Abstract
Background: Studies have shown that circulating tumor DNA (ctDNA) indicates a poor prognosis in ovarian cancer. In this study, meta-analysis was used to assess the relationship between ctDNA and the prognosis of patients with epithelial ovarian cancer. Methods: The clinical trials included in this study were obtained via a search of PubMed, the Cochrane Library, the Web of Science and Embase between the period of establishment and March 2020. We selected clinical studies using qualitative or quantitative ctDNA methods to analyse the prognosis of ovarian epithelial cancer, screened the studies according to the determined inclusion and exclusion criteria, and used the modified JADAD score scale and NOS scale for evaluation, with OS (overall survival) and PFS (progression-free survival) as end events. The Cochrane Evaluation Tool was used to evaluate the quality of the randomized controlled trials. Stata 15.0 software was used to combine the effect ratio (hazard ratio, HR) and its 95% confidence interval (CI). In addition, a source analysis of ctDNA specimens, an analysis of ctDNA detection methods and a subgroup and sensitivity analysis of FIGO staging were performed. Results: A total of 8 studies were included in this meta-analysis, and ctDNA was found to be an independent risk factor for patients with epithelial ovarian cancer (OS: HR = 2.36, 95% CI [1.76,3.17], P < .001; PFS: HR = 2.51, 95% CI [1.83,3.45]). Conclusions: The results of our analysis suggested that ctDNA is a potential biomarker that can be used to evaluate the prognosis of patients with ovarian cancer.
Keywords: circulating tumor DNA, ovarian cancer, prognosis, meta-analysis
Introduction
Ovarian cancer is one of the three major gynaecological malignancies. According to the latest statistics in China, approximately 51 000 women suffered from ovarian cancer in 2014, and 23 000 died from the disease. Based on the data of Chinese cancer patients from 2003 to 2015, the 5-year survival rate of ovarian cancer patients (only 39%) has not changed significantly in the past 10 years, 1 and the 5-year recurrence rate has reached 70%. 2 Although surgery and chemotherapy for ovarian cancer are becoming increasingly advanced, there has been no significant improvement in the prognosis of ovarian cancer patients, and there has been no major change in their prognosis. Precise treatment and full-course management are the most recent approaches to ovarian cancer. With the development of targeted chemotherapy and targeted drugs, genetic testing and sequencing technology have become the basis for precise treatment. Therefore, identifying biomarkers related to treatment response and prognosis may help improve the clinical prognosis of ovarian cancer patients.
Currently, histopathological examination is considered the gold standard for ovarian cancer diagnosis; however, this method is invasive, and it is difficult to detect tumor changes via repeat sample acquisition. HE4 and CA125 are widely used for the early diagnosis of ovarian cancer as well as for monitoring and prognostic assessment. However, HE4 and CA125 cannot reflect tumor changes at the genetic level because tumors are heterogeneous, and tumor cells continue to develop, leading to metastasis, recurrence, and drug resistance. Circulating tumor DNA (ctDNA) is derived from primary tumors or metastatic lesions and can be isolated from plasma or serum by means of non-invasive methods. It can be used for early diagnosis and for monitoring the treatment response, detecting drug resistance and predicting clinical results.
It has been reported that ctDNA persists in patients with ovarian cancer after surgery, suggesting a poor clinical prognosis and its higher sensitivity than CA125.3–7 A series of evidence-based medical research analyses showed that ctDNA has positive significance for the diagnosis and prognosis of prostate cancer, non-small cell cancer, breast cancer and other tumors.8–13 Zhou et al. 14 found that ctDNA has positive significance for the early diagnosis of ovarian cancer through systematic review and multivariate analysis. However, to date, there have been no meta-analyses exploring the prognostic value of ctDNA for ovarian cancer. This article focuses on this aspect and provides a scientific basis for accelerating the clinical application of ctDNA.
Materials and Methods
My systematic review protocol has been submitted to the INPLASY register and has been published on the INPLASY website (DOI number: 10.37766/inplasy2021.8.0038; INPLASY202180038).
A comprehensive literature search was performed using the PubMed, Embase, Web of Science and Cochrane Library databases between the period of establishment and March, 2020. The search strategies are shown in Tables 1–4. This meta-analysis is performed based on the related items of the Preferred Reporting Items for Systematic Reviews and Meta-analysis statement (PRISMA statement). 15
Table 1.
Search Strategies of Pub Med Database.
| #1 | prognosis[Title/Abstract] OR overall survival[Title/Abstract] OR OS[Title/Abstract] OR progression free survival[Title/Abstract] OR PFS[Title/Abstract] OR Prognoses[Title/Abstract] OR Prognostic Factors[Title/Abstract] OR Factor, Prognostic[Title/Abstract] OR Factors, Prognostic[Title/Abstract] OR Prognostic Factor[Title/Abstract] OR Prognostic Factor[Title/Abstract] |
| #2 | Progression-Free Survival [Mesh] |
| #3 | #1 AND #2 |
| #4 | Neoplasm, Ovarian[Title/Abstract] OR Ovarian Neoplasm[Title/Abstract] OR Ovary Neoplasms[Title/Abstract] OR Ovary Cancer[Title/Abstract] OR Cancer, Ovary[Title/Abstract] OR Cancers, Ovary[Title/Abstract] OR Ovary Cancers[Title/Abstract] OR Ovarian Cancer[Title/Abstract] OR Cancer, Ovarian[Title/Abstract] OR Ovarian Cancers[Title/Abstract] OR epithelial ovarian cancer"[Title/Abstract] |
| #5 | Ovarian Neoplasms [Mesh] |
| #6 | #4 AND #5 |
| #7 | DNA,Circulating Tumor [Title/Abstract] OR Tumor DNA, Circulating[Title/Abstract] OR Cell-Free Tumor DNA[Title/Abstract] OR Cell Free Tumor DNA[Title/Abstract] OR Cell Free Tumor DNA[Title/Abstract] OR Tumor DNA, Cell-Free[Title/Abstract] OR ctDNA[Title/Abstract] |
| #8 | Circulating Tumor DNA[Mesh] |
| #9 | #7 AND #8 |
| #10 | #3 AND #6 AND #9 |
Table 4.
Cochrane Library Database.
| #1 | (prognosis):ti,ab,kw OR (overall survival):ti,ab,kw OR (os):ti,ab,kw OR (progression free survival):ti,ab,kw OR (pfs):ti,ab,kw |
| #2 | (ovary tumor):ti,ab,kw OR (ovarian neoplasms):ti,ab,kw OR (ovary cancer):ti,ab,kw OR (ovary cancers):ti,ab,kw OR (ovarian cancer):ti,ab,kw |
| #3 | (ctdna):ti,ab,kw OR (circulating tumor dna):ti,ab,kw OR (cell free tumor dna):ti,ab,kw |
| #4 | #1 AND #2 AND #3 |
Table 2.
Search Strategies of Embase Database.
| #1 | prognosis:ab,ti OR ‘overall survival’:ab,ti OR os:ab,ti OR ‘progression free survival’:ab,ti OR pfs:ab,ti |
| #2 | ‘ovary tumor’:ab,ti OR ‘ovarian neoplasms’:ab,ti OR ‘ovary cancer’:ab,ti OR ‘ovary cancers’:ab,ti OR ‘ovarian cancer’:ab,ti |
| #3 | ctdna:ab,ti OR ‘circulating tumor dna’:ab,ti OR ‘cell free tumor dna’:ab,ti |
| #4 | #1 AND #2 AND #3 |
Table 3.
Web of Science Database.
| #1 | TS = ([prognosis] OR [overall survival] OR [os] OR [progression free survival] OR [pfs]) |
| #2 | TS = ([ovary tumor] OR [ovarian neoplasms] OR [ovary cancer] OR [ovary cancers] OR [ovarian cancer]) |
| #3 | TS = ([ctdna] OR [circulating tumor dna] OR [cell free tumor dna]) |
| #4 | #1 AND #2 AND #3 |
Inclusion and Exclusion Criteria
Inclusion criteria
(1) Clinical research on ctDNA detection and ovarian cancer prognosis, including overall survival (OS) and progression-free survival (PFS); (2) patients with epithelial ovarian cancer confirmed by histopathology; (3) peripheral blood samples; and (4) prospective and retrospective research.
Exclusion criteria
(1) Case reports, editorials, meeting abstracts, and reviews; (2) repeated publications (in these cases, we selected the version with the most samples); or (3) required outcome indicators that could not be obtained directly or could not be calculated. For documents with unavailable full texts or incomplete data, we tried to contact the authors to obtain relevant data.
Data Extraction and Quality Assessment
The data included information on the first author, country, publication date, FIGO stage, blood type, peripheral blood test method, overall survival (OS), progression-free survival (PFS) and risk ratio (HR). Randomized controlled trials were evaluated via a modified JADAD 16 scale. This modified JADAD scale was generated according to the random sequence of the literature, allocation concealment, blindness, and whether it described the study participants' withdrawal details. Studies with a total score of 4 to 7 were classified as high-quality research, and those with a total score of 1 to 3 were classified as low-quality research. Retrospective studies were evaluated using the Newcastle-Ottawa Scale (NOS), 17 which has three categories, study selection, comparability, and outcome, consisting of a total of 9 stars; 1 to 3 stars indicated low quality, 4 to 6 stars indicated medium quality, and 7 to 9 stars indicated high quality.
Statistical Analysis
The meta-analysis was performed using Stata 15.0. OS and PFS were used as the main evaluation indicators, and subgroup analysis was performed based on the source of ctDNA specimens, ctDNA detection methods, and FIGO staging. The I2 test was used to assess the heterogeneity between studies. 18 If the studies were homogenous (P ≥ .1 and I2 ≤ 50%), the fixed effect model was used for combined analysis; if the studies were heterogeneous (P < .1, I2 > 50%), sensitivity analysis or subgroup analysis was used to find the source of the heterogeneity. When the heterogeneity was still large, the random effect model was used, or the results were discarded and descriptive analysis was used. When the number of individual outcome indicators included in the literature was more than 10, the publication bias of each indicator was analysed using a funnel chart and Begg's bias test. 19
Results
Search Results
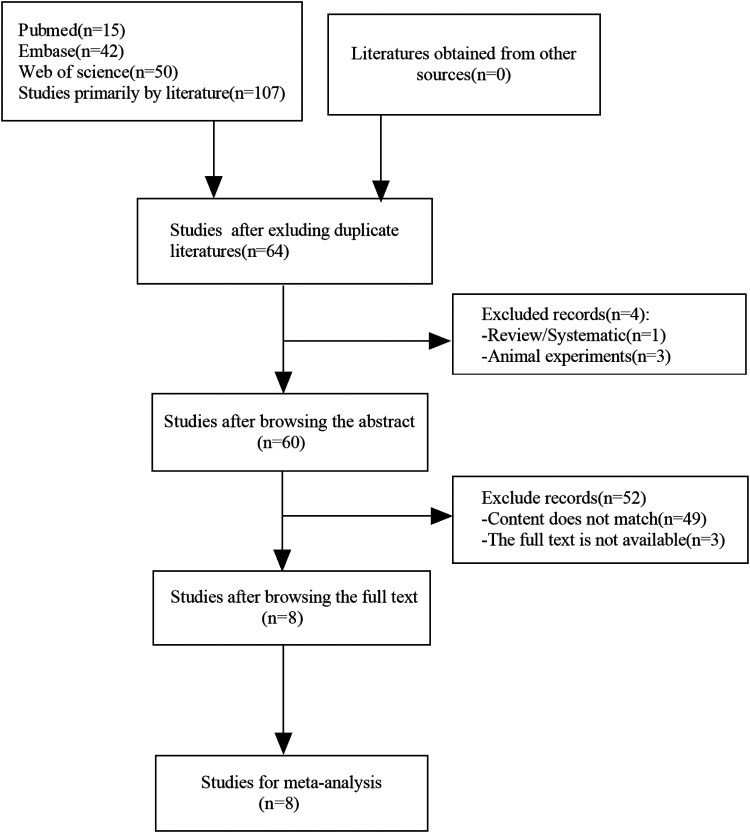
A total of 107 related documents were retrieved from the database, and 64 articles remained after excluding duplicates. After a full-text review, 8 studies with a total of 627 patients were included. The search and selection process are shown in Figure 1.
Figure 1.
Articles searching flow chart.
Characteristics of the Included Studies
The main indicators of all studies were OS and PFS, and the 8 included studies were published between 2012 and 2020. All of these 8 studies discussed ctDNA and ovarian cancer FIGO staging.4,20–26 Five articles were retrospective studies,4,21,22,24,25 and 3 articles were randomized control studies.20,23,26 In 6 articles, the ctDNA test specimens were plasma (426 patients),4,20,23–26 and in 2 articles, the test specimens were serum (201 patients).21,22 Five studies evaluated ctDNA by real-time PCR (531 patients),20–22,25,26 and 3 studies evaluated ctDNA by digital PCR (96 patients).4,23,24 The basic characteristics of the included studies are shown in Table 5.
Table 5.
Basic Characteristic of Included Studies.
| Author | Year | Research type | Study area | Number of patients | Sourceof DNA | Detection method | FIGO staging |
|---|---|---|---|---|---|---|---|
| HONG NO 21 | 2012 | Retrospective | Korea | 36 | Serum | Real time-PCR | III/IV |
| Steffensen 24 | 2014 | Retrospective | Denmark | 144 | Plasma | Real time-PCR | I-IV |
| Parkinson 4 | 2016 | Retrospective | UK | 40 | Plasma | Digital PCR | III/IV |
| Vanderstichele 25 | 2019 | RCT | European | 119 | Plasma | Real time-PCR | III/IV |
| Kalavska 19 | 2018 | RCT | Slovakia | 67 | Plasma | Real time-PCR | I-IV |
| Meng 20 | 2019 | Retrospective | China + Germany | 165 | Serum | Real time-PCR | I-IV |
| Rusan 22 | 2020 | RCT | Denmark | 24 | Plasma | Digital PCR | III/IV |
| Steffensen 23 | 2018 | Retrospective | Denmark | 32 | Plasma | Digital PCR | III/IV |
Quality Assessments
Three articles in this included study were randomized controlled trials and were evaluated using a modified JADAD scale. All of them were high-quality studies (4 points) (Table 6). The remaining 5 studies were retrospective studies and were evaluated using NOS. All of these studies were high-quality studies (7-8 points) (Table 7).
Table 6.
Quality Assessments of Randomized Controlled Trials.
| Author | Random allocation | Concealed distribution scheme | Blinding | Withdrawal | Jadad score |
|---|---|---|---|---|---|
| Vanderstichele 25 | Randomized method (details unknown) | Unknown | Not mentioned | Known | 3 |
| Kalavska 19 | Randomized method (details unknown) | Unknown | Not mentioned | Known | 3 |
| Rusan 22 | Randomized method (details unknown) | Unknown | Not mentioned | Known | 3 |
Table 7.
Quality Assessments of Retrospective Research.
Meta-Analysis Results
Overall Survival (OS)
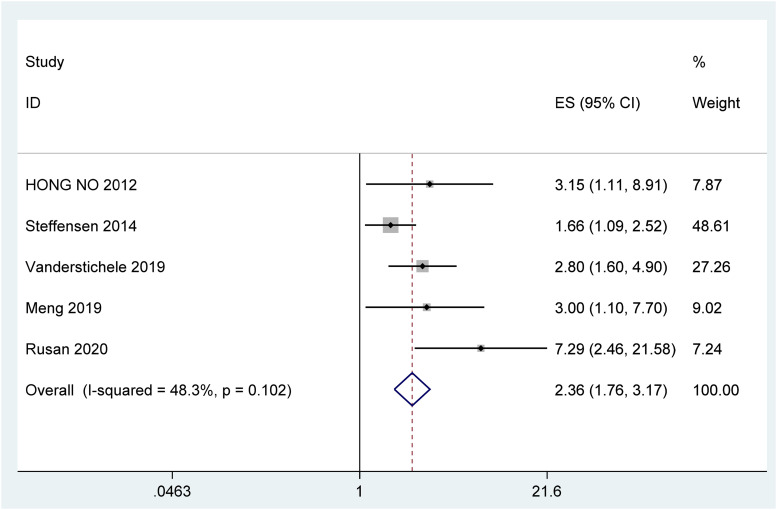
The heterogeneity test (I2 = 48.3%<50%, P = 0.102 > 0.1) suggested that there was no obvious heterogeneity between the studies, with a combined fixed effects HR = 2.36 (95% CI [1.76, 3.17], P < .001) (Figure 2), indicating that ctDNA is significantly associated with decreased OS in patients with ovarian cancer.
Figure 2.
Forest plot of the association between ctDNA and OS in patients with Ovarian cancer.
Progression-Free Survival (PFS)
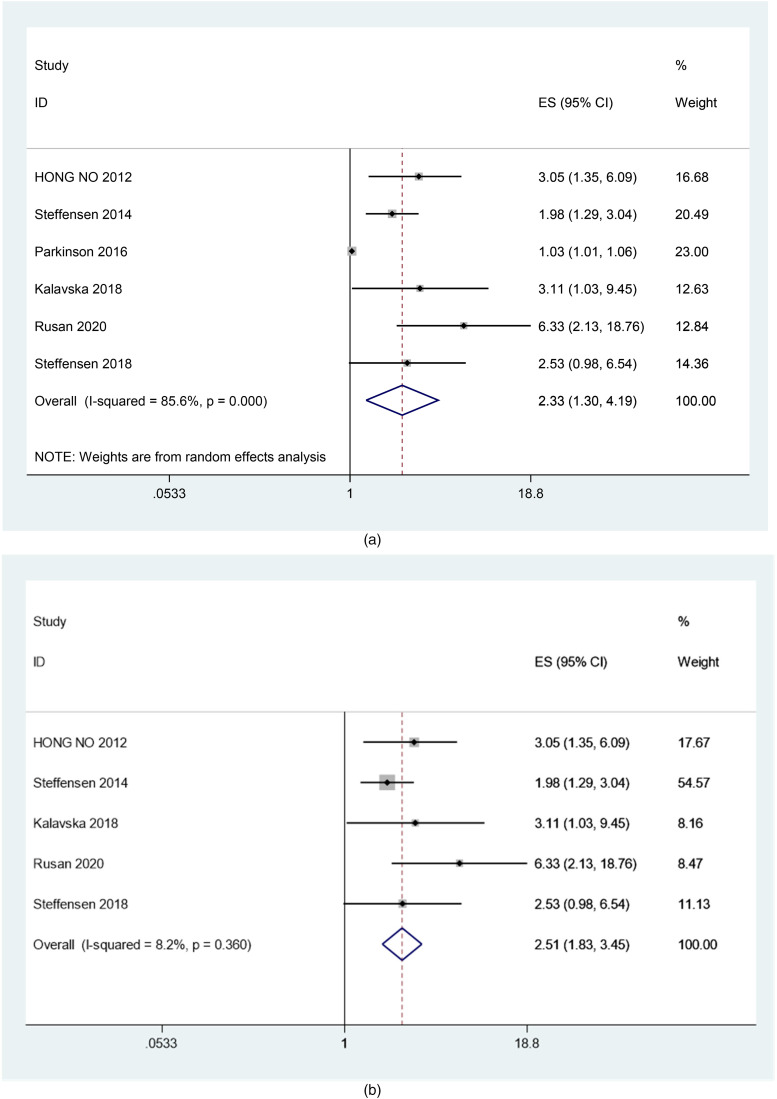
The heterogeneity test (I2 = 85.6%, P < .001) suggested that there was strong heterogeneity between the studies (Figure 3A). After exclusion of the article by Parkinson (2016) from the sensitivity analysis, the heterogeneity was significantly reduced (I2 = 8.2%, p = 0.360), with a combined fixed effects HR = 2.51 (95% CI [1.83, 3.45], P < .001) (Figure 3B), indicating that ctDNA is significantly associated with PFS reduction in ovarian cancer patients.
Figure 3.
(A) Forest plot of the association between ctDNA and PFS in patients with Ovarian cancer (before sensitivity analysis). (B) Forest plot of the association between ctDNA and PFS in patients with Ovarian cancer (after sensitivity analysis).
Subgroup Analysis
Subgroup analysis of Os
Relationship between the origin of ctDNA specimens and Os of ovarian cancer
The results of the relationship between ctDNA origin and OS were significant because there no heterogeneity was found in the serum group (I2 = 0%, p = 0.947), with a combined fixed effects HR = 3.07 (95% CI [1.51, 6.25], p = 0.002). There was a certain heterogeneity in the plasma group (I2 = 71.9%, p = 0.029), with a combined random effects HR = 2.78 (95% CI [1.40, 5.53], p = 0.003) (Table 8, Figure S1). These results were significant, indicating that serum-derived ctDNA has a strong relationship with the decrease in the OS of ovarian cancer patients.
Table 8.
Subgroup Analysis of OS and PFS.
| Subgroup | Number of studies | Pooled HR (95%CI) | Heterogeneity |
|---|---|---|---|
| OS | |||
| Origin of ctDNA | |||
| Serum | 2 | 3.07(1.51,6.25) | I2 = 0.0%, p = 0.947 |
| Plasma | 3 | 2.78(1.40, 5.53) | I2 = 71.9%,p = 0.029 |
| FIGO stage | |||
| III/IV | 3 | 3.37(2.15, 5.28) | I2 = 15.9%,p = 0.305 |
| I-IV | 2 | 1.82(1.24, 2.68) | I2 = 16.6%, p = 0.274 |
| Detection methods | |||
| Real time-PCR | 4 | 2.16(1.60, 2.93) | I2 = 8.7%,p = 0.350 |
| Digital PCR | 1 | 7.29(2.46, 21.59) | / |
| PFS | |||
| Origin of ctDNA | |||
| Serum | 1 | 3.0 (1.44, 6.48) | / |
| Plasma | 4 | 2.41 (1.70, 3.42) | I2 = 25.9%,p = 0.256 |
| FIGO stage | |||
| III/IV | 3 | 3.41(2.03, 5.72) | I2 = 0.0%, p = 0.426 |
| I-IV | 2 | 2.10(1.41, 3.13) | I2 = 0.0%, p = 0.456 |
| Detection methods | |||
| Real time-PCR | 3 | 2.28(1.60, 3.24) | I2 = 0%, p = 0.524 |
| Digital PCR | 2 | 3.76(1.84, 7.69) | I2 = 35.5%, p = 0.213 |
Relationship between FIGO stage and Os of ovarian cancer
The results of the relationship between FIGO and OS were significant because no heterogeneity was observed in the III/IV group (I2 = 15.9%, p = 0.305), with a combined fixed effects HR = 3.37 (95% CI [2.15, 5.28], P < .001). There was also no heterogeneity in the I-IV group (I2 = 16.6%, p = 0.274), with a combined fixed effects HR = 1.82 (95% CI [1.24, 2.68], p = 0.009) (Table 8, Figure S2). These results were significant, indicating that ctDNA of III/IV has a strong relationship with the reduction in the OS of patients with ovarian cancer. It can be speculated that there is a certain correlation between ctDNA and tumor stage.
Relationship between ctDNA detection methods and Os of ovarian cancer
The results for the relationship between detection method and OS showed that there was no heterogeneity in the real-time PCR group (I2 = 8.7%, p = 0.350), with a combined fixed effects HR = 2.16 (95% CI [1.60, 2.93], P < .001), and this result was significant. The combined effect of digital PCR showed a HR = 7.29 (95% CI [2.46, 21.59], P < .001) (Table 8, Figure S3), and this result was significant. Thus, it can be considered that digital PCR has a stronger relationship with the reduction in the OS of patients with ovarian cancer than real-time PCR. We speculated that this may be due to the higher positivity rate of digital PCR than real-time PCR for ctDNA detection.
Subgroup analysis of PFS
Relationship between the origin of ctDNA specimens and PFS of ovarian cancer
The results for the relationship between ctDNA origin and PFS showed that the combined effect in the serum group had an HR = 3.05 (95% CI [1.44, 6.48], p = 0.004), and these results were significant. There was no obvious heterogeneity in the plasma group (I2 = 25.9%, p = 0.256), with a combined fixed effects HR = 2.41 (95% CI [1.70, 3.42], P < .001) (Table 8, Figure S4), so the results were significant. This result indicates that both serum and plasma-derived ctDNA are strongly associated with the reduction in the PFS of ovarian cancer patients and that serum-derived ctDNA has a strong relationship with PFS in patients with ovarian cancer.
The relationship between FIGO staging and PFS of ovarian cancer
The results for the relationship between FIGO staging and PFS were significant because they showed that there was no heterogeneity in the III/IV group (I2 = 0.0%, p = 0.426), with a combined fixed effects HR = 3.41 (95% CI [2.03, 5.72], p = 0.49). The results were also significant because there was no heterogeneity in the I-IV group (I2 = 0%, p = 0.456), with a combined fixed effects HR = 2.10 (95% CI [1.41, 3.13], P < .001) (Table 8, Figure S5), indicating that the ctDNA of III/IV is relatively whole and has a stronger relationship with the decrease in PFS of patients with ovarian cancer. Therefore, it can be speculated that there is a certain correlation between ctDNA and tumor stage.
Relationship between ctDNA detection methods and PFS of ovarian cancer
The results for the relationship between the detection method and PFS showed that there was no heterogeneity in the real-time PCR group (I2 = 0%, p = 0.524), with a combined fixed effects HR = 2.28 (95% CI [1.60, 3.24], P < .001), which indicated significance. There was still no obvious heterogeneity in the digital PCR group (I2 = 35.5%, p = 0.213), with a combined effects HR = 3.76 (95% confidence interval [1.84, 7.69], P < .001), which also indicated significance(Table 8, Figure S6). These results suggest that digital PCR results have a stronger relationship with the reduction in PFS of patients with ovarian cancer than real-time PCR results.
Sensitivity Analysis
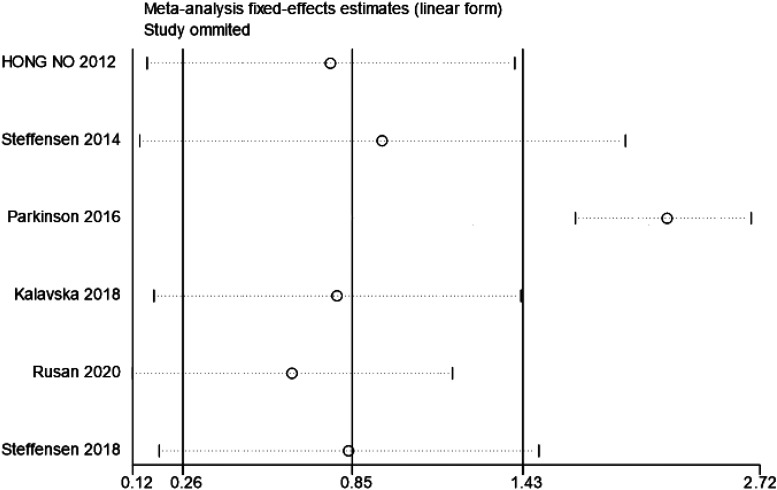
A sensitivity analysis was performed by removing documents in sequence, and the results revealed that the research by Parkinson (2016) had a great impact on the results, so this study was eliminated (Figure 4).
Figure 4.
The sensitivity analysis for ctDNA and PFS in patients with Ovarian cancer.
Detection of Publication Bias
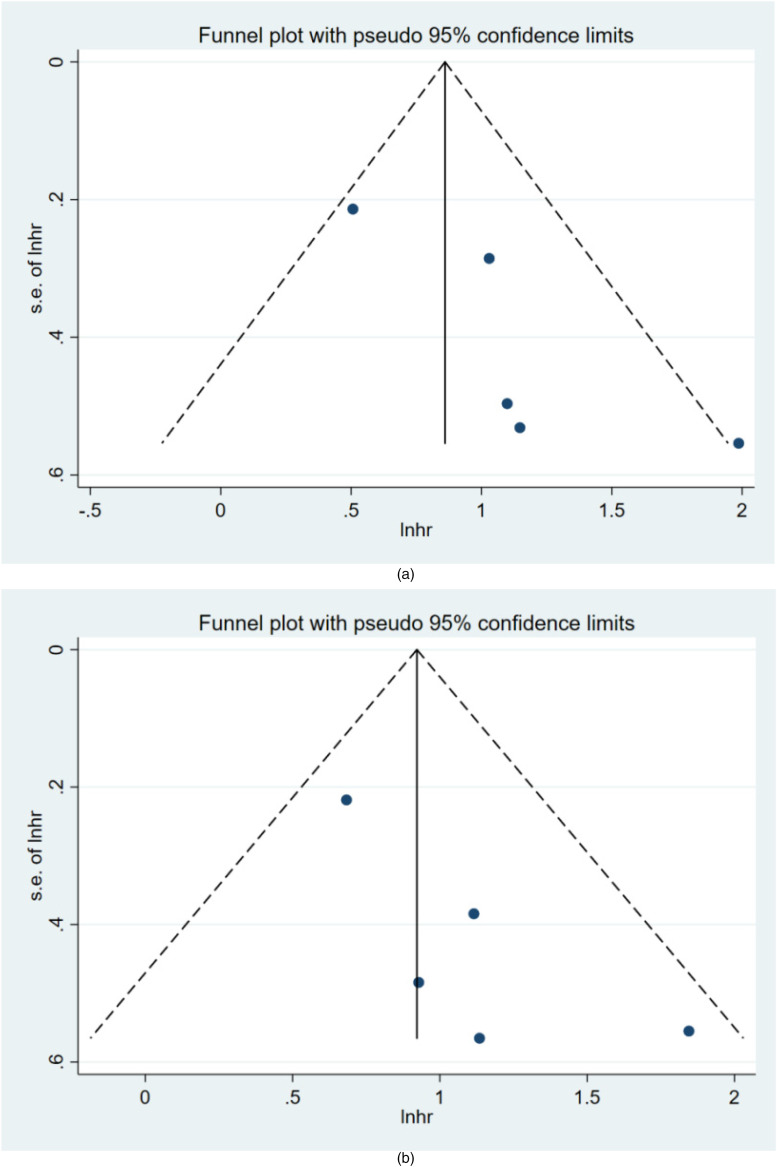
Funnel plots and Begg's test were used to evaluate the publication bias of the included studies in this study. The results of Begg's bias test, which was based on funnel plots, showed that there was no publication bias in the included literature for OS (z = 1.22, p = 0.221) (Figure 5A) or PFS (z = 0.73, p = 0.462) (Figure 5B).
Figure 5.
Funnel plots of publication bias for OS (A) and PFS (B) in patients with Ovarian cancer.
Discussion
Characteristics of Circulating Tumor DNA
The amount of ctDNA released is related to the tumor load, and the concentration of ctDNA in the serum of cancer patients is positively correlated with protease activity. The activity of proteases increase, leading to the degradation of the extracellular matrix and promoting the invasion and metastasis of tumor cells. CtDNA carries tumor-related genetic changes, such as point mutations, deletions, and copy number variations. With every 1 cm3 increase in tumor volume, the proportion of mutant alleles in plasma increases by approximately 0.08%, and 6 mutation copies per mL of plasma are added. 4 As a readily available liquid biomarker and a potential substitute for the entire tumor genome, ctDNA may play a valuable role in predicting the prognosis of ovarian cancer patients.
The Effect of Circulating Tumor DNA on the Prognosis of Ovarian Cancer
Several studies have explored the effectiveness of ctDNA as a tool for the screening and early diagnosis of ovarian cancer. The application of ctDNA in prognostic predictions is also gaining increasing attention. Related studies have confirmed that high levels of ctDNA in patients with ovarian cancer are associated with their poor prognosis.3,6,7,27 However, the results of these studies have not been systematically evaluated. A recent meta-analysis showed the relationship between ctDNA and solid tumors through a systematic review of electronic databases. In the multivariate analysis, the detection of ctDNA was significantly associated with poor OS (HR = 2.70; 95% CI [2.02-3.61], P < .001). Additionally, in the univariate analysis, there was a statistically significant correlation between high levels of total ctDNA and poor OS (HR = 1.91; 95% CI [1.59-2.29], P < .001) (OR = 2.82; 95% CI [1.93-4.13], P < .001). The above results indicate that the presence of high levels of total ctDNA and ctDNA is related to the poor survival rate of patients with solid tumors. 28 The prognostic value of ctDNA for breast cancer, lung cancer, and rectal cancer has been supported by research-based evidence, illustrating its clinical significance.10,12,13 To the best of our knowledge, this is the first meta-analysis to report the value of ctDNA in predicting the prognosis of ovarian cancer patients and provides new clinical practice guidelines for future research.
A total of 672 patients with ovarian cancer were included in this study, and those with high levels of ctDNA had poor OS and PFS. Patients with ovarian cancer are usually treated by surgery to remove the lesion; however, for advanced cancer patients, whether ovarian cancer cytoreductive surgery can be achieved, that is, R0 resection status, is closely related to the prognosis of the patient. Moreover, the sensitivity of current detection methods is unsatisfactory, so tumor recurrence cannot be detected at an earlier stage. Fortunately, ctDNA levels are significantly correlated with ovarian cancer tumor volume 4 and have been found to offer an earlier indication of treatment response than radiological methods. 29 Under stable conditions, if the patient's tumor load is 100 g (approximately 3 × 1010 cells), approximately 3.3% of ctDNA will be released into the bloodstream every day. 30 Patients with a high tumor burden may have even more ctDNA released into the blood. After treatment, ctDNA levels decline rapidly as the tumor regresses. 31 Therefore, the level of peripheral blood ctDNA can reflect tumor burden, can be used as a substitute for tumor burden and can become a valuable predictive prognostic marker.
There have been many clinical studies on targeted therapy for ovarian cancer, and PARP inhibitors are currently the first-line maintenance therapy for ovarian epithelial cancer. Approximately 25% of high-grade serous ovarian cancers have BRCA mutations, and 50% of these patients will develop homologous recombination defect (HRD)-positive cancers that include BRCA1/2 mutations; however, BRCA1/2 mutations are detected in only 22% of cases. Patients with BRCA mutations (systemic and germline) can obtain the most benefit from PARP inhibitor treatment, and some patients with HRDs can also benefit from PARP inhibitors according to their sensitivity to PARP inhibitors, as follows: BRCA germline mutation > BRCA systemic mutation > HRD > platinum sensitivity > other. Targeted therapy based on molecular characteristics has an important impact on the treatment strategy for nonovarian cancer. Gene mutation analysis is a commonly used predictive biomarker for patients with ovarian cancer who are receiving targeted therapy. In addition, some patients develop drug resistance, and cells with strong drug resistance survive and possess novel mutations. The current mutation analysis is mostly based on tumor tissue, which has many limitations. It is not feasible to perform a real-time evaluation of treatment resistance through repeated surgery or biopsy. Considering these limitations, ctDNA, which can be obtained in a non-invasive manner, is a reliable biomarker for monitoring the efficacy of targeted drugs and the overall survival and progression-free survival of patientsThere have been many clinical studies on targeted therapy for ovarian cancer, and PARP inhibitors are currently the first-line maintenance therapy for ovarian epithelial cancer. Approximately 25% of high-grade serous ovarian cancers have BRCA mutations, and 50% of these patients will develop homologous recombination defect (HRD)-positive cancers that include BRCA1/2 mutations; however, BRCA1/2 mutations are detected in only 22% of cases. Patients with BRCA mutations (systemic and germline) can obtain the most benefit from PARP inhibitor treatment, and some patients with HRDs can also benefit from PARP inhibitors according to their sensitivity to PARP inhibitors, as follows: BRCA germline mutation > BRCA systemic mutation > HRD > platinum sensitivity > other. Targeted therapy based on molecular characteristics has an important impact on the treatment strategy for nonovarian cancer. Gene mutation analysis is a commonly used predictive biomarker for patients with ovarian cancer who are receiving targeted therapy. In addition, some patients develop drug resistance, and cells with strong drug resistance survive and possess novel mutations. The current mutation analysis is mostly based on tumor tissue, which has many limitations. It is not feasible to perform a real-time evaluation of treatment resistance through repeated surgery or biopsy. Considering these limitations, ctDNA, which can be obtained in a non-invasive manner, is a reliable biomarker for monitoring the efficacy of targeted drugs and the overall survival and progression-free survival of patients.32,33
Heterogeneity is an important issue in meta-analyses. The results of the heterogeneity analysis in this research showed that there was no heterogeneity among the 5 articles included in the OS study. The 6 articles included in the PFS study showed strong heterogeneity. To investigate the potential sources of heterogeneity, heterogeneity analysis was performed in each subgroup according to ctDNA source, FIGO staging, and ctDNA detection method, and the results showed that the factors in these subgroups did not have a significant impact on heterogeneity. This indicated that the study design did not have a prognostic impact, the accuracy of the judgement had a substantial impact, and the influencing factors were unclear. To continue to explore the sources of heterogeneity, a sensitivity analysis was conducted based on the results of the subgroup analysis.
The results showed that Parkinson's research had large heterogeneity. After removing this study, the heterogeneity test was conducted again and indicated no heterogeneity, which may be related to the experimental design of the study, sample size, or evaluation criteria, among other factors. Begg's bias test of OS and PFS revealed that there was no publication bias in this study and that the results of our meta-analysis were reliable.
Circulating Tumor DNA and Clinical FIGO Staging of Ovarian Cancer
The prognosis of malignant tumors of various systems, including ovarian cancer, is clearly related to the clinical stage. The later the clinical stage is, the worse the prognosis. It has been reported that ctDNA is related to tumor staging. Bettegowda et al. 34 evaluated the ability of ctDNA to detect tumors in 640 patients with different cancer types and stages. Compared with that in stage I patients, the median concentration of ctDNA in stage IV patients increased by 100 times. In advanced pancreatic cancer, ovarian cancer, colorectal cancer, bladder cancer, oesophagogastric cancer, breast cancer, melanoma, liver cancer, and head and neck cancer, ctDNA can be detected in 75% of patients, but in early primary brain cancer, kidney cancer, prostate cancer or thyroid cancer, the detection rate is less than 50%. A quantitative analysis of tumor mutations in each patient showed that patients with stage I disease had fewer than 10 copies per 5 mL of plasma. In contrast, patients with advanced prostate ovarian cancer or colorectal cancer had a median plasma concentration of 100 to 1000 copies per 5 mL. Later clinical stage of the cancer and higher levels of ctDNA are indicative of a worse prognosis. This study analysed the FIGO staging subgroups, and the results showed that the I-IV group had a lower HR value than the III/IV group (1.82 vs 3.37, 2.10 vs 2.44), indicating that the ctDNA of advanced ovarian cancer in the III/IV group is strongly associated with poor OS and PFS. Therefore, it can be speculated that there is a certain correlation between ctDNA and tumor stage. Despite this correlation, the level of ctDNA is actually related to many factors, such as differences in tumor histology, affecting, the rate of cell apoptosis, and in tumor vascularization, affecting the release of ctDNA into the blood.
Detection Methods for Circulating Tumor DNA
The low tumor load in patients with early or relapsed ovarian cancer, as well as post treatment, are consistent with low levels of ctDNA, which means that successful detection of low-level ctDNA is another challenge. Diamandis and Fiala 35 found that the level of ctDNA extracted from 10 mL of blood was very low, and when the tumor diameter fell below 10 mm, not even one mutant DNA copy could be detected. The subgroup analysis of this study showed that the ability to detect ctDNA via digital PCR was associated with lower OS and PFS in ovarian cancer patients. This may be because digital PCR could have a higher positivity rate for ctDNA detection than real-time PCR. Bettegowda et al. 34 used digital PCR to detect ctDNA in patients with advanced metastatic tumors, and the detection rate was 75%. Among patients with localized tumors, 73% had colorectal cancers, 57% had gastroesophageal cancers, 48% had pancreatic cancers, and 50% had breast cancers.
CtDNA detection methods include digital PCR (dPCR), droplet digital PCR (ddPCR) and BEAMing technology. The limitation of the above detection methods is that only known mutations can be detected. Next-generation sequencing (NGS) can detect unknown genes and detect multiple genes at the same time, making it particularly suitable for the detection of ctDNA because the number of DNA samples required is small. The strategies for ctDNA analysis can be divided into three categories 36 : single point or multiple PCR, target region capture sequencing, and whole genome sequencing (WGS). The hotspot mutation analysis of allele-specific PCR is very sensitive; targeted sequencing can detect ctDNA when the allele frequency is less than 0.1%; WGS can only detect 5%-10% fragments of allele mutations. Thus, it is not suitable for early disease research and is more suitable for the detection of tumor-related copy number variations.
Sample Source
The DNA content may differ greatly between serum and plasma. Morgan et al. 37 asserted that plasma-derived ctDNA can affect serum specimens. The main reason is that the coagulation process of leukocyte lysis or exogenous genomic DNA will reduce the proportion of ctDNA.38,39 This difference may affect the detection of ctDNA. The subgroup analysis in Boxuan et al.'s meta-analysis of the diagnostic significance of ctDNA in patients with ovarian cancer suggested that ctDNA in plasma is less diagnosable than that in serum. 40 Some studies have pointed out that due to the large amount of immune cell contamination in serum, it is better to use plasma as a specimen. 36 The results of the subgroup analysis of ctDNA specimens in this study indicated that serum-derived ctDNA is significantly associated with poor OS and PFS in patients with ovarian cancer. As the results of the above studies are inconsistent, the current level of evidence is insufficient determine the most reliable source of ctDNA detection.
Limitations
This study has some limitations that should be mentioned. The small number of studies that could be included in the meta-analysis may have resulted in poor stability in the analysis results. More studies need to be included in the future to enhance these results. First, our analysis was based on the available literature. There was a lack of single-person data, so single-data meta-analysis could not be performed. The methodological differences in the original literature also had an impact on our results. Second, the analysis of PFS in this study suggested that there was strong heterogeneity. When subgroup analysis was performed, there was no change in the heterogeneity between groups. Thus, this heterogeneity may be partly derived from other variations, such as country and language. Third, the current research on ctDNA lacks uniform standards in terms of specimen extraction, sample collection time, sample type, and detection method. The standardization of future ctDNA research would be conducive to comparing different experimental results and would make the experiments highly repeatable. Fourth, some studies were excluded because data could not be extracted or HR data were not provided, which could have affected the results. Among the 8 articles included, only the study by Steffensen et al. 25 provided data on the response to chemotherapy, so the utility of ctDNA in monitoring the response to chemotherapy warrants further exploration.
Conclusions
For the first time, a meta-analysis was conducted to explore the prognostic value of ctDNA in ovarian cancer, and the results of this meta-analysis suggest that ovarian cancer patients with high levels of ctDNA have a poor prognosis. In view of the limitations of this study, we believe that further large-scale prospective studies are needed to verify the prognosis and predictive value of ctDNA in ovarian cancer patients.
Supplemental Material
Supplemental material, sj-docx-1-tct-10.1177_15330338211043784 for The Prognostic Value of Circulating Tumor DNA in Ovarian Cancer: A Meta-Analysis by Yuanyuan Lu and Li Li in Technology in Cancer Research & Treatment
Supplemental material, sj-doc-2-tct-10.1177_15330338211043784 for The Prognostic Value of Circulating Tumor DNA in Ovarian Cancer: A Meta-Analysis by Yuanyuan Lu and Li Li in Technology in Cancer Research & Treatment
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical Approval: Our study did not require an ethical board approval because it did not contain human or animal trials.
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6(5):e555-ee67. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong DK, Alvarez RD, Bakkum-Gamez JN, et al. NCCN guidelines insights: ovarian cancer, version 1.2019. J Natl Compr Canc Netw. 2019;17(8):896-909. [DOI] [PubMed] [Google Scholar]
- 3.Harris FR, Kovtun IV, Smadbeck J, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkinson CA, Gale D, Piskorz AM, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton CA, Hacker NF, Clark SJ, et al. DNA Methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109(1):129-139. [DOI] [PubMed] [Google Scholar]
- 6.Bondurant AE, Huang Z, Whitaker RS, et al. Quantitative detection of RASSF1A DNA promoter methylation in tumors and serum of patients with serous epithelial ovarian cancer. Gynecol Oncol. 2011;123(3):581-587. [DOI] [PubMed] [Google Scholar]
- 7.Giannopoulou L, Mastoraki S, Buderath P, et al. ESR1 Methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol Oncol. 2018;150(2):355-360. [DOI] [PubMed] [Google Scholar]
- 8.Yin C, Luo C, Hu W, et al. Quantitative and qualitative analysis of circulating cell-free DNA can be used as an adjuvant tool for prostate cancer screening: a meta-analysis. Dis Markers. 2016(2016):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai B, Liu H, Huang Y, et al. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016;7(28):44583-44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Shao F, Wu X, et al. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysis. Lung Cancer. 2010;69(2):225-231. [DOI] [PubMed] [Google Scholar]
- 12.Cargnin S, Canonico PL, Genazzani AA, et al. Quantitative analysis of circulating cell-free DNA for correlation with lung cancer survival: a systematic review and meta-analysis. J Thorac Oncol. 2017;12(1):43-53. [DOI] [PubMed] [Google Scholar]
- 13.Fan G, Zhang K, Yang X, et al. Prognostic value of circulating tumor DNA in patients with colon cancer: systematic review. PLoS One. 2017;12(2):e0171991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q, Li W, Leng B, et al. Circulating cell free DNA as the diagnostic marker for ovarian cancer: a systematic review and meta-analysis. PLoS One. 2016;11(6):e0155495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Open Med. 2009;3(3):e123-30. [PMC free article] [PubMed] [Google Scholar]
- 16.Jadad AR, Moore RA, Carroll D, et al. Assessing the quality of reports of randomized clinical trials: is blinding necessary?. Control Clin Trials. 1996;17(1):1-12. [DOI] [PubMed] [Google Scholar]
- 17.Wells G SB, O’Connell D, Peterson J, Welch V, Losos M, Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrando-mised studies in meta-analyses 2013. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 18.Colditz GA, Burdick E, Mosteller F. Heterogeneity in meta-analysis of data from epidemiologic studies: a commentary. Am J Epidemiol. 1995;142(4):371-382. [DOI] [PubMed] [Google Scholar]
- 19.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088-1101. [PubMed] [Google Scholar]
- 20.Kalavska K, Minarik T, Vlkova B, et al. Prognostic value of various subtypes of extracellular DNA in ovarian cancer patients. J Ovarian Res. 2018;11(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng X, Schwarzenbach H, Yang Y, et al. Circulating mitochondrial DNA is linked to progression and prognosis of epithelial ovarian cancer. Transl Oncol. 2019;12(9):1213-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.No JH, Kim K, Park KH, et al. Cell-free DNA level as a prognostic biomarker for epithelial ovarian cancer. Anticancer Res. 2012;32(8):3467-3471. [PubMed] [Google Scholar]
- 23.Rusan M, Andersen RF, Jakobsen A, et al. Circulating HOXA9-methylated tumour DNA: a novel biomarker of response to poly (ADP-ribose) polymerase inhibition in BRCA-mutated epithelial ovarian cancer. Eur J Cancer. 2020(125):121-129. [DOI] [PubMed] [Google Scholar]
- 24.Steffensen KD, Andersen RF, Jakobsen AKM. Methylated circulating tumor DNA as a potential marker of PARP inhibitor efficiency in BRCA mutated ovarian cancer patients. J Clin Oncol. 2018;36(15):5540-5540. [Google Scholar]
- 25.Steffensen KD, Madsen CV, Andersen RF, et al. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer. 2014;50(15):2611-2618. [DOI] [PubMed] [Google Scholar]
- 26.Vanderstichele A, Concin N, Busschaert P, et al. TP53 Mutations in cell-free DNA as early markers of therapeutic response in platinum-resistant relapsed ovarian cancer (PROC): a prospective translational analysis of the phase II GANNET53 clinical trial. Int J Gynecol Cancer. 2019;29:A11. [Google Scholar]
- 27.Wimberger P, Roth C, Pantel K, et al. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. Int J Cancer. 2011;128(11):2572-2580. [DOI] [PubMed] [Google Scholar]
- 28.Ocaña A, Díezgonzález L, Garcíaolmo DC, et al. Circulating DNA and survival in solid tumors. Cancer Epidemiol Biomarkers Prev. 2016;25(2):399-406. [DOI] [PubMed] [Google Scholar]
- 29.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368-16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szpechcinski A, Chorostowska-Wynimko J, Kupis W, et al. Quantitative analysis of free-circulating DNA in plasma of patients with resectable NSCLC. Expert Opin Biol Ther. 2012(12):S3-S9. [DOI] [PubMed] [Google Scholar]
- 32.Thomsen CB, Andersen RF, Steffensen KD, et al. Delta tocotrienol in recurrent ovarian cancer. A phase II trial. Pharmacol Res. 2019;141:392-396. [DOI] [PubMed] [Google Scholar]
- 33.Piskorz AM, Lin KK, Morris J, et al. Feasibility of monitoring response to the PARP inhibitor rucaparib with targeted deep sequencing of circulating tumor DNA (ctDNA) in women with high grade ovarian carcinoma on the ARIEL2 trial. Eur J Cancer. 2016;(69):S123-S12S. [Google Scholar]
- 34.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diamandis EP, Fiala C. Can circulating tumor DNA be used for direct and early stage cancer detection?. F1000Res. 2017;6:2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223-238. [DOI] [PubMed] [Google Scholar]
- 37.Morgan SR, Whiteley J, Donald E, et al. Comparison of KRAS mutation assessment in tumor DNA and circulating free DNA in plasma and Serum samples. Clin Med Insights Pathol. 2012;(5):15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee TH, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41(2):276-282. [DOI] [PubMed] [Google Scholar]
- 39.Page K, Powles T, Slade MJ, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci. 2006;1075:313-317. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Pu K, Ge L, et al. Diagnostic significance assessment of the circulating cell-free DNA in ovarian cancer: an updated meta-analysis. Gene. 2019;714:143993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tct-10.1177_15330338211043784 for The Prognostic Value of Circulating Tumor DNA in Ovarian Cancer: A Meta-Analysis by Yuanyuan Lu and Li Li in Technology in Cancer Research & Treatment
Supplemental material, sj-doc-2-tct-10.1177_15330338211043784 for The Prognostic Value of Circulating Tumor DNA in Ovarian Cancer: A Meta-Analysis by Yuanyuan Lu and Li Li in Technology in Cancer Research & Treatment