Abstract
Accurate predictions from models based on physical principles are the ultimate metric of our biophysical understanding. While there has been stunning progress towards structure prediction, quantitative prediction of enzyme function has remained challenging. Realizing this goal will require large numbers of quantitative measurements of rate and binding constants, and the use of these ground-truth datasets to guide the development and testing of these quantitative models. Ground truth data more closely linked to the underlying physical forces are also desired. Here we describe technological advances that enable both types of ground truth measurements. These advances allow classic models to be tested, provide novel mechanistic insights, and place us on the path toward a predictive understanding of enzyme structure and function.
Keywords: enzymology, energetic landscapes, ensembles, functional prediction, enzyme design
Introduction
Scientists can now predict and design protein structure with ångström accuracy, a triumph culminating from decades of experimental and computational efforts [1,2]. In this perspective, we describe why the approaches that have been so successful in protein design are unlikely to lead to analogously predictive models for protein function, and we introduce concepts and experimental approaches that address these limitations and move us toward the ultimate goals of accurately and quantitatively predicting and designing function.
Structure broadly and deeply informs our understanding of function—consider the striking visualizations of motor proteins that have revealed the lever arms of myosin, dynein, and kinesin and their ATP-dependent power strokes [3], and the myriad of proteins whose shape is integral to their function, like the β-clamp that encircles DNA to enhance polymerase processivity [4]. Nevertheless, more than structure is needed to describe, understand, and quantitatively predict function. Indeed, many proteins with the same fold differ in function, quantitively and even qualitatively [5].
Function involves a series of states, such as the conformations through the myosin reaction cycle or the states in chemical reactions catalyzed by enzymes (substrate binding (E•S), transition state (E•S‡), product complex (E•P), and release (E + P) to regenerate free enzyme ready for another round of catalysis). A minimal description of protein function therefore requires describing these states and determining the rate and equilibrium constants that define their transition probabilities and relative populations, respectively.
But still more is required to understand and ultimately predict and design new functions—an ability to specify the functional consequences of sequence changes. Enzymes are large, with residues beyond the active site required for function, minimally to fold into and stabilize the correct binding and active site configurations [6]. But regions far from the active site can also have considerable functional consequences [7], as evidenced by allosteric modulation [8,9] and remote mutational effects frequently identified in high-throughput screens [10–12]. To find and describe which residues, sets of residues, and substructures affect function, as well as the particular aspects of function that are affected, we need approaches to systematically interrogate all residues and to determine the effects of perturbing them through each step of the enzyme’s reaction cycle. In other words, we need to measure many rate and equilibrium constants for many mutants. Here, we highlight a breakthrough approach that allows these measurements to be made.
In principle, with sufficient empirical data, machine and deep learning approaches could be employed to provide accurate, predictive models of enzyme function. However, sequence space is vast—so vast that nature has only sampled a miniscule corner of it [13]. And whereas pairwise residue information is largely sufficient to predict protein structure [14], enzyme function is much more complex, with multiple distal residues exerting functional effects on one another. These effects correspond mathematically to higher-order terms to account for the effects from combinations of multiple residues, and are thusterms that are needed to quantitatively describe function. Because of this complexity, vast amounts of data would likely be required to define the relationship between sequence and function, and we suspect that the scale of data needed to predict enzyme function via machine- and deep-learning approaches may greatly exceed what is measurable, even with recent breakthroughs.
In contrast, physics-based approaches are scalable, so that simple rules can be used to describe the behavior of arbitrarily complex systems [15,16]. These models relate atomic forces and motions, captured by the preferred substates in conformational ensembles, to thermodynamic and kinetic constants. From the perspective of statistical mechanics, the constants that define function arise from the energy landscapes that define an ensemble of enzyme substates and the transition and reaction probabilities for each substate, represented mathematically in Figure 1 [17,18].
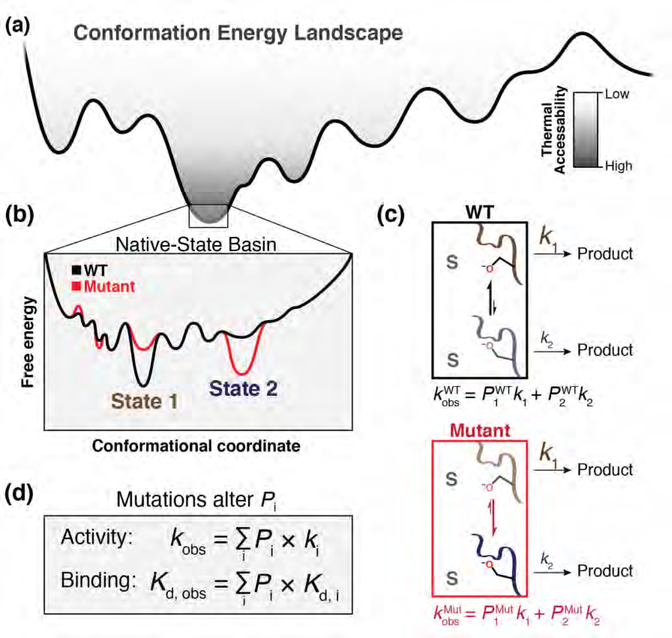
Figure 1. Enzymes function on a high-dimensional energy landscape.
(a) Enzymes form a set of states specified by energy wells on a free energy landscape, with dimensionality defined by the thousands of degrees of freedom from each rotatable bond of each residue. (b) An example ensemble of near-energy substates in which State 1 (brown) and State 2 (blue) lie within the lowest-energy basin (“Native-State Basin”) and equilibrate at room temperature. These substates have different intrinsic reactivities, reflecting barrier heights along the reaction coordinate that differ when projected from different conformational coordinates. (c) A mutation that does not affect intrinsic reactivity (red) can nevertheless diminish catalysis by redistributing equilibrium populations in the native-state basin to favor less-active conformers (i.e., with higher free energy barriers to reaction; State 2 vs. State 1). Mathematically, the apparent rate of the WT (kobsWT) and mutant (kobsMut) toward a substrate (S) is the probability-weighted (occupancy-weighted) sum of the intrinsic rate constants of each microscopic substate. In this two-state example, k1 > k2, so kobsWT > kobsMut. (d) Observed enzymatic activity is the summed activity of each microscopic substate (ki) weighted by its fractional occupancy (Pi). Mutations can alter the free energy landscape and change apparent activity (kobs), or binding (Kd), by altering Pi, as shown in the figure, or by altering the reaction barrier (not shown).
Thus, we need to go beyond structures to ensembles, and beyond structure–function relationships to ensemble–function relationships, and we will need to use experimental determinations of these relationships to test and build the physics-based models needed to quantitatively and accurately predict enzyme function. Here, we describe emerging X-ray crystallographic approaches that can provide this needed ensemble information.
Current design efforts yield enzymes that require cycles of randomization and selection to begin to approach natural enzymes. Perhaps the tortoise rather than the hare is needed to win this race [19], wherein large-scale quantitative and in-depth data are first collected and used to test and build models that will ultimately have the accuracy to predict and design enzyme function. We expect accurate enzyme functional prediction to remain a grand challenge of 21st century biophysics—it is a still-distant goal. Systematic blind tests of models built from large-scale quantitative data provide a promising, and perhaps necessary, path forward.
1. Ground truths are needed for model development
To develop and establish a model, “ground truths” are needed. Ground truths are experimental data in a form that can be predicted by and thus compared to a model; without ground truths there is no way to definitively test a model.
The most sophisticated models in enzymology combine quantum mechanics and molecular mechanics (QM/MM) and have been used to predict reaction rate constants [20,21]. However, in nearly all instances the rate constants predicted by QM/MM were already measured, and thus do not represent actual predictions that foretell a future event and that are incontrovertibly independent of the existing experimental findings [22]. The importance of predictions prior to measurement is underscored by the fact that the inability of protein folding models of the 1970s–90s to predict structures was not apparent until they were challenged with truly blinded predictions (CASP, Critical Assessment of Structure Prediction); [1,23,24]). Ultimately, the algorithms and models that accurately predict structure were built using information from the large number of solved structures in the Protein Data Bank (PDB) [25], mining this information, and combining it with vast information from sequence conservation along with simplified energy potentials or rules derived via machine or deep learning [14,26–29,2].
Analogously, we need many measurements of kinetic and thermodynamic constants as ground truths to build and test predictive models of enzyme function, but current approaches are severely limited in their ability to deliver these essential data. Changes in residues throughout an enzyme can affect kinetic and thermodynamic constants and combined changes will often not give additive effects; from a mathematical perspective, this property corresponds to a need for many terms in a model that can predict the effects from all sequence changes. With only a handful of measurements, there will not be enough data to constrain the model—from a simple algebraic standpoint, one needs the number of measurements to equal or exceed the number of variables in an equation to solve for those variables. While we are unlikely to ever obtain sufficient measurements to fully define all of the variables of a master equation for function, we need many measurements to guide model development, and then many predictions from these models—followed by many additional quantitative measurements—to provide a robust test of the models.
Structural ensembles can provide orthogonal ground truths. The relative occupancy of different conformational states reflects a balance of physical forces and thus provides ground truths that can be used as tests of models that account for these forces. Of further value, each ensemble provides a wealth of data—the distribution of states for each residue and around each backbone and sidechain bond, as well as information about their hydrogen bonds and electrostatic and van der Waals interactions. In contrast, average structures can be predicted without these “details” being accurate, as evidenced by the rather simple force models present in successful Rosetta structural prediction algorithms [26,30].
The sections that follow describe recent advances in obtaining these ground truths for enzyme function.
2. The need for quantitative enzymology at scale
Recognizing the need for an immense amount of data to describe and understand enzymes and their function, high-throughput approaches have been used to interrogate up to ~106 sequence variants in parallel. In particular deep mutational scanning (DMS) approaches have been applied to all possible single mutants for dozens of unique proteins [31,32].
Some DMS studies report the effects of mutations in a particular protein on organismal fitness, a convolution of multiple factors [33,34]. These experiments can also be designed to report more specific aspects of function, including catalytic efficiency, substrate specificity, stability, and interaction with binding partners [10,11,35–39]. While valuable, and sometimes of immediate practical benefit, these functional readouts still represent a convolution of contributing factors. For example, for observed catalytic function a mutant enzyme down 100-fold in catalysis can be 99% unfolded, 99% partitioned to an alternative misfolded state, have a misaligned catalytic residue, have a binding interaction removed, or exhibit some combination of these factors. Thus, these functional readouts fall short of delivering the needed ground truths.
At the other end of the spectrum, traditional enzymology provides kinetic and thermodynamic constants that describe the catalytic cycle and have been combined with incisive mechanistic probes (e.g., alternative substrates, isotope effects, etc.) to provide deep mechanistic insights. However, these approaches are only feasible for a small number of variants of each enzyme. Past efforts to quantify properties of many variants in T4 lysozyme, pyruvate kinase, and β-glucosidase B underscore that data for many mutants can be collected when heroic means are employed [40–43]. But even in these cases, the time and cost to carry out the additional measurements required to probe the mechanistic origins of the observed effects would be prohibitive.
It is hard to identify a discipline that has not been transformed at one time or another by a breakthrough technology. Here, technology was needed to efficiently provide the rich and quantitative data of traditional enzymology at a much larger scale for many variants and multiple enzymes.
Quantitative enzymology on a chip
Advances in microfluidics provided the opportunity to marry the strengths of traditional enzymology with automated high-throughput data collection and bring enzymology into the genomic age [44–46]. High-throughput microfluidic enzyme kinetics (HT-MEK; Figure 2) allows 1500 enzyme variants to be produced, purified, and subjected to multiple quantitative assays in days, at a miniscule fraction of the cost of traditional approaches [47].
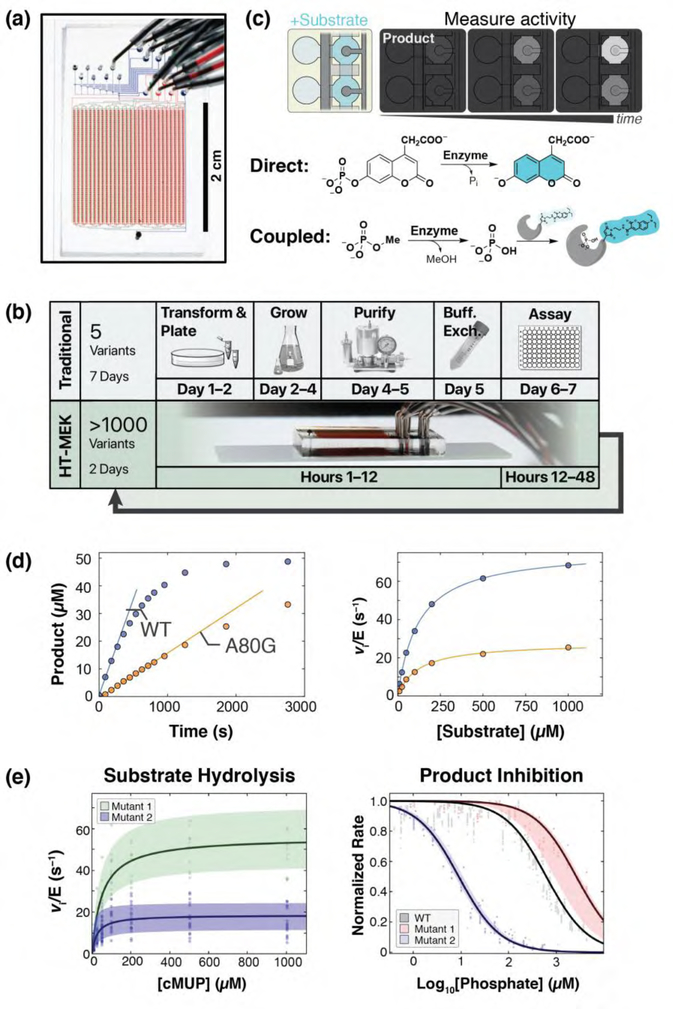
Figure 2. Application of HT-MEK to study catalysis and inhibition.
(a) HT-MEK uses a valved microfluidic device containing >1500 chambers to (b) in vitro express, purify, and assay 1500 enzyme variants in days. Iteratively varying substrates, inhibitors, and conditions give kinetic and thermodynamic parameters of function. (c) Reactions are performed by introducing fluorogenic substrate (light blue) into reaction chambers and synchronously exposing surface-immobilized enzyme to the substrate with pneumatic valving. Product (darker blue) is quantified by fluorescence over time, using direct (fluorogenic) or indirect (coupled) assays. The assays shown are for phosphatase activity or for any enzyme that directly or indirectly generates inorganic phosphate. (d) Example on-chip fluorogenic substrate turnover curves for the PafA phosphatase with fit initial rates (left) and Michaelis-Menten curves (right) for WT (blue) and mutant (orange) PafA variants. A per-chamber standard curve is used to convert fluorescence to product concentration. (e) Many replicates (chambers) for each variant (Mutant 1: Y74V, Mutant 2: Y112G) over multiple chips are used to calculate bootstrap errors on fit Michaelis-Menten (left) and competitive inhibition (right) parameters (reproduced from Markin, Mokhtari, et al. (2021) [47]).
Figure 2 outlines how HT-MEK experiments are performed. HT-MEK uses a microfluidic device with chambers aligned to a DNA microarray of 1500 isolated variant plasmids (Figure 2a). Expression and purification of enzyme variants is carried out in parallel, so that all 1500 enzymes can be purified, quantified, recruited to antibody-patterned surfaces, and ready for assay in hours (Figure 2b). Pneumatically controlled valves allow the user to protect the enzyme from flow-induced shear forces while the expression solution is removed and an assay solution containing substrate is added to the chambers, followed by opening of the valves to simultaneously initiate reactions in all chambers (Figure 2c). Product formation is quantified over time via fluorescence, either directly using a fluorogenic substrate or indirectly using a coupled assay (Figure 2c). Once complete, reaction product is flowed out and a new substrate stock is flowed in so that a series of assays can be performed iteratively. Figure 2d shows example Michaelis-Menten and inhibition curves obtained in HT-MEK experiments.
Each HT-MEK device can be used to carry out tens of reactions and a single researcher can fabricate tens of devices in a day. These properties make it possible to carry out hundreds of assays—with multiple substrates, inhibitors, and solution conditions, as used traditionally in mechanistic enzymology— but to do so across thousands of enzyme variants and to do so in a few weeks. Thus, the properties of HT-MEK allow measurement of many kinetic and thermodynamic constants that provide valuable information about an enzyme and can serve as ground truths for model testing.
For PafA [48], our test case, we obtained >6000 kinetic and thermodynamic constants from >650,000 kinetic measurements for 1036 mutants. HT-MEK provides a wide dynamic range, ~105 in rate for PafA, which allows measurement of large active site effects and reaction rates for non-cognate substrates. High measurement precision is obtained with rigorous error estimates using bootstrapping, which is possible because of the many replicates acquired within each HT-MEK assay.
In addition to providing a large number of ground truth measurements that can be used to evaluate and guide the development of quantitative models, the initial PafA data provided extensive mechanistic information not previously accessible. The observation that mutations at most of the 526 PafA positions altered one or more kinetic and thermodynamic parameter underscores the need for measurements throughout an enzyme to map its function. Further revealing and displaying the intricacy of enzyme function, different sets of residues affected different reaction steps and underlying catalytic mechanisms as well as folding, as illustrated in the functional maps of Figure 3. The largest mutational effects were seen at the active site and directly around it, as expected, but effects extended from the active site all the way to the enzyme surface, with large effects many ångströms from the active site and different remote regions affecting different aspects of function (Figure 3a). We do not think that these effects could have been predicted a priori using current approaches. Regardless, researchers with predictive algorithms can now use those algorithms on any of the multiple of enzymes amenable to HT-MEK, so that we can determine the algorthm’s predictive power.
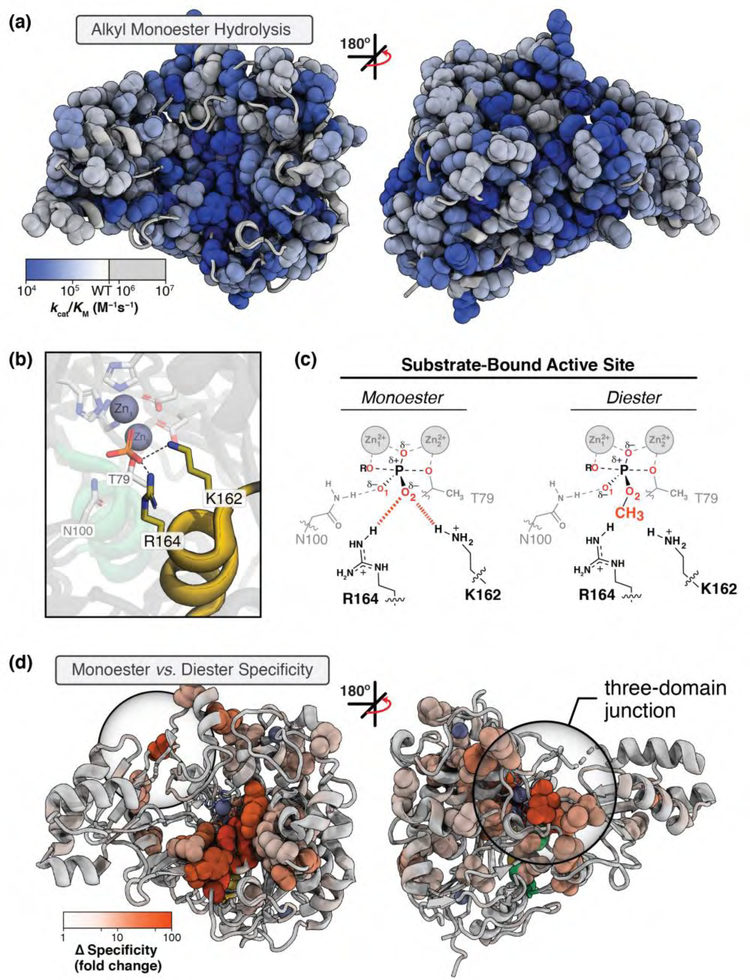
Figure 3. Spatial patterns of PafA mutational effects on specific functions.
(a) Alkyl phosphate hydrolysis (kcat/KM) for glycine and valine substitutions at each position in PafA. Significant effects are shown as spheres, color-coded by the size of the effect. (b) PafA active site (PDBID: 5TJ3) showing contacts between covalently-bound phosphate (T79) and active site K162 and R164 (yellow sticks). (c) Schematic of PafA active site residue contacts to bound phosphate monoester (left) vs diester (right) substrates. (d) Substrate specificity effects upon valine and glycine mutation of PafA positions, visualized as in a. Large mutational effects (spheres) cluster primarily around R164 and K162, extending distally to a three-domain junction (circled) more than 20 Å away. Panels a, c, and d are adapted from Markin, Mokhtari, et al. (2021) [47].
Consider, as an example, the active site arginine and lysine residues that contact one of the substrate phosphoryl oxygen atoms and are responsible for reaction specificity for phosphate monoesters over diesters (which are substrates of related superfamily members; [48–52]) (Figure 3b and c). While mutation of most residues contacting these active site residues diminished specificity, a majority of the affecting residues were remote, including residues at the junction of three auxiliary domains (Figure 3d; auxiliary domains are structural regions present in subsets of Alkaline Phosphatase superfamily members [52]). The auxiliary domain junction sits ~20 Å from the active site and on the opposite side of the enzyme from the catalytic pocket, but nevertheless exhibits mutational effects of up to 60-fold [47]. These observations suggest that the auxiliary domains and their positioning are critical for catalytic function by the active site arginine and lysine, but we would not have predicted these or other remote effects for mutations throughout PafA.
One would be tempted to conclude, in the absence of data to the contrary, that most distal functional effects arose from destabilizing the active enzyme. With HT-MEK (and related approaches under development), we can independently assay folding [47,53]. We found that none of the PafA effects arose from equilibrium unfolding (PafA is a secreted enzyme and is highly stable). Nevertheless, our ability to assay PafA with multiple substrates and under multiple expression and reaction conditions allowed us to uncover the presence of an unanticipated long-lived misfolded state of the enzyme. Without accounting for the effects of misfolding and unfolding on observed reaction rates, functional models cannot be unambiguously made or tested.
As noted above, an immediate challenge is to predict distal effects for multiple enzymes and to use HT-MEK to determine what is correctly predicted, quantitatively or qualitatively. We can also directly use data from HT-MEK to aid enzyme engineering at a practical level. Functional maps generated by HT-MEK can inform where mutations should be made to enhance the chance of altering and tuning specific functional parameters. In addition, HT-MEK can rapidly assess human alleles to reveal the biophysical bases of mutations associated with disease and to inspire new and precise therapeutic strategies. For instance, the discovery of surface residues (through mutation) allosterically linked to function may allow druggable enzyme activation as well as inhibition [54].
3. Conformational ensembles for evaluating and building physical and catalytic models
Ultimately, we want to predict kinetic and thermodynamic constants for a particular reaction with a specified sequence. We can measure many of these values via HT-MEK as ground truths and compare them to values predicted by models, but these functional constants are emergent properties that result from the enzyme’s underlying physical properties. We would like to have ground truths more closely connected to the enzyme’s physical properties; these measurements would provide more direct tests to evaluate and improve physics-based models. In particular, we want to know an enzyme’s conformational landscape and how this varies with bound ligands and through the enzyme’s reaction, and we want to determine the affinities and reactivities of the states that constitute the landscape (Figure 1a to c).
The value of and need for ensemble information to understand protein folding and function has been recognized for decades [17,55,56]. For catalysis, the clear evidence for remote effects—from allosteric ligands and post-translational modifications—and efforts to understand how enzymes so efficiently navigate their reaction paths have led to a panoply of functional models that invoke dynamics (e.g., [57–61]). Experimentally, NMR provides relevant information about the rates of transitions between conformational states and information (e.g., order parameters) on the relative conformational freedom of residues (e.g., [62,63]). However, detailed atomic information that provides more direct tests of models, such as the extent and direction of motion and which motions are coupled or independent, is difficult to obtain via NMR. Fortunately, emerging X-ray crystallographic approaches can provide extensive and detailed information about conformational ensembles that can be more directly related to predictions from physics-based models.
A key technological breakthrough in X-ray crystallography was cryo-freezing crystals to reduce their susceptibility to radiation damage and make crystal handling more reliable. Indeed at least 90% of more than 150,000 protein X-ray structural models in the PDB were obtained under cryogenic conditions (diffraction source temperature ≤ 125 K) [25]. Nevertheless, X-ray structures can be obtained at ambient temperatures as well, conditions that do not quench a protein’s inherent dynamic motions [64]. Ambient or “room-temperature” (RT) X-ray crystallography requires high resolution (typically sub-1.5 Å) to provide reliable and extensive information about conformational heterogeneity at the atomic level and requires larger-than-average crystals to limit X-ray damage during data collection. Fortunately, many enzymes of interest yield crystals of the desired size and quality, and recent methodological improvements allow RT X-ray crystallography to be broadly implemented [65].
Conformational ensembles can also be generated from cryo X-ray structures, by combining multiple static structures into a so-called pseudo-ensemble [66,67]. In brief, it is assumed that individual cryogenic X-ray structures of proteins sharing the same or highly-similar sequences (e.g., with one or a few mutations) provide conformers trapped in different low-energy wells on the protein’s energy landscape, so that combining many cryo-structures can approximate the protein’s accessible ensemble of states (for pseudo-ensemble computational tools, see refs [68–71]). Although motions are restricted and some are changed upon freezing, several lines of evidence and direct comparisons support a close correspondence of the flexibility within pseudo-ensembles and the motions present at ambient temperatures [67,72].
Pseudo-ensembles and RT data are complimentary—the latter providing the most reliable information about conformational heterogeneity and the former retaining information about correlated motions within the constituent conformers. These approaches have provided insights into multiple systems, including HRas GTPase, protein tyrosine phosphatase, proline isomerase, soybean lipoxygenase, β-lactamase, dihydrofolate reductase, isocyanide hydratase, herpes virus protease, and designed and laboratory-evolved kemp eliminases [73–83]. The most extensive X-ray ensemble data to be collected and analyzed is for ketosteroid isomerase (KSI; Figure 4a). For KSI, pseudo-ensembles and high-resolution RT ensembles have been obtained for complexes representing the states in the enzyme’s reaction cycle, for KSIs from two different species, and for WT and mutant KSIs [72].
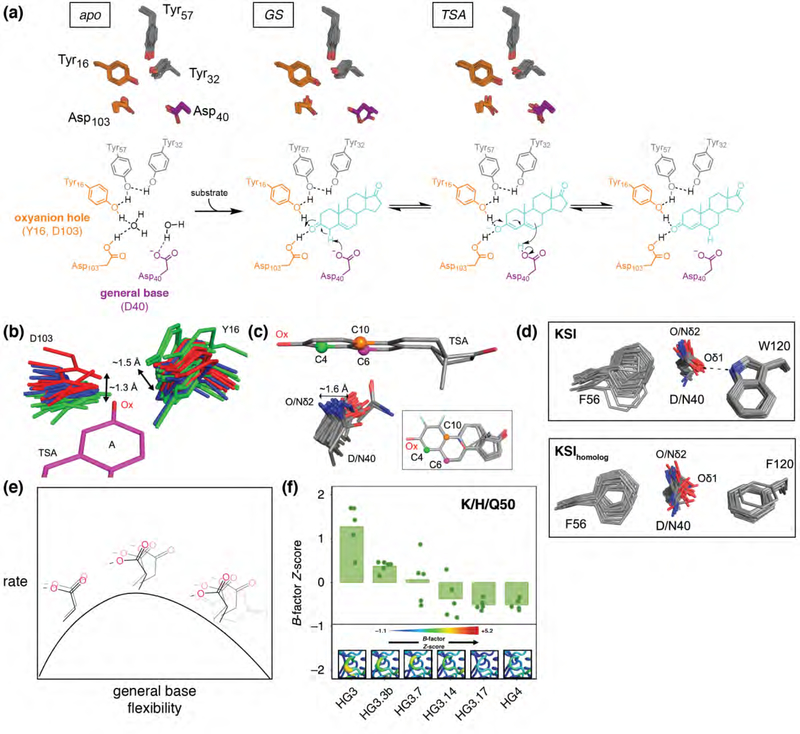
Figure 4. Ensemble crystallography provides mechanistic insights toward enzyme catalysis and engineering.
(a) KSI reaction mechanism and ensemble crystallography of multiple states in the reaction cycle. Active site residues from experimental RT multi-conformer models are shown for apo, ground state bound (GS), and transition state analog bound (TSA) KSI. (b, c) Pseudo-ensembles for KSI oxyanion hole residues Y16 and D103 (b) and the general base (D40, WT, or mutant D40N) (c), derived from cryo structures containing bound transition state analogs (TSA, equilenin and phenols). Bound TSAs were superimposed, aligned on phenol (“A”) rings, and shown as a single structure for simplicity. (d) Pseudo-ensemble for the KSI general base (D40, or mutant D40N) and neighboring residues for KSI (F56, W120) and a KSIhomolog (F56, F120). KSI and KSIhomolog kcat measurements for both the WT and the position 120 reciprocal mutants (KSI W120F; KSIhomolog F120W) demonstrate a 4-fold (KSI) and 8-fold (KSIhomolog) rate increase for phenylalanine vs. tryptophan containing variants. (e) Cartoon depiction of the relationship between KSI rate and general base (D40) flexibility. (f) Increase in active site rigidity during directed evolution of a designed Kemp eliminase (from HG3 to HG4). B-factor Z-scores are plotted for each heavy atom of the position 50 side chain (K, H, or Q). Structural views of the Kemp eliminase residues 87–90 loop from room-temperature crystallography, with B-factor Z-scores colored according to legend and increasing with sausage plot thickness (inset). Panels b to e adapted from Yabukarski et al. (2020) [72]; panel f adapted from Broom et al. [83].
Insights from KSI ensembles
The function of a catalytic residue depends not only on its presence in the vicinity of the substrate, but on the adoption of conformational states with the correct distance and orientation to the substrate and/or other residues. Indeed, positioning is universally invoked or assumed in descriptions of enzyme catalysis, but without ensemble information we cannot know the nature and extent of this positioning. Ensemble information is also needed to understand the motion inherent in all chemical reactions, minimally to go from van der Waals distance to form a bond, a change on the order of 1 Å, and how or whether this is affected by the enzyme environment. Furthermore, many enzymes catalyze multi-step reactions and use the same functional groups in different poses to carry out successive reaction steps. Again, ensemble information is needed to understand how enzymes navigate these challenges.
KSI, a steroid isomerase, abstracts a proton from its steroid substrate with a general base, transferring the proton to a different position of the resulting intermediate to give the more stable conjugated product, using an oxyanion hole to stabilize negative charge accumulation on the intermediate (Figure 4a). Oxyanion holes for serine proteases have been suggested to contribute catalytically via ground state destabilization, by forming sub-optimal, geometrically-constrained hydrogen bonds that sit out of the plane of the ground state sp2 oxygen [84–87]. The KSI ensembles reveal motions of the oxyanion hole hydrogen bond donors on the scale of ~1 Å and an absence of discrimination between the sp2 and sp3 oxygen electronic configurations (Figure 4b). Instead, the oxyanion hole seems to provide catalysis by forming hydrogen bonds that are stronger than those to water in solution [6,72,88–90].
Extensive site-directed mutagenesis studies revealed an astounding effective molarity of 103–105 M for the KSI aspartate general base [91]. While the simplest explanation for this large catalytic effect is precise positioning, positioning at multiple sites would be required to accommodate KSI’s multiple substrates and successive reaction steps. As above, KSI ensembles allowed this model to be tested, revealing a broad distribution of general base positions (Figure 4c), as needed to abstract and donate protons at multiple positions and indicating that alternative models are needed to account for the highly efficient observed general base catalysis [91–93]. In addition, the flexibility in the oxyanion hole, while precluding ground state destabilization, contributes to the conformational plasticity of the general base and substrate with respect to one another (Figure 4b and c).
One might expect there to be a balance between positioning and flexibility: clearly too much flexibility of the KSI general base would hamper catalysis while too-restricted positioning could as well, by limiting access to the multiple states needed to carry out the full reaction cycle. Remarkably, functional results and ensembles for wild type and mutant KSIs provide evidence for the balance: an aromatic-anion interaction provides greater flexibility of the general base than a hydrogen bond as well as faster reaction, whereas mutations in the general base loop that disorder it substantially impair catalysis (Figure 4d and e).
In addition, comparisons of how conformational ensembles of KSI side chains change from mutations in nearby residues provides information about the balance of forces, including conformational entropy, that determine where and how much the oxyanion hole tyrosine is positioned [72] (Figure 4b). Effects like these will provide rich testing grounds for force fields in physics-based models. Finally, the observation that, at least in this case, ensemble rearrangements remain local suggests at least some limitation to the complexity of models needed to account for energy landscapes of enzymes (Figure 1a and b; see also “The complexity of functional models” below).
Rules for enzyme design from ensemble crystallography
A major challenge is to understand why the performance of de novo enzyme designs falls short of natural enzymes [94–96], and how to rationally engineer solutions that Nature (or researchers) discover through evolution. Early de novo design of Kemp eliminases (KE) yielded some success [97], but the same fold rate enhancement, or more, for the eliminase reaction is achieved in the active site of KSI, an enzyme evolved to carry out different chemistry with different substrates. This result suggests that designed KEs accomplish only coarse positioning against a general base within a binding pocket [98]. With laboratory-evolved improvements (17 substitutions), an ~105-fold increase in kcat/KM was achieved [99]. Recent work from the Chica and Fraser groups sought to understand the mechanistic bases for these improvements, interrogating four variants along the mutational trajectory via room-temperature crystallography [83,100]. This effort revealed that apo-state catalytic elements rigidified along the mutational trajectory, favoring catalytically-preorganized poses, consistent with classical proposals for origins of catalysis from positioning of substrates and catalytic groups (Figure 4f) [93,101,102].
More generally, crystallographic ensembles can be used to test models of catalysis that attempt to link motions or positioning to function, identifying the types and scales of motions that may be relevant to progress along the reaction coordinate. This ability allows the structure-function paradigm to be supplanted by ensemble-function analyses.
Ensemble measurements versus the reaction coordinate
The X-ray ensemble approaches described above are needed to relate structure to energetics and function but also have limitations. Most centrally, they provide information about the “lower levels” of the enzyme’s conformational landscape. States that are uphill by >~2 kcal/mol, representing <5% of the total population, are unlikely to be observed. This limitation alone is not severe, as one can decipher much of the underlying energetics by having lots of data—as is provided by X-ray ensembles—in the ~0–2 kcal/mol energy range. But what is missing is information about the conformations and motions as one climbs further toward the reaction’s transition state. Transition states are by definition fleeting states, lasting <1 picosecond, and highly improbable. To assess what happens at these rarified regions of the energy landscape, and whether the data obtained closer to the base of these mountains is adequate to model reaction coordinate, we need additional ground truths for these transient, high-energy states. While such information cannot (yet) be obtained in high throughput, several methods exist to provide this critical information.
The highest time resolution structural data use laser pulses to initiate a process and serial high resolution X-ray data collection. X-ray free-electron lasers (XFEL) and cutting-edge synchrotron sources capture crystallographic snapshots at ambient temperatures by supplying intense femtosecond X-ray pulses [103,104]. Tenboer et al. used nanosecond laser pulses in conjunction with XFEL crystallography to isomerize the double bond of the photoactive yellow protein chromophore and to measure protein and chromophore conformational changes after time delays of 10 ns and 1 μs. These experiments revealed the significant side chain displacements associated with photocycle transients at high resolution [105]. Schlichting and coworkers used a 150 fs laser pulse to dissociate CO from myoglobin and were able to follow in real time the synchronous non-equilibrium picosecond oscillations of the heme ring that arise from the CO dissociation energy and dissipate on the order of 10–100 ps [106]. Vibrational spectroscopy, while not directly measuring atomic positions, is particularly powerful because frequencies can be assigned to specific bonds located within proteins or bound reactants and can provide information about changes in the strength and properties of those interactions. Dyer and Callender used temperature-jump infrared spectroscopy on the microsecond timescale to identify multiple distinct and non-interconverting substrate binding conformations with different reactivities in lactate dehydrogenase, providing a detailed energetic map of reaction trajectories unavailable with traditional methods [107]. Vibrational spectroscopy, while not directly measuring atomic positions, is particularly powerful because frequencies can be assigned to specific bonds located within proteins or bound reactants and can provide information about changes in the strength and properties of those interactions. It may also be possible to carry out time-resolved vibrational studies on enzyme libraries in high throughput. A critical next step will be to apply these synchronized approaches more broadly so that motions and transitions are not averaged among the population of molecules in the crystal [82].
4. The complexity of functional models
Residues are functionally, and thus energetically, interdependent. This interconnection is most simply appreciated by recognizing that without the “right” residues surrounding the catalytic and binding residues those active site residues do not yield significant catalysis, and vice versa [108]. Consequently, descriptions of “residue function” cannot be made from single-mutant variants alone. The extent of residue connectivity—how many residues affect the function of a particular residue and by how much—defines the complexity of the model that is needed to mathematically describe an enzyme’s function [109,110]. While this complexity is likely to vary—for enzymes with different folds, with allostery, etc.—we want to know the scale and range of this complexity as it will dictate the form of models and how they are developed and tested [111,112].
We also know that residues are not fully interdependent (epistatic), as if this were the case any single mutation would shatter the active site and fully abolish activity. Classical mutational studies have found some regions, including active sites, with limited energetic dependencies among sets of three or fewer residues [113–115]. In one striking case, a single residue change was predominantly responsible for improved activity in psychrophiles versus improved stability in thermophiles [116]. Phylogenetic comparisons, verified experimentally, identified two nearby residue changes needed to fully shift stability and activity, but with much smaller effects.
Phylogenetic comparisons across many enzyme families containing psycrophilic, mesophilic, and thermophilic variants suggested instances of limited epistasis in temperature adaptation [116]. Most of these covarying sets of residues corresponded primarily to pairs of residues that correlate with temperature adaptation in and likely confer function within divergent sequence backgrounds of a given enzyme family. These observations suggest substantial simplifications in residue interdependences and model complexity ion many cases. Nevertheless, there are also larger co-occurring sets, and our initial HT-MEK experiments in PafA have also identified functional interconnections among tens of residues in regions extending from the active site to the surface.
Experimentally, even measuring effects of all possible triple-mutant substitutions within a single small (100 residue) enzyme is intractable, as it would require >109 variants. Instead, it will be necessary to prioritize higher-order mutants most likely to be informative, guided by maps of enzyme architecture identified in initial HT-MEK surveys and additional phylogenetic information such as that described to understand temperature adaptation [117]. This is an area that will likely require innovative ideas and rigorous tests to identify paths towards predictive models. The length of those paths and the difficulty in traversing them will be determined by how rapidly partially-predictive physics-based models can be developed and used as guides.
Conclusion and outlook
Accurate quantitative prediction of protein and enzyme functions from primary sequence is the ultimate litmus test of our biophysical understanding. But as other disciplines have experienced, breakthroughs frequently arrive later than hoped, and only at the nexus of deep need, technical ability, an empirical or theoretical framework, and a systematic and sustained effort.
The need for accurate and general quantitative models of functional prediction is clear—they would transform medicine, industry, and biotechnology. But what remains uncertain is whether we possess the requisite theoretical and technological foundation. Where we stand is likely to be clarified only through careful tests of current general predictive models and algorithms, particularly physics-based ones, using blinded comparisons to large-scale empirical measurements. With HT-MEK and crystallographic ensembles, we are starting to acquire the needed quality and quantity of data to compare with predictions.
Most immediately, HT-MEK can be used to study new enzymes, as it can be applied to any enzyme with a direct or coupled fluorogenic assay. HT-MEK, and extensions to it currently under development, can assay protein stability as well as function, and important applications include dissecting the functional effects of human alleles that do or may cause disease; providing foundational information to guide protein engineering and design efforts using current approaches; and combining enzymes to efficiently engineer metabolic pathways.
We envision a future wherein large-scale mutational studies are routinely performed with HT-MEK. In this future, analysis, interpretation, and modeling are the rate-limiting steps of advancing protein biochemistry, instead of experimentation. To drive future advances in these models, conformational ensembles will provide ground-truth atomic positions and motions and describe how these are altered in mutants of differing functions. Ensembles can be assembled for many enzymes from the vast data available in the PDB [72], and ensemble information can now be readily attained for new enzyme complexes and variants through advances in room temperature data collection [65].
We suspect that a “critical assessment of quantitative protein function” will be needed. While blind protein function prediction challenges exist, these contests typically use in vivo coarse phenotypic data (e.g., whether or not particular mutations are deleterious) as targets, reflecting the absence of and need for large-scale in vitro quantitative measurements of specific functional parameters [118,119]. A new effort would involve solicitation of large-scale quantitative functional datasets ahead of publication and creation of objective metrics for success that scale across different parameters and sizes of datasets. We look forward to contributing to this effort by basic scientists, engineers, and theoreticians.
Finally, with deep functional data in hand, it will also be possible to extend predictive models to the systems level, connecting the basic enzyme properties responsible for selective advantages observed in DMS experiments and in natural and laboratory evolution. Here comparisons of effects predicted from metabolic models for particular kinetic perturbations with experimental measurements of fitness and of metabolite levels will provide a path to develop robust models to understand cellular metabolism and to engineer new metabolic pathways.
Acknowledgements
We dedicate this article to the memory of Dan Tawfik who recently and tragically passed away. Dan’s creativity and enthusiasm was an inspiration to us, and many others. We thank Craig Markin, Siyuan Du, and members of the Herschlag and Fordyce laboratories for helpful discussions and review. This work was supported with funding from a National Institutes of Health (NIH) R01 grant (GM064798), Gordon and Betty Moore Foundation grant, and Ono Pharma Foundation Breakthrough Initiative Prize awarded to D.H. and P.M.F, and a National Science Foundation (NSF) grant (MCB-1714723) awarded to D.H. D.A.M. acknowledges support from the Stanford Medical Scientist Training Program and a Stanford Interdisciplinary Graduate Fellowship (Anonymous Donor) affiliated with Stanford ChEM-H. P.M.F. is a Chan Zuckerberg Biohub Investigator.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.CASP13 proceedings. Proteins 2019, 87:1007–1388. [Google Scholar]
- 2.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, et al. : Improved protein structure prediction using potentials from deep learning. Nature 2020, 577:706–710. [DOI] [PubMed] [Google Scholar]
- 3.Kato Y, Miyakawa T, Tanokura M: Overview of the mechanism of cytoskeletal motors based on structure. Biophys Rev 2018, 10:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indiani C, O’Donnell M: The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol 2006, 7:751–761. [DOI] [PubMed] [Google Scholar]
- 5.Keskin O, Nussinov R: Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng Des Sel 2005, 18:11–24. [DOI] [PubMed] [Google Scholar]
- 6.Kraut DA, Sigala PA, Pybus B, Liu CW, Ringe D, Petsko GA, Herschlag D: Testing electrostatic complementarity in enzyme catalysis: hydrogen bonding in the Ketosteroid Isomerase oxyanion hole. PLoS Biol 2006, 4:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Goodey NM: Catalytic contributions from remote regions of enzyme structure. Chem Rev 2011, 111:7595–7624. [DOI] [PubMed] [Google Scholar]
- 8.Hilser VJ, Wrabl JO, Motlagh HN: Structural and energetic basis of allostery. Annu Rev Biophys 2012, 41:585–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nussinov R, Tsai C-J: Allostery in disease and in drug discovery. Cell 2013, 153:293–305. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds KA, McLaughlin RN, Ranganathan R: Hot spots for allosteric regulation on protein surfaces. Cell 2011, 147:1564–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrenbeck EE, Azouz LR, Whitehead TA: Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded. Nat Commun 2017, 8:15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, Russ WP, Benkovic SJ, Ranganathan R: Surface sites for engineering allosteric control in proteins. Science 2008, 322:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dryden DTF, Thomson AR, White JH: How much of protein sequence space has been explored by life on Earth? J R Soc Interface 2008, 5:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C: Protein 3D structure computed from evolutionary sequence variation. PLoS ONE 2011, 6:e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau KF, Dill KA: A lattice statistical mechanics model of the conformational and sequence spaces of proteins. Macromolecules 1989, 22:3986–3997. [Google Scholar]
- 16.Adcock SA, McCammon JA: Molecular dynamics: survey of methods for simulating the activity of proteins. Chem Rev 2006, 106:1589–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frauenfelder H, Sligar S, Wolynes P: The energy landscapes and motions of proteins. Science 1991, 254:1598–1603. [DOI] [PubMed] [Google Scholar]
- 18.Wei G, Xi W, Nussinov R, Ma B: Protein ensembles: how does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem Rev 2016, 116:6516–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aesop Gibbs L (Eds): Aesop’s fables. Oxford University Press; 2008. [Google Scholar]
- 20.Claeyssens F, Harvey JN, Manby FR, Mata RA, Mulholland AJ, Ranaghan KE, Schütz M, Thiel S, Thiel W, Werner H-J: High-accuracy computation of reaction barriers in enzymes. Angew Chem Int Ed 2006, 45:6856–6859. [DOI] [PubMed] [Google Scholar]
- 21.Mulholland AJ: Chemical accuracy in QM/MM calculations on enzyme-catalysed reactions. Chem Cent J 2007, 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevenson Angus, Lindberg Christine A.: New Oxford American Dictionary (3 ed.). Oxford University Press; 2010. [Google Scholar]
- 23.Wooley JC, Ye Y: A Historical Perspective and Overview of Protein Structure Prediction. In Computational Methods for Protein Structure Prediction and Modeling. Edited by Xu Y, Xu D, Liang J. Springer; New York; 2007:1–43. [Google Scholar]
- 24.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A: Critical assessment of methods of protein structure prediction (CASP) - round x: critical assessment of structure prediction. Proteins 2014, 82:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman HM: The Protein Data Bank. Nucleic Acids Res 2000, 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das R, Baker D: Macromolecular modeling with Rosetta. Annu Rev Biochem 2008, 77:363–382. [DOI] [PubMed] [Google Scholar]
- 27.Kamisetty H, Ovchinnikov S, Baker D: Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci U S A 2013, 110:15674–15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.AlQuraishi M: End-to-end differentiable learning of protein structure. Cell Systems 2019, 8:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhlman B, Bradley P: Advances in protein structure prediction and design. Nat Rev Mol Cell Biol 2019, 20:681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, DiMaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, et al. : The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput 2017, 13:3031–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fowler DM, Fields S: Deep mutational scanning: a new style of protein science. Nat Methods 2014, 11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esposito D, Weile J, Shendure J, Starita LM, Papenfuss AT, Roth FP, Fowler DM, Rubin AF: MaveDB: an open-source platform to distribute and interpret data from multiplexed assays of variant effect. Genome Biol 2019, 20:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr TN, Thornton JW: Epistasis in protein evolution: Epistasis in Protein Evolution. Protein Sci 2016, 25:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mehlhoff JD, Stearns FW, Rohm D, Wang B, Tsou E-Y, Dutta N, Hsiao M-H, Gonzalez CE, Rubin AF, Ostermeier M: Collateral fitness effects of mutations. Proc Natl Acad Sci U S A 2020, 117:11597–11607. •• Cellular fitness effects of mutations in protein-coding genes can arise from defects unrelated to the gene’s specific function, like misfolding, aggregation, and stress induction. The authors found that for TEM-1 β-lactamase, these “collateral” mutational effects can occur as often or more than effects on intrinsic function or abundance, suggesting fitness-based deep mutational scanning data can reflect phenotypes with limited linkage to specific function.
- 35.Romero PA, Tran TM, Abate AR: Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc Natl Acad Sci U S A 2015, 112:7159–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris DT, Wang N, Riley TP, Anderson SD, Singh NK, Procko E, Baker BM, Kranz DM: Deep mutational scans as a guide to engineering high affinity T cell receptor interactions with peptide-bound major histocompatibility complex. J Biol Chem 2016, 291:24566–24578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandaru P, Shah NH, Bhattacharyya M, Barton JP, Kondo Y, Cofsky JC, Gee CL, Chakraborty AK, Kortemme T, Ranganathan R, et al. : Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife 2017, 6:e27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamer YT, Gaszek IK, Abdizadeh H, Batur T, Reynolds K, Atilgan AR, Atilgan C, Toprak E: High-order epistasis in catalytic power of dihydrofolate reductase gives rise to a rugged fitness landscape in the presence of trimethoprim selection. Mol Biol Evol 2019, 36:1533–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson S, Zhang Y, Ingle C, Reynolds KA, Kortemme T: Altered expression of a quality control protease in E. coli reshapes the in vivo mutational landscape of a model enzyme. eLife 2020, 9:e53476. •• Examined the impacts of altered cellular contexts on the mutational sensitivity of DHFR using deep mutational scanning. The authors observed that presence or absence of cellular quality control machinery (Lon protease) substantially altered mutational effects with cell fitness as a readout, and that apparently advantageous variants become deleterious when Lon was present. These findings suggest widespread activity-stability tradeoffs that can be masked or elicited depending on the specific strains and conditions used when performing deep mutational scanning.
- 40.Baase WA, Liu L, Tronrud DE, Matthews BW: Lessons from the lysozyme of phage T4. Protein Sci 2010, 19:631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Q, Fenton AW: Whole-protein alanine-scanning mutagenesis of allostery: A large percentage of a protein can contribute to mechanism. Hum Mutat 2017, 38:1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlin DA, Caster RW, Wang X, Betzenderfer SA, Chen CX, Duong VM, Ryklansky CV, Alpekin A, Beaumont N, Kapoor H, et al. : Kinetic characterization of 100 glycoside hydrolase mutants enables the discovery of structural features correlated with kinetic constants. PLOS ONE 2016, 11:e0147596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlin DA, Hapig-Ward S, Chan BW, Damrau N, Riley M, Caster RW, Bethards B, Siegel JB: Thermal stability and kinetic constants for 129 variants of a family 1 glycoside hydrolase reveal that enzyme activity and stability can be separately designed. PLOS ONE 2017, 12:e0176255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maerkl SJ, Quake SR: A systems approach to measuring the binding energy landscapes of transcription factors. Science 2007, 315:233–237. [DOI] [PubMed] [Google Scholar]
- 45.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR: De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol 2010, 28:970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aditham AK, Markin CJ, Mokhtari DA, DelRosso N, Fordyce PM: High-throughput affinity measurements of transcription factor and DNA Mutations reveal affinity and specificity determinants. Cell Syst 2021, 12:112–127.e11. [DOI] [PubMed] [Google Scholar]
- 47. Markin CJ, Mokhtari DA, Sunden F, Appel MJ, Akiva E, Longwell SA, Sabatti C, Herschlag D, Fordyce PM: Revealing enzyme functional architecture via high-throughput microfluidic enzyme kinetics. bioRxiv 2020, doi: 10.1101/2020.11.24.383182. •• Development and application of a high-throughput platform for simultaneous protein expression, purification, and kinetic analysis for 1500 enzyme variants. This study revealed enzyme functional architecture by obtaining and comparing multiple catalytic and binding parameters for mutations at every position of a highly-proficient phosphate monoesterase.
- 48.Sunden F, AlSadhan I, Lyubimov AY, Ressl S, Wiersma-Koch H, Borland J, Brown CL, Johnson TA, Singh Z, Herschlag D: Mechanistic and evolutionary insights from comparative enzymology of phosphomonoesterases and phosphodiesterases across the alkaline phosphatase superfamily. J Am Chem Soc 2016, 138:14273–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien PJ, Herschlag D: Functional interrelationships in the alkaline phosphatase superfamily: phosphodiesterase activity of Escherichia coli alkaline phosphatase. Biochemistry 2001, 40:5691–5699. [DOI] [PubMed] [Google Scholar]
- 50.Zalatan JG, Fenn TD, Brunger AT, Herschlag D: Structural and functional comparisons of Nucleotide Pyrophosphatase/Phosphodiesterase and Alkaline Phosphatase: implications for mechanism and evolution. Biochemistry 2006, 45:9788–9803. [DOI] [PubMed] [Google Scholar]
- 51.Wiersma-Koch H, Sunden F, Herschlag D: Site-directed mutagenesis maps interactions that enhance cognate and limit promiscuous catalysis by an alkaline phosphatase superfamily phosphodiesterase. Biochemistry 2013, 52:9167–9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sunden F, AlSadhan I, Lyubimov A, Doukov T, Swan J, Herschlag D: Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution. J Biol Chem 2017, 292:20960–20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atsavapranee B, Stark CD, Sunden F, Thompson S, Fordyce PM: Fundamentals to function: quantitative and scalable approaches for measuring protein stability. Cell Systems 2021, Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdel-Magid AF: Allosteric modulators: an emerging concept in drug discovery. ACS Med Chem Lett 2015, 6:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frauenfelder H, Petsko GA, Tsernoglou D: Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature 1979, 280:558–563. [DOI] [PubMed] [Google Scholar]
- 56.Hartmann H, Parak F, Steigemann W, Petsko GA, Ponzi DR, Frauenfelder H: Conformational substates in a protein: structure and dynamics of metmyoglobin at 80 K. Proc Natl Acad Sci U S A 1982, 79:4967–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammes GG, Benkovic SJ, Hammes-Schiffer S: Flexibility, diversity, and cooperativity: pillars of enzyme catalysis. Biochemistry 2011, 50:10422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz SD: Protein dynamics and the enzymatic reaction coordinate. In Dynamics in Enzyme Catalysis. Edited by Klinman J, Hammes- Schiffer S. Springer; Berlin Heidelberg; 2013:189–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanoian P, Liu CT, Hammes-Schiffer S, Benkovic S: Perspectives on electrostatics and conformational motions in enzyme catalysis. Acc Chem Res 2015, 48:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohen A: Role of dynamics in enzyme catalysis: substantial versus semantic controversies. Acc Chem Res 2015, 48:466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warshel A, Bora RP: Perspective: Defining and quantifying the role of dynamics in enzyme catalysis. J Chem Phys 2016, 144:180901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmer AG: NMR characterization of the dynamics of biomacromolecules. Chem Rev 2004, 104:3623–3640. [DOI] [PubMed] [Google Scholar]
- 63.Alderson TR, Kay LE: NMR spectroscopy captures the essential role of dynamics in regulating biomolecular function. Cell 2021, 184:577–595. [DOI] [PubMed] [Google Scholar]
- 64.Halle B: Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc Natl Acad Sci U S A 2004, 101:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Doukov T, Herschlag D, Yabukarski F: Instrumentation and experimental procedures for robust collection of X-ray diffraction data from protein crystals across physiological temperatures. J Appl Crystallogr 2020, 53:1493–1501. • Reports experimental strategies to facilitate the collection of high resolution (<1.5 Å) x-ray structures at above-room temperatures, up to 363 K. Applied to proteinase K, thaumatin, and lysozyme crystals, this approach is generalizable to crystallographic study of other enzymes at physiological temperatures and can be readily implemented at other beamlines.
- 66.Zoete V, Michielin O, Karplus M: Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J Mol Biol 2002, 315:21–52. [DOI] [PubMed] [Google Scholar]
- 67.Best RB, Lindorff-Larsen K, DePristo MA, Vendruscolo M: Relation between native ensembles and experimental structures of proteins. Proc Natl Acad Sci U S A 2006, 103:10901–10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monzon AM, Rohr CO, Fornasari MS, Parisi G: CoDNaS 2.0: a comprehensive database of protein conformational diversity in the native state. Database 2016, 2016:baw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Z, Jaroszewski L, Iyer M, Sedova M, Godzik A: FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res 2020, 48:W60–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang S, Krieger JM, Zhang Y, Kaya C, Kaynak B, Mikulska-Ruminska K, Doruker P, Li H, Bahar I: ProDy 2.0: increased scale and scope after 10 years of protein dynamics modelling with Python. Bioinformatics 2021, doi: 10.1093/bioinformatics/btab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grant BJ, Skjærven L, Yao X: The BIO3D packages for structural bioinformatics. Protein Sci 2021, 30:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yabukarski F, Biel JT, Pinney MM, Doukov T, Powers AS, Fraser JS, Herschlag D: Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray–derived conformational ensembles. Proc Natl Acad Sci U S A 2020, 117:33204–33215. •• Obtains conformational ensembles from “room temperature” cryogenic X-ray diffraction data for wild type and mutant ketosteroid isomerase in complexes representative of the states through its reaction cycle. Ensemble-function analysis is applied to evaluate catalytic models and the forces underlying positioning of active site residues.
- 73.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T: Hidden alternative structures of proline isomerase essential for catalysis. Nature 2009, 462:669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fraser JS, van den Bedem H, Samelson AJ, Lang PT, Holton JM, Echols N, Alber T: Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc Natl Acad Sci U S A 2011, 108:16247–16252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keedy DA, van den Bedem H, Sivak DA, Petsko GA, Ringe D, Wilson MA, Fraser JS: Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR. Structure 2014, 22:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dellus-Gur E, Elias M, Caselli E, Prati F, Salverda MLM, de Visser JAGM, Fraser JS, Tawfik DS: Negative epistasis and evolvability in TEM-1 β-Lactamase—The thin line between an enzyme’s conformational freedom and disorder. J Mol Biol 2015, 427:2396–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keedy DA, Kenner LR, Warkentin M, Woldeyes RA, Hopkins JB, Thompson MC, Brewster AS, Van Benschoten AH, Baxter EL, Uervirojnangkoorn M, et al. : Mapping the conformational landscape of a dynamic enzyme by multitemperature and XFEL crystallography. eLife 2015, 4:e07574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Acker TM, Gable JE, Bohn M-F, Jaishankar P, Thompson MC, Fraser JS, Renslo AR, Craik CS: Allosteric inhibitors, crystallography, and comparative analysis reveal network of coordinated movement across human herpesvirus proteases. J Am Chem Soc 2017, 139:11650–11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Offenbacher AR, Hu S, Poss EM, Carr CAM, Scouras AD, Prigozhin DM, Iavarone AT, Palla A, Alber T, Fraser JS, et al. : Hydrogen–deuterium exchange of lipoxygenase uncovers a relationship between distal, solvent exposed protein motions and the thermal activation barrier for catalytic proton-coupled electron tunneling. ACS Cent Sci 2017, 3:570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keedy DA, Hill ZB, Biel JT, Kang E, Rettenmaier TJ, Brandão-Neto J, Pearce NM, von Delft F, Wells JA, Fraser JS: An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering. eLife 2018, 7:e36307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hu S, Offenbacher AR, Thompson EM, Gee CL, Wilcoxen J, Carr CAM, Prigozhin DM, Yang V, Alber T, Britt RD, et al. : Biophysical characterization of a disabled double mutant of soybean lipoxygenase: the “undoing” of precise substrate positioning relative to metal cofactor and an identified dynamical network. J Am Chem Soc 2019, 141:1555–1567. • Reports room-temperature crystallography, EPR and hydrogen-deuterium exchange for soybean lipoxygenase. By comparing ensembles of WT and active site mutants, the authors identified an altered microenvironment (motions and interactions) around the iron cofactor that impairs optimal substrate binding for hydrogen transfer.
- 82.Dasgupta M, Budday D, de Oliveira SHP, Madzelan P, Marchany-Rivera D, Seravalli J, Hayes B, Sierra RG, Boutet S, Hunter MS, et al. : Mix-and-inject XFEL crystallography reveals gated conformational dynamics during enzyme catalysis. Proc Natl Acad Sci U S A 2019, 116:25634–25640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Broom A, Rakotoharisoa RV, Thompson MC, Zarifi N, Nguyen E, Mukhametzhanov N, Liu L, Fraser JS, Chica RA: Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico. Nat Commun 2020, 11:4808. •• Determines the mechanistic basis for catalytic improvements provided by directed evolution of a de novo designed Kemp eliminase. Ensemble crystallography was used to identify more favorable positioning of active site residues along the mutational trajectory of evolved variants.
- 84.Robertus JD, Kraut J, Alden RA, Birktoft JJ: Subtilisin. Stereochemical mechanism involving transition-state stabilization. Biochemistry 1972, 11:4293–4303. [DOI] [PubMed] [Google Scholar]
- 85.Kraut J: Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem 1977, 46:331–358. [DOI] [PubMed] [Google Scholar]
- 86.Kamerlin SCL, Chu ZT, Warshel A: On catalytic preorganization in oxyanion holes: highlighting the problems with the gas-phase modeling of oxyanion holes and illustrating the need for complete enzyme models. J Org Chem 2010, 75:6391–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simón L, Goodman JM: Hydrogen-bond stabilization in oxyanion holes: grand jeté to three dimensions. Org Biomol Chem 2012, 10:1905. [DOI] [PubMed] [Google Scholar]
- 88.Sigala PA, Kraut DA, Caaveiro JMM, Pybus B, Ruben EA, Ringe D, Petsko GA, Herschlag D: Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole. J Am Chem Soc 2008, 130:13696–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sigala PA, Fafarman AT, Schwans JP, Fried SD, Fenn TD, Caaveiro JMM, Pybus B, Ringe D, Petsko GA, Boxer SG, et al. : Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site. Proc Natl Acad Sci U S A 2013, 110:E2552–E2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pinney MM, Natarajan A, Yabukarski F, Sanchez DM, Liu F, Liang R, Doukov T, Schwans JP, Martinez TJ, Herschlag D: Structural coupling throughout the active site hydrogen bond networks of Ketosteroid Isomerase and Photoactive Yellow Protein. J Am Chem Soc 2018, 140:9827–9843. •• Describes simple molecular mechanisms accounting for thermo-adaptation of activity and stability in a mesophilic and a thermophilic ketosteroid isomerase (KSI), almost fully accounted for by a single residue change at the active site. Through phylogenetic comparisons of ~2000 classes of homologous enzymes across ~5800 organisms, the authors found 1000s of examples of parallel changes in temperature adaptation, indicative of adaptive changes from small numbers of residues and limited epistasis.
- 91.Lamba V, Yabukarski F, Pinney M, Herschlag D: Evaluation of the catalytic contribution from a positioned general base in Ketosteroid Isomerase. J Am Chem Soc 2016, 138:9902–9909. [DOI] [PubMed] [Google Scholar]
- 92.Jindal G, Warshel A: Misunderstanding the preorganization concept can lead to confusions about the origin of enzyme catalysis. Proteins 2017, 85:2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Menger FM, Nome F: Interaction vs preorganization in enzyme catalysis. A dispute that calls for resolution. ACS Chem Biol 2019, 14:1386–1392. [DOI] [PubMed] [Google Scholar]
- 94.Lassila JK, Baker D, Herschlag D: Origins of catalysis by computationally designed retroaldolase enzymes. Proc Natl Acad Sci U S A 2010, 107:4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hilvert D: Design of protein catalysts. Annu Rev Biochem 2013, 82:447–470. [DOI] [PubMed] [Google Scholar]
- 96.Kiss G, Çelebi-Ölçüm N, Moretti R, Baker D, Houk KN: Computational enzyme design. Angew Chem Int Ed 2013, 52:5700–5725. [DOI] [PubMed] [Google Scholar]
- 97.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. : Kemp elimination catalysts by computational enzyme design. Nature 2008, 453:190–195. [DOI] [PubMed] [Google Scholar]
- 98.Lamba V, Sanchez E, Fanning LR, Howe K, Alvarez MA, Herschlag D, Forconi M: Kemp eliminase activity of Ketosteroid Isomerase. Biochemistry 2017, 56:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blomberg R, Kries H, Pinkas DM, Mittl PR, Grutter MG, Privett HK, Mayo SL, Hilvert D: Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 2013, 503:418–21. [DOI] [PubMed] [Google Scholar]
- 100.Otten R, Pádua RAP, Bunzel HA, Nguyen V, Pitsawong W, Patterson M, Sui S, Perry SL, Cohen AE, Hilvert D, et al. : How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 2020, doi: 10.1126/science.abd3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Page MI, Jencks WP: Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci U S A 1971, 68:1678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Menger FM: On the source of intramolecular and enzymatic reactivity. Acc Chem Res 1985, 18:128–134. [Google Scholar]
- 103.Chapman HN: X-Ray Free-Electron Lasers for the Structure and Dynamics of Macromolecules. Annu Rev Biochem 2019, 88:35–58. [DOI] [PubMed] [Google Scholar]
- 104.Orville AM: Recent results in time resolved serial femtosecond crystallography at XFELs. Current Opinion in Structural Biology 2020, 65:193–208. [DOI] [PubMed] [Google Scholar]
- 105.Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, et al. : Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science 2014, 346:1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barends TRM, Foucar L, Ardevol A, Nass K, Aquila A, Botha S, Doak RB, Falahati K, Hartmann E, Hilpert M, et al. : Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science 2015, 350:445–450. [DOI] [PubMed] [Google Scholar]
- 107.Reddish MJ, Peng H-L, Deng H, Panwar KS, Callender R, Dyer RB: Direct Evidence of Catalytic Heterogeneity in Lactate Dehydrogenase by Temperature Jump Infrared Spectroscopy. J Phys Chem B 2014, 118:10854–10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kraut DA, Carroll KS, Herschlag D: Challenges in enzyme mechanism and energetics. Annu Rev Biochem 2003, 72:517–571. [DOI] [PubMed] [Google Scholar]
- 109.Poelwijk FJ, Krishna V, Ranganathan R: The context-dependence of mutations: A linkage of formalism. PLOS Comput Biol 2016, 12:e1004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang G, Anderson DW, Baier F, Dohmen E, Hong N, Carr PD, Kamerlin SCL, Jackson CJ, Bornberg-Bauer E, Tokuriki N: Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme. Nat Chem Biol 2019, 15:1120–1128. • Employs ancestral reconstruction and high-order mutagenesis to understand the recently-evolved phosphotriesterase activity of methyl-parathion hydrolase (MPH). Only five spatially clustered mutations (none at the active site) converted a dihydrocoumarin hydrolase to an MPH. Higher-order epistasis accounted for only <1% (5-fold) of the ~1000-fold net MPH activity gain, suggesting promiscuous activity can be greatly enhanced without strongly epistatic networks, but evolutionary trajectories were constrained by epistasis among intermediates.
- 111.Sailer ZR, Harms MJ: Molecular ensembles make evolution unpredictable. Proc Natl Acad Sci U S A 2017, 114:11938–11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Morrison AJ, Wonderlick DR, Harms MJ: Ensemble epistasis: thermodynamic origins of non-additivity between mutations. bioRxiv 2020, doi: 10.1101/2020.10.14.339671. •• Presents an analytical framework for predicting and understanding epistasis based on conformational ensembles. The authors find that the presence of multiple protein substates with differing functional properties, along with differential effects on substate occupancy upon mutation, can account for widespread epistasis in biology.
- 113.Carter P, Wells JA: Dissecting the catalytic triad of a serine protease. Nature 1988, 332:564. [DOI] [PubMed] [Google Scholar]
- 114.Wells JA: Additivity of mutational effects in proteins. Biochemistry 1990, 29:8509–8517. [DOI] [PubMed] [Google Scholar]
- 115.Sunden F, Peck A, Salzman J, Ressl S, Herschlag D: Extensive site-directed mutagenesis reveals interconnected functional units in the alkaline phosphatase active site. eLife 2015, 4:e06181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pinney MM, Mokhtari DA, Akiva E, Yabukarski F, Sanchez DM, Liang R, Doukov T, Martinez TJ, Babbitt PC, Herschlag D: Parallel molecular mechanisms for enzyme temperature adaptation. Science 2021, 371:eaay2784. [DOI] [PubMed] [Google Scholar]
- 117. Russ WP, Figliuzzi M, Stocker C, Barrat-Charlaix P, Socolich M, Kast P, Hilvert D, Monasson R, Cocco S, Weigt M, et al. : An evolution-based model for designing chorismate mutase enzymes. Science 2020, 369:440. • Direct coupling analysis, using first and second-order coupling terms from an alignment of extant chorismate mutase orthologs, was used as a generative model for non-natural sequences. More than 1000 artificial designed mutases were assayed with a competitive growth and deep-sequencing protocol, and nearly half (up to 80% sequence divergence) were apparently active, underscoring the importance of low-order coupling terms in specifying enzyme function, at least for this enzyme that is at the lower end in the range of catalytic rate enhancements.
- 118.Zhou N, Jiang Y, Bergquist TR, Lee AJ, Kacsoh BZ, Crocker AW, Lewis KA, Georghiou G, Nguyen HN, Hamid MN, et al. : The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol 2019, 20:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andreoletti G, Pal LR, Moult J, Brenner SE: Reports from the fifth edition of CAGI: The Critical Assessment of Genome Interpretation. Hum Mutat 2019, 40:1197–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]