Abstract
Cellular identity and physiologic function in mammary epithelial cells and in many breast cancers flow from the action of a network of master transcriptional regulators including Estrogen Receptor alpha, GATA3 and FOXA1. The last decade has seen the completion of multiple large sequencing projects focusing on breast cancer. These massive compendia of sequence data have provided a wealth of new information linking mutation in these transcription factors to alterations in tumor biology and transcriptional program. The emerging details on mutation in cancer, and direct experimental exploration of hypotheses based upon it, are now providing a wealth of new information on the roles played by Estrogen Receptor alpha, GATA3 and FOXA1 in regulating gene transcription and how their combined action contributes to a network shaping cell function in both physiologic and disease states.
The mammary gland undergoes a complex set of developmental transitions associated with puberty, pregnancy, lactation, and menopause. This tissue must respond to acute physiologic signals to activate the developmental programs dictating elaboration of a complex structure destined for milk production and reverse this program upon cessation of lactation leading to gland involution. This program is orchestrated by the reproductive hormones estrogen and progesterone which act through a network of transcription factors including Estrogen Receptor alpha (ESR1), GATA3, and FOXA1 [1–3]. Current models implicate FOXA1 and GATA3 respectively in preparation of genomic loci for ligand-activated ESR1 through local remodeling of chromatin architecture [2,4,5]. Indeed, Gata3, Foxa1 and Esr1 are all required for normal development of the mammary gland and elaboration of ductal structures in mice [3,6,7]. Pathologic co-option of this network occurs with high frequency in breast cancer where approximately 2/3 of breast cancers express ESR1 and are reliant on estrogens for growth [8].
With broad declines in the costs associated with next-generation sequencing projects, considerable attention has turned to cancer sequencing. In the context of breast cancer, researchers have reported the mutational and transcriptional signatures of invasive breast cancer in multiple cohorts [9–12], the landscape of lobular breast cancer [13,14], and mutational and transcriptional signatures associated with endocrine resistant breast cancer [15]. These studies provide a wealth of information on the mutational status of the Estrogen Receptor/GATA3/FOXA1 transcriptional network and they associate mutation status with transcriptome and outcome data. Recent biochemical and molecular analysis of GATA3 and FOXA1 have added substantially to our understanding of how these factors function and are regulated in the context of breast cancer.
Estrogen Receptor alpha (ESR1) mutations in breast cancer
Estrogen receptor alpha (ESR1) has the domain architecture typical of steroid hormone receptors with a central DNA binding domain flanked by an amino terminal activation domain and a C-terminal ligand binding/activation domain. Multiple structures of ESR1 domains have been solved providing atomic level understanding of protein function [16]. ESR1 is a relatively infrequent target of mutation in breast cancer with missense mutations occurring in around 3% of cases and amplification occurring at a similar level [10,12,13,15]. ESR1 missense mutations are found in both lobular and ductal carcinomas; truncating mutations are rarely observed. Two locations in ESR1 are hotspots for missense mutation – tyrosine 537/aspartic acid 538 and glutamic acid 380. Mutation of Y537 has been known for decades; mutation of this residue in the ligand binding domain alters interaction with the p160 family of coactivators, leading to loss of ligand dependence [17,18]. Likewise, mutation of E380 is proposed to render the receptor independent of ligand for activation, making it constitutively active [16]. Overall, these missense mutations in ESR1 are frequently found in metastatic ESR1 positive tumors refractory to endocrine therapy where they are largely considered to favor a conformation of the receptor that resembles the agonist bound state (Table 1) [19,20]. The structural basis and implications of mutations of ESR1 in cancer have been recently reviewed extensively elsewhere [16] and will not be further considered here.
Table 1.
Summary of mutations in ESR1
| GENE | MUTATION TYPE | FUNCTIONAL DOMAIN | PREDICTED IMPACT |
|---|---|---|---|
| ESR1 | Missense | Ligand Binding | Reinforce transcriptional program |
GATA3 mutations in breast cancer
GATA3 was initially discovered in the immune system as a regulator of the T-cell receptor [21]. Subsequent studies have identified roles for GATA3 in cell-type specification in multiple systems. GATA3 plays an integral role in establishment and development of the mammary gland where it is expressed solely in the luminal epithelial compartment. Deletion of Gata3 in the mammary gland leads to severe developmental perturbations with failure to specify the luminal epithelial cell fate [3]. In breast cancer cells, GATA3 participates in a reciprocal regulatory loop with ESR1; each binds to cis-acting regulatory DNA of the other and regulates its expression [1]. In addition, GATA3 can function as an initiating, or pioneering, transcription factor in breast cancer cells, enabling downstream function of both ESR1 and FOXA1 [4].
GATA3 is a prototypical zinc finger transcription factor with 2 GATA family zinc fingers providing sequence specific DNA recognition (Figure 1). GATA3 is characterized by 2 transcriptional activation domains at the amino terminus and a short C-terminal region of unknown function [22]. Mutation of GATA3 in breast cancer was discovered prior to the advent of large-scale sequencing studies [23]. Genomic sequencing projects have indicated that GATA3 is mutated with high frequency in breast cancer, with 10 to 15% of breast tumors harboring GATA3 mutation [9,10]. The majority of GATA3 mutations are found in luminal cancers. In most cases, mutation occurs on a single allele with the remaining wildtype allele also being expressed. Interestingly, mutations, which are largely frameshift, truncation, or splice site mutations, are found throughout the zinc fingers and carboxyl terminus (See Figure 1). The wide distribution of mutations, as opposed to concentration at one or a few residues, leads to the prediction that most GATA3 mutations in breast cancer are loss of function [24].
Figure 1. Mutations in GATA3 in breast cancer.
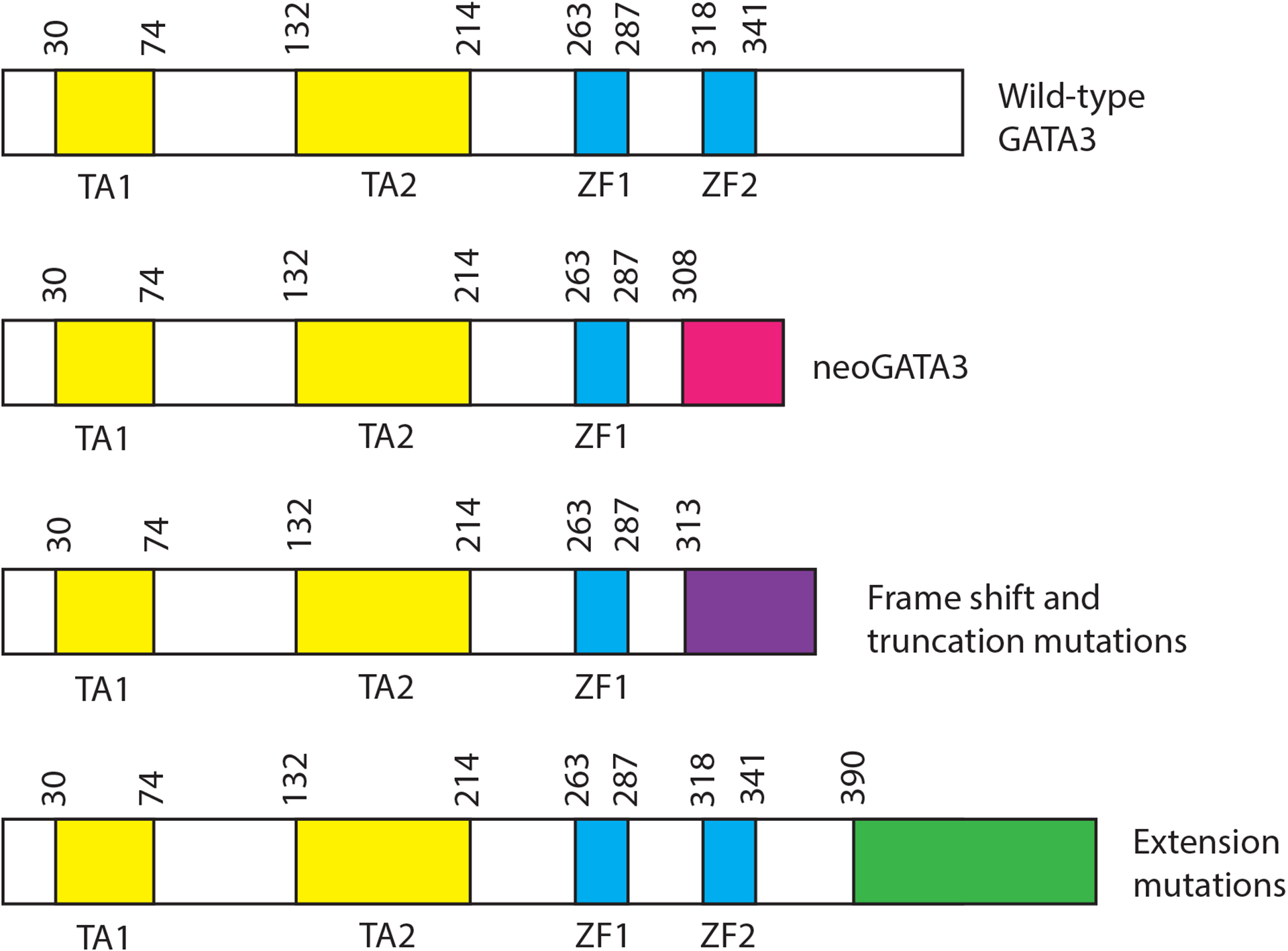
The figure illustrates the location of the major classes of GATA3 mutation found in breast cancers. Locations are given by amino acid numbers above each illustration. Features of the mutations are discussed in the text. TA = transactivation domain; ZF = zinc finger
The most frequently observed mutation in GATA3 in breast cancer is loss of a splice acceptor at the 5’ end of exon 5. Exon 5 encodes the second zinc finger and this class of mutations typically results in a frameshift and truncation with complete loss of amino acids 308–444. The shift to an alternative reading frame results in addition of 44 novel amino acids onto the first 307 amino acids of GATA3 – a feature referred to as neoGATA3 [25]. NeoGATA3 is observed exclusively in ESR1 positive tumors where it is associated with favorable outcome and younger age of onset. Exogenous expression of neoGATA3 in ESR1 positive breast cancer cell lines perturbs the cell cycle stimulatory impact of estrogens and perturbs the ability of progesterone to cause cell cycle arrest. These impacts on the ESR1 transcriptional program are consistent with losses of function, although gain of function imparted by the addition of 44 novel amino acids is also possible.
A large number of GATA3 mutations, more than 200 in the TCGA [13], Metabric [12] or MSK [15] cohorts, involve frameshift mutations starting at S390 and extending to the C-terminus, with a hotspot located at Proline 408. Interestingly, these mutations predominantly shift to the same reading frame which results in loss of the C-terminal portion of GATA3 and extension of the protein by approximately 60 amino acids encoded by the alternative reading frame. These mutations have been referred to as extension mutations [26,27] and they are associated with significantly shorter disease free survival in one cohort [9,26] but not in another [10,26]. When modeled in an ESR1 negative cell line, extension mutations result in alteration of expression of many transcripts and increased sensitivity to drugs inhibiting the histone methyltransferases G9a and GLP [26]. It is currently unclear whether the extension mutations impact the ESR1/GATA3/FOXA1 transcriptional network as the cellular model utilized was ESR1 negative.
The third major class of GATA3 mutations in breast cancer are frameshift and truncation mutations located from A313 to S369. Most of these mutations impact zinc finger 2 or amino acids adjacent to zinc finger 2, causing loss of key residues involved in DNA recognition [27,28]. Two highly utilized models of ESR1 positive cancer, MCF7 and KPL1, harbor such mutations [23,29]. Truncation in zinc finger 2 has been associated with poor prognosis, as compared to all other GATA3 mutations [27]. A cellular model of ZF2 mutant GATA3 in which one allele is mutated while the other retains wild-type sequence displays altered growth characteristics, dramatic remodeling of the genomic landscape of GATA3, FOXA1 and ESR1 localization, alterations in the transcriptome, dramatic declines in expression of progesterone receptor and changes in growth properties [27,30]. Transgenic mice expressing a ZF2 truncation mutation (R335fs) in the mammary gland exhibit increased ductal branching while a cellular model of the same mutation results in increased growth in 3D culture in the absence of estrogens [31].
In summary, breast cancer specific mutations in GATA3 impact several regions of the protein, mostly through frameshift and truncation (Figure 1, Table 2). The truncation mutations that have been studied extensively suggest that the presence of one mutant allele and one wild type allele in the same cell results in some loss of binding at loci bound by GATA3, FOXA1 and ESR1 in wild type cells [30], consistent with loss of function. In contrast, other loci exhibit novel binding by these three transcription factors consistent with gain of function. Biologically, the impact of GATA3 mutation on expression of progesterone receptor noted in both splice site [25] and truncation mutations [27] may explain some clinical characteristics of tumors bearing these mutations such as early age of onset.
Table 2.
Summary of mutations in GATA3
| GENE | MUTATION TYPE | FUNCTIONAL DOMAIN | PREDICTED IMPACT |
|---|---|---|---|
| GATA3 | Splice | Zinc Finger 2 | Decreased dosage of wt GATA3 (decreased Progesterone Receptor) |
| Gain of function? | |||
| Frame shift and truncation | Zinc Finger 2 | Decrease dosage of wt GATA3 (decreased Progesterone Receptor) | |
| Novel DNA binding pattern | |||
| Extension | C-terminus | Unknown |
FOXA1 mutations in breast cancer
FOXA1 belongs to a family of proteins with a common DNA interaction surface termed the forkhead domain. This domain consists of three alpha helices, three beta strands and a pair of loops or wings. Structural data indicates that the forkhead domain occupies the major groove of DNA with the wings making specific DNA contacts in a manner that resembles linker histone [18]. FOXA1 co-localizes with ESR1 at a large number of genomic sites and its depletion results in crippled genomic binding of ESR1 [2,32]. FOXA1 mutation is not observed with the same frequency as GATA3 mutation and appears to be lower in primary sites than in metastatic ones [13]. Mutations are observed in both ductal and lobular carcinoma and they frequently co-occur in lobular carcinoma with mutations in CDH1 [13,14]. At the molecular level, both missense and truncation mutations are observed in breast cancer, hotspots for missense mutation occur in the forkhead domain while truncations are largely C-terminal to the forkhead domain. Missense hotspots include sites in helix 1 (I176), between helix 3 and beta strand 2 (D226), beta strand 3 (S242/Y243 insertion/deletion mutations) and in wing 2 (H247, S250).
Functional analysis of breast cancer specific mutations in FOXA1 is in a developing stage. Arruabarrena-Aristorena and colleagues recently analyzed a set of mutations via exogenous expression in MCF7 [33]. Mutations in Wing 2 of the forkhead domain (H247Y, S250F, F266L) conferred a growth advantage when estrogens were limiting and exhibited increased genomic binding upon estrogen stimulation, leading to an enhanced estrogen response at the transcriptome level. The beta strand 3 hotspot mutation (S242/Y243 ins/del) conferred genomic localization to a substantial number of novel binding sites, suggesting altered genomic site selection. These ‘gained’ binding sites were associated with a novel transcriptome, including genes involved in proliferation such as Myc target genes. These analyses suggest that the FOXA1 mutations in Wing 2 consolidate the ESR1/GATA3/FOXA1 transcriptional program while mutations in beta strand 3 have novel gains of function (Table 3).
Table 3.
Summary of mutations in FOXA1
| GENE | MUTATION TYPE | FUNCTIONAL DOMAIN | PREDICTED IMPACT |
|---|---|---|---|
| FOXA1 | Missense | Wing 2 (Forkhead) | Reinforce transcriptional program |
| Missense | β strand 3 (Forkhead) | Novel DNA binding pattern |
Closing thoughts
Detailed analysis of mutations identified in breast cancer genomic sequencing projects has begun to reveal new information on the ESR1 transcriptional network (Table 1, Table 2, Table 3). Mutations in the receptor itself largely confer ligand independence for transcriptional activation, consolidating the transcriptional program. Mutations in GATA3, while still under investigation, lead to specific losses of function (including decreased expression of progesterone receptor) at some loci while exhibiting gains of function at others. Mutations in FOXA1, also still an active research area, lead to consolidation of the ESR1 transcriptional program in some cases and to gains of function in others. In most cases, there are compelling hypotheses or strong structural data to facilitate interpretation of the findings.
Current models describing the ESR1 transcriptional network suggest that multiple transcription factors, minimally ESR1, FOXA1 and GATA3 cooperate to produce active enhancers that modulate gene expression [5]. However, the identity of the transcription factor(s) that act as initiator of new enhancer formation and principal shaper of productive chromatin interaction by the network is currently a matter of some debate in the field [4,6,27,34–36]. It seems useful, then, to consider whether cancer-specific mutations in the ESR1 network can address this question. ESR1 mutations largely render the receptor independent of ligand for transcriptional activation [16], thereby altering the dynamics of receptor interaction with DNA rather than altering specificity. FOXA1 mutations, while also infrequent in primary tumors, include mutations in the Forkhead domain which mediates interaction with chromatin. Mutations in Wing 2 of the Forkhead domain reinforce the estrogen-induced transcriptional program and are suggested to stabilize the interaction of FOXA1 with DNA [33]. Like the case with ESR1, these mutations suggest that altering the kinetics of interaction of one component with DNA can stabilize the transcriptional output of the network, consistent with a central role for transcription factor dynamics. Finally, multiple mutations in GATA3 directly impact the second zinc finger which contributes to specificity. Zinc Finger 2 mutations clearly exhibit losses of function, they also exhibit gains of function, redirecting all three network factors to new loci in the genome [4,27,30]. In this respect, the cancer-specific mutations of GATA3 and in beta strand 3 of FOXA1 are reminiscent of activated IL6 signaling in breast cancer that also alters localization of ESR1 and FOXA1 [37]. Taken together, the behavior of cancer-specific mutations in this network seems to indicate that multiple factors, including GATA3 and FOXA1, can function at specific loci to direct action of the remaining network members, with other network inputs (including aberrant signaling) also shaping the enhancer repertoire utilized by ER-α. Further, the data suggest that the dynamic nature of the interaction of these transcription factors with the chromatin fiber is a critical determinant in regulating downstream gene expression.
Many challenges remain in the quest to understand the consequences of cancer-specific mutations. Data integration at multiple levels, including patient metadata as well as molecular data, is limiting for many analyses. Current model systems for ESR1 positive tumors have limitations with known mutations in GATA3 [23] or ARID1a [29] confounding experiments in MCF7 and T47D, respectively. Further, experimental analyses that rely on exogenous expression may not accurately model the impact of loss of one wild-type allele on gene regulation. Finally, lack of structural data on GATA3 outside the zinc fingers impairs the ability to interpret some mutational data. Solutions to these, and other, experimental issues will lead to new principles on how the ESR1/GATA3/FOXA1 transcriptional network regulates cell growth, differentiation, and function.
Highlights:
Mutations in estrogen receptor found in endocrine therapy refractive breast cancer stabilize the ligand-bound conformation
Mutations in the forkhead domain of FOXA1 reinforce the estrogen mediated transcriptional program
Mutations in GATA3 lead to loss and gains of function
Acknowledgements
The authors apologize to many colleagues whose work could not be cited here for space reasons. This work was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (ES101965 to P.A.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare no conflicts of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M: Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 2007, 67:6477–6483. [DOI] [PubMed] [Google Scholar]
- 2.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS: FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011, 43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouros-Mehr H, Kim JW, Bechis SK, Werb Z: GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol 2008, 20:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theodorou V, Stark R, Menon S, Carroll JS: GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 2013, 23:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong SL, Li G, Loh SL, Sung WK, Liu ET: Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 2011, 7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, et al. : FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development 2010, 137:2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korach KS: Insights from the study of animals lacking functional estrogen receptor. Science 1994, 266:1524–1527. [DOI] [PubMed] [Google Scholar]
- 8.Siersbaek R, Kumar S, Carroll JS: Signaling pathways and steroid receptors modulating estrogen receptor alpha function in breast cancer. Genes Dev 2018, 32:1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas N: Comprehensive molecular portraits of human breast tumours. Nature 2012, 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. : The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486:346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. : Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. : The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016, 7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. : Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmedt C, Zoppoli G, Gundem G, Pruneri G, Larsimont D, Fornili M, Fumagalli D, Brown D, Rothe F, Vincent D, et al. : Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol 2016, 34:1872–1881. [DOI] [PubMed] [Google Scholar]
- 15.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, et al. : The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018, 34:427–438 e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S: Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer 2018, 18:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS: Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol 1996, 10:1388–1398. [DOI] [PubMed] [Google Scholar]
- 18.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA: Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry 1997, 36:14897–14905. [DOI] [PubMed] [Google Scholar]
- 19.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. : ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013, 45:1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, Smith A, Wilson J, Morrow C, Wong WL, et al. : Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov 2017, 7:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM: Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J 1991, 10:1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho IC, Tai TS, Pai SY: GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol 2009, 9:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, et al. : Mutation of GATA3 in human breast tumors. Oncogene 2004, 23:7669–7678. [DOI] [PubMed] [Google Scholar]
- 24.Adomas AB, Grimm SA, Malone C, Takaku M, Sims JK, Wade PA: Breast tumor specific mutation in GATA3 affects physiological mechanisms regulating transcription factor turnover. BMC Cancer 2014, 14:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hruschka N, Kalisz M, Subijana M, Grana-Castro O, Del Cano-Ochoa F, Brunet LP, Chernukhin I, Sagrera A, De Reynies A, Kloesch B, et al. : The GATA3 X308_Splice breast cancer mutation is a hormone context-dependent oncogenic driver. Oncogene 2020, 39:5455–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This manuscript provides detailed analysis of the clinical features associated with splice site mutation in GATA3 in cancer. Additional functional studies in breast cancer cell lines indicate defects in estrogen and progesterone signaling.
- 26.Mair B, Konopka T, Kerzendorfer C, Sleiman K, Salic S, Serra V, Muellner MK, Theodorou V, Nijman SM: Gain- and Loss-of-Function Mutations in the Breast Cancer Gene GATA3 Result in Differential Drug Sensitivity. PLoS Genet 2016, 12:e1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaku M, Grimm SA, Roberts JD, Chrysovergis K, Bennett BD, Myers P, Perera L, Tucker CJ, Perou CM, Wade PA: GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun 2018, 9:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The manuscript describes an analytical scheme for classifying GATA3 mutant breast cancers. Functional analysis of a model generated by genome engineering suggests zinc finger 2 mutation leads to an altered transcriptome and a substantial decrease in progesterone receptor levels.
- 28.Bates DL, Chen Y, Kim G, Guo L, Chen L: Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol 2008, 381:1292–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. : The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takaku M, Grimm SA, De Kumar B, Bennett BD, Wade PA: Cancer-specific mutation of GATA3 disrupts the transcriptional regulatory network governed by Estrogen Receptor alpha, FOXA1 and GATA3. Nucleic Acids Res 2020, 48:4756–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper utilizes an engineered breast cancer cell line bearing a GATA3 zinc finger 2 mutation to demonstrate that mutation of one allele of GATA3 significantly redistributes ESR1 and FOXA1 in the genome.
- 31.Emmanuel N, Lofgren KA, Peterson EA, Meier DR, Jung EH, Kenny PA: Mutant GATA3 Actively Promotes the Growth of Normal and Malignant Mammary Cells. Anticancer Res 2018, 38:4435–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. : Genome-wide analysis of estrogen receptor binding sites. Nat Genet 2006, 38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 33.Arruabarrena-Aristorena A, Maag JLV, Kittane S, Cai Y, Karthaus WR, Ladewig E, Park J, Kannan S, Ferrando L, Cocco E, et al. : FOXA1 Mutations Reveal Distinct Chromatin Profiles and Influence Therapeutic Response in Breast Cancer. Cancer Cell 2020, 38:534–550 e539. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper provides comprehensive description of the correlation of FOXA1 mutation with clinical parameters in breast cancer. Functional studies of various mutants are conducted in breast cancer cell lines revealing a class of mutations in Wing 2 of the forkhead domain that reinforce the estrogen stimulated transcriptional program as well as mutations elsewhere in the forkhead domain that alter DNA binding specificity of FOXA1.
- 34.Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, et al. : Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 2016, 165:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glont SE, Chernukhin I, Carroll JS: Comprehensive Genomic Analysis Reveals that the Pioneering Function of FOXA1 Is Independent of Hormonal Signaling. Cell Rep 2019, 26:2558–2565 e2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paakinaho V, Swinstead EE, Presman DM, Grontved L, Hager GL: Meta-analysis of Chromatin Programming by Steroid Receptors. Cell Rep 2019, 28:3523–3534 e3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siersbaek R, Scabia V, Nagarajan S, Chernukhin I, Papachristou EK, Broome R, Johnston SJ, Joosten SEP, Green AR, Kumar S, et al. : IL6/STAT3 Signaling Hijacks Estrogen Receptor alpha Enhancers to Drive Breast Cancer Metastasis. Cancer Cell 2020, 38:412–423 e419. [DOI] [PMC free article] [PubMed] [Google Scholar]


