Abstract
RAG1/2 (RAG) is an RNH-type DNA recombinase specially evolved to initiate V(D)J gene rearrangement for generating the adaptive immune response in jawed vertebrates. After decades of frustration with little mechanistic understanding of RAG, the crystal structure of mouse RAG recombinase opened the flood gates in early 2015. Structures of three different chordate RAG recombinases including protoRAG, and the evolutionarily preceding Transib transposase have been determined in complex with various DNA substrates. Biochemical studies along with the abundant structural data have shed light on how RAG has evolved from an ordinary transposase to a specialized recombinase in initiating gene rearrangement. RAG has also become one of the best characterized RNH-type recombinases, illustrating how a single active site can cleave the two antiparallel DNA strands of a double helix.
Keywords: V(D)J recombination, asymmetric DNA recognition, transposition
From transposition to V(D)J recombination
RAG1/2 recombinase (RAG), which initiates the V(D)J gene rearrangement during development of the adaptive immune system in jawed vertebrates, is a member of the RNH-transposase family [1]. The RNH-type transposases that are characterized by their RNase H-like catalytic center include bacterial Tn10 (conferring antibiotic tetracycline resistance), bacteriophage Mu, HIV integrase, Drosophila P element, Tc1/mariner, Hermes and piggyBac [2,3,4*,5*]. Each RNH-type transposase can bind a pair of terminal inverted repeat sequences (TIRs) and catalyze transposition by the simple “cut-and-paste” mechanism [6,7] (Fig. 1a). RAG also recognizes a pair of recombination signal sequences (RSS, equivalent to TIRs) and makes double-strand cleavage at the borders of RSS and a flanking sequence, which can be more or less random. Unlike DNA transposases, the principal function of RAG is not transposition but this cleavage, which enables the flanking V, D or J gene segments to be selected from large arrays and joined imprecisely by non-homologous end-joing (NHEJ) to form the repertoire of diverse antibodies and antigen receptors [1] (Fig. 1b). Transposition by RAG is observed in vitro [8,9], but rare in vivo [10,11].
Fig. 1.
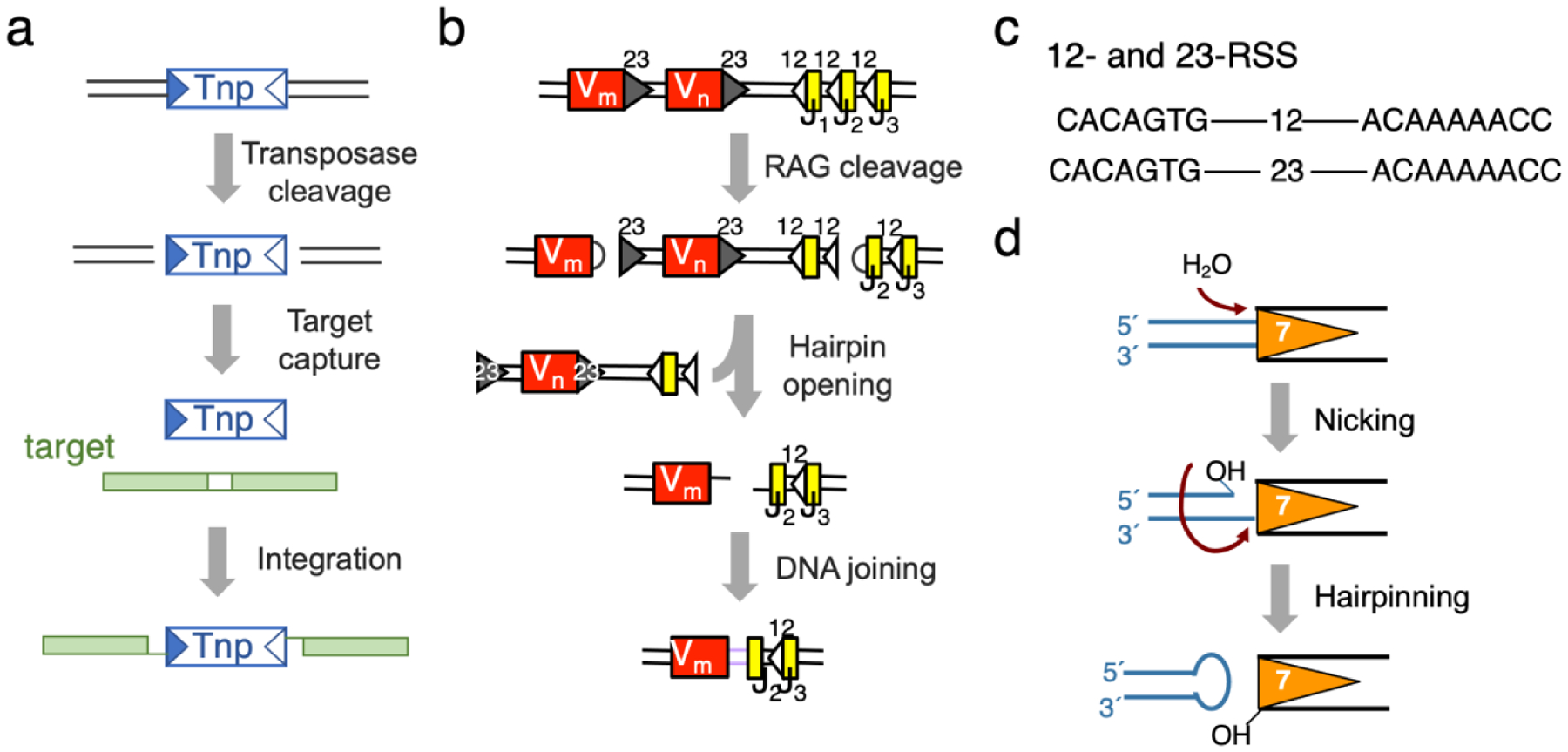
Diagram of V(D)J recombination versus DNA transposition. (a) Transposition and transposons (cut-and-paste). Transposase (Tnp) recognizes TIRs (triangles) and transposes transposon DNA to a new location (green target). (b) V(D)J recombination. κ chain is used as an example to illustrate the cleavage of a pair of V and J segments (Vm bordered by 23RSS and J2 bordered by 12RSS) by RAG. The hairpin products are processed and joined by the NHEJ pathway.
(c) Sequence of 12- and 23-RSS. (d) DNA double-strand cleavage reactions catalyzed by RAG.
Successful V(D)J gene rearrangement requires pairing and joining of two different kinds of gene segments, V and J, V and D or D and J. Selection of gene segments is achieved by the stringent pairing of two kinds of RSS [12]. Each RSS contains a 7 bp (heptamer) and 9 bp (nonamer) conserved sequence, but with a 12 or 23 bp spacer in between, hence known as 12- and 23-RSS [13] (Fig. 1c). The two gene segments to be joined are always flanked by RSS DNAs with different spacers, and RAG adheres to this 12/23 rule [14]. In contrast, many transposons have slightly different left and right TIRs, which allow modulations of the transposition event, but transposases in most cases can cleave a pair of identical TIRs [2,3,5*,15,16]. Furthermore, different from stochastic events of transposition, V(D)J recombination is highly regulated and cell-type and development-stage specific. For regulatory purposes, beyond the essential catalytic core domains [17], RAG1 contains the RING domain and can be auto-ubiquitylated, and RAG2 contains a PHD domain for its recruitment to the histone H3K4Me3 sites, as well as a phosphorylation site for its degradation [1].
All transposases and recombinases are molecular acrobats; each enzyme must be capable of catalyzing two or more different reactions consecutively and undergoing conformational changes in the process. For example, an RNH-type transposase needs to hydrolyze a phosphodiester bond in one DNA strand and use the resulting 3′-OH as a nucleophile to either cleave the second strand by forming a hairpin, or integrate the transposon end into a target DNA [17,18] (Fig. 1a, d). RAG recombinase provides the first example of how a single active site is configured to accommodate DNA strands of different polarities and catalyze reactions of different chemical natures.
Features of RAG that insure asymmetric DNA pairing
RAG is composed of two RAG1 subunits, each containing the RNH-like catalytic center characterized by three acidic residues (DDE), and two RAG2 subunits with the β-propeller fold [1]. Based on the protein sequences and reaction chemistry, Hermes transposases are identified as homologs, and Transib the precursor, of RAG1 [19], but these transposases lack a RAG2-like partner. In 2015, RAG2 homologs (RAG2L) were finally found, forming protoRAG with RAG1L in sea urchin, oyster, starfish and lancelet, but these organisms lack V(D)J recombination [20,21]. Recently reported structures of Transib, protoRAG, zebrafish and mouse RAG and their complexes with substrate DNA [22*,23**,24**,25**,26**,27*,28**,29**,30*] (Table 1) shed a great deal of light on the evolution and reaction chemistry of these fascinating DNA enzymes.
Table 1.
Structures of the RAG recombinase, protoRAG and Transiba
| apo | PRC | NFC | HFC | SEC/TEC/TCC | STC | |
|---|---|---|---|---|---|---|
| Mouse RAG | 4WWX (3.2 Å) |
6CIK (3.15 Å)
6OEM (3.6 Å)/20030 |
6OER (3.3 Å)/20033
6OEP (3.7 Å)/20034b |
5ZE0 (2.75 Å)
5ZE1 (3.0 Å) 6CG0 (3.17 Å)/7470 |
5ZE2 (3.3 Å)c
6XNZ (3.8 Å)/22274d |
6XNY (2.9 Å)/22273
6OES (3.1 Å)/20036 |
| Zebrafish RAG | 6DBT (4.3 Å)/7849 |
6DBL (5.0 Å)/7845
6DBR (4.0 Å)/7848b |
6DBI (3.4 Å)/7843 | 3JBX (3.4 Å)/6487e | ||
| protoRAG | 7043 (5.3 Å)g | 6B40 (4.3 Å)/7046 | ||||
| Transib | 6PQN (3.0 1Å) | 6PQU (3.3 Å)/20453 | 6PQX (4.6 Å)/20455 | 6PQY (4.2 Å)/20456f | 6PRS (3.3 Å)/20457 |
PDB accession codes of crystal structures are colored in orange; PDB and EMDB accession codes of cryoEM structures are colored blue.
The model consists of a half of PRC and a half of NFC.
SEC (signal end complex) structure before the hairpin products are release.
TCC (target capture complex) with hairpin product replaced by target DNA for transposition.
SEC with the hairpins products replaced by blunt-end DNAs.
TEC (transposon end complex) structure, equivalent to SEC and with hairpin products released.
The structure coordinates are not available, and cryoEM map is deposited in EMDB.
Despite 400 million years of evolutionary separation, zebrafish and mouse both have adaptive immune systems, and their RAG proteins and RAG-DNA complexes are nearly identical (Fig. 2). The catalytic core of RAG1 contains the nonamer-binding domain (NBD), DNA-binding and dimerization domain (DDBD), pre-RNH (preR), RNH, Zn-binding modules (ZnC2 and ZnH2 or ZnB) inserted in the RNH domain, and the C-terminal domain (CTD) after RNH (Fig. 3). Two RAG1 subunits form a Y-shaped dimer via domain swapping between NBDs and dimerization of DDBD in the stem. The catalytic centers are located in the middle of the Y arms, and RAG2 caps each arm (Fig. 3a–c). RAG is thus a dimer of RAG1/2 heterodimers, and the two halves bind 12- and 23-RSS DNAs asymmetrically. The RSS DNAs bind the lower 2/3 of the Y-shaped RAG (below the active site), while the flanking DNA (V, D or J) segment binds the upper 1/3 and is clamped by RAG2 and ZnH2 (Fig. 3e).
Fig. 2.

Structure-based sequence alignment of RAG, protoRAG and Transib. (a) mouse and zebrafish RAG1, lancelet (Bb) RAG1L, and Transib alignment. (b) RAG2 and RAG2L alignment.
Fig. 3.
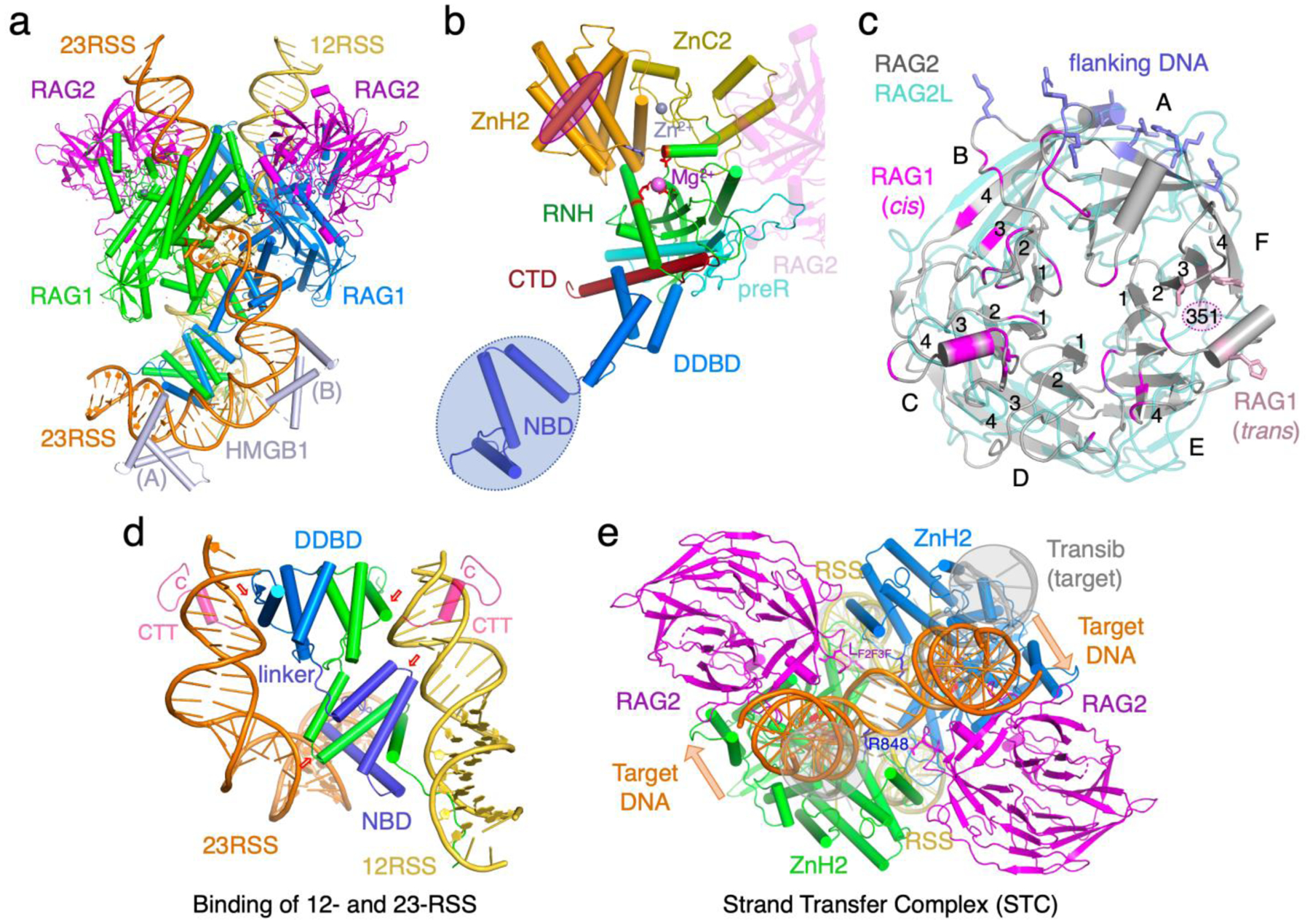
RAG and RAG-DNA complex structures. (a) The HFC structure is an example of the fully functional RAG complex (PDB: 5ZE0). The three catalytic carboxylates are shown as red sticks. The heptamer and nonamer sequences are shown with sugars and bases, while the rest of DNAs are shown as tube-and-ladder. (b) Structure of one RAG1 subunit with each domain color-coded and labeled. The interface with RAG2 in cis is marked by the partial structure of RAG2 (semi transparent), and the interface with RAG2 in trans is outlined by a magenta oval. (c) RAG2 is shown in grey with the three interfaces (DNA, cis and trans RAG1) highlighted in blue, magenta and pink, respectively. The protoRAG2L, which is superimposed (pale teal), differs from RAG2 in the β propeller structure, particularly in blades F and A, where interactions with RAG1 (in trans) and DNA occur. (d)A zoom-in view of the asymmetric RSS DNA-binding by the NBD and DDBD domains. The nonamers and 12-bp and 23-bp spacers (yellow and orange) sandwich the RAG1 dimer (blue and green) (PDB: 5ZE0). Two pairs of small red arrows mark equivalent regions in the dimeric RAG1. The CTT of protoRAG1L (PDB: 6B40, pink and superimposed onto the RAG structure) wraps around each DNA. (e) The RAG STC structure (PDB: 6OES). The flanking DNA of the integration target (orange) is superimposable with the coding flanks in HFC. The extra twists imposed by RAG2 are indicated by the orange arrows. R838 (RAG1) and LF2F3 (RAG2) next to the integration sites are shown in sticks and labeled. In the absence of RAG2, the target DNA in the Transib STC (PDB:6PR5, shown in semi-transparent grey and highlighted in grey circles) has no extra twist and is wider apart in the flanks. As the two sides of the target DNA are of different lengths, one is more obvious than the other. The highly distorted central 3 bp between two integration sites (in the center of this view) are well superimposed between RAG and Transib.
The NBD domains and RAG2, which occupy the extremities of the Y-shaped structure, are absent in the ancestral transposases and Transib, and their appearance is associated with V(D)J recombination. RAG2L in protoRAG is rather different from RAG2 in sequence and structure and is also devoid of the C-terminal 180 residues of regulatory functions [21] (Fig. 2b). Several long loops in RAG1 and RAG2 form an extended interface (Fig. 2, 3), which makes the two an integral entity. RAG2 appears to have the following three roles: binding the flanking gene segment in either the minor or major groove [22*], bridging protein interactions across the two Y-arms [24**], and regulating RAG activity and degradation [31–33].
NBD forms a domain-swapped dimer, and each NBD chain links two nonamers of an RSS pair like a shoelace threading back and forth (Fig. 3d). Adjacent to it, DDBD, which is conserved from Transib to RAG, binds the heptamer adjacent to the 12/23 spacer. NBD and DDBD are connected by a 6-aa linker spanning less than 20 Å. The constraints of binding the nonamer (by NBD) and heptamer (by DDBD) on the same side of each RSS DNA, which requires their separation by integral helical turns (12 and 23 bp or ~35 and 70 Å), and of binding two RSS DNAs, result in asymmetric tilting of the NBD dimer and the need for one short and one long DNA spacer. To accommodate the 6-aa linker between NBD and DDBD, both the 12- and 23-RSS are bent, 60° and 150° respectively. HMGB (1 or 2) is found to stabilize the severe DNA bending [22*,34].
Deletion of either NBD from RAG1 or the nonamers from RSS DNAs has little impact on RAG-DNA complex formation, but inactivates DNA cleavage completely [22*]. Structures of RAG at different stages of DNA cleavage indicate that the association of NBD and nonamer anchors the protein-DNA complex and enables the large conformational changes. In the absence of NBD, such as in protoRAG and Transib, a C-terminal tail (CTT) appended to CTD wraps around the DNA and thus stabilizes DNA for cleavage [28**] (Fig. 3d). But without NBD, protoRAG and Transib pair and cleave symmetric TIRs [28**,29**]. Although sequence differences in the shared RNH domain may contribute to the asymmetric RSS DNA cleavage [28**], the NBD domain and its association with the nonamer dictate the 12/23-rule in V(D)J recombination.
Conformational changes and reaction chemistry of RAG recombination
RAG undergoes a series of conformational changes in the process of binding a pair of RSS DNAs (pre-reaction complex, PRC), then first strand cleavage (nick-forming complex, NFC), and finally second strand cleavage (hairpin-forming complex, HFC) (Fig. 1d). Structures of mouse RAG, having the best resolutions and being in agreement with results from zebrafish, protoRAG and Transib, are used here to illustrate atomic details.
Compared to the apo-protein form, RAG opens two arms by swinging out its ZnH2 domains and tilting NBD domains to bind 12- and 23-RSS DNAs. But in PRC, the “wrong” strand is near the catalytic center, and the active site is incomplete (Fig. 4a) [22*]. The third catalytic carboxylate (E962 or its equivalent) is 10Å away from the other two (D600 and D708). To bring the “right” strand into the catalytic center, each DNA has to unwind by nearly 180°, while forming a “DNA zipper”, in which the two antiparallel strands are in a plane and joined by interdigitating base stacking [24**] (Fig. 4b). Unwinding also effectively “lengthens” the DNA, thus placing the scissile phosphate in the active site, which in turn promotes repositioning of E962 and capture of two Mg2+ in the fully formed active site [24**]. DNA unwinding and zipper formation occur at the 2nd and 3rd bp of the the totally conserved CAC heptamer sequence. Alternating purine and pyrimidine (AC, AT or GT) predispose DNA to distortion and unwinding [24**,35*]. Accompanying the DNA transformation from PRC to NFC, the NBD and nonamers tilt toward the 23RSS side by 5°, and the ZnH2 domain undergoes a ~5Å movement in the direction opposite the DNA unwinding. The joint movements stabilize the DNA zipper in NFC.
Fig. 4.
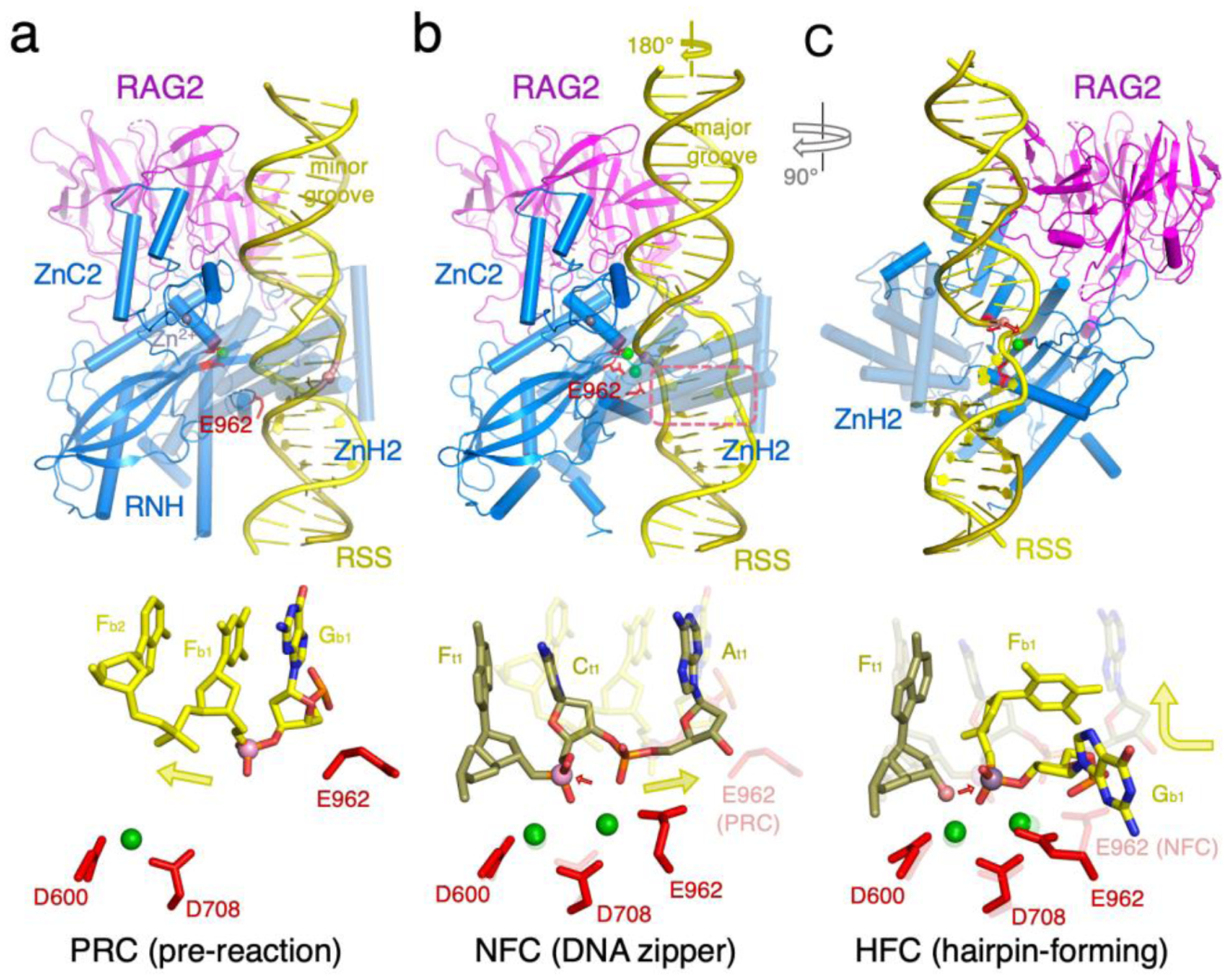
Double-strand DNA cleavage by RAG. (a) One RAG arm bound to the heptamer and flanking DNA in the PRC (PDB: 5OEM). RAG2 mainly interacts with the DNA minor groove. The wrong DNA strand (yellow color) is near the active site, and the active site is incompletely formed with E962 far away. A zoom-in view of the active site is shown below. The heptamer is labeled according to the sequence, and the flanking DNA is labeled as “F”. For both, base positions are labeled in subscript. (b) The same region in the NFC structure (PDB: 6OER). The DNA is unwound by 180°, so RAG2 interacts mainly with the major groove. The 2nd and 3rd base pairs in the heptamer form a zipper (ourlined in a pink box). The strand (olive color) to be nicked is placed in the fully formed active site. In the zoom-in view below, the superimposed PRC structure is semi-transparent. (c) The same region in the HFC (PDB: 6CG0 and 5ZE1) is shown after a 90° rotation. The hydrolysis product of the first (olive) strand, 3′-OH (shown as a pink ball), is poised to cleave the second strand (yellow). The three catalytic carboxylates are shown as red sticks, and green spheres are Ca2+ or Mn2+ ions. In the zoom-in view of the active site, the superimposed NFC structure is semi-transparent.
To place the second DNA strand in the active site for cleavage and hairpin formation (HFC), the two arms of RAG undergo a scissor-like 12–14° closing motion, pivoting around the NBD-DDBD domains[24**]. Accompanying the large movement, the 6-residue linker between the NBD and DDBD on the 23RSS side stretches by >2Å. Meanwhile, the nucleophile 3′-OH generated in the hydrolysis reaction remains in the active site (Fig. 4c). The second strand to be cleaved, whose polarity is opposite the first one, is bent 90° at the scissile phosphate to fit in the active site (Fig. 4c). Accompanying the large protein and DNA movement, E962 is reconfigured by a change of rotamer conformation in HFC to accommodate the different DNA substrate. In HFC, heptamer recognition is again mainly based on the deformed structure of AC and TG dinucleotides with only two to three specific hydrogen bonds between protein and DNA [22*].
With mouse and zebrafish RAG, formation of the unusual DNA zipper appears to be a bottleneck in the cleavage process [24**,27*]. NFC is a weakly populated state even when one of the two RSS DNAs is nicked; pairs of intact and singly nicked RSS DNAs predominantly assume the PRC state [22*,24**]. NFC structures can be “isolated” only during image processing by the cryoEM technique. HFC, however, readily forms if both RSS DNAs are nicked. For instance, crystal structures of mouse HFC were determined up to 2.75Å resolution, and the hairpin-forming process was recorded in crystallo [22*]. In the absence of RAG2 and RAG2L, the structure of Transib’s NFC is still unresolved, and HFC appears unstable. Even in the presence of two nicked DNAs, Transib predominantly forms PRC [29**].
RAG2 and ZnH2 may prevent transposition
In accordance with the rarity of transposition events in vivo, mouse RAG efficiently catalyzes the disintegration reaction, which is the reversal of transposition (integration) [25**]. In contrast, transposases are usually inefficient at disintegration [16,36]. The structure of the strand-transfer complex (STC), in which two RSS ends are inserted into a target DNA 5 bp apart and RAG is poised to disintegrate them, is superimposable with the HFC structure [25**,30*]. The severely distorted structure of the target DNA suggests why RAG may be a poor transposase. The 5 bp target DNA is bent 85° twice 1 bp away from each target-RSS junction, and thus assuming a “U” shape. The central 5 bp between RSS insertions has a dramatically expanded major groove and extremely narrow minor groove [25**,30*]. The interactions between the flanking DNA and RAG2 lead to addition “arm” twists of the sharply bent target DNA (Fig. 3e). In STC complexes of most transposases, target DNAs are often bent and have expanded major groove for transposition to take place, but the severe bending often occurs right at the integration site (target-TIR junction) preventing disintegration, and not 1 bp away [25**].
RAG2, which binds the distal ends of a target DNA and contacts both RAG1 subunits in the same (cis) and the other Y arm (trans), determines the overall shape of the target DNA. In addition, ZnH2 (particularly R848) and LF2F3 of RAG2 contact and stabilize the sharply kinked DNA near the integration sites (Fig. 3e). Deletion of 4 residues at the tip of LF2F3 indeed increases transposition by 2–3 fold while having no effect on DNA cleavage by mouse RAG [30*]. The results confirm that the severely bent and twisted target DNA is a barrier to transposition. Unexpectedly, mutation of R848 at the DNA kink to Ala accelerates all reactions catalyzed by mouse RAG [25**], but disintegration is enhanced less than the forward reaction. We suspect that R848 serves as a molecular brake, with the result of reduced transposition. Biochemical data from the Schatz group point to further inhibition of transposition by the C-terminal regulatory region of RAG2 beyond the catalytic core [28**].
Interestingly, Transib’s STC is superimposable with mouse STC in the RSS (TIR) regions and the 5-bp between two integration sites. But Transib’s flanking DNAs are wider apart, and the target DNA appears more smoothly bent with no additional twists (Fig. 3e). In the absence of RAG2, the flanking DNA and associated ZnB of Transib are more flexible than the DNAs bound to RAG, and Transib releases the flanking DNA readily after hairpin formation [29**]. In protoRAG, RAG2L is loosely attached to RAG1L in cis with a small interface and doesn’t contact RAG1L in trans. As a result, the two flanking DNAs are further apart than those bound to mouse RAG [28**] (Fig. 3e). RAG2L also has a much shorter LF2F30 loop (Fig. 2) and is unlikely to reach to the integration sites to stabilize the severely distorted target DNA. We suspect that the structural rigidity imposed by RAG2 may further inhibit transposition.
Conclusion
The detailed structural and biochemical studies of the RAG recombinase and its predecessors provide an elegant case study of evolutionary processes. Stage by stage, alterations have repurposed a biological process to a completely different function, while maintaining the same basic chemistry throughout. It is also striking that a standard RNH catalytic site is used to cut the strand of DNA opposite to that first bound by forcing the DNA to unwind, and that a second DNA motion is required to allow the hairpinning reaction in the same site. Further adaptations will undoubtedly become apparent when the regulatory domains are incorporated in the overall structure.
Acknowledgement
This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases intramural grants DK036167 (M.G.); DK036147 and DK036144 (W.Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no conflict of interest.
References
- 1.Schatz DG, Swanson PC: V(D)J recombination: mechanisms of initiation. Annu Rev Genet 2011, 45:167–202. [DOI] [PubMed] [Google Scholar]
- 2.Montano SP, Rice PA: Moving DNA around: DNA transposition and retroviral integration. Curr Opin Struct Biol 2011, 21:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hickman AB, Voth AR, Ewis H, Li X, Craig NL, Dyda F: Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase. Nucleic Acids Res 2018, 46:10286–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *4.Ghanim GE, Kellogg EH, Nogales E, Rio DC: Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts. Nat Struct Mol Biol 2019, 26:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the unique structural features and biochemical validation of how P element transposase recognizes the terminal inverted repeats with the assistance of GTP.
- *5.Chen Q, Luo W, Veach RA, Hickman AB, Wilson MH, Dyda F: Structural basis of seamless excision and specific targeting by piggyBac transposase. Nat Commun 2020, 11:3446. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structures of how the piggyBac transposase asymmetrically binds two identical TIR DNA ends and specifically inserts them into a DNA target containg AATT sequence.
- 6.Craig NL: Unity in transposition reactions. Science 1995, 270:253–254. [DOI] [PubMed] [Google Scholar]
- 7.Arinkin V, Smyshlyaev G, Barabas O: Jump ahead with a twist: DNA acrobatics drive transposition forward. Curr Opin Struct Biol 2019, 59:168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiom K, Melek M, Gellert M: DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell 1998, 94:463–470. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal A, Eastman QM, Schatz DG: Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 1998, 394:744–751. [DOI] [PubMed] [Google Scholar]
- 10.Reddy YV, Perkins EJ, Ramsden DA: Genomic instability due to V(D)J recombination-associated transposition. Genes Dev 2006, 20:1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterji M, Tsai CL, Schatz DG: Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol 2006, 26:1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S, Okazaki K, Sakano H: Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science 1987, 238:1134–1138. [DOI] [PubMed] [Google Scholar]
- 13.Ramsden DA, Baetz K, Wu GE: Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res 1994, 22:1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapkouski M, Chuenchor W, Kim MS, Gellert M, Yang W: Assembly Pathway and Characterization of the RAG1/2-DNA Paired and Signal-end Complexes. J Biol Chem 2015, 290:14618–14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rousseau P, Loot C, Turlan C, Nolivos S, Chandler M: Bias between the left and right inverted repeats during IS911 targeted insertion. J Bacteriol 2008, 190:6111–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montano SP, Pigli YZ, Rice PA: The mu transpososome structure sheds light on DDE recombinase evolution. Nature 2012, 491:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gellert M: V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 2002, 71:101–132. [DOI] [PubMed] [Google Scholar]
- 18.Craigie R: Nucleoprotein Intermediates in HIV-1 DNA Integration: Structure and Function of HIV-1 Intasomes. Subcell Biochem 2018, 88:189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapitonov VV, Jurka J: RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol 2005, 3:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapitonov VV, Koonin EV: Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct 2015, 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, et al. : Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell 2016, 166:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Kim MS, Chuenchor W, Chen X, Cui Y, Zhang X, Zhou ZH, Gellert M, Yang W: Cracking the DNA Code for V(D)J Recombination. Mol Cell 2018, 70:358–370 e354. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the highest resolution structures of RAG recombinase and detailed protein-DNA interactions and intermediates of hairpin formation.
- **23.Kim MS, Lapkouski M, Yang W, Gellert M: Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 2015, 518:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first crystal structure of hetero-tetrameric RAG catalytic core. This study defines the architecture and domain structures of mouse RAG and lays the foundation for cryoEM analysis.
- **24.Chen X, Cui Y, Best RB, Wang H, Zhou HZ, Yang W, Gellert M: Cutting antiparallel DNA strand in a single active site. Nat Struct Mol Biol 2020, 27:119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper includes the cryoEM structures of mouse RAG in PRC, NFC and HFC and reveals conformational changes, formation of the DNA zipper, and active site rearrangements in the process of cleaving two antiparallel DNA strands.
- **25.Chen X, Cui Y, Wang H, Zhou ZH, Gellert M, Yang W: How mouse RAG recombinase avoids DNA transposition. Nat Struct Mol Biol 2020, 27:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first RAG STC structure (DNA strand transfer and disintegration state). This study reveals the cooperation of RAG1 and RAG2 in distoring the target DNA to suppress DNA transposition and the role of R848 in favoring disintegration.
- **26.Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H: Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell 2015, 163:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first struture of a RAG recombinase complexed with 12- and 23-RSS DNAs. This study also shows the power of cryoEM in characterizing heterogeneous macromolecular assemblies.
- *27.Ru H, Mi W, Zhang P, Alt FW, Schatz DG, Liao M, Wu H: DNA melting initiates the RAG catalytic pathway. Nat Struct Mol Biol 2018, 25:732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the NFC and PRC structures of zebrafish RAG.
- **28.Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, et al. : Transposon molecular domestication and the evolution of the RAG recombinase. Nature 2019, 569:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structures of lancelet protoRAG in nick- and hairpin-forrming complexes and two-tiered control evolved from protoRAG to RAG to avoid transposition.
- **29.Liu C, Yang Y, Schatz DG: Structures of a RAG-like transposase during cut-and-paste transposition. Nature 2019, 575:540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive series of Transib structures from apo protein to STC.
- *30.Zhang Y, Corbett E, Wu S, Schatz DG: Structural basis for the activation and suppression of transposition during evolution of the RAG recombinase. EMBO J 2020, 39:e105857. [DOI] [PMC free article] [PubMed] [Google Scholar]; Structural, mutagenic and biohemical studies pinpoint the regions in RAG1 and RAG2 that suppress transposition.
- 31.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. : RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 2007, 450:1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W: The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A 2007, 104:18993–18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin WC, Desiderio S: Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science 1993, 260:953–959. [DOI] [PubMed] [Google Scholar]
- 34.van Gent DC, Hiom K, Paull TT, Gellert M: Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J 1997, 16:2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Rubio-Cosials A, Schulz EC, Lambertsen L, Smyshlyaev G, Rojas-Cordova C, Forslund K, Karaca E, Bebel A, Bork P, Barabas O: Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance. Cell 2018, 173:208–220 e220. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first report of interdigitating base stacking when DNA is unwound by ~60°.
- 36.Maertens GN, Hare S, Cherepanov P: The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 2010, 468:326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]


