Dear Editor,
Targeted protein degradation (TPD) represents a promising research field that has quickly attracted attention and efforts from both academia and pharmaceutical industry. TPD technology uses small molecules to induce the degradation of target proteins by harnessing the ubiquitin–proteasome system. PROTAC (PROteolysis Targeting Chimeras)1–3 and MG (Molecular Glue)4,5 are two major modes of TPD. PROTACs comprise three parts, including a ligand for binding a target protein, another ligand for recruiting an E3 ligase, and a linker, which helps anchoring the target protein to the E3 ubiquitin ligase, to promote its ubiquitination and subsequent proteasomal degradation. Similar to PROTACs, MGs can also cause ubiquitination and degradation of a target protein. In contrast to PROTACs, MG molecules are small-molecular-weight compounds that trigger a compact protein–protein interaction between a target protein and an E3 ubiquitin ligase. Both PROTACs and MGs have their own advantages and limitations. For example, PROTACs are more suitable for rational design, and a broad range of targets have been successfully degraded using the PROTAC approach. However, PROTACs are often relatively large and sometimes suffer from insufficient druggability. In contrast, MG molecules have a clear advantage in their smaller molecular weights and better druggability. However, the design of MGs is rather difficult due to limited understanding of the controlling factors, and there have been few successful cases.6,7 Therefore, we planned to design molecules with the characteristics of these two techniques to combine their strengths in this study.
The BTK (Bruton Tyrosine Kinase) protein, a tyrosine kinase and key regulator of the BCR pathway, is overactivated in a variety of lymphoma cells.8 In our previous work, we achieved efficient degradation of wild-type and mutant BTK proteins via PROTAC.9,10 Although the inhibitory effect of the degraders L18I and P13I on BTK-mutant lymphoma is clearly better than that of ibrutinib, both BTK inhibitors and PROTAC degraders show very limited efficacy for refractory DLBCL (diffuse large B cell lymphoma)11 and AML (acute myeloid leukemia).12 Achieving an ideal therapeutic effect on DLBCL and AML upon the inhibition or degradation of BTK alone is difficult. GSPT1 (G1 to S phase transition 1) is a translation termination factor that recognizes the termination codon by binding eRF1, forcing the proteins to dissociate from the ribosome after translation.13 Downregulation of GSPT1 can cause the abnormal expression of key proteins, inhibit proliferation or induce apoptosis in diverse tumor cells. It was recently proven that GSPT1 can be degraded by MGs.14,15 Therefore, we propose that the concurrent degradation of GSPT1 may enhance the therapeutic potential of BTK degraders in DLBCL, thereby eliminating the practical barriers of PROTACs in these types of lymphoma.
Herein, as a proof of concept, we report a dual-mechanism degrader that can concurrently chemically induce the degradation of BTK and GSPT1 by merging the PROTAC and MG strategies. The representative double-mechanism degrader, GBD-9, efficiently degraded BTK and GSPT1 by recruiting the E3 ligase cereblon (CRBN) in various DLBCL and AML cell lines. Moreover, GBD-9 effectively inhibited cancer cell growth. Our work provides a new example for the design of double-mechanism degraders with the characteristics of both MG and PROTACs (Fig. 1a, b) and expands the indications of PROTACs targeting BTK, which may have broader clinical application prospects.
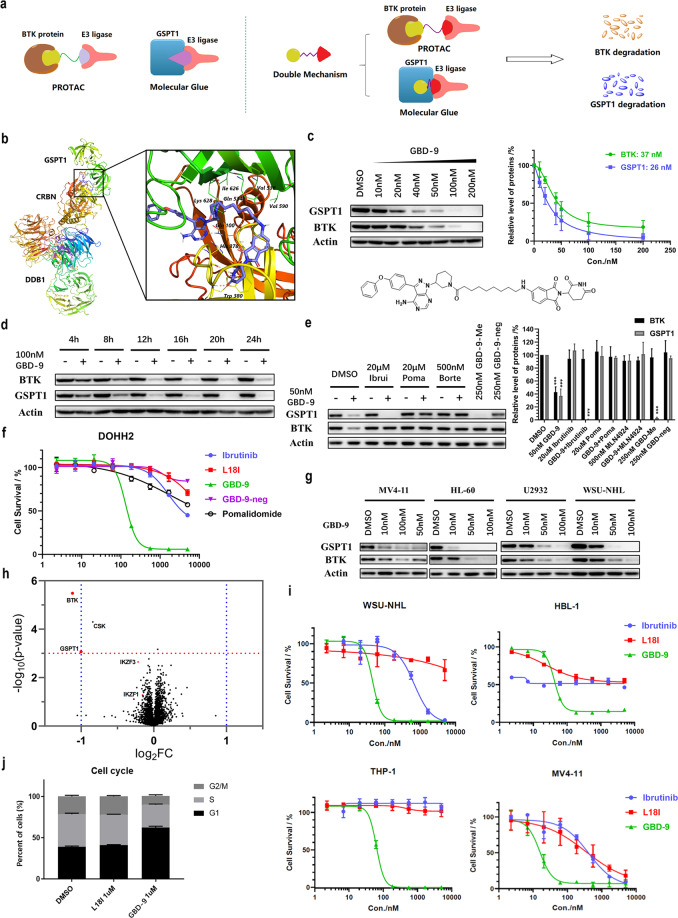
Fig. 1. Schematic representation and characterization of the double-mechanism degrader GBD-9.
a A brief illustration of the PROTAC, MG and double-mechanism degrader developed in the study. b Molecular modeling of the binding of GBD-9 (Lyons blue) with GSPT1 and CRBN. An available structure of the GSPT1–CRBN complex with the ligand CC-885 (PDB code: 5HXB) was used. c Immunoblot analysis of BTK, GSPT1 and β-actin in DOHH2 cells after treatment with the double-mechanism degrader GBD-9 at the indicated concentrations for 24 h. d Immunoblot analysis of BTK, GSPT1 and β-actin in DOHH2 cells treated with 100 nM GBD-9 for the indicated durations. e Immunoblot analysis of BTK, GSPT1 and β-actin in DOHH2 cells after treatment with GBD-9 (50 nM) or GBD-9 along with ibrutinib (20 µM), pomalidomide (20 µM), or MLN-4924 (500 nM) for 24 h. Poma, pomalidomide; Ibrui, ibrutinib. “+” refers to treatment with GBD-9; “−” refers to treatment without GBD-9. f In vitro study of the inhibitory effects of the BTK degrader L18I, GBD-9, ibrutinib, pomalidomide, and negative control (GBD-9-neg) on the DOHH2 cell line. In 96-well plates, 5000 cells were incubated in each well at 37 °C for 72 h. Cell viability was determined by CCK-8 assay. g Immunoblot analysis of BTK, GSPT1 and β-actin in MV4-11, HL-60, U2932 and WSU-NHL cells. h Proteomics analysis. DOHH2 cells were treated with 300 nM GBD-9 for 8 h before proteomics measurement. i In vitro study of the inhibitory effects of the BTK degrader L18I, ibrutinib and GBD-9 on different DLBCL (WSU-NHL, HBL-1) and AML (THP-1, MV4-11) cell lines. j GBD-9 induced G1 phase arrest in DOHH2 cells. Cells were treated with different concentrations of L18I and GBD-9 for 24 h and then stained with propidium iodide. The cell cycle distribution was assessed using flow cytometry. Quantification of western blot in c and e was performed by grayscale analysis; the ratios were normalized to control (DMSO). *P < 0.05, **P < 0.01, ***P < 0.001 (treated groups versus control group (DMSO), independently). The gels are representative of, minimally, three biologically independent experiments (c, d, e, g). The error bars represent the means ± SD of three biological duplicates. The significance analysis was conducted by two-tailed unpaired Student’s t-test.
Since the molecular weights of PROTACs are much larger than those of MGs, the difficulty in integrating the PROTAC and MG techniques lies in balancing their features. Based on our previous works on the development of BTK PROTACs9,10 and recent reports on MG molecules targeting GSPT1, our strategy is to design and optimize the characteristics required to target GSPT1 in BTK degraders. Unlike the complexes induced by PROTACs, the spatial distances between CRBN and target proteins in the complexes induced by MG molecules are very small, and thus the molecules need to be as short as possible to stabilize the interaction between the target proteins and CRBN (Fig. 1a). Therefore, we designed a library of molecules with a shorter linker based on the original BTK degrader and tried to change the type of linkers or connecting sites of the CRBN ligand, hoping to find a degrader that retained the efficiency of PROTACs and concurrently possessed the features of MGs. The L18I linker contains 16 atoms, and we further shortened it to 6–12 atoms (Supplementary information, Table S1, such as GBD-2, 4, 6, 8, 10, 12, 14); because a shorter linker can limit the molecular conformation and affect the efficiency, we also designed molecules with meta-substituted pomalidomide, which has a more stretched conformation (Supplementary information, Table S1, such as GBD-3, 5, 7, 9, 11, 13, 15).
Next, we simulated the binding of these designed lead molecules with GSPT1 and CRBN based on the CRBN–DDB1–CC-885–GSPT1 cocrystal structure. The docking and MD (molecular dynamics) simulation results revealed that a few of the designed PROTAC molecules may be suitable as potential MGs. Based on the simulation results, we synthesized the designed molecules and evaluated their degradation efficiencies in the DLBCL cell line DOHH2. The results indicated that linkers with 7–9 carbons could induce the concurrent degradation of BTK and GSPT1. Treatment with all these degraders at 50 nM for 24 h could induce > 70% degradation of the BTK and GSPT1 proteins; among them, a representative degrader named GBD-9, has the greatest effect on both targets, degrading BTK and GSPT1 by 80% and 90%, respectively. As displayed in Fig. 1b, the docking of GBD-9 in the CRBN–DDB1–CC-885–GSPT1 cocrystal structure may provide a deeper insight into MG design (see Supplementary information for details about docking analysis and SAR study).
As screening results showed that GBD-9 possessed the best degradation efficiency, we conducted further mechanistic and activity studies with this degrader. In the DLBCL cell line DOHH2, GBD-9 at 50 nM exhibited strong degradation activity against the BTK and GSPT1 proteins and could downregulate both target proteins to < 20% of baseline level (Fig. 1c). The degradation of BTK and GSPT1 occurred quite fast, taking only 4 h under treatment with 100 nM GBD-9 (Fig. 1d). To confirm that GBD-9 degrades BTK and GSPT1 independently via the PROTAC and MG mechanisms, ligands at both ends of GBD-9 were added to the treatment. When ibrutinib was used to occupy the binding pocket of BTK, the degradation activity of GBD-9 against BTK was abolished, but it could still degrade GSPT1, which showed that the degradation of GSPT1 does not completely depend on binding to the ibrutinib-containing end. This was confirmed by the binding mode in the CRBN–DDB1–CC-885–GSPT1 structure. When pomalidomide was added to block the binding pocket of CRBN, as expected, GBD-9 could not degrade BTK and simultaneously lost its effect on GSPT1. When treated with GBD-9-neg (see Supplementary information, Table S2 for the structure), in which the pomalidomide-containing end of GBD-9 was ethylated, the same result was observed. This indicated that the pomalidomide end played an important role in the formation of the ternary complex induced by GBD-9, regardless of whether the target was BTK or GSPT1. The results above verified that when GBD-9 acted as a PROTAC molecule to induce BTK degradation, the ligands at both ends were required to concurrently bind the target proteins and CRBN. Nevertheless, when the molecule induced the degradation of GSPT1, it functioned as a complete MG, and therein, the pomalidomide end made the greater contribution. When MLN-4924 was added to inhibit the activation of E1 ubiquitin-activating enzyme, neither BTK nor GSPT1 was degraded, indicating that GBD-9 induces the degradation of target proteins through the ubiquitin–proteasome system. Furthermore, CRBN–DDB1, GSPT1 and BTK were purified for the pull-down assay; the CRBN–DDB1 protein was apparently pulled down by tagged GSPT1 or BTK after incubation with GBD-9 (Supplementary information, Fig. S6b, c). These results illustrated the formation of ternary complex and engagement of these proteins in the process of degradation induced by GBD-9. To further establish that the downregulation of BTK was not caused by the degradation of GSPT1, we methylated the ibrutinib end of GBD-9, which eliminated its binding affinity to BTK, producing GBD-9-Me (see Supplementary information, Table S2 for the structure). GBD-9-Me could only induce the degradation of GSPT1 and had no effect on the protein level of BTK, which indicated that the degradations of GSPT1 and BTK occur through two independent processes (Fig. 1e). Collectively, our results demonstrated that GBD-9 acts both as a PROTAC molecule to induce the degradation of BTK and as a MG to degrade GSPT1.
In view of the neo-substrates of CRBN and multi-targets of ibrutinib that possibly become the off-targets of GBD-9, we confirmed the selectivity of the degrader through TMT-labeled quantitative proteomics assay. GBD-9 displayed fair selectivity on GSPT1 and BTK, which were the main proteins to be downregulated (Fig. 1h). It is worth mentioning that no degradation of IKZF1/3 or CK1α was detected, which was further verified by immunoblot analysis (Supplementary information, Fig. S5d).
Both ibrutinib and L18I inhibited the proliferation of DLBCL cells to some extent but did not kill the cells, and the GSPT1 degrader CC-90009 is not so efficient in this type of cells either, limiting their clinical applications. In this work, GBD-9, which degrades BTK and GSPT1 at the same time, significantly reduced the survival ratio of DOHH2 cells even at low concentrations. Compared with ibrutinib, GBD-9 showed a greatly enhanced inhibitory effect on the proliferation of DOHH2 cells (IC50 = 133 nM), but the negative control (GBD-9-neg) had no obvious inhibitory effect, which confirmed that the activity of GBD-9 is derived from degrading instead of inhibiting BTK (Fig. 1f). Additionally, compared with the single BTK degrader L18I, GBD-9 induced G1 cell cycle arrest, which was mainly manifested as ~20% increase in the proportion of cells at G1 phase (Fig. 1j). In addition to ibrutinib and L18I, GBD-9 induced the apoptosis of DOHH2 cells, in which the antiapoptotic proteins BCL-2, MCL-1, and XIAP were downregulated and caspase-3 was cleaved (Supplementary information, Fig. S5c). The above data clearly confirmed that GBD-9 is superior to ibrutinib and L18I in inhibiting cell survival through double-mechanism degradation. Moreover, the anti-proliferative effect of GBD-9 was much better than those of the single degraders L18I and CC-90009, but equal to the effect of combination of the two molecules (Supplementary information, Fig. S4c, d). The conclusion can be drawn that both degradations of BTK and GSPT1 contribute to the inhibitory activity of GBD-9 in DLBCL cell lines. In addition to the DOHH2 cell line, other GCB-DLBCL and AML cell lines were tested. We found that GBD-9 stably and concurrently degraded BTK and GSPT1 in the tested DLBCL and AML cell lines (Fig. 1g). As shown in Fig. 1i, compared with ibrutinib or L18I, GBD-9 exhibited much stronger inhibitory effect against various lymphomas (see Supplementary information, Fig. S2 for more details).
In summary, for the first time, we have developed a degrader that targets the BTK and GSPT1 proteins through a unique double mechanism. Our study provides insights into the design of degraders integrating characteristics of both MG and PROTAC. Moreover, by combining PROTAC and MG mechanisms, GBD-9 retained the ability of PROTACs to degrade BTK and also functioned as a MG to degrade GSPT1. As a result, GBD-9 illustrated much greater anti-proliferative effects on a variety of DLBCL and AML cell lines than ibrutinib, and may have the potential to overcome the inability of BTK inhibitors to treat DLBCL and AML patients.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81622042, 81773567), and National Major Scientific and Technological Project (2020YFE0202200 and 2018ZX09711001). We thank Fang Fang, Xiuyun Sun, Liguo Wang (Tsinghua University) for helpful support and suggestions. We thank Tianbai Zhong (Tsinghua University) for kindly providing cell lines. We thank Can Zhu, Yi Shi, and Ya Tan (Tsinghua University) for kind help in the construction of knockdown cell line. We also thank Profs. Haiteng Deng and Chongchong Zhao in Protein Chemistry and Proteomics Facility at Tsinghua University Technology Center for Protein Research for mass spectrometry analysis of proteins.
Author contributions
YR, ZY, and YS conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. ZY and YS performed most of the experiments and bioinformatic study. ZN assisted with synthesis of compounds and the experiment of cell proliferation inhibition. CY purified the proteins and performed the experiments of pull-down and iSCAMs. YT and YL constructed the CRBN-knockdown cell line. HL directed the experimental design of protein purification and pull-down assay.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD025609.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zimo Yang, Yonghui Sun.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00533-6.
References
- 1.Sakamoto, K. M. et al. Proc. Natl. Acad. Sci. USA98, 8554–8559 (2001).
- 2.Sun, X. et al. Signal Transduct. Target. Ther.4, 64 (2019). [DOI] [PMC free article] [PubMed]
- 3.Schapira, M., Calabrese, M. F., Bullock, A. N & Crews, C. M. Nat. Rev. Drug Discov.18, 949–963 (2019). [DOI] [PubMed]
- 4.Tan, X. et al. Nature446, 640–645 (2007).
- 5.Han, T. et al. Science356, eaal3755 (2017).
- 6.Krönke, J. et al. Nature523, 183–188 (2015).
- 7.Faust, T. B. et al. Nat. Chem. Biol.16, 7–14 (2020). [DOI] [PMC free article] [PubMed]
- 8.Davis, R. E. et al. Nature463, 88–92 (2010). [DOI] [PMC free article] [PubMed]
- 9.Sun, Y. et al. Cell Res.28, 779–781 (2018). [DOI] [PMC free article] [PubMed]
- 10.Sun, Y. et al. Leukemia33, 2105–2110 (2019). [DOI] [PubMed]
- 11.Wilson, W. H. et al. Nat. Med.21, 922–926 (2015). [DOI] [PMC free article] [PubMed]
- 12.Cortes, J. E., Jonas, B. A., Graef, T., Luan, Y. & Stein, A. S. Clin. Lymphoma Myeloma Leuk.19, 509–515.e1 (2019). [DOI] [PubMed]
- 13.Chauvin, C., Salhi, S. & Jean-Jean, O. Mol. Cell. Biol.27, 5619–5629 (2007). [DOI] [PMC free article] [PubMed]
- 14.Matyskiela, M. E. et al. Nature535, 252–257 (2016). [DOI] [PubMed]
- 15.Yang, J. et al. J. Med. Chem.62, 9471–9487 (2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD025609.