Abstract
The development of genetically-encoded tools to record and manipulate neurons in vivo has greatly increased our understanding of how neuronal activity affects behavior. Recent advances enable the use of these tools in species not typically considered genetically tractable. This progress is revolutionizing neuroscience in general, and insect neuroethology in particular. Here we cover the latest innovations and some of their applications in phylogenetically-diverse insect species. We discuss the importance and implications of these approaches for both basic and translational research. We focus on genetically- and virally-encoded tools used for calcium imaging, optogenetics, and synaptic silencing. Finally, we discuss potential future developments of universally applicable, modular, and user-friendly genetic toolkits for neuroethological studies of insect behavior.
Introduction
The last few decades have produced rapid progress in the control of gene expression and manipulation in specific neural tissues and neuron types. The main drivers behind these advances have been traditional genetically-tractable animal species such as the vinegar fly Drosophila melanogaster, the zebrafish Danio rerio, the mouse Mus musculus, and the nematode Caenorhabditis elegans. Their success largely derived from the fruitful integration of classical genetics and molecular biology. By borrowing components of organisms like bacteria, yeast, algae, fungi, and jelly fish scientists engineered new molecular tools to probe the nervous system with high spatial and temporal resolution [1,2]. Subsequently, these tools have greatly advanced the mapping of neural circuits to behavior.
Independently, neuroethological approaches have advanced our understanding of brain function by integrating behavioral, neurophysiological and computational methods in a comparative and ecological framework. Such studies have focused on a wide range of behaviors in vertebrate and invertebrate species, e.g., vocal learning in songbirds [3], auditory sound localization in barn owls [4], the electrical jamming avoidance response of weakly electric fish [5], olfactory processing in several insect species [6], including honeybees [7] and grasshoppers [8], sound discrimination in crickets and grasshoppers [9,10], pheromone scent tracking in silk moths [11,12], and air displacement sensing in the cercal system of crickets [13]. This approach was designed to provide a comprehensive explanation of behavior summarized by four distinct levels of analysis: (i) the function of a particular behavior, (ii) its phylogenetic origin, and, most importantly, (iii) its neurobiological implementation, and (iv) development. Tinbergen, one of the founders of modern ethology along with von Frisch and Lorenz, proposed this scheme [14–17]. The merging of these ethological principles and neurobiological methods led to the field of neuroethology that favors the selection of a diverse set of ‘specialists’ for studying specific behaviors [18].
Extending genetic methods to such species would greatly expand our perspective and usher in a new era of comparative neuroethological research. This type of transgenesis – the process of integrating exogenous or modified genes, i.e., transgenes, into the genome of a recipient organism – might have seemed a distant dream only a few years ago. However, it is now starting to be broadly applied based on the ever-decreasing costs of sequencing and the recent development of precise genome editing tools. Here, we review the techniques at play and specific examples of their applications to insect neurons, focusing on work not covered extensively earlier [19].
Approaches to transgenesis
With the exception of Drosophila until recently, transgenic manipulations of most insect species completely depended on the use of modified transposons such as piggyBac [20]. However, the rapid development of CRISPR/Cas9-dependent genome editing [21,22], derived from a bacterial antiviral acquired immunity system, is blurring the boundaries between ‘genetically tractable’ and ‘non-genetically tractable’ model systems (CRISPR stands for ‘clustered regularly interspaced short palindromic repeats’, a component of this adaptive immune system, and Cas9 for ‘CRISPR associated nuclease, type 9’). Both transposon- and CRISPR/Cas9-mediated genome transformations usually depend on the injection of nucleic acids/proteins into embryos to target the future germline cells thus resulting in heritable transgene integration (alternative delivery routes for CRISPR/Cas9 are described below). The main difference between these techniques is that transposons are typically inserted semi-randomly (see below) while CRISPR/Cas9 can target any specific site in the genome, giving it higher precision and flexibility. A hybrid natural system of CRISPR/Cas9-guided transposition was also recently discovered [23,24].
The piggyBac transposon.
Natural transposons are mobile elements that can extract themselves from their location in the genome and re-insert at another random location. First discovered in maize [25], such sequences exist in many organisms including insects. The piggyBac transposon consists of two 13 base pair (bp) long inverted terminal repeat sequences flanking a gene that encodes a transposase enzyme [20]. When the transposase is expressed, it will recognize the inverted terminal repeat sequences, excise the transposon and then reinsert it at a random position in the genome, but preferentially into TTAA sequences. Such a transposon can be used for genetic engineering by replacing the genetic material between the two inverted terminal repeat sequences with a transgene of interest. The transposase enzyme required for inserting the modified transposon into the genome of the host organism is provided separately, e.g., by a co-injected helper plasmid or an mRNA that encodes it (Fig. 1).
Figure 1: Basic elements of transposon transgenesis.

(a) Structure of a natural transposon. Transposons have two essential components: inverted terminal repeats (1) that flank a transposase (2) encoding gene. The transposase recognizes the inverted repeats, excises the sequence and transpose the element to a new place in the genome, typically in a recognizable target sequence. ORF: open reading frame.
(b) Binary transposition systems. Scientists have engineered natural transposons into a binary transposition system, where one element has the inverted repeats flanking genetic cargo that replaces the transposase coding sequence to move these genetic components into a genome. These immobile elements can only move in the presence of a transposase-producing element that lacks the inverted repeats and therefore cannot move into a genome itself but can move inverted repeat surrounded cargo elements when they are co-injected into early embryos targeting their future germ line for integration events.
(c) Three step transposition process. A transposase in the form of a helper plasmid or produced by expression from a natural transposon will recognize the inverted terminal repeat sequences, bend the DNA to bring the transposon ends together before excising the element creating double strand breaks that are repaired. The transposase bound to the excised DNA brings the element to a new target site and catalyze an integration event moving the DNA into a new location.
CRISPR/Cas9.
Genome editing with this tool involves an engineered single guide RNA (sgRNA) that recognizes a specific target sequence in the genome, and a nuclease capable of double strand breaks, Cas9 [26]. The combination of sgRNA and Cas9 directs the double strand break to the specific location encoded by the sgRNA sequence, thus allowing to select with high specificity the targeted region for genome editing. The double strand break is then repaired by the cellular machinery through non-homologous end-joining (NHEJ), which will sometimes introduce a small mutation that can inactivate the targeted gene. Alternatively, the double strand break can be mended by homology directed repair (HDR) from a template provided along with the sgRNA/Cas9 enzyme complex and used to introduce a transgene at the targeted location. With two different sgRNAs, a repair template can be used to replace existing sequences with any transgene of interest (Fig. 2).
Figure 2: Crispr/Cas9 mediated genome editing.

An engineered single guide RNA (brown sequence) binds to the Cas9 enzyme and recognizes a specific sequence (green sequence) in the target DNA complementary to the guide RNA and immediately adjacent to the PAM sequence (red). The Cas9 enzyme will generate a double strand break four base pairs (bp) from the PAM sequence. The double strand break will be repaired by one of two mechanisms: 1) non-homologous end-joining (NHEJ, left pathway), which can cause a small insertion or deletion (InDel) if repair is incorrect, or 2) homology directed repair (HDR, right pathway) which requires a repair template with homology on both ends immediately adjacent to the cut site. Through homologous recombination anywhere in the flanking sequence the repair template can integrate a transgene sequence (surrounded by the homology arms) into the cut site. This pathway allows knocking DNA into the genome. If two guide RNAs are used, the repair process can replace an existing sequence in the genome with the transgene sequence.
Transgenic payloads
The transgenes delivered by transposon or CRISPR/Cas9 editing fall into three general categories briefly discussed below: reporters, effectors, and components of binary expression systems, which have revolutionized the use of reporters and effectors. An extensive list of these categories and their use in Drosophila has been covered elsewhere [2].
Reporter proteins include fluorescent proteins that when expressed under the control of a specific promoter will allow visualization of the cells in which the promoter is active. These include green fluorescent protein (GFP) and its engineered modifications that fluoresce at different wavelengths as well as others such as the red marker tdTomato [27]. In some cases, these fluorescent proteins can be targeted to specific cellular sub compartments such as the nucleus, the lysosome, or the cell membrane [28,29]. Another widely used reporter is the GCaMP family of calcium indicators (e.g., GCaMP6s, where ‘6’ is the generation number, and ‘s’ denotes a slow, high sensitivity version) [30,31]. These indicators change their fluorescence with intracellular calcium concentration and thus allow indirect monitoring of neuronal activity since changes in calcium concentration are typically caused by changes in membrane potential through the opening and closing of calcium channels. Genetically encoded voltage indicators also exist but are more difficult to use [32].
Among effector proteins, channel rhodopsin is a blue light-gated cation channel which enables positive ion flow into neurons when open [33,34]. This channel and its red-shifted variants make it possible to excite neurons with a light pulse and directly perturb their activity in resolved time windows. Inhibitory versions that target chloride/proton pumps or anion channels exist as well [35–37]. Tetanus toxin light chain (TeTxLC) is another effector protein, which blocks neuronal activity-evoked synaptic transmission (see below) [38].
Binary expression systems allow the flexible use of reporters and effectors expressed in a tissue specific manner. The first of these, GAL4-UAS, was originally developed, and remains ubiquitously used, in Drosophila [39]. The yeast GAL4 gene encodes a transcription activator that binds to the DNA upstream activating sequence (UAS) thereby driving expression of genes downstream of UAS. Thus, when GAL4 is under control of a cell- or tissue-specific promoter it drives expression of a gene downstream of UAS encoding a reporter or effector protein in a cell- or tissue-specific manner. The GAL4-UAS system was systematically improved in Drosophila by modifying different features of the construct to optimize expression [40,41]. By maintaining independent tissue specific promoter-GAL4 lines and UAS-reporter/effector lines, a simple cross allows to express any of these reporters or effectors in the desired tissue. The tissue specificity of GAL4 expression depends on the promoter that drives GAL4 and can derive from a specific gene or a cloned promoter or enhancer element [42]. The Q system (QF2-QUAS) is another, independent binary expression system derived from the bread fungus Neurospora crassa [43,44]. This system was also developed in Drosophila where its use with GAL4/UAS added to the sophistication of genetic manipulations [43]. Just as for GAL4-UAS, the QF2 transcription factor under expression of an endogenous promoter activates genes cloned downstream of a QUAS promoter. LexA-LexAop is the third available binary system in Drosophila, that originated from the λ phage [45]. For these binary expression systems, methods to further restrict expression to specific neuronal populations or within selected temporal windows sometimes exist. For example, the repressor GAL80 expressed in a tissue specific manner suppresses the activity of GAL4 [46]. Similarly, QS is a suppressor of the Q system that can be inactivated within a selected temporal window by administration of an exogenous drug [43]. A similar innovation for GAL4 drivers not yet widely adopted is the drug inducible variant, called GeneSwitch, which also allows temporal control of the driver in addition to tissue specificity [47,48].
Transposon-based transgenesis
Transposon-based genetic engineering has a long history in Drosophila [49,50] but its most commonly used transposon, the P-element, never worked in non-drosophilid insects [51]. The piggyBac transposon described above was discovered in the cabbage looper moth and first used to transform the Mediterranean fruit fly [52]. Later, it was shown to work in many other insects [53], including the silk moth Bombyx mori [54].
A subsequent powerful application of piggyBac transformation in Bombyx was its use in adapting the GAL4-UAS expression system [55]. Further improvements similar to those developed in Drosophila were exploited in Bombyx, leading to reliable expression of GFP, GCaMP, and TeTxLC. These tools helped refine the understanding of how the sex pheromone bombykol impacts male mating behavior, and how ecdysone and insulin-like hormones affect development in a sex-specific manner [56].
CRISPR/Cas9-dependent gene knock-out and knock-in
The semi-random integration pattern of transposons across the genome does not allow targeting specific DNA sequences for modification. Targeted loss-of-function mutations are the bread-and-butter of CRISPR/Cas9. They have been used in a range of insect species to answer questions related to many aspects of their biology, including the role of sensory perception in behavior [19]. One example sheds light on an intriguing phenotype well-known in grasshoppers [57]. Several grasshopper species possess the ability to transition from their usually solitarious existence to a gregarious one, where they aggregate in swarms that migrate collectively [58,59]. This phenomenon, known as locust phase polyphenism, carries important economical and human implications as locust swarming repeatedly devastates the crops and livelihood of African, Asian, and South American countries and sometimes even large parts of these continents. The mechanisms that regulate the transition from the solitarious to the gregarious phase are not fully understood but rely in part on olfaction. One potential aggregation pheromone is 4-vinylanisole (4VA). Behavioral experiments confirmed that 4VA is attractive to Locusta migratoria and emitted by gregarious animals in a density-dependent manner. The olfactory receptor OR35 detects 4VA and its knock-out by CRISPR/Cas9 triggered NHEJ resulted in lost attraction to 4VA in Or35−/− mutants. The attraction of Locusta migratoria to 4VA was confirmed in the wild, suggesting that it might be used for locust pest control. Another application of CRISPR/Cas9 mediated gene knock-out revealed the existence of a new pheromone receptor unrelated to similar previously described receptors in moths. This suggests that a class of pheromone receptors may have evolved independently at least twice in this insect family [60].
In mosquitoes, CRISPR/Cas9 gene knock-out was rendered more efficient by endogenous expression of Cas9 through insertion of this transgene using piggyBac. Lines expressing Cas9 endogenously may allow the development of mosquito population control strategies [61]. One application of gene knock-out in this species has shown that the mechanisms for ammonia sensing, a component of human sweat, likely differ in vinegar flies and mosquitos [62].
HDR-mediated knock-in of genetic material in generic insects is still rare but that is rapidly changing. One example illustrating the power of this approach, particularly when combined with targeted RNA sequencing (RNAseq) analysis, comes from the mosquito Aedes aegypti [63]. The choice between blood and nectar feeding is well-regulated in female mosquitos but the mechanism of this regulation is poorly understood. Using a combination of tools delivered with piggyBac transposons or CRISPR/Cas9, the mechanism of these feeding modalities is now better understood. The Gr4a receptor locus in Ae. aegypti encodes the gustatory receptor most closely related to sugar-detecting receptors of Drosophila. A knock-in of QF2 in this locus was combined with a piggyBac integrated QUAS-TrpV1 co-expressing a red fluorescent protein (Fig. 3). Red fluorescence showed that Ae. aegypti’s Gr4a is expressed in the labium and the legs, the two major taste appendages of insects. Activation with capsaicin, the chemical trigger of TrpV1, was sufficient to drive the knock-in animals to consume water but not blood, suggesting that nectar feeding is initiated when Gr4a expressing neurons are activated. Blood feeding in female mosquitos is done with a syringe-like appendage called the stylet used to pierce the skin of their prey and operated like a straw to suck up blood. The stylet is surrounded by chemosensory neurons that may play a role in blood detection. A knock-in of QF2 in the pan-neuronal Brp locus (see also [64]) combined with a piggyBac integrated QUAS-GCaMP6s (see also [65]) showed that these neurons respond to blood. They also respond to sugar, salt, sodium bicarbonate (NaHCO3) and ATP, a mixture triggering consumption of blood in females [66]. By exposing each of these components separately and evaluating Ca2+ responses, five clusters of neurons were detected. A comparative RNAseq analysis of the female stylet identified two putative ionotropic receptor encoding genes, Ir7a and Ir7f that may play a role in detecting the chemical components triggering blood engorgement. A QF2 knock-in targeting these genes showed that they are expressed by a small subset of non-overlapping stylet neurons, and only in females. Ca2+ responses showed that Ir7a expressing neurons respond to NaHCO3 while Ir7f expressing neurons respond to the whole mix, thus likely representing ‘integrator’ neurons. Interestingly, neither of these neuron clusters responded to nectar-specific sugars.
Figure 3: Genetic engineering strategies in non-model system insects.
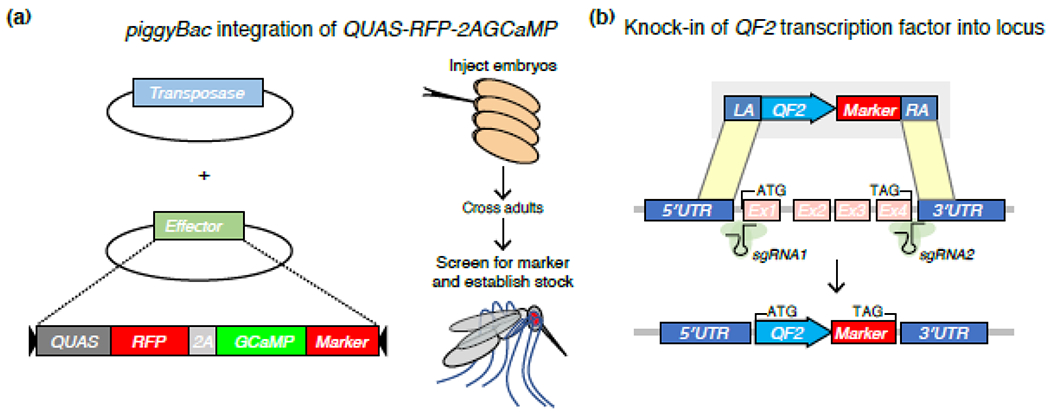
(a) Transgenesis with piggyBac. Embryos are injected with a mixture of helper plasmid, providing the transposase source, and the effector plasmid. The effector plasmid is a transposon that lacks the transposase but still has the inverted repeats that can be recognized by the transposase and therefore integrate in the genome at random locations typically in a TTAA sequence. Injected fertilized eggs will undergo transgenic integrations at low frequency in the cells that will become the future germline of the embryo. Adults that emerge from the injected embryos are crossed to animals of the opposite sex and screened for transformed offspring. The transgenic offspring can be recognized because the effector plasmid that integrates in the genome contains a screenable marker. The screenable marker typically is a fluorescent protein that is expressed with an eye specific promoter making the eyes of transgenic animals fluorescent [91]. The example effector shown here contains a QUAS response element that will respond to the QF2 transcriptional activator that is generated in a separate animal. The example effector expresses a Ca2+ indicator, GCaMP, that will respond with an increase in fluorescence when Ca2+ levels in the cell increase [92]. To mark the GCaMP expressing cells, a 2nd fluorescent protein can be expressed from the same QUAS response element (e.g., red fluorescent protein, RFP). Both proteins are produced from the same transcript because they are linked by a viral 2A sequence which leads to bond skipping during translation at the penultimate amino acid of the 2A sequence separating the 2 protein from each other [93].
(b) Crispr/Cas9 mediated knock-in. The cartoon shows a knock-in strategy of the QF2 transcriptional activator in a gene with four exons that will be replaced with the QF2 sequence driving expression of QF2 in the spatio-temporal pattern of the gene that is replaced. To generate the knock-in, embryos are injected as in the piggyBac strategy in (a), but instead of a transposase, the Cas9 enzyme is injected with sgRNAs (both can also be provided as plasmids expressing these sequences) that will target the Cas9 enzyme to the two recognition sites cutting the gene out of its location. By providing a repair template plasmid that is co-injected, the gene can be replaced with the QF2 sequence. The replacement happens through HDR via two ~1kbp flanking homology arms that have identical sequences to those around the sgRNA cut sites in the host DNA (left arm, LA and right arm, RA). To screen the injected embryos for transgenic transformation, surviving adults are crossed as above and screened for the marker that again indicates a successful transgenic event. The event is also molecularly verified to make the sure the integration occurred as intended.
Another knock-in application of CRISPR/Cas9 in mosquitoes suggests that their olfactory system may be organized unconventionally, with multiple olfactory receptors co-expressed in single olfactory neurons. Further, convergence of olfactory receptor neurons expressing different receptors in the antennal lobe may not be as strictly segregated as documented earlier in such widely divergent species as mice and vinegar flies (Younger et al., bioRxiv doi: 10.1101/2020.11.07.368720). Similar observations have now also been reported in Drosophila (Task et al., bioRxiv doi: 10.1101/2020.11.07.355651).
Comparing transposon and CRISPR/Cas9 mediated transgenesis
As is clear from the above discussed examples, transposon mediated transgenesis has long been widely used. Its major advantages are broad applicability [53] and relatively high efficiency, even in insects difficult to transform genetically [67]. Its major weakness is that it is not targeted. Most transposons integrate randomly into the genome, with some bias. As a result, transposons can accidentally inactivate a gene causing an unintended phenotype. In Drosophila transposons were in fact used early on to mutagenize genes and rapidly identify the resulting mutation [68,69]. In addition to potential mutagenicity, the integrated material is subject to position effects, which can affect expression of the transgene because of the genomic context of the integration site [70]. This makes it hard to compare effectors inserted at different sites.
These problems have been bypassed in Drosophila by using a viral integration system based on the bacteriophage integrase φC31 and its complementary DNA docking sites, attP and attB [71]. The attP docking site is first randomly inserted in the genome through transposition. The different strains generated with specific docking sites [72] can then be used to integrate effectors using φ031, a plasmid with the complementary attB site, and the cloned payload. Different transgenes integrated in the same site can be compared because they are subject to the same genomic context. This site-specific integration system is now also used in mosquitos [73].
In principle, Crispr/Cas9 based genome-editing addresses all the disadvantages of transposons. However, it can lead to off target effects despite its precision [74]. In addition, the efficiency of HDR mediated integration is still not as high as random or site-directed integration. In the long run, as Crispr/Cas9 transgenesis becomes more efficient and reliable, it may reduce the role of transposition in genome modification or even usurp transposition as in a recently developed hybrid method [23,24].
Other delivery routes for Crispr/Cas9 genome editing
As mentioned earlier, the molecular components necessary for genome editing are typically injected into an early embryo to target its future germline (but see [61], discussed above). The editing cocktail is often entirely plasmid-based, but it can be a mixture of gRNA, Cas9 protein or RNA and a donor plasmid for repair. When targeted microinjection is difficult such as for insects with small embryos, an alternate method exploits receptor-mediated endocytosis in the ovaries. In this approach, the gene-editing cargo is combined with a ligand derived from yolk protein precursors. The molecular cargo is then delivered to the ovaries of females, so it can be deposited in oocytes during yolk formation (vitellogenesis). This technique referred to as Receptor-Mediated Ovary Transduction of Cargo (ReMOT Control) has been demonstrated in several insects including jewel wasps (Nasonia vitripennis [75]), mosquitos [76] and whiteflies (Bemisia tabaci [77]). An adaptation of the ReMOT control approach was also proposed in insect models where yolk protein precursors are not known or are difficult to synthesize. In this approach, delivery to ovaries is achieved using nanosphere peptides that encapsulate ribonucleoproteins [75].
Optogenetics through viral transfection in generic insects
Transient transfection of insect neurons using recombinant viruses represents an alternative to germline transgenic transformation. Although not yet implemented in insects, this method is often combined with complex transgenesis in mice [78]. Its advantage is that it does not involve genome transformation, making it simpler to implement. The disadvantage is that it is limited in its specificity and reproducibility, which are mainly determined by the injection site and the number of viral particles injected into the animal. The Sindbis virus has been used to study development of butterfly wings and of beetle embryos [79]. Recently, the Semliki forest virus, an alpha virus closely related to Sindbis, was used in grasshoppers to deliver channelrhodopsin [80]. This technique allowed channelrhodopsin expression in excitatory neurons presynaptic to an identified collision-detecting visual neuron of the grasshopper optic lobe, the lobula giant movement detector (LGMD) [81]. Subsequent stimulation through pulses of laser light elicited vigorous spiking in the LGMD, thus demonstrating that optogenetics can be implemented in non-genetic model insects using virus-mediated delivery (Fig. 4).
Figure 4: Viral transfection of insect neurons using Semliki Forest Virus (SFV).

(a) The main effector plasmid codes for the non-structural SFV proteins one to four (SFVnsP1-4), for an effector protein such as channelrhodopsin (ChR) and a fluorescent marker (Venus) under control of a ubiquitous promoter (CMV). Additionally, the SFV structural proteins are provided in a distinct helper plasmid so as not to be packaged during viral production, resulting in the fabrication of replication-deficient virions.
(b) The plasmids are linearized and transcribed before being electroporated in BHK-21 cells. The assembled SFV particles are collected for subsequent in vivo experiments.
(c) The virus is injected through the eye into the brain of live grasshoppers fed with retinal. After a few days the animals are used to image stained neurons and stimulate them using cyan light.
This allows trans-synaptic stimulation of the LGMD neuron.
Conclusions and outlook
The combination of high throughput genomic approaches with genome-editing technologies is allowing unprecedented genetic manipulation in generic insects. Tools developed in traditional genetic models are now adapted to these non-traditional insect models. We highlighted examples where this approach revealed novel mechanisms previously poorly understood. These approaches will likely expand into more animals, exploiting their unique biological features to investigate novel aspects of brain function and behavior.
A comprehensive understanding of behavior requires integration at many levels. First, the neurons and circuits that control behavior must be identified. Then, we must understand how activity in these circuits affects behavior and identify the genes required for circuit development or function. Tissue specific transcriptome data, temporal expression profiles, and single cell atlases will help find genes that could be targets for editing and functional studies. A reference genome is also important to make the best use of such profiling data.
Many of these future developments will have translational and societal benefits. For example, mosquitos are vectors for a range of diseases [82]. Manipulating their genomes and learning about their behaviors will help control the spread of diseases associated with them. Honey bees are critical for agriculture and understanding their biology could potentially save entire industries [83]. Locusts cause devastating plagues that regularly destroy the food supply of entire countries, often underdeveloped [84]. Understanding the mechanisms switching on gregarious swarming will only be possible through an integrated approach dependent on all the techniques discussed above.
What are some of the challenges we need to overcome to make these new approaches usable in a broad range of species [73]? One problem not yet fully resolved is tissue specific expression. In Drosophila many thousands of GAL4 lines with different promoters and enhancers are available [41] to drive the expression of effectors allowing to image neurons or manipulate their function. Such large collections, maintained independently by entities like the Bloomington Drosophila Stock Center (https://bdsc.indiana.edu/), simply do not exist for other insects. Identifying and testing similar promoters in other species will need to be optimized. For this purpose, genome sequences and cell specific expression atlases will be necessary. A virally mediated approach may help test putative promoters before making the effort of modifying the genome of an animal. Some genes, for example the rate limiting enzymes in the synthesis of serotonin and dopamine, have narrow expression patterns so a knock-in in their locus may be one of the easier expression tools to generate (sects. 5.9 and 5.10 of [85]). Neuropeptides are also expressed in limited neuronal populations (sects. 5.13 - 5.27 of [85]) and could be attractive targets since they play key roles in regulating basic biological functions and behavior, such as feeding and reproduction.
Another problem is the efficiency of Crispr/Cas9 transgenesis. In mosquitos for example, thousands of embryos must be injected to find a successful Crispr/Cas9-mediated transgenic HDR event, in contrast to piggyBac-mediated transgenesis. One solution is to use a selection rather than a screening approach. By coupling a selection marker to the transgene, only transformed animals will survive, eliminating the need to examine all offspring for the presence of the screening marker, often a fluorescent protein expressed in the eye. This approach was recently developed in Drosophila and reduces the pool of animals screened from several hundreds to just a few [86]. The 3xP3 enhancer widely used for screening works well across many insects [87], showing robust expression in the eyes, but few other marker enhancers exist. For selection, an enhancer that is expressed early in development is necessary and several promoters, either viral [88] or constitutive [89], could work for this purpose. Developing new enhancers for marker expression would nevertheless also be useful. Another solution to low transgenic efficiency may be to simplify the delivery cocktail. Adding all the required components into a single plasmid increased the rate of transgenesis in microorganisms [90] and may work similarly in Drosophila (Dierick, unpublished results).
While still challenging, advances in insect transgenesis will likely transform both basic and applied insect research, including studies of neural function and behavior. They will finally enable us to comprehensively address Tinbergen’s four questions and provide new avenues for understanding brain function and behavior from a comparative, evolutionary perspective. As illustrated by the examples discussed above, these advances can shed light on all of Tinbergen’s questions when combined with techniques such as behavior, electrophysiology, imaging, or phylogenetic and RNAseq analysis. This new road is promising to further blur the lines between traditional genetic model systems and those that were largely refractory to genetic analysis.
Highlights.
Species-independent insect transgenesis is rapidly advancing
Genetically-encoded tools allow to manipulate and record neuronal activity in vivo
Viruses offer a transient somatic alternative to transgene germline transformations
Mosquitoes, silk moths, and locusts are among recently transformed insects
Acknowledgments
Funding: This work was supported by the National Science Foundation [grant numbers 1453022, 1607518, 1707221, 1754264, 2021795] and the National Institutes of Health [grant numbers RO1GM109938, RO1MH107474].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Dr. Herman Dierick, Dep. Human and Molecular Genetics, Baylor College of Medicine
Dr. Yehuda Ben-Shahar, Dep. of Biology, Washington University in St. Louis
Dr. Baranidharan Raman, Dep. of Bioengineering, Washington University in St. Louis
Dr. Fabrizio Gabbiani, Dep. of Neuroscience, Baylor College of Medicine
References
- 1.del Valle Rodríguez A, Didiano D, Desplan C: Power tools for gene expression and clonal analysis in Drosophila. Nat Methods 2011, 9:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venken KJT, Simpson JH, Bellen HJ: Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 2011, 72:202–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Garcia-Oscos F, Dinh D, Roberts TF: Inception of memories that guide vocal learning in the songbird. Science 2019, 366:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grothe B: How the Barn Owl Computes Auditory Space. Trends in Neurosciences 2018, 41:115–117. [DOI] [PubMed] [Google Scholar]
- 5.Zupanc GKH, Bullock TH: Walter Heiligenberg: the jamming avoidance response and beyond. J Comp Physiol A 2006, 192:561–572. [DOI] [PubMed] [Google Scholar]
- 6.Hansson BS, Stensmyr MC: Evolution of Insect Olfaction. Neuron 2011, 72:698–711. [DOI] [PubMed] [Google Scholar]
- 7.Stopfer M, Bhagavan S, Smith BH, Laurent G: Impaired odour discrimination on desynchronization of odour-encoding neural assemblies. Nature 1997, 390:70–74. [DOI] [PubMed] [Google Scholar]
- 8.Saha D, Leong K, Li C, Peterson S, Siegel G, Raman B: A spatiotemporal coding mechanism for background-invariant odor recognition. Nat Neurosci 2013, 16:1830–1839. [DOI] [PubMed] [Google Scholar]
- 9.Ronacher B, Hennig RM, Clemens J: Computational principles underlying recognition of acoustic signals in grasshoppers and crickets. J Comp Physiol A 2015, 201:61–71. [DOI] [PubMed] [Google Scholar]
- 10.Kostarakos K, Hedwig B: Pattern recognition in field crickets: concepts and neural evidence. J Comp Physiol A 2015, 201:73–85. [DOI] [PubMed] [Google Scholar]
- 11.Namiki S, Iwabuchi S, Pansopha Kono P, Kanzaki R: Information flow through neural circuits for pheromone orientation. Nat Commun 2014, 5:5919. [DOI] [PubMed] [Google Scholar]
- 12.Sakurai T, Namiki S, Mitsuno H, Kanzaki R: 11 - Pheromone detection and processing in the silkmoth Bombyx mori. In Insect Pheromone Biochemistry and Molecular Biology (Second Edition). Edited by Blomquist GJ, Vogt RG. Academic Press; 2021:329–354. [Google Scholar]
- 13.Baba Y, Ogawa H: Cereal System-Mediated Antipredator Behaviors. In The Cricket as a Model Organism: Development, Regeneration, and Behavior. Edited by Horch HW, Mito T, Popadić A, Ohuchi H, Noji S. Springer Japan; 2017:211–228. [Google Scholar]
- 14.Bateson P, Laland KN: Tinbergen’s four questions: an appreciation and an update. Trends in Ecology & Evolution 2013, 28:712–718. [DOI] [PubMed] [Google Scholar]
- 15.Bateson P, Laland KN: On current utility and adaptive significance: a response to Nesse. Trends in Ecology & Evolution 2013, 28:682–683. [DOI] [PubMed] [Google Scholar]
- 16.Nesse RM: Tinbergen’s four questions, organized: a response to Bateson and Laland. Trends in Ecology & Evolution 2013, 28:681–682. [DOI] [PubMed] [Google Scholar]
- 17.Tinbergen N: On aims and methods of Ethology. Zeitschrift für Tierpsychologie 1963, 20:410–433. [Google Scholar]
- 18.Konishi M: Neuroethology: What is it? In Encyclopedia of Animal Behavior. . Elsevier; 2010:562–565. [Google Scholar]
- 19.Mansourian S, Fandino RA, Riabinina O: Progress in the use of genetic methods to study insect behavior outside Drosophila. Current Opinion in Insect Science 2019, 36:45–56. [DOI] [PubMed] [Google Scholar]
- 20.Elick TA, Bauser CA, Fraser MJ: Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica 1996, 98:33–41. [DOI] [PubMed] [Google Scholar]
- 21.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. : Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doudna JA, Charpentier E: The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346. [DOI] [PubMed] [Google Scholar]
- 23.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH: Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 2019, 571:219–225. [DOI] [PubMed] [Google Scholar]
- 24.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F: RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019, 365:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClintock B: The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences of the United States of America 1950, 36:344.--. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F: Genome engineering using the CRISPR-Cas9 system. Nature Protocols 2013, 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA: Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiological Reviews 2010, 90:1103–1163. [DOI] [PubMed] [Google Scholar]
- 28.Lee T, Luo L: Mosaic Analysis with a Repressible Cell Marker for Studies of Gene Function in Neuronal Morphogenesis. Neuron 1999, 22:451–461. [DOI] [PubMed] [Google Scholar]
- 29.Gerdes HH, Kaether C: Green fluorescent protein: applications in cell biology. FEBS Lett 1996, 389:44–47. [DOI] [PubMed] [Google Scholar]
- 30.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. : Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baird GS, Zacharias DA, Tsien RY: Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 1999, 96:11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang HH, St-Pierre F: Genetically Encoded Voltage Indicators: Opportunities and Challenges. J Neurosci 2016, 36:9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P: Channelrhodopsin-1: a light-gated proton channel in green algae. Science 2002, 296:2395–2398. [DOI] [PubMed] [Google Scholar]
- 34.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E: Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A 2003, 100:13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL: NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349:647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Wang L-P, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. : Multimodal fast optical interrogation of neural circuitry. Nature 2007, 446:633–639. [DOI] [PubMed] [Google Scholar]
- 37.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, et al. : High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 2010, 463:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ: Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 1995, 14:341–351. [DOI] [PubMed] [Google Scholar]
- 39.Brand AH, Perrimon N: Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118:401–415. [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer BD, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM: Refinement of Tools for Targeted Gene Expression in Drosophila. Genetics 2010, 186:735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer BD, Truman JW, Rubin GM: Using translational enhancers to increase transgene expression in Drosophila. PNAS 2012, 109:6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gnerer JP, Venken KJT, Dierick HA: Gene-specific cell labeling using MiMIC transposons. Nucleic Acids Res 2015, 43:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potter CJ, Tasic B, Russler EV, Liang L, Luo L: The Q System: A Repressible Binary System for Transgene Expression, Lineage Tracing, and Mosaic Analysis. Cell 2010, 141:536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riabinina O, Luginbuhl D, Marr E, Liu S, Wu MN, Luo L, Potter CJ: Improved and expanded Q-system reagents for genetic manipulations. Nature Methods 2015, 12:219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai S-L, Lee T: Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci 2006, 9:703–709. [DOI] [PubMed] [Google Scholar]
- 46.Suster ML, Seugnet L, Bate M, Sokolowski MB: Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis 2004, 39:240–245. [DOI] [PubMed] [Google Scholar]
- 47.Roman G, Endo K, Zong L, Davis RL: P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A 2001, 98:12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osterwalder T, Yoon KS, White BH, Keshishian H: A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A 2001, 98:12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin GM, Spradling AC: Genetic transformation of Drosophila with transposable element vectors. Science 1982, 218:348–353. [DOI] [PubMed] [Google Scholar]
- 50.Engels WR: The origin of P elements in Drosophila melanogaster. Bioessays 1992, 14:681–686. [DOI] [PubMed] [Google Scholar]
- 51.Handler AM, Gomez SP, O’Brochta DA: A functional analysis of the P-element gene-transfer vector in insects. Arch Insect Biochem Physiol 1993, 22:373–384. [DOI] [PubMed] [Google Scholar]
- 52.Handler AM, McCombs SD, Fraser MJ, Saul SH: The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. PNAS 1998, 95:7520–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Handler AM: Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochemistry and Molecular Biology 2002, 32:1211–1220. [DOI] [PubMed] [Google Scholar]
- 54.Tamura T, Thibert C, Royer C, Kanda T, Eappen A, Kamba M, Kômoto N, Thomas J-L, Mauchamp B, Chavancy G, et al. : Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nature Biotechnology 2000, 18:81.–. [DOI] [PubMed] [Google Scholar]
- 55.Imamura M, Nakai J, Inoue S, Quan GX, Kanda T, Tamura T: Targeted Gene Expression Using the GAL4/UAS System in the Silkworm Bombyx mori. Genetics 2003, 165:1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hara C, Morishita K, Takayanagi-Kiya S, Mikami A, Uchino K, Sakurai T, Kanzaki R, Sezutsu H, Iwami M, Kiya T: Refinement of ectopic protein expression through the GAL4/UAS system in Bombyx mori: application to behavioral and developmental studies. Scientific Reports 2017, 7:11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo X, Yu Q, Chen D, Wei J, Yang P, Yu J, Wang X, Kang L: 4-Vinylanisole is an aggregation pheromone in locusts. Nature 2020, doi: 10.1038/s41586-020-2610-4. [DOI] [PubMed] [Google Scholar]; ** This paper uses a combination of behavior, electrophysiology and genome engineering to unravel the role of olfaction in locust swarming behavior. Specifically, it identifies and characterizes an aggregation pheromone that might prove useful in locust swarm control.
- 58.Waloff N, Popov GB: Sir Boris Uvarov (1889-1970): The Father of Acridology. Annu Rev Entomol 1990, 35:1–26. [Google Scholar]
- 59.Simpson SJ, Sword GA: Locusts. Current Biology 2008, 18:R364–R366. [DOI] [PubMed] [Google Scholar]
- 60.Bastin-Héline L, de Fouchier A, Cao S, Koutroumpa F, Caballero-Vidal G, Robakiewicz S, Monsempes C, Francis M-C, Ribeyre T, Maria A, et al. : A novel lineage of candidate pheromone receptors for sex communication in moths. eLife 2019, 8:e49826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M, Bui M, Yang T, Bowman CS, White BJ, Akbari OS: Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc Natl Acad Sci U S A 2017, 114:E10540–E10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye Z, Liu F, Ferguson ST, Baker A, Pitts RJ, Zwiebel LJ: Ammonium transporter AcAmt mutagenesis uncovers reproductive and physiological defects without impacting olfactory responses to ammonia in the malaria vector mosquito Anopheles coluzzii. Insect Biochem Mol Biol 2021, 134:103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jové V, Gong Z, Hoi FJH, Zhao Z, Sorrells TR, Carroll TS, Prakash M, McBride CS, Vosshall LB: Sensory Discrimination of Blood and Floral Nectar by Aedes aegypti Mosquitoes. Neuron 2020, 108:1163–1180.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The study provides the first detailed response characterization of sexually dimorphic chemosensory neurons present in the needle-like stylet organ used to feed a blood meal. Stylet-specific driver lines were generated to identify non-overlapping subsets of neurons either expressing an ionotropic receptor sensitive to an individual component of the blood mixture (Ir7a expressing neurons activated by NaHCO3) or expressing Ir7f that was activated only by the whole cocktail.
- 64.Zhao Z, Tian D, McBride CS: Development of a pan-neuronal genetic driver in Aedes aegypti mosquitoes. Neuroscience; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matthews BJ, Younger MA, Vosshall LB: The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. Elife 2019, 8:e43963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galun R, Oren N, Zecharia M: Effect of plasma components on the feeding response of the mosquito Aedes aegypti L. to adenine nucleotides. Physiological Entomology 1984, 9:403–408. [Google Scholar]
- 67.Schulte C, Theilenberg E, Miiller-Borg M, Gempe T, Beye M: Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). PNAS 2014, 111:9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooley L, Kelley R, Spradling A: Insertional mutagenesis of the Drosophila genome with single P elements. Science 1988, 239:1121–1128. [DOI] [PubMed] [Google Scholar]
- 69.Cooley L, Berg C, Kelley R, McKearin D, Spradling A: Identifying and cloning Drosophila genes by single P element insertional mutagenesis. Prog Nucleic Acid Res Mol Biol 1989, 36:99–109. [DOI] [PubMed] [Google Scholar]
- 70.Mayer LR, Diegelmann S, Abassi Y, Eichinger F, Pflugfelder GO: Enhancer trap infidelity in Drosophila optomotor-blind. Fly (Austin) 2013, 7:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Groth AC, Fish M, Nusse R, Calos MP: Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 2004, 166:1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bischof J, Maeda RK, Hediger M, Karch F, Basler K: An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 2007, 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews BJ, Vosshall LB: How to turn an organism into a model organism in 10 ‘easy’ steps. Journal of Experimental Biology 2020, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang S-J, Qiao H-H, Wang X, Hu Q, et al. : Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep 2014, 9:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaverra-Rodriguez D, Dalla Benetta E, Heu CC, Rasgon JL, Ferree PM, Akbari OS: Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol Biol 2020, 29:569–577. [DOI] [PubMed] [Google Scholar]; * Chaverra-Rodriguez et al. (2020) In this study, the authors demonstrate efficient delivery of CRISPR/Cas 9 ribonucleoprotein complex to jewel wasp ovaries thereby avoiding the need for microinjection-based delivery. Specific peptides (P2C and Branched Amphiphilic Peptide Capsules) were used to facilitate uptake of the gene editing cargo into the ovaries. The authors report both somatic and germline gene editing using this approach.
- 76.Chaverra-Rodriguez D, Macias VM, Hughes GL, Pujhari S, Suzuki Y, Peterson DR, Kim D, McKeand S, Rasgon JL: Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun 2018, 9:3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heu CC, McCullough FM, Luan J, Rasgon JL: CRISPR-Cas9-Based Genome Editing in the Silverleaf Whitefly (Bemisia tabaci). The CRISPR Journal 2020, 3:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murphey DK, Herman AM, Arenkiel BR: Dissecting inhibitory brain circuits with genetically-targeted technologies. Front Neural Circuits 2014, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis DL, DeCamillis MA, Brunetti CR, Haider G, Kassner VA, Selegue JE, Higgs S, Carroll SB: Ectopic gene expression and homeotic transformations in arthropods using recombinant Sindbis viruses. Curr Biol 1999, 9:1279–1287. [DOI] [PubMed] [Google Scholar]
- 80.Wang H, Dewell RB, Ehrengruber MU, Segev E, Reimer J, Roukes ML, Gabbiani F: Optogenetic manipulation of medullary neurons in the locust optic lobe. Journal of Neurophysiology 2018, 120:2049–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper uses an arbovirus, the Semliki Forest virus to efficiently transfect grasshopper neurons in vivo. Application of this technical tool demonstrates the feasibility of optogenetic neuronal activation using viral delivery vectors in non-genetic insect models.
- 81.Dewell RB, Gabbiani F: Biophysics of object segmentation in a collision-detecting neuron. eLife 2018, 7:e34238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carey AF, Carlson JR: Insect olfaction from model systems to disease control. PNAS 2011,108:12987–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klein A-M, Boreux V, Fornoff F, Mupepele A-C, Pufal G: Relevance of wild and managed bees for human well-being. Current Opinion in Insect Science 2018, 26:82–88. [DOI] [PubMed] [Google Scholar]
- 84.Sword GA, Lecoq M, Simpson SJ: Phase polyphenism and preventative locust management. Journal of Insect Physiology 2010, 56:949–957. [DOI] [PubMed] [Google Scholar]
- 85.Burrows M: The Neurobiology of an Insect Brain. Oxford University Press; 1996. [Google Scholar]
- 86.Matinyan N, Karkhanis MS, Gonzalez Y, Jain A, Saltzman A, Malovannaya A, Sarrion-Perdigones A, Dierick HA, Venken KJT: Multiplexed drug-based selection and counterselection genetic manipulations in Drosophila. Cell Reports 2021, 36:109700. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper optimized the use of selection and counterselection markers to significantly reduce the workload of transgenesis in Drosophila and also generated a cloning toolbox to make the transfer of these tools to other insect species easier.
- 87.Horn C, Schmid BGM, Pogoda FS, Wimmer EA: Fluorescent transformation markers for insect transgenesis. Insect Biochemistry and Molecular Biology 2002, 32:1221–1235. [DOI] [PubMed] [Google Scholar]
- 88.Kandul NP, Liu J, Hsu AD, Hay BA, Akbari OS: A drug-inducible sex-separation technique for insects. Nat Commun 2020, 11:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miyata Y, Tokumoto S, Sogame Y, Deviatiiarov R, Okada J, Cornette R, Gusev O, Shagimardanova E, Sakurai M, Kikawada T: Identification of a novel strong promoter from the anhydrobiotic midge, Polypedilum vanderplanki, with conserved function in various insect cell lines. Sci Rep 2019, 9:7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fang Y, Cui L, Gu B, Arredondo F, Tyler BM: Efficient Genome Editing in the Oomycete Phytophthora sojae Using CRISPR/Cas9. Current Protocols in Microbiology 2017, 44. [DOI] [PubMed] [Google Scholar]
- 91.Horn C, Offen N, Nystedt S, Häcker U, Wimmer EA: piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 2003, 163:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Broussard GJ, Liang R, Tian L: Monitoring activity in neural circuits with genetically encoded indicators. Front Mol Neurosci 2014, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD: E unum pluribus: multiple proteins from a self-processing polyprotein. Trends in Biotechnology 2006, 24:68–75. [DOI] [PubMed] [Google Scholar]


