52]), Set2 (PDB id 6NZO, nucleosome only 6PX1 [49]), and Sox2/Oct4 at SHL-6 (PDB id 6T90, nucleosome only 6T93 [54]); translocation as depicted for UV-DDB with THF2 lesion at -3 position (PDB id 6R91, nucleosome only 6R94 [62]); and bulging with Sox11 (PDB id 6T7A, nucleosome only 6T79 [74]) and Sox2/Oct4 at SHL+6 (PDB id 6YOV, nucleosome only 6T93 [54]).
To date, the most commonly observed DNA conformational change upon protein-nucleosome interaction is unwrapping of entry/exit DNA. The entry/exit DNA is known to spontaneously unwrap [47] and such DNA end breathing is observed in single particle cryo-EM analysis of nucleosomes [48]. However, several structures demonstrate that nucleosome binding proteins can interact with DNA ends and/or linker DNA to stabilize an unwrapped conformation to elicit specific effects. Examples include H3 K36 methyltransferases Set2 and NSD3 that gain access to their targeted lysine upon DNA unwrapping [49,50], remodelers like SWR1 that unwrap DNA to facilitate histone exchange [51], RNA PolII with elongation factors Elf1 and Spt4/5 transcribing through a nucleosome [52], the inner kinetochore CCAN complex [53], and pioneer transcription factors Sox2 and Oct4, when their sequence recognition motifs are imbedded in positions near the DNA entry/exit site [54]. Interestingly, DNA bulging is induced by Sox2 and Oct4, instead of unwrapping, when motifs are positioned at internal locations. Frequently observed DNA unwrapping, contrasts with nucleosome-bound structures of linker histones [7,55–60] and the H3 K4 demethylase LSD1 [61], which engage linker DNA and stabilize a fully wrapped DNA conformation. A more unique DNA rearrangement is observed for the DNA damage recognition complex UV-DDB. While binding to surface exposed lesions in nucleosomal DNA without altering the nucleosome structure, UV-DDB recognizes occluded lesions by engaging and stabilizing a translocated DNA conformation that relocalizes the lesion to an accessible, exposed position [62]. One noteworthy mechanism of nucleosomal DNA recognition is used by PARP2, whose contacts do not cause major DNA conformational changes, but rather bridges two nucleosomes through recognition of DNA ends to function in double strand break repair [63,64]. As more structures are solved especially with native DNA sequences, we anticipate that these and other DNA conformational changes will be commonplace.
Perspective
Despite the availability of so many new structures of chromatin factors and enzymes in complex with the nucleosome, we still lack structures for many unique classes of nucleosome-binding proteins. Moreover, most structures have been solved with artificially stable nucleosome positioning DNA sequences and histones with one, or most often, no histone modifications. Pivoting toward structures with native DNA sequences and established patterns of histone modifications can capture chromatin complexes in a more physiological context. We also need more information about the dynamics of chromatin complexes. Increasingly, cryo-EM data analysis allows dynamic motions of macromolecular complexes to be inferred from conformational heterogeneity. It is clear that most chromatin factors and enzymes engage nucleosomes through flexible and dynamic interactions. Selecting a highly homogeneous subset of particles may enable high resolution reconstructions, but neglects the true dynamic nature of the complexes. Finally, while the accessibility of cryo-EM has provided much needed structural insights, comprehensive functional characterization has lagged behind. While less flashy than increasingly ambitious structures, a greater emphasis on careful mechanistic investigation is necessary to fully leverage the rapidly growing set of structures towards biomedical innovations.
Acknowledgements
This work was supported by National Institutes of Health grant NIH GM127034 to S.T. and NIH GM133498 to R.K.M. and a grant from the Pew-Stewart Scholars for Cancer Research program to R.K.M..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
The authors declare no conflict of interest.
References
- 1.McGinty RK, Tan S: Recognition of the nucleosome by chromatin factors and enzymes. Curr Opin Struct Biol 2016, 37:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM: The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 2006, 311:856–861. [DOI] [PubMed] [Google Scholar]
- 3.Makde RD, England JR, Yennawar HP, Tan S: Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 2010, 467:562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato H, Jiang J, Zhou B-R, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y: A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 2013, 340:1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGinty RK, Henrici RC, Tan S: Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 2014, 514:591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armache K-J, Garlick JD, Canzio D, Narlikar GJ, Kingston RE: Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 2011,334:977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou B-R, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y: Structural Mechanisms of Nucleosome Recognition by Linker Histones. Molecular Cell 2015, 59:628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P: Structural basis for retroviral integration into nucleosomes. Nature 2015, 523:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC: The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface 2013, 10:20121022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGinty RK, Tan S: Nucleosome Structure and Function. Chem. Rev. 2015, 115:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389:251–260. [DOI] [PubMed] [Google Scholar]
- 12.Skrajna A, Goldfarb D, Kedziora KM, Cousins EM, Grant GD, Spangler CJ, Barbour EH, Yan X, Hathaway NA, Brown NG, et al. : Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res 2020, 48:9415–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper reports an unbiased proteomics screen of the nucleosome interactome. The authors show that the acidic patch is the major hot spot for nucleosome disk binding and identify more that 250 proteins that directly or indirectly require the acidic patch for nucleosome binding.
- 13.Pierce SB, Stewart MD, Gulsuner S, Walsh T, Dhall A, McClellan JM, Klevit RE, King M-C: De novo mutation in RING1 with epigenetic effects on neurodevelopment. Proc Natl Acad Sci USA 2018, 115:1558–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer JA, Spangler CJ, Strauss JD, Cesmat AP, Liu P, McGinty RK, Zhang Q: Structural basis of nucleosome-dependent cGAS inhibition. Science 2020, 370:450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathare GR, Decout A, Glüick S, Cavadini S, Makasheva K, Hovius R, Kempf G, Weiss J, Kozicka Z, Guey B, et al. : Structural mechanism of cGAS inhibition by the nucleosome. Nature 2020, 587:668–672. [DOI] [PubMed] [Google Scholar]; • This is one of six studies establishing the structural basis for inhibition of cGAS by the nucleosome. Nucleosome binding prevents cGAS from inappropriate immune pathway activation by self-DNA.
- 16.Zhao B, Xu P, Rowlett CM, Jing T, Shinde O, Lei Y, West AP, Liu WR, Li P: The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature 2020, 587:673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, Hornung V, Hopfner K-P: Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature 2020, 587:678–682. [DOI] [PubMed] [Google Scholar]
- 18.Cao D, Han X, Fan X, Xu R-M, Zhang X: Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res 2020, 30:1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, Hirai S, Funabiki H, Kurumizaka H: Structural basis for the inhibition of cGAS by nucleosomes. Science 2020, 370:455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson CJ, Baird MR, Hsu A, Barbour EH, Koyama Y, Borgnia MJ, McGinty RK: Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase. Cell Rep 2019, 26:1681–1690.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worden EJ, Hoffmann NA, Hicks CW, Wolberger C: Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell 2019, 176:1490–1501.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This is one of five articles reporting the structure of human Dot1L bound to the nucleosome. The structure shows how Dot1L binds to the acidic patch and makes direct hydrophobic interactions with ubiquitin attached to H2B K120, providing a mechanistic understanding of a classic histone crosstalk pathway.
- 22.Valencia-Sánchez MI, De loannes P, Wang M, Vasilyev N, Chen R, Nudler E, Armache J-P, Armache K-J: Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Molecular Cell 2019, 74:1010–1019.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao T, Jing W, Hu Z, Tan M, Cao M, Wang Q, Li Y, Yuan G, Lei M, Huang J: Structural basis of the crosstalk between histone H2B monoubiquitination and H3 lysine 79 methylation on nucleosome. Cell Res 2019, 29:330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He S, Wu Z, Tian Y, Yu Z, Yu J, Wang X, Li J, Liu B, Xu Y: Structure of nucleosome-bound human BAF complex. Science 2020, 367:875–881. [DOI] [PubMed] [Google Scholar]; • The authors solve the structure of the BAF remodeling complex bound to the nucleosome. Malfunction of the BAF complex is causative in several types of cancer.
- 25.Wagner FR, Dienemann C, Wang H, Stützer A, Tegunov D, Urlaub H, Cramer P: Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 2020, 579:448–451. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study reports the structure of the nucleosome-bound RSC remodeling complex and illustrates important mechanistic insights into how nucleosome binding defines its function.
- 26.Ye Y, Wu H, Chen K, Clapier CR, Verma N, Zhang W, Deng H, Cairns BR, Gao N, Chen Z: Structure of the RSC complex bound to the nucleosome. Science 2019, 366:838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worden EJ, Zhang X, Wolberger C: Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. elife 2020, 9:e53199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu PL, Shi H, Leonen C, Kang J, Chatterjee C, Zheng N: Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS. Molecular Cell 2019, 76:712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This article reports the structure of COMPASS bound to unmodified and ubiquitylated nucleosomes to show how ubiquitin reverses enzymatic autoinhibition.
- 29.Ho C-H, Takizawa Y, Kobayashi W, Arimura Y, Kimura H, Kurumizaka H: Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase. Life Sci Alliance 2021,4:e202000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witus SR, Burrell AL, Farrell DP, Kang J, Wang M, Hansen JM, Pravat A, Tuttle LM, Stewart MD, Brzovic PS, et al. : BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat Struct Mol Biol 2021, 28:268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De loannes P, Leon VA, Kuang Z, Wang M, Boeke JD, Hochwagen A, Armache K-J: Structure and function of the Orc1 BAH-nucleosome complex. Nat Commun 2019, 10:2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valencia-Sánchez MI, De loannes P, Wang M, Truong DM, Lee R, Armache J-P, Boeke JD, Armache K-J: Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Science 2021, 371:eabc6663. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors solve the structure of the yeast Dot1p protein in complex with a ubiquitylated nucleosome. This structure shows a mechanism for nucleosome binding that is divergent from the human ortholog.
- 33.Dann GP, Liszczak G, Bagert JD, Müiller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et al. : ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang T, Yang Y, Song X, Wan X, Wu B, Sastry N, Horbinski CM, Zeng C, Tiek D, Goenka A, et al. : PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Molecular Cell 2021, 81:1276–1291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinty RK, Tan S: Histone, Nucleosome and Chromatin Structure. In Fundamentals of Chromatin. Edited by Workman JL. Springer; 2014:1–28. [Google Scholar]
- 36.Xue H, Yao T, Cao M, Zhu G, Li Y, Yuan G, Chen Y, Lei M, Huang J: Structural basis of nucleosome recognition and modification by MLL methyltransferases. Nature 2019, 573:445–449. [DOI] [PubMed] [Google Scholar]; • Structures of MLL1 and MLL3 core catalytic complexes bound to the nucleosome described in this study suggest a role for H2B ubiquitylation in stabilizing a productive complex as well as unique modes of regulation of the catalytic subunits of the MLL complexes by an accessory subunit.
- 37.Farnung L, Vos SM, Wigge C, Cramer P: Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 2017, 550:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundaramoorthy R, Hughes AL, El-Mkami H, Norman DG, Ferreira H, Owen-Hughes T: Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. elife 2018, 7:e35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farnung L, Ochmann M, Cramer P: Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations, elife 2020, 9:e56178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Li M, Xia X, Li X, Chen Z: Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 2017, 544:440–445. [DOI] [PubMed] [Google Scholar]
- 41.Armache J-P, Gamarra N, Johnson SL, Leonard JD, Wu S, Narlikar GJ, Cheng Y: Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. elife 2019, 8:e46057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan L, Wu H, Li X, Gao N, Chen Z: Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat Struct Mol Biol 2019, 26:258–266. [DOI] [PubMed] [Google Scholar]
- 43.Fang Q, Chen P, Wang M, Fang J, Yang N, Li G, Xu R-M: Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome. elife 2016, 5:e11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C: Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 2016, 351:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, Faini M, Toso D, Aebersold R, Nogales E: JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 2021, 371:eabc3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner K-P: Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 2018, 556:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, Levitus M, Bustamante C, Widom J: Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 2005, 12:46–53. [DOI] [PubMed] [Google Scholar]
- 48.Bilokapic S, Strauss M, Halic M: Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol 2018, 25:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bilokapic S, Halic M: Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat Commun 2019, 10:3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Tian W, Yuan G, Deng P, Sengupta D, Cheng Z, Cao Y, Ren J, Qin Y, Zhou Y, et al. : Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 2021, 590:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willhoft O, Ghoneim M, Lin C-L, Chua EYD, Wilkinson M, Chaban Y, Ayala R, McCormack EA, Ocloo L, Rueda DS, et al. : Structure and dynamics of the yeast SWR1-nucleosome complex. Science 2018, 362:eaat7716. [DOI] [PubMed] [Google Scholar]
- 52.Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine S-I: Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 2019, 363:744–747. [DOI] [PubMed] [Google Scholar]; • This study describes a structural mechanism for decreased stalling of RNA polymerase II at defined nucleosome positions when in complex with elongation factors. The elongation factors prevent nucleosome interactions that contribute to pausing and reorient the polymerase for forward progression through nucleosomal DNA.
- 53.Yan K, Yang J, Zhang Z, McLaughlin SH, Chang L, Fasci D, Ehrenhofer-Murray AE, Heck AJR, Barford D: Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature 2019, 574:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The authors present a structure of the inner kinetochore CCAN complex on a centromeric nucleosome providing key mechanistic insights into kinetochore assembly at the centromere.
- 54.Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, Bunker RD, Schenk AD, Graff-Meyer A, Pathare GR, et al. : Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 2020, 368:1460–1465. [DOI] [PubMed] [Google Scholar]
- 55.Zhou B-R, Feng H, Kale S, Fox T, Khant H, de Val N, Ghirlando R, Panchenko AR, Bai Y: Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Molecular Cell 2021, 81:166–182.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Vogirala VK, Soman A, Berezhnoy NV, Liu ZB, Wong ASW, Korolev N, Su C-J, Sandin S, Nordenskiöld L: Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome. Sci Rep 2021, 11:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adhireksan Z, Sharma D, Lee PL, Davey CA: Near-atomic resolution structures of interdigitated nucleosome fibres. Nat Commun 2020, 11:4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou B-R, Jiang J, Ghirlando R, Norouzi D, Sathish Yadav KN, Feng H, Wang R, Zhang P, Zhurkin V, Bai Y: Revisit of Reconstituted 30-nm Nucleosome Arrays Reveals an Ensemble of Dynamic Structures. J Mol Biol 2018, 430:3093–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Pápai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, et al. : Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Molecular Cell 2017, 66:384–397.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adhireksan Z, Sharma D, Lee PL, Bao Q, Padavattan S, Shum WK, Davey GE, Davey CA: Engineering nucleosomes for generating diverse chromatin assemblies. Nucleic Acids Res 2021, doi: 10.1093/nar/gkab070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S-A, Zhu J, Yennawar N, Eek P, Tan S: Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate. Molecular Cell 2020, 78:903–914.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsumoto S, Cavadini S, Bunker RD, Grand RS, Potenza A, Rabl J, Yamamoto J, Schenk AD, Schübeler D, Iwai S, et al. : DNA damage detection in nucleosomes involves DNA register shifting. Nature 2019, 571:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This article describes a mechanism to explain the recognition of damaged DNA that is occluded due its position in the nucleosome. Using thorough structural and biochemical characterization, the authors show that UV-DDB binds to and stabilizes a translocated nucleosome that repositions a DNA lesion to an accessible position.
- 63.Bilokapic S, Suskiewicz MJ, Ahel I, Halic M: Bridging of DNA breaks activates PARP2-HPF1 to modify chromatin. Nature 2020, 585:609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study reports the structural basis for PARP2 function at DNA double strand breaks. PARP2 engages DNA ends from two separate nucleosomes, bridging the ends for ligation and stimulating PARP2 signaling activity.
- 64.Gaullier G, Roberts G, Muthurajan UM, Bowerman S, Rudolph J, Mahadevan J, Jha A, Rae PS, Luger K: Bridging of nucleosome-proximal DNA double-strand breaks by PARP2 enhances its interaction with HPF1. PLoS ONE 2020, 15:e0240932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou B-R, Yadav KNS, Borgnia M, Hong J, Cao B, Olins AL, Olins DE, Bai Y, Zhang P: Atomic resolution cryo-EM structure of a native-like CENP-A nucleosome aided by an antibody fragment. Nat Commun 2019, 10:2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu T-H, Liu M, Zhou XE, Liang G, Zhao G, Xu HE, Melcher K, Jones PA: Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature 2020, 586:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lesbats P, Serrao E, Maskell DP, Pye VE, O’Reilly N, Lindemann D, Engelman AN, Cherepanov P: Structural basis for spumavirus GAG tethering to chromatin. Proc Natl Acad Sci USA 2017, 114:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehmann LC, Bacic L, Hewitt G, Brackmann K, Sabantsev A, Gaullier G, Pytharopoulou S, Degliesposti G, Okkenhaug H, Tan S, et al. : Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch. Cell Rep 2020, 33:108529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien J-P, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D: The structural basis of modified nucleosome recognition by 53BP1. Nature 2016, 536:100–103. [DOI] [PubMed] [Google Scholar]
- 70.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ: Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol 2002, 319:1097–1113. [DOI] [PubMed] [Google Scholar]
- 71.Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE: Structure of the Human Core Centromeric Nucleosome Complex. Curr Biol 2019, 29:2625–2639.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J-H, Bollschweiler D, Schäfer T, Huber R: Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies. Sci Adv 2021, 7:eabd4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson MD, Renault L, Maskell DP, Ghoneim M, Pye VE, Nans A, Rueda DS, Cherepanov P, Costa A: Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer. Nat Commun 2019, 10:4189. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A structure of the prototypical PFV viral integrase in action on the nucleosome establishes a mechanism for DNA repositioning to drive retroviral integration into nucleosomal DNA.
- 74.Dodonova SO, Zhu F, Dienemann C, Taipale J, Cramer P: Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020, 580:669–672. [DOI] [PubMed] [Google Scholar]


