Abstract
Precision oncology emphasizes genotyping as one of the mainstays of oncological decision‐making. The core information element exchanged between the laboratory and the oncologist is the precise mutation. Specifically, it is the written representation typically in the form of a variant description at the DNA or protein level. These annotations can be confusing, and many commercial laboratories have abandoned DNA‐level annotations. Here we present a complex double‐point mutation to illustrate a situation where a formally “correct” reporting nomenclature can obscure clinically relevant and potentially clinically actionable information.
Key Points
The Human Genome Variation Society (HGVS) currently recommends that “two variants separated by one or more nucleotides should be described individually and not as a combined ‘delins’ (deletion‐inserion).”
There remains confusion about the appropriate nomenclature to report variants and the significance of these variants among clinicians. It is the clinically integrated molecular‐genetic interpretation that will help clinicians make informed decisions to improve patient care.
Keywords: Biomarker • IDH • Variant annotation
Short abstract
With the increasingly popular use of next‐generation sequencing for tumor genotyping, molecular pathologists encounter variants that are on the same allele and close enough to be combined and annotated together. This article reports a case of acute myeloid leukemia with two in cis IDH2 variants, illustrating the importance of correct variant annotation as good communication practices between molecular laboratories and oncologists.
Patient Story
A 62‐year‐old man with a history of Klinefelter syndrome and essential thrombocythemia on hydroxyurea and aspirin presented with pancytopenia. As blood counts remained low despite discontinuation of hydroxyurea, a bone marrow biopsy was performed and showed a hypercellular marrow at 90% cellularity with 10%–15% blasts (Fig. 1A). Bone marrow aspirate showed 13.8% blasts by morphology and 17% myeloid blasts by flow cytometry (CD33 dim/−, CD13+, MPO−, CD117+, CD34+, HLA‐DR+ cells). Cytogenetics showed a karyotype of 47,XXYc[20]. A diagnosis of myeloproliferative neoplasm with emerging blast phase was made. The patient shortly progressed to acute myeloid leukemia (AML) after a repeat bone marrow biopsy showed 29% blasts. A next‐generation sequencing panel with n=103 genes was performed on a bone marrow aspirate sample. The panel revealed variants predicted to result in DNMT3A Arg882His, RUNX1 Arg204Gln, MPL Trp515Leu, and two single nucleotide variants in the IDH2 gene—c.502A>G and c.514A>G (ENST00000330062.3)—each with an allele frequency of 31% (Fig. 1).
Figure 1.
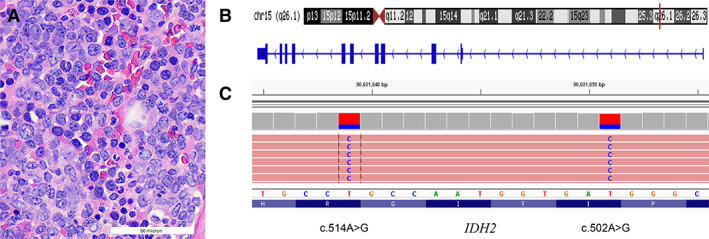
(A): Microphotograph of the bone marrow biopsy showing hypercellular marrow with an increase of blasts. (B, C): Visual exploration of the two in cis transition mutations in IDH2 exon 4 identified in this patient. Note: IDH2 is encoded in antisense direction.
Abbreviation: bp, base pair.
Molecular Tumor Board
The two IDH2 variants are 12 nucleotides apart and present on the same allele (in cis) (Fig. 1B, 1C). This finding can be viewed as two separate events affecting the same allele or as a single complex deletion/insertion (delins) variant. One way of reporting the latter (i.e., the combined variant) would be c.502_514delATCACCATTGGCAinsGTCACCATTGGCG, which translates to p.Ile168_Arg172delinsValThrIleGlyGly (Fig. 2). Without manual review of the variant (i.e., pile‐up; Integrative Genomics Viewer, IGV, Broad Institute, Boston, MA) or realization that the delins annotation may not be optimal to indicate the underlying (actionable) amino acid changes, the variant might be interpreted as “of unknown significance.” Importantly, without an interpretative comment or direct communication with the oncologist, such unusual findings may be interpreted by the oncologist as “variants of unknown significance (VUS).” In this case, the pathology team was in direct correspondence with the patient's oncologist and clarified that the combined annotation included the canonical IDH2 amino acid alteration at position 172. After correspondence with the oncologist, we reported two missense mutations, Ile168Val and Arg172Gly, and explained in a comment that their orientation was in cis. IDH2 Arg172 is a well‐known oncogenic hotspot that can be therapeutically actionable by conferring sensitivity to IDH2 inhibitors [1].
Figure 2.
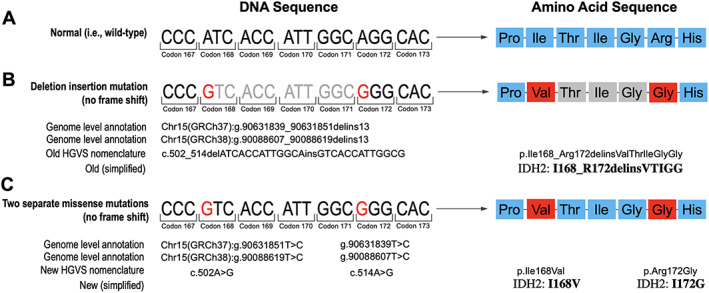
Illustration of the two alternative ways of variant annotation and reporting; wild‐type sequence for reference (A). The annotation as a deletion/insertion is formally correct yet difficult to interpret (B). The traditionally “wrong” annotation as two separate substitutions is more accessible to clinicians (C).Abbreviation: HGVS, Human Genome Variation Society.
Patient Update
The patient received 10 months of systemic chemotherapy with decitabine and venetoclax after initial diagnosis, followed by matched unrelated allogeneic donor stem cell transplantation (allo‐SCT). The patient was subsequently enrolled in a clinical trial evaluating post–allo‐SCT maintenance therapy with enasidenib, an IDH2 inhibitor, for IDH2‐mutant AML.
Discussion
The case illustrates several important aspects. First, there remains confusion about the appropriate nomenclature to report variants and the significance of these variants among clinicians. The Human Genome Variation Society (HGVS) developed and maintains comprehensive nomenclature recommendations that serve as an international standard for reporting of human variants and information exchange [2]. HGVS used to recommend annotation of combined variants as ‘delins’. However, this has recently been revised (https://varnomen.hgvs.org/recommendations/DNA/variant/delins/). The current version (version 20.05) recommends that “two variants separated by one or more nucleotides should be described individually and not as a ‘delins.’” The explicit reason for the change was that bioinformatics tools are not sophisticated enough to combine variants into the appropriate delins annotation [3]. We contend, however, that improved variant calling software is available [4]. Second, this case illustrates that clinically relevant information may sometimes hide in plain sight and that appropriate interpretation may require contextual knowledge and/or direct consultation with the molecular pathologist. It is also important for laboratories to recognize the complexity of some results and directly communicate their significance with the clinician taking care of the patient. Third, when encountering two or more variants in close proximity, one should (a) review the pile‐up for the orientation (phasing) of the variants (cis vs. trans) and (b) consider “unmasking” of clearly pathogenic mutations at the protein level (in particular when considering use of a converged, grouped, or combined nomenclature). Fourth, HGVS guidelines and other online resources (e.g., ClinVar, PubMed, Google) are critically important in completing these tasks and also in differentiating polymorphisms, germline variants, and somatic variants [5]. Fifth, at this time, it remains unclear whether the efficacy of specific inhibitors and their possible side effects are altered by additional nearby variants. Thus, correlative studies that take co‐occurring variants into account are urgently needed, and a unified nomenclature will certainly help clarify a comprehensive interpretation. Finally, from a practical perspective, we make the following suggestions: (a) use the nomenclature to include clearly pathogenic variants whenever possible because it helps the clinician to extract relevant information, (b) add a clarifying comment, and (c) actively seek consultation with the clinician instead of writing the commonly used phrase “clinical correlation recommended.” It is the clinically integrated molecular‐genetic interpretation that will help clinicians make informed decisions to improve patient care.
Glossary of Genomic Terms and Nomenclature
CD: cluster of differentiation
DNMT3A: DNA (cytosine‐5)‐methyltransferase 3A
HLA: human leukocyte antigen
IDH2: isocitrate dehydrogenase (NADP(+)) 2
MPO: myeloperoxidase
MPL: myeloproliferative leukemia virus oncogene
RUNX1: runt‐related transcription factor 1
Author Contributions
Conception/design: Ying‐Chun Lo, Jochen K. Lennerz
Provision of study material or patients: Rupa Narayan, Valentina Nardi
Collection and/or assembly of data: Ying‐Chun Lo, Jochen K. Lennerz
Data analysis and interpretation: Ying‐Chun Lo, Valentina Nardi, Jochen K. Lennerz
Manuscript writing: Ying‐Chun Lo, Rupa Narayan, Jochen K. Lennerz
Final approval of manuscript: Ying‐Chun Lo, Rupa Narayan, Valentina Nardi, Jochen K. Lennerz
Disclosures
Ying‐Chun Lo: Takeda Pharmaceutical Company Limited (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
References
- 1. Stein EM, DiNardo CD, Pollyea DA et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. den Dunnen JT, Dalgleish R, Maglott DR et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 2016;37:564–569. [DOI] [PubMed] [Google Scholar]
- 3. Deans ZC, Fairley JA, den Dunnen JT et al. HGVS nomenclature in practice: An example from the United Kingdom National External Quality Assessment Scheme. Hum Mutat 2016;37:576–578. [DOI] [PubMed] [Google Scholar]
- 4. Schmidt RJ, Macleay A, Le LP. VarGrouper: A bioinformatic tool for local haplotyping of deletion‐insertion variants from next‐generation sequencing data after variant calling. J Mol Diagn 2019;21:384–389. [DOI] [PubMed] [Google Scholar]
- 5. Li MM, Datto M, Duncavage EJ et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


