Abstract
Introduction
Freshly isolated uncultured adipose tissue-derived stromal cells (u-ADSCs), containing miscellaneous cells like the relatively abundant mesenchymal stem cells, are attractive for repair and regenerative therapy. However, the detailed characteristics and therapeutic efficacy of u-ADSCs obtained from disease-affected hosts are unknown. We compared the properties of u-ADSCs obtained from wild-type mice and from a mouse model of non-alcoholic steatohepatitis (NASH).
Methods
The NASH model was established by feeding C57BL/6J mice an atherogenic high-fat diet for 4 (NASH (4w)) or 12 weeks (NASH (12w)), followed by the isolation and characterization of u-ADSCs. Wild-type u-ADSCs or NASH-derived u-ADSCs were administered to mice with NASH cirrhosis, followed by analyses of hepatic inflammatory cells, antigen profiles, fibrosis, and gene expression.
Results
Wild-type u-ADSCs and NASH-derived u-ADSCs did not show marked differences in surface antigen profiles. In NASH (4w) u-ADSCs, but not NASH (12w) u-ADSCs, the frequencies of the leukocyte markers CD11b, CD45, and CD44 were elevated; furthermore, we observed an increase in the M1/M2 macrophage ratio only in NASH (12w) u-ADSCs. Only in NASH-4w u-ADSCs, the expression levels cell cycle-related genes were higher than those in u-ADSCs. Wild-type u-ADSCs administered to mice with NASH-related cirrhosis decreased the infiltration of CD11b+, F4/80+, and Gr-1+ inflammatory cells, ameliorated fibrosis, and had a restorative effect on liver tissues, as determined by gene expression profiles and the NAFLD activity score. The therapeutic effects of NASH (4w) u-ADSCs and NASH (12w) u-ADSCs on NASH-related cirrhosis were highly similar to the effect of wild-type u-ADSCs, including reductions in inflammation and fibrosis.
Conclusions
NASH-derived u-ADSCs, similar to wild-type u-ADSCs, are applicable for reparative and regenerative therapy in mice with NASH.
Keywords: Non-alcoholic fatty liver disease, Adipose tissue, Stromal cells, Mesenchymal stem cells
Abbreviations: u-ADSCs, uncultured adipose tissue-derived stromal cells; NASH, nonalcoholic steatohepatitis; NAFLD, nonalcoholic fatty liver disease; MSCs, mesenchymal stem cells; AT-HF, atherogenic high-fat; NASH (4w) u-ADSCs, NASH (4 weeks)-derived u-ADSCs; NASH (12 w) u-ADSCs, NASH (12 weeks)-derived u-ADSCs; HICs, hepatic inflammatory cells; FCM, flow cytometry; qRT-PCR, quantitative real-time polymerase chain reaction; AST, aspartate aminotransferase; LD, lactate dehydrogenase; NAS, NAFLD activity score
Highlights
-
•
Uncultured adipose tissue-derived stromal cells (u-ADSCs) in regenerative therapy.
-
•
Nonalcoholic steatohepatitis (NASH) mice model was established.
-
•
We confirmed the efficacy of u-ADSCs for treatment of cirrhotic mice.
-
•
We studied the NASH mouse model-derived u-ADSCs for treatment of cirrhotic mice.
-
•
NASH-u-ADSCs and wild-type u-ADSCs are anti-inflammatory and effective for cirrhosis.
1. Introduction
Chronic liver diseases are commonly characterized by persistent hepatic inflammation with the development of hepatic fibrosis, ultimately leading to serious conditions, including liver cirrhosis [1]. Nonalcoholic steatohepatitis (NASH) is a chronic liver disease associated with metabolic syndrome and obesity [2]. Fatty deposits on the liver are common in nonalcoholic fatty liver disease (NAFLD). Around 80–90% of patients with NAFLD do not progress to steatohepatitis; however, the remaining patients suffer from persistent chronic steatohepatitis and 10–15% of patients develop liver cirrhosis [3]. The detailed mechanisms by which chronic hepatitis progresses to cirrhosis is unknown; furthermore, effective treatments have not been established. Freshly isolated uncultured adipose tissue-derived stromal cells (u-ADSCs) have attracted substantial attention for repair and regenerative therapy, as they are a source of abundant mesenchymal stem cells (MSCs) [4], which are pluripotent somatic stem cells able to differentiate into diverse cell lineages [5]. Moreover, u-ADSCs possess strong immunomodulatory properties, capable of suppressing harmful inflammation [6]. Therefore, u-ADSCs are promising for repairing and/or regenerating injured organs, especially under harmful inflammation [7]. Moreover, u-ADSCs can be obtained in an autologous manner from subcutaneous tissues [8]. However, u-ADSCs obtained from individuals with NASH have not been comprehensively characterized, and comparative analyses of their therapeutic efficacy with that of u-ADSCs from unaffected individuals are needed. In this study, using a mouse model of steatohepatitis, we characterized uncultured ADSCs of mice with progression to non-alcoholic steatohepatitis after 4 weeks of an atherogenic high-fat (AT-HF) diet (NASH (4w)) or mice with late NASH/early cirrhosis after 12 weeks of an AT-HF diet (NASH (12w)) [9]. We observed similar gene expression profiles in u-ADSCs derived from mice with NASH and wild-type u-ADSCs, with some differences, including the upregulation of pathways related to cell cycling and B cell proliferation in NASH-u-ADSCs. Despite these differences, the therapeutic efficacy of NASH-derived u-ADSCs was maintained in a mouse model of NASH, including an immunosuppressive effect and effective reduction in fibrosis.
2. Methods
2.1. Establishment of NASH model mice, isolation of adipose tissue-derived stromal cells, and treatments
The Institutional Review Board of Kanazawa University approved the procedures for the care and use of laboratory animals for this study (approval no. AP-163728). Murine models of NASH were established by feeding C57BL/6J female mice (8–12 weeks old; Charles River Laboratories Japan, Yokohama, Japan) an AT-HF diet (Oriental Yeast Co., Tokyo, Japan) as previously reported [10]. Adipose tissue-derived stromal cells were isolated as previously described [6]. Briefly, immediately after euthanasia with carbon dioxide or isoflurane (Wako Pure Chemical Industries, Osaka, Japan), the inguinal adipose tissues were isolated from 15- to 20-week-old wild-type C57BL/6J female mice or NASH model mice fed an AT-HF diet for 4 or 12 weeks. Inflammation in the liver is already enhanced at these two time points, while fibrosis progression is observed starting from 12 weeks of AT-HF diet [9]. The tissues were digested by using type I collagenase (Worthington Biochemical Corporation, Lakewood, NJ, USA), the stromal fraction was separated from the fatty components by centrifugation, and samples were washed with PBS. The resultant u-ADSCs, NASH (4w)-derived u-ADSCs (NASH (4w) u-ADSCs), and NASH (12w)-derived u-ADSCs (NASH (12w) u-ADSCs) were used directly for experiments. NASH-cirrhosis model mice were kept under anesthesia with 0.3 mg/kg medetomidine (1 mg/mL Domitor; Nippon Zenyaku Kogyo, Koriyama, Japan), 4 mg/kg midazolam (10 mg/2 mL Dormicum; Astellas Pharma, Tokyo, Japan), and 5 mg/kg butorphanol (5 mg/mL Vetorphale; Meiji Seika Pharma, Tokyo, Japan). Then, u-ADSCs, NASH (4w) u-ADSCs, or NASH (12w) u-ADSCs (1 × 106 or 7.5 × 105 cells) were suspended in 100 μL of PBS and injected into the splenic subcapsule at weeks 24 and 26 of AT-HF diet feeding. Blood was collected from mice on week 28 from the venous plexus within the retroorbital space. The serum was separated from the clotted components of blood by centrifugation, and aspartate aminotransferase (AST) and lactate dehydrogenase (LD) levels were determined as described previously [11]. Liver tissues were obtained on week 28 and used for gene expression and immunohistochemical analyses as well as for the estimation of the NAFLD activity score (NAS); we calculated the NAS as the sum of scores for steatosis (score range: 0–3; hematoxylin and eosin (HE) images analyzed at ×100 magnification), lobular inflammation (score range: 0–3; HE images analyzed at ×200 magnification), and hepatocyte ballooning (score range: 0–2; HE images analyzed at ×100 magnification). Hepatic inflammatory cells (HICs) were isolated by the homogenization of a part of the liver, digestion with type I collagenase, washing with PBS, and separation with Percoll® PLUS (GE Healthcare, Chicago, IL, USA). The HICs were then analyzed by flow cytometry (FCM).
2.2. Flow cytometry
Freshly isolated u-ADSCs, NASH (4w) u-ADSCs, NASH (12w) u-ADSCs or HICs were incubated in PBS with 2% v/v FBS (FCM buffer; Gibco, Life Technologies) for 10 min at 4 °C with FcR Blocking Reagent (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Cells were washed with FCM buffer and then incubated with the following fluorescence-labeled antibodies as per the manufacturers' protocols: FITC anti-mouse CD11b (clone: M1/70), PE anti-mouse CD11b (clone: M1/70), FITC anti-mouse CD4 (clone: RM4-5), APC anti-mouse CD8a (clone: 53–6.7), PE anti-mouse CD29 (clone: HM β1-1), PerCP anti-mouse CD45 (clone: 30-F11), APC anti-mouse CD44 (clone: IM7), FITC anti-mouse CD31 (clone: MEC 13.3; BD Pharmingen, San Jose, CA, USA), PE anti-mouse CD90 (clone: G7) (Novus Biologicals, Littleton, CO, USA), PE anti-mouse CD31 (clone: 390) (Beckman Coulter, Brea, CA, USA), PE anti-mouse CD34 (clone: HM34), PE anti-mouse CD73 (clone: TY/11.8) (BioLegend, San Diego, CA, USA), APC anti-mouse CD105 (clone: MJ7/18), APC anti-mouse Ly-6C (clone: 1G7.G10), PE anti-mouse Gr-1 (clone: RB6-8C5), APC anti-mouse Gr-1 (clone: RB6-8C5; Miltenyi Biotec, Tokyo, Japan), APC anti-mouse Egr2 (clone: erongr2; M2 macrophage marker [12,13]), FITC anti-mouse CD38 (clone: 90; M1 macrophage marker [12]; Thermo Fisher Scientific, Waltham, MA, USA). Cell suspensions were processed by using the FACSCalibur® or BD Accuri™ C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed using FlowJo® V10 (Tree Star, Ashland, OR, USA).
2.3. Histological and immunohistochemical analyses
Liver tissues were fixed in IHC Zinc Fixative® (BD Pharmingen) for paraffin embedding. Sectioning, deparaffinization, epitope retrieval, and peroxidase/protein blocking steps were performed as described previously [11]. The HE staining involved the application of hematoxylin, followed by a rinse in weak acid solution to remove excess of hematoxylin, then the tissues were counterstained with eosin. Regarding the immunohistochemical staining, the following primary antibodies were used for staining as per the manufacturers' protocols: rat anti-mouse CD11b (clone: M1/70), Gr-1 (clone: RB6-8C5), CD4 (clone: H129.19), CD8a (clone: 53–6.7) (BD Pharmingen), F4/80 (clone: BM8; Invitrogen, Life Technologies, Camarillo, CA, USA), HRP goat polyclonal mouse serum Albumin, rabbit polyclonal Collagen I, rabbit polyclonal alpha smooth muscle Actin (Abcam, Cambridge, UK), rabbit anti-mouse Ki67 (clone: SP6; Novus Biologicals). Tissues were incubated with antibodies in 1% BSA/PBS and maintained overnight at 4 °C. Histofine Simple Stain Mouse MAX PO® (Nichirei Corporation, Tokyo, Japan) was used for antibody detection. Hydrogen peroxide/imidazole-HCl buffer was applied, followed by the DAB substrate solution (DAKO ChemMate™ EnVision Kit, Dako, Kyoto, Japan) and hematoxylin for counterstaining and visualization. Paraffin sections of livers were stained with Heidenhain's AZAN trichrome stain; to quantify fibrosis, images were captured with the BIOREVO BZ-9000® microscope (Keyence, Osaka, Japan), and fibrotic area was calculated by using the ImageJ software (NIH, Bethesda, MD, USA).
2.4. Quantitative real-time PCR
Isospin Cell & Tissue RNA (Nippon Gene, Tokyo, Japan) was used for RNA isolation as per the manufacturer's protocol. Single-stranded cDNA was synthesized by reverse transcriptase (Applied Biosystems, Waltham, MA, USA). cDNA was mixed with qPCR MasterMix Plus® (Eurogentec, Seraing, Belgium) and the TaqMan® Gene Expression Assay hydrolysis probe (Applied Biosystems) for the detection of Alb, Col1a1, Acta2, Cyp27a1, Ldha, and Tat. The Applied Biosystems® 7900HT Fast Real-Time PCR System was used for quantitative real-time PCR (qRT-PCR) as per the manufacturer's protocol.
2.5. DNA microarray analysis
A DNA microarray analysis was performed as described previously [14]. Briefly, purified RNAs were amplified and labeled with Cy3 by using the Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA, USA). Complementary RNA was hybridized to the Whole Mouse Genome 4 × 44K Array (Agilent Technologies). The Agilent G2505B DNA Microarray Scanner was used to read the microarray. BRB ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html) was used for the gene expression analysis. Filtering and normalization were performed as follows, unless otherwise stated: 1) filters were set to exclude spots with intensities below 1.0 or greater than 1 × 106, and values for replicate spots within an array were averaged; 2) normalization factors were determined by setting the specific target percentile to 50 and target intensity to 500. Gene class comparison and gene set comparison were performed for the evaluation of differentially expressing genes and gene sets containing the most differentially expressed genes. Furthermore, a list of MetaCore pathway maps (MetaCore from Clarivate Analytics, Philadelphia, PA, USA) was obtained following the enrichment analysis of differentially expressing genes identified in the preliminary class comparison.
2.6. Statistical analysis
Data are expressed as means ± SD, unless otherwise stated. Origin, Version 2020 (OriginLab Corporation, Northampton, MA, USA) was used to generate plots and for data analysis. All comparisons were evaluated by Student's t-tests, unless otherwise stated. Statistical significance was set to p < 0.05.
3. Results
3.1. Characterization of NASH mouse-derived u-ADSCs
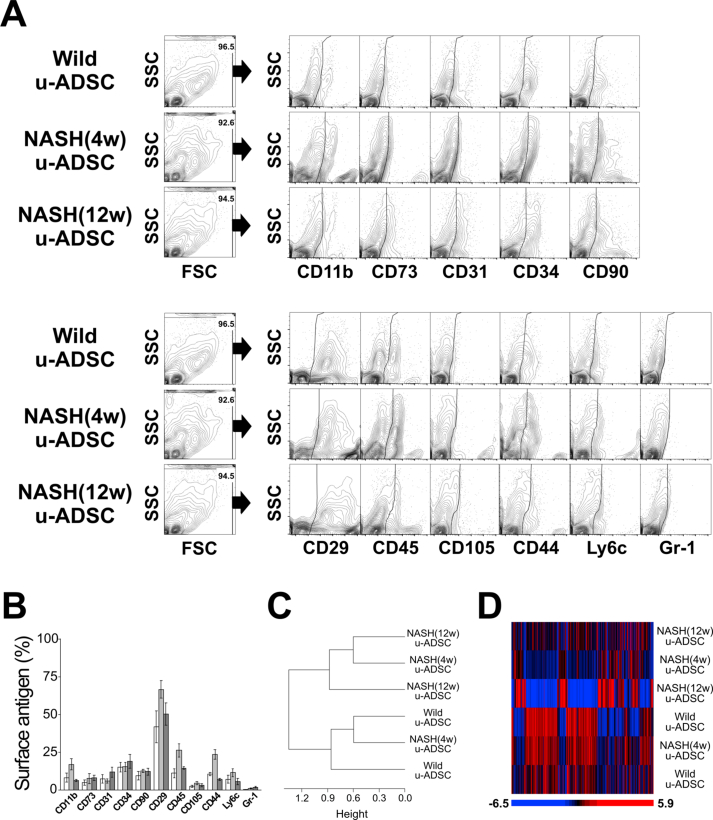
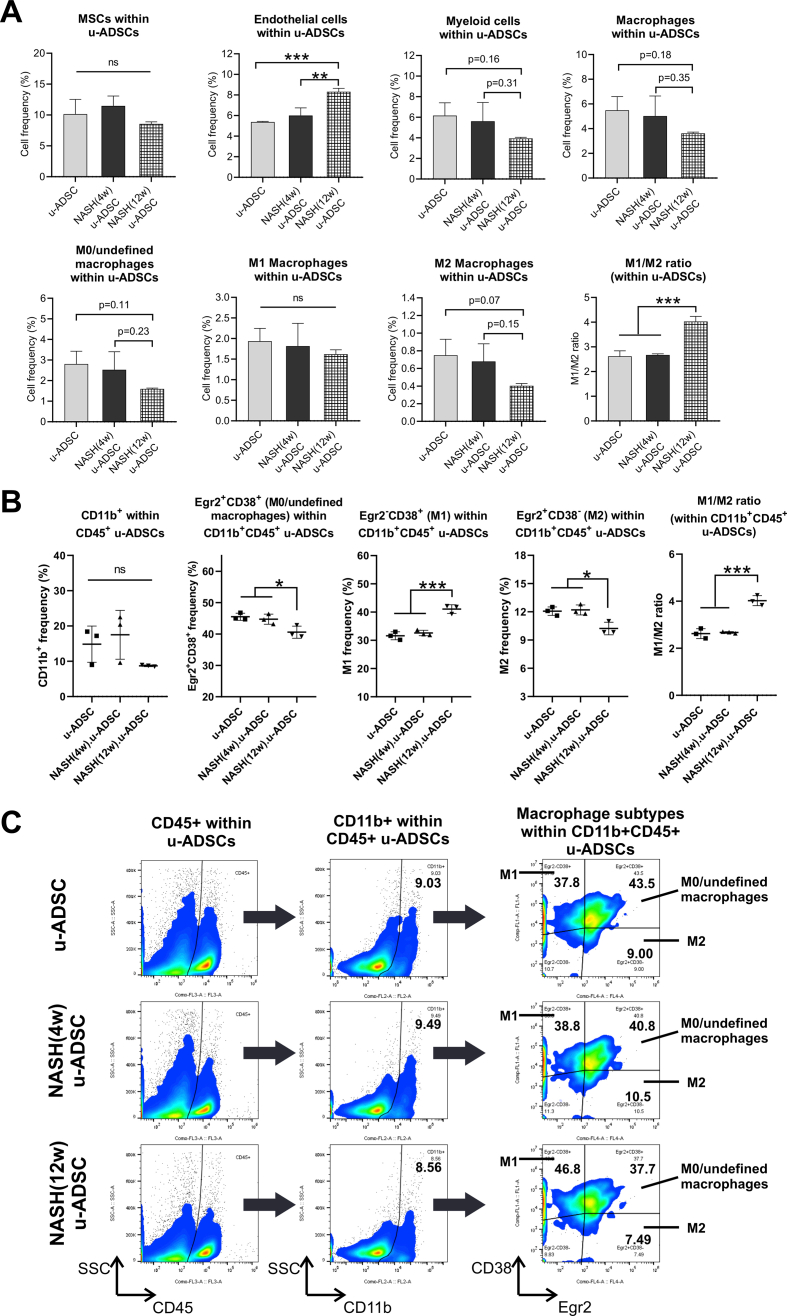
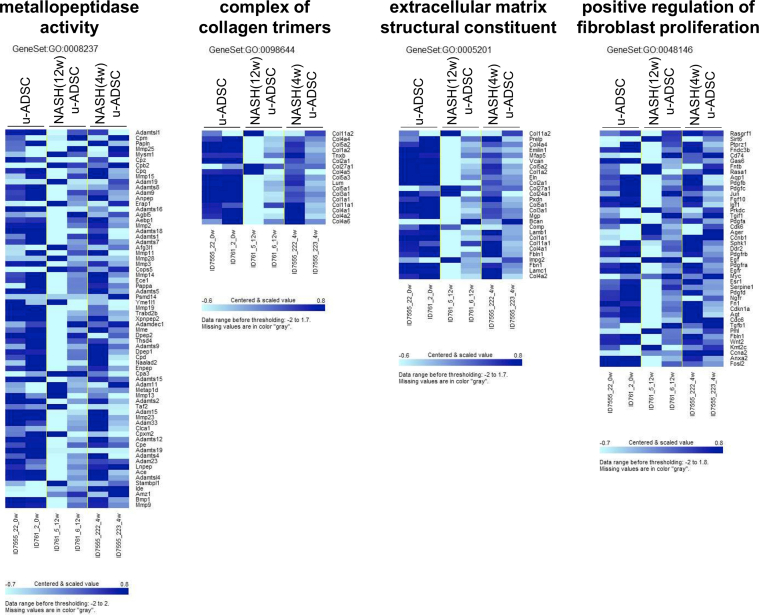
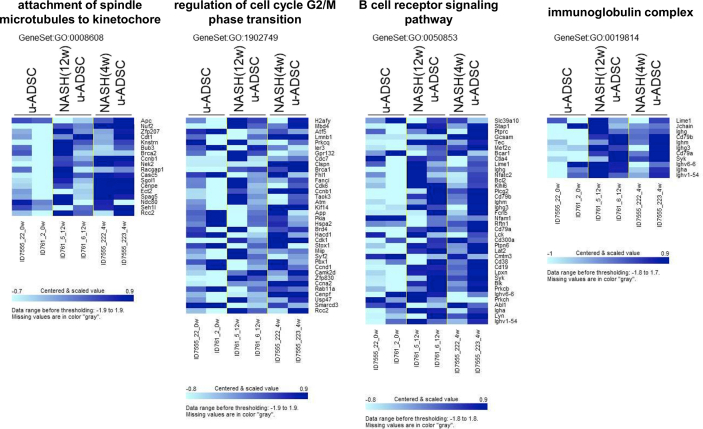
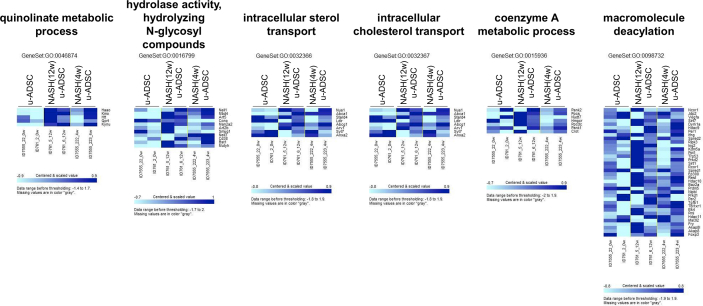
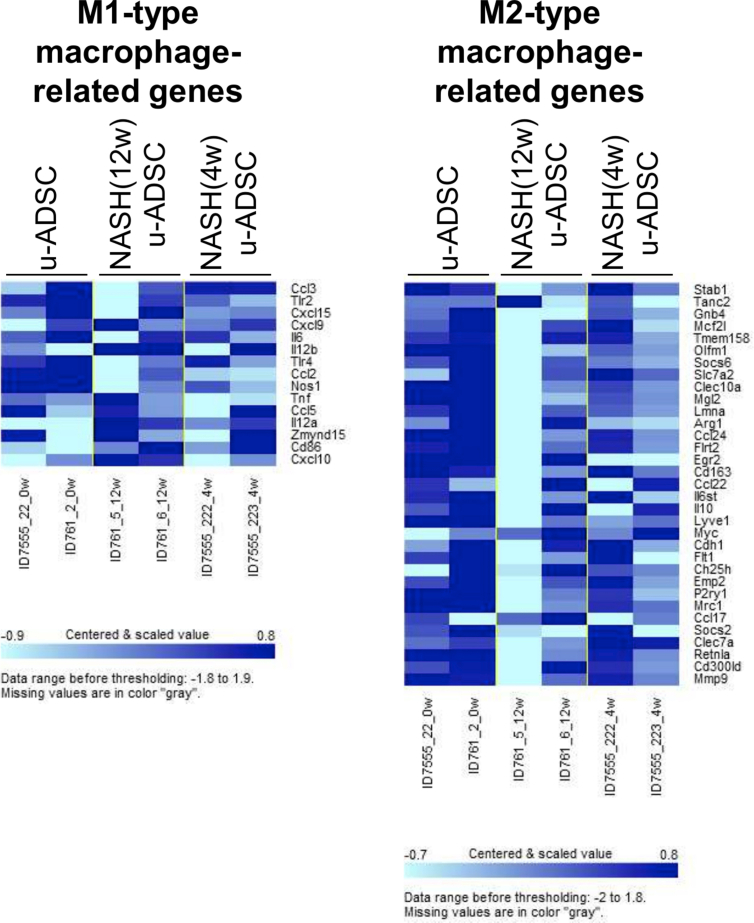
We characterized NASH mouse-derived u-ADSCs after 4 and 12 weeks of AT-HF diet feeding. Surface antigens for both u-ADSCs and NASH-u-ADSCs were verified by FCM. The frequencies of CD29, CD44, CD90, CD34, CD73, and CD105 were similar in all cell populations, confirming the presence of tissue-resident MSCs. CD31, related to endothelial cells, and CD11b, CD45, and Ly6c, related to myeloid and lymphoid cells, had overall frequencies of 5%–25%. Gr-1, a neutrophil marker, was not detected (Fig. 1A and B). Thus, we did not detect a substantial difference between wild-type u-ADSCs, NASH (4w) u-ADSCs, and NASH (12w) u-ADSCs with respect to MSC markers, except for slight increases in frequency of CD11b+, CD45+, and CD44+ cells in NASH (4w) u-ADSCs. These results indicated that there were slightly higher frequencies of myeloid and lymphoid cells in the adipose tissue after 4 weeks of an atherogenic diet, with subsequent decreases at 12 weeks, when late NASH/early cirrhosis is reached. Furthermore, we calculated the frequencies of MSCs (CD45−CD31−CD29+CD44+), endothelial cells (CD45−CD31+), myeloid cells (CD45+CD11b+), macrophages, and subsets of macrophages within u-ADSCs, NASH (4w) u-ADSCs, and NASH (12w) u-ADSCs (Supplementary Fig. S1A). We did not detect differences in the frequency of MSCs, however, we observed an increased frequency of endothelial cells and decreased frequencies of myeloid cells and macrophages in NASH (12w) u-ADSCs when compared to other groups. With respect to macrophage subsets within u-ADSCs, we observed reductions in M0/undefined (CD45+CD11b+CD38+Egr2+) and M2 (CD45+CD11b+CD38−Egr2+) macrophages within NASH (12w) u-ADSCs when compared to wild-type u-ADSCs and NASH (4w) u-ADSCs, while no difference in the frequency of M1 (CD45+CD11b+CD38+Egr2−) macrophages between groups was observed (Supplementary Fig. S1A). When we analyzed the frequencies of macrophage subsets in greater detail, within the CD45+CD11b+ myeloid cells, we observed a stronger decrease in M0/undefined and M2 macrophages as well as a steeper increase in M1 macrophages in NASH (12w) u-ADSCs; consequently, the M1/M2 macrophage ratio was higher in NASH (12w) u-ADSCs than in other groups (Supplementary Fig. S1B and S1C). The wild-type u-ADSCs, NASH (4w) u-ADSCs, and late NASH/early cirrhosis (12w) u-ADSCs were also subjected to a comparative gene expression analysis. By an unsupervised clustering analysis of expression levels of all filtered 5777 gene probes, we distinguished between wild-type u-ADSCs and NASH (12w) u-ADSCs, whereas NASH (4w) u-ADSCs did not form a single distinct cluster (Fig. 1C and D). Furthermore, we performed a gene set enrichment analysis of u-ADSCs, NASH (12w) u-ADSCs, and NASH (4w) u-ADSCs (Supplementary Table S1). Wild-type u-ADSCs were enriched for various terms in the Gene Ontology (GO) biological processes category, including metallopeptidase activity, complex of collagen trimers, extracellular matrix structural constituent and positive regulation of fibroblast proliferation (Supplementary Fig. S2); the GO biological processes upregulated in NASH (12w) u-ADSCs or NASH (4w) u-ADSCs were related to cell cycle, BCR signaling pathway and immunoglobulin complex (Supplementary Fig. S3), while the GO biological processes upregulated only in NASH (12w) u-ADSCs, when compared to u-ADSCs and NASH (4w) u-ADSCs, were related to metabolic processes, molecular modifications and intracellular transport of sterol and cholesterol (Supplementary Fig. S4). The observed gene sets of GO biological processes were closely related to u-ADSC characteristics; for example, the regulation of cytokine biosynthetic process, somatic stem cell division or regulation of hematopoietic progenitor cell differentiation, did not result differentially expressed (data not shown). Furthermore, we verified the expression of genes associated to M1-and M2-type macrophages [12]; no clear difference was observed regarding the M1-type macrophages-related genes between groups, while a downregulation of genes related to M2-type macrophages was observed in NASH (12w) u-ADSCs when compared to u-ADSCs or NASH (4w) u-ADSCs (Supplementary Fig. S5); these results further confirm a shift towards a higher M1/M2 macrophage ratio within the NASH (12w) u-ADSCs when compared to u-ADSCs or NASH (4w) u-ADSCs.
Fig. 1.
Characterization of u-ADSCs, NASH (4w) u-ADSCs, and NASH (12w) u-ADSCs. Freshly isolated u-ADSCs were isolated from inguinal subcutaneous adipose tissue of wild-type C57BL/6J female mice or C57BL/6J female mice fed an atherogenic diet for either 4 weeks or 12 weeks. (A) Scatter plots of flow cytometry results using fluorescence-conjugated antibodies to surface antigens (results are representative of three independent experiments). (B) Frequencies of wild-type u-ADSCs (white bar), NASH (4w) u-ADSCs (light grey bar), and NASH (12w) u-ADSCs (dark grey bar) expressing surface markers; bars: mean ± SD (n = 3). (C, D) RNA was isolated from u-ADSCs, NASH (4w) u-ADSCs, and NASH (12w) u-ADSCs for a DNA microarray analysis. (C) Dendrogram depicting an unsupervised clustering analysis of 5777 filtered genes; genes were excluded if: 1) fewer than 20% of expression values showed fold change of at least 1.5 in either direction from the median value for the gene, 2) the p-value for differences in log intensity was greater than 0.05, 3) the percent of missing spot values among the arrays exceeded 5%. (D) Heatmap depicting expression results for 5777 filtered genes for each sample.
3.2. Effect of wild-type u-ADSC treatment on hepatic inflammatory cell infiltration and fibrosis
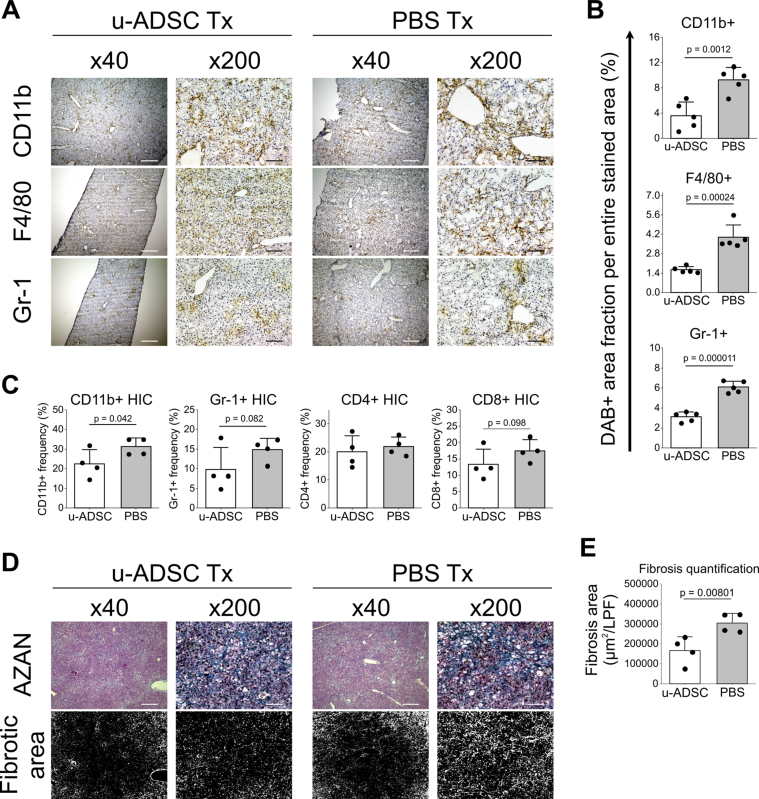
The u-ADSCs are a heterogeneous cell population with repair, regenerative, and immunomodulatory properties, they contain not only mesenchymal stem cells, but fibroblasts, endothelial cells, leukocytes, and pericytes [6,15]. Therefore, we evaluated whether the administration of u-ADSCs derived from wild-type mice had a therapeutic effect on mice with NASH cirrhosis. Previous studies have shown that the single administration of u-ADSCs is effective in a murine model of acute hepatitis [6]; in this study, we administered u-ADSCs twice to a mouse model of chronic NASH at weeks 24 and 26 of AT-HF diet feeding, in order to obtain a consistent and stable effect. At week 28, liver tissues and HICs were analyzed. Treatment with u-ADSCs reduced the infiltration of inflammatory CD11b+, F4/80+, and Gr-1+ cells within the liver tissues (Fig. 2A and B). Furthermore, we observed that the major inflammatory cells influenced by u-ADSC treatment were CD11b+ and Gr-1+ cells among HICs separated from all liver cells by density gradient centrifugation (Fig. 2C). No obvious difference was detected in the infiltration of CD4+ or CD8+ cells (Fig. 2C). A profound decrease in fibrosis was observed in NASH mice that received u-ADSC treatment (Fig. 2D and E).
Fig. 2.
Therapeutic effect of u-ADSC administration on a mouse model of NASH. C57Bl/6J female mice were fed an AT-HF diet, followed by u-ADSC injection (1 × 106 cells) into the splenic subcapsule of mice with NASH-cirrhosis on week 24 and 26 of the AT-HF diet. Liver tissues were sampled on week 28. (A) DAB staining of CD11b+, F4/80+, and Gr-1+ cells in livers treated with u-ADSCs or untreated livers (PBS). The DAB+ area was quantified by using ImageJ (B; area analyzed = 1.576 mm2, bars: mean ± SD; n = 5). (C) HICs were isolated, and frequencies of CD11b+, Gr-1+, CD4+, and CD8+ HICs quantified by flow cytometry (bars: mean ± SD; n = 4). (D) AZAN-stained sections with the relative fibrotic area and (E) quantification of fibrosis (bars: mean ± SD; n = 4). (A, D) Bars: 500 μm for × 40 magnification, 100 μm for ×200 magnification. Data are from the same experiment repeated twice.
3.3. Restorative effect of u-ADSCs on impaired liver tissues
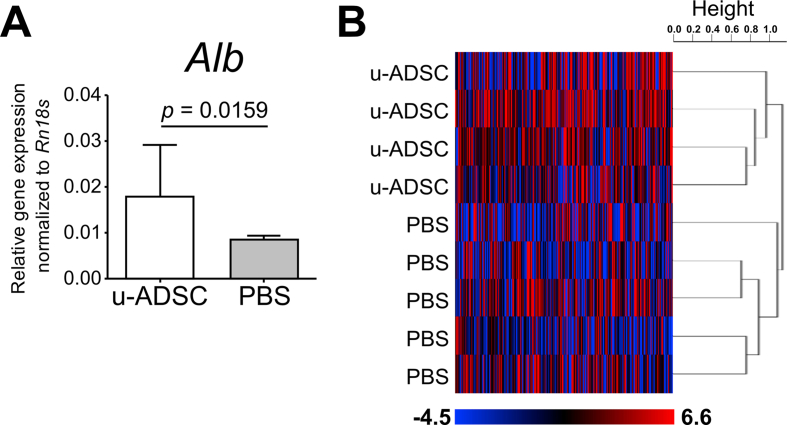
We further analyzed gene expression in liver tissues of mice treated with wild type u-ADSCs. We confirmed that u-ADSC treatment increased the expression of albumin (Fig. 3A). A comprehensive DNA microarray analysis of liver tissues revealed two complete clusters corresponding to u-ADSC treatment and the control (Fig. 3B). Among genes meeting the differential expression criteria in the microarray analysis, we identified 6950 genes that were significantly upregulated in liver tissues treated with u-ADSCs when compared to controls. We evaluated these genes by a MetaCore enrichment analysis (Table 1). The biological maps included mostly development-related pathways, restoration of transport mechanism and CFTR folding, cytoskeleton remodeling, chemotaxis SDF-1/CXCR4 signaling-dependent, as well as IL-6 signaling-related pathways, suggesting that gene expression patterns in the liver reflected the reparative/restorative effect of wild-type u-ADSCs.
Fig. 3.
Gene expression analysis of livers of NASH mice treated with u-ADSCs. The u-ADSCs (1 × 106 cells) were injected into the splenic subcapsule of NASH mice on week 24 and 26 of AT-HF diet feeding. Control mice received PBS. Liver tissues were sampled on week 28. RNA was isolated from liver tissues for qRT-PCR and DNA microarray analyses. (A) Expression of albumin assessed by qRT-PCR; bars represent mean ± SE, u-ADSCs: n = 4, PBS: n = 5. (B) Heatmap and dendrogram depicting an unsupervised clustering analysis of 8148 filtered genes whose expression for which the p-value of the log-ratio was lower than 0.05, and genes were excluded if the percent of missing spot values among arrays exceeded 1%; centered correlation and average linkage was used for clustering.
Table 1.
List of MetaCore pathways obtained by an enrichment analysis of 6950 upregulated genes in livers treated with u-ADSCs (downregulated in untreated livers (PBS)) identified in a preliminary class comparison (parametric p < 0.05).
| # | Maps | Total | p value | FDR | In Data | Network Objects from Active Data |
|---|---|---|---|---|---|---|
| 1 | Development_VEGF signaling via VEGFR2 - generic cascades | 93 | 3.060E-11 | 4.505E-08 | 38 | RhoA, Pyk2(FAK2), PI3K cat class IA, NCK1, CCL2, H-Ras, PAK2, MEK3(MAP2K3), TCF7L2 (TCF4), ROCK1, eIF4E, ERK1/2, VEGF-A, c-Raf-1, VEGFR-2, SOS, MNK1, p90Rsk, Paxillin, IP3 receptor, Shc, p120GAP, PKC, PI3K reg class IA, Vinculin, PKC-alpha, alpha-V/beta-3 integrin, HSP27, MEK2(MAP2K2), FAK1, IKK-gamma, CREB1, Neurofibromin, ERK2 (MAPK1), AKT(PKB), HSP90, Calcineurin B (regulatory), NF-AT2(NFATC1) |
| 2 | CFTR folding and maturation (normal and CF) | 24 | 1.511E-10 | 1.005E-07 | 17 | PARP-1, GANAB, UGCGL1, EDEM, HSP105, HSP90 alpha, CFTR, Hdj-2, Sti1, HSP90 beta, Aha1, HSPBP1, HSP40, Calnexin, HSP70, BAG-3, p23 co-chaperone |
| 3 | Chemotaxis_Lysophosphatidic acid signaling via GPCRs | 129 | 2.657E-10 | 1.005E-07 | 45 | RhoA, E-cadherin, cPKC (conventional), PLD1, PI3K cat class IA (p110-beta), Tcf(Lef), H-Ras, TRAF6, ROCK1, MKL2, ERK1/2, PKC-zeta, c-Raf-1, G-protein alpha-q/11, PKC-epsilon, TRIP6, CD36, Paxillin, IP3 receptor, LPAR2, MLCP (reg), ROCK, PI3K reg class IA (p85), Caspase-7, PAK, PDZ-RhoGEF, PKC, PLC-beta, Vinculin, ATF-2, DIA1, HAS2, alpha-V/beta-3 integrin, FasR(CD95), FAK1, CREB1, Bcl-XL, MEK1/2, Rho GTPase, N-CoR, JNK(MAPK8-10), MKL1, AKT(PKB), p53, Elk-1 |
| 4 | Transport_Clathrin-coated vesicle cycle | 71 | 2.731E-10 | 1.005E-07 | 31 | Syntaxin 6, Rab-4, VTI1A, Myosin I, Rabaptin-5, HIP1, Rab11-FIP1, PIP5KIII, Rab-8, Eps15, SNX9, VAMP4, Dynamin-2, Actin, YKT6, GS15, RAB9P40, Rab-7, Rab-11A, Endophilin B1, PLEKHA8 (FAPP2), Rab11-FIP2, VPS45A, PREB, BIN1 (Amphiphysin II), EEA1, Clathrin, Syntaxin 16, Myosin Vb, Clathrin heavy chain, VAMP2 |
| 5 | Development_The role of GDNF ligand family/RET receptor in cell survival, growth and proliferation | 92 | 1.837E-09 | 5.408E-07 | 35 | RhoA, PI3K cat class IA, B-Raf, NCK1, IRS-1, H-Ras, ATF-1, ERK1/2, VEGF-A, c-Raf-1, SOS, Paxillin, IP3 receptor, CDK2, Shc, ROCK, PI3K reg class IA (p85), MLC2, CREM (activators), SHP-2, alpha-V/beta-3 integrin, N-Ras, CaMK II alpha, VAV-2, FAK1, IKK-gamma, CREB1, Bcl-XL, MEK1/2, Cyclin D1, JNK2(MAPK9), NF-kB, c-FLIP, AKT(PKB), Elk-1 |
| 6 | Cytoskeleton remodeling_FAK signaling | 57 | 2.384E-09 | 5.848E-07 | 26 | RhoA, PI3K cat class IA, Cyclin D3, TRAF3, H-Ras, DOCK1, ERK1/2, p190RhoGAP, VEGF-A, c-Raf-1, VEGFR-2, SOS, G-protein alpha-q/11, FARP2, Paxillin, IP3 receptor, Talin, Shc, PI3K reg class IA, PLC-beta, PTEN, RIPK1, MEK2(MAP2K2), FAK1, AKT(PKB), Elk-1 |
| 7 | Immune response_TCR alpha/beta signaling pathway | 97 | 9.339E-09 | 1.922E-06 | 35 | STIM1, STIM2, B-Raf, CD45, TRAF3, TRAF2, H-Ras, TRAF6, ERK1/2, CABIN1, c-FLIP(Long), E2N(UBC13), c-Raf-1, Ca(II)channel L-type, Csk, Pannexin-1, Caspase-8, GRAP2, IP3 receptor, MEF2D, ORAI1, PKC-theta, PAG, PKC-alpha, CD3, CalDAG-GEFII, P2X1, RIPK1, CFLAR p43, IKK-gamma, MEK1/2, AKT(PKB), UEV1A, Calcineurin B (regulatory), NF-AT2(NFATC1) |
| 8 | IL-6 signaling in breast cancer cells | 53 | 1.175E-08 | 1.922E-06 | 24 | N-cadherin, E-cadherin, PI3K cat class IA, IP10, NF-kB1 (p50), H-Ras, ERK1/2, c-Raf-1, SOS, HSD17B1, MDR1, STAT1, Shc, PI3K reg class IA, SHP-2, Fascin, IL-6, Bcl-XL, MEK1/2, AKT2, Cyclin D1, AKT(PKB), Jagged1, Mcl-1 |
| 9 | DNA damage_ATR activation by DNA damage | 53 | 1.175E-08 | 1.922E-06 | 24 | RAD17, HUWE1, ERK1/2, Histone H2AX, Rad50, PRMT5, RAD9A, CINP, HSP90 beta, p90Rsk, RAD9, p53BP1, EGLN3, MRN complex, TTI1, Casein kinase II, NEK1, TELO2, TTI2, ETAA1, PP5, USP20 (VDU2), Nibrin, DBC1 |
| 10 | Chemotaxis_SDF-1/CXCR4-induced chemotaxis of immune cells | 79 | 2.754E-08 | 4.055E-06 | 30 | RhoA, Pyk2(FAK2), PI3K cat class IA, VCAM1, CD45, CALDAG-GEFI, NCK1, PLD1, ICAM1, PKA-cat alpha, PAK2, ROCK1, ERK1/2, PKC-zeta, PKA-reg (cAMP-dependent), Csk, JAK3, Paxillin, Talin, Shc, PI3K reg class IA (p85), PAK, PLC-beta, Vinculin, CD3, CD3 zeta, FAK1, MEK1/2, AKT(PKB), SFK |
| 11 | Immune response_B cell antigen receptor (BCR) pathway | 110 | 3.105E-08 | 4.154E-06 | 37 | STIM1, VCAM1, B-Raf, NCK1, NF-kB1 (p50), K-RAS, ICAM1, ETS1, H-Ras, PIP5KIII, MEK3(MAP2K3), ERK1/2, c-Raf-1, PP2A catalytic, BCAP, SOS1, PIP5KI, IP3 receptor, CDK6, Shc, PI3K reg class IA (p85), ORAI1, Rb protein, ATF-2, CalDAG-GEFII, N-Ras, VAV-2, MEK2(MAP2K2), GSK3 alpha/beta, IKK-gamma, Bcl-XL, MEK1/2, NF-kB, AKT(PKB), Calcineurin B (regulatory), Elk-1, NF-AT2(NFATC1) |
| 12 | G-protein signaling_K-RAS regulation pathway | 25 | 5.626E-08 | 6.902E-06 | 15 | B-Raf, CALDAG-GEFI, K-RAS, c-Raf-1, SOS, A-Raf-1, FTase, KAP3, Shc, PI3K cat class IA (p110-alpha), PKC-theta, RASSF1, Rap1GDS1, CalDAG-GEFII, RASGRF1 |
| 13 | Regulation of degradation of deltaF508-CFTR in CF | 39 | 9.216E-08 | 1.044E-05 | 19 | HSC70, HSP105, CFTR, HDAC6, Hdj-2, Sti1, SAE1, Dynein 1, cytoplasmic, heavy chain, Aha1, HSP27, HSPBP1, MJD (ataxin-3), BAG-2, UBE2J1, UBE2D1, HSP90, CHIP, HSP70, Derlin1 |
| 14 | Development_WNT/Beta-catenin signaling in the nucleus | 62 | 9.938E-08 | 1.045E-05 | 25 | TRRAP, TNIK, BAF53, PLD1, Tcf(Lef), TCF7L2 (TCF4), TBP, BRG1, MED1, PYGO2, BCL9/B9L, MEF2, TLE, DOT1, Kaiso, UBR5, hSPT3, RUNX, Pitx2, Casein kinase II, SET8, JNK(MAPK8-10), SMARCA5, EP400, RUVBL1 |
| 15 | Development_Positive regulation of STK3/4 (Hippo) pathway and negative regulation of YAP/TAZ function | 71 | 1.309E-07 | 1.258E-05 | 27 | RhoA, GCGR, E-cadherin, PKA-cat alpha, INADL, Alpha-1 catenin, 14-3-3, AMPK alpha subunit, PKA-reg (cAMP-dependent), MARKK, c-Raf-1, PP2A cat (alpha), Alpha-catenin, ZO-2, MOBKL1A, WW45, LATS1, Schwannomin (NF2), LKB1, STK4, RASSF1, FasR(CD95), Mol1b, Skp2/TrCP/FBXW, Beta-2 adrenergic receptor, Angiomotin (AMOT), PKA-cat (cAMP-dependent) |
| 16 | IL-6 signaling in multiple myeloma | 51 | 1.394E-07 | 1.258E-05 | 22 | PI3K cat class IA, K-RAS, ERK1/2, VEGF-A, c-Raf-1, VEGFR-2, SOS, STAT1, CDK6, Shc, PI3K reg class IA (p85), Rb protein, SHP-2, N-Ras, MEK2(MAP2K2), IL-6, Bcl-XL, Cyclin D1, JNK2(MAPK9), AKT(PKB), p53, Mcl-1 |
| 17 | Development_Fetal brown fat cell differentiation | 55 | 1.453E-07 | 1.258E-05 | 23 | PI3K cat class IA, SREBP1 (nuclear), IRS-1, H-Ras, ERK1/2, PKC-zeta, PKA-reg (cAMP-dependent), c-Raf-1, SOS, IGF-2 receptor, FASN, STAT5B, TR-beta, Prolactin receptor, SREBP1 precursor, Shc, PI3K reg class IA (p85), MEK2(MAP2K2), CREB1, AKT(PKB), PPAR-gamma, PKA-cat (cAMP-dependent), C/EBPalpha |
Abbreviations: u-ADSC, uncultured adipose tissue-derived stromal cells; PBS, phosphate-buffered saline; VEGF, vascular endothelial growth factor; CFTR, cystic fibrosis transmembrane conductance regulator; CF, cystic fibrosis; GPCRs, G protein-coupled receptors; GDNF, glial cell-derived neurotrophic factor; RET, rearranged during transfection; FAK, focal adhesion kinase; TCR, T-cell receptor; ATR, Rad3-related; SDF1, stromal cell-derived factor 1; CXCR-4, C-X-C chemokine receptor type 4.
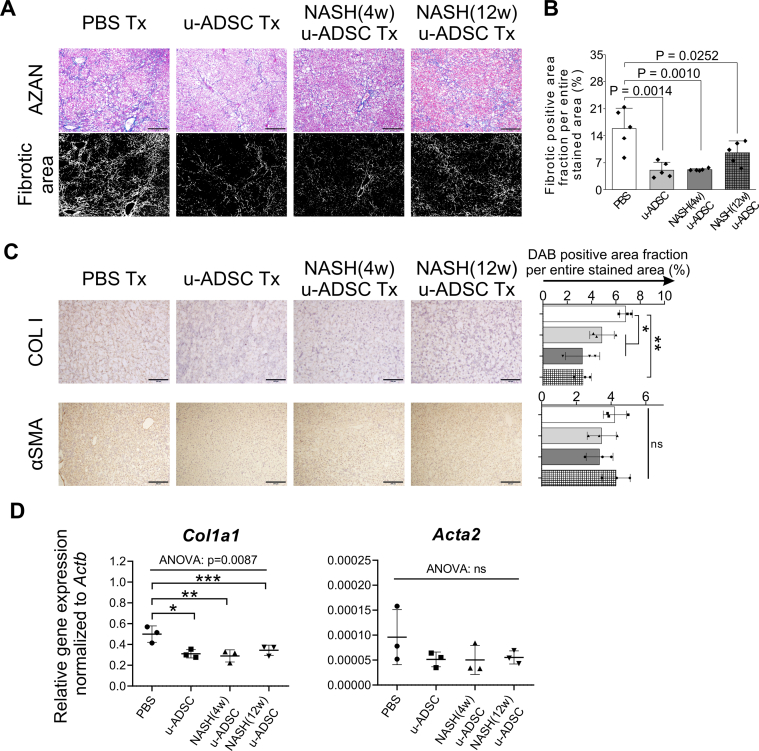
3.4. Immunosuppressive and restorative effect of NASH u-ADSCs on mice with NASH-cirrhosis
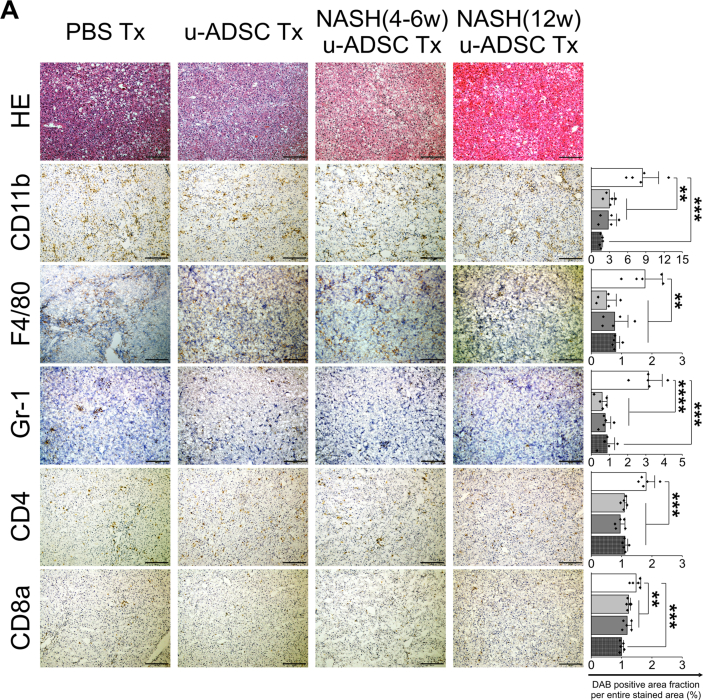
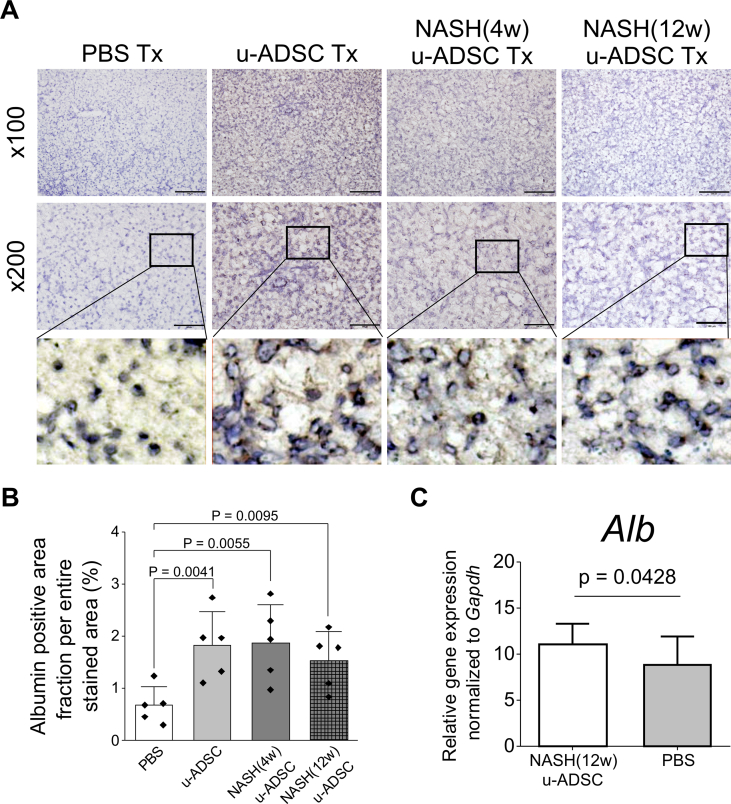
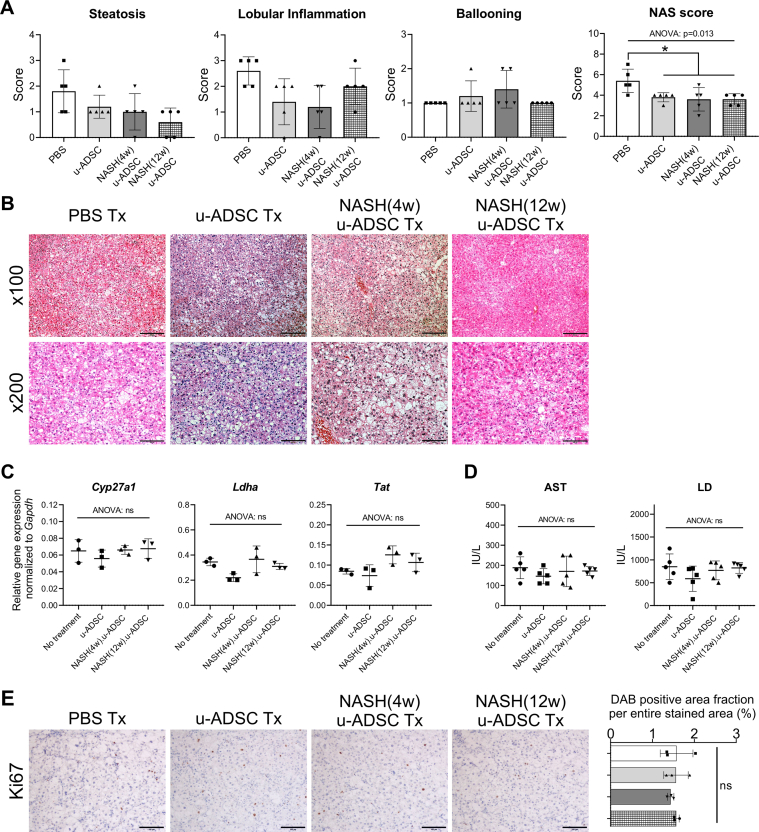
We observed that NASH mouse-derived u-ADSCs and wild-type u-ADSCs showed similar surface antigen expression as well as gene expression profiles, except for subtle increases in CD44 and CD11b cells and cell cycle activity in NASH (4w) u-ADSCs. We next assessed whether NASH (4w) u-ADSCs or NASH (12w) u-ADSCs had a therapeutic effect on mice with NASH-cirrhosis after treatment at weeks 24 and 26 of AT-HF diet feeding. On week 28, immunohistochemical staining of liver tissues was performed to evaluate inflammatory cells, fibrosis, and albumin expression. All treatments reduced the hepatic infiltration of inflammatory CD11b+, F4/80+, and Gr-1+ cells, which were prominent in NASH mice, and only a slight decrease was detected in the infiltration of CD4+ or CD8+ cells (Fig. 4A). Liver fibrosis was reduced by all treatments (Fig. 5A and B). Furthermore, the distribution and expression levels of collagen I, a key interstitial matrix main component, decreased; meanwhile α-SMA and Acta2 were not greatly affected by treatments (Fig. 5C and D). Furthermore, the NAS results confirmed that all treatments resulted in a global improvement in livers (Supplementary Figs. S6A and S6B), we noticed that the degree of improvement in individual items of the NAS score resulted different depending on the types of u-ADSCs used for the treatment; specifically, wild-type u-ADSCs and NASH (4w) u-ADSCs had a stronger effect at reducing lobular inflammation, conversely, NASH (12w) u-ADSCs resulted more efficacious at reducing steatosis; no major difference was observed in hepatocyte ballooning between treatments. Albumin expression levels also indicated an improvement in response to all treatments (Fig. 6A and B). Moreover, as determined by qRT-PCR, NASH (12w) u-ADSCs treatment induced the Albumin expression when compared to control (Fig. 6C), although we did not observe increases in the expression of Cyp27a1, Ldha, or Tat (Supplementary Fig. S6C). We also analyzed serum markers to assess liver enzymes and found that AST and LD levels were not affected by treatments (Supplementary Fig. S6D). Furthermore, we did not detect a difference in levels of Ki67, cell proliferation marker, among treatments (Supplementary Fig. S6E). Collectively, freshly isolated ADSCs obtained from wild-type and NASH mice were effective against NASH cirrhosis.
Fig. 4.
Therapeutic effect of u-ADSC, NASH (4w) u-ADSC, or NASH (12w) u-ADSC administration on a mouse model of NASH. Wild-type u-ADSCs, NASH (4w) u-ADSCs, or NASH (12w) u-ADSCs (7.5 × 105 cells) were administered into the splenic subcapsule of mice with NASH-cirrhosis on week 24 and 26 of AT-HF diet. Liver tissues were sampled on week 28. (A) Hematoxylin and eosin-stained sections and DAB staining of CD11b+, F4/80+, Gr-1+, CD4+, and CD8a cells in livers treated with PBS (control), u-ADSCs, NASH (4w) u-ADSCs, or NASH (12w) u-ADSCs. Bars: 200 μm, ×100 magnification. The DAB+ area, identifying cell infiltration was quantified by using ImageJ (area analyzed = 1.576 mm2, bars: mean ± SD; n = 5 independent biological replicates); white bar: No treatment (PBS); light grey bar: u-ADSCs; dark grey bar: NASH (4w) u-ADSCs; dark grey/hatched bar: NASH (12w) u-ADSCs.
Fig. 5.
Anti-fibrotic effects of u-ADSC, NASH (4w) u-ADSCs, and NASH (12w) u-ADSCs on NASH progression. Wild-type u-ADSCs, NASH (4w) u-ADSCs, or NASH (12w) u-ADSCs (7.5 × 105 cells) were administered to the splenic subcapsule of mice with NASH-cirrhosis on week 24 and 26 of AT-HF diet feeding; liver tissues were sampled on week 28. (A) AZAN-stained fibrotic area was identified using ImageJ in liver tissues. Magnification: ×100. Bars: 200 μm. (B) Quantification of fibrosis (n = 5, three different areas were analyzed for each biological sample); white bar: no treatment (PBS); light grey bar: u-ADSCs; dark grey bar: NASH (4w) u-ADSCs; dark grey/hatched bar: NASH (12w) u-ADSCs. Bars represent mean ± SD. (C) Immunohistochemical analysis of liver tissues for the detection of Collagen I (magnification: ×200; bars: 100 μm) and α-SMA (magnification: ×100; bars: 200 μm), and relative quantification of the DAB+ area; the Kruskal–Wallis test and post hoc Conover test were used for statistical analysis; n = 3, three different areas were analyzed for each biological sample, each area analyzed = 1.576 mm2; white bar: no treatment (PBS); light grey bar: u-ADSCs; dark grey bar: NASH (4w) u-ADSCs; hatched bar: NASH (12w) u-ADSCs. Bars represent means ± SD. (D) Expression levels of fibrosis-related genes were assessed by qRT-PCR; Y-axis represents the relative gene expression normalized to Actb; n = 3. Data are expressed as means ± SD; one-way ANOVA followed by the Tukey's HSD post hoc tests were performed for comparisons; ∗p < 0.05, ∗∗p < 0.01.
Fig. 6.
Effect of NASH u-ADSCs on a mouse model of NASH. (A) Albumin-positive cells in livers treated with no treatment (PBS), u-ADSCs, NASH (4w) u-ADSCs or NASH (12w) u-ADSCs, along with a zoomed-in area showing the subcellular localization of Albumin within hepatocytes; bars: 200 μm for ×100 magnification, 100 μm for ×200 magnification. The DAB+ area, identifying albumin-producing cells (A), was quantified by using ImageJ (B; area analyzed = 1.576 mm2, n = 5); white bar: No treatment (PBS); light grey bar: u-ADSCs; dark grey bar: NASH (4w) u-ADSCs; dark grey/hatched bar: NASH (12w) u-ADSCs; bars represent means ± SD. (C) Expression levels of albumin in liver tissues assessed by qRT-PCR; bars represent mean ± SD, NASH (12w) u-ADSCs: n = 8, PBS: n = 3, each replicate is an independent biological sample.
4. Discussion
Despite drastic advances in the treatment for chronic viral hepatitis [[16], [17], [18]], new therapies for liver diseases based on underlying causes are still needed [1]. NASH is emerging as a major cause of chronic liver disease requiring effective treatments to prevent progression to liver cirrhosis [19]. Stromal cells of adipose tissues have beneficial reparative/regenerative effects, since they contain somatic pluripotent MSCs [4], which are thought to be advantageous owing to their pluripotency as well as their immunomodulatory effects for the treatment of various injured organs, including cirrhotic livers [6]. Furthermore, autologous cells are relatively accessible and easy to obtain [20] from a patient's own adipose tissue; thus, the use of the freshly isolated autologous adipose tissue-derived stromal cells for reparation/regeneration therapy for injured organs is a practical approach [8,[21], [22], [23], [24]]. However, it is necessary to characterize freshly isolated u-ADSCs from affected hosts and to determine their effects on cirrhotic liver. We examined various surface antigens expressed on u-ADSCs. The frequencies of cells expressing MSC markers were nearly identical for wild-type and NASH-derived u-ADSCs, except for CD44 and CD11b antigens in NASH (4w) u-ADSCs. Furthermore, in a detailed analysis of the subfraction of macrophages, we observed by flow cytometry an increase in the M1/M2 macrophage ratio only within NASH (12w) u-ADSCs. Accordingly, it was also observed a downregulation of M2-type macrophage-related genes within the entire population of NASH (12w) u-ADSCs when compared to u-ADSCs or NASH (4w) u-ADSCs, while no distinct difference was seen regarding the M1-type macrophage-related genes between the entire populations of u-ADSCs, NASH (4w) u-ADSCs and NASH (12w) u-ADSCs; consequently, gene expression results also suggest a higher M1/M2 macrophage ratio within the NASH (12w) u-ADSCs when compared to other treatments. In our mouse model of NASH, 4 weeks of AT-HF diet initiated inflammation in the liver. Adipose tissues of the host, affected by smoldering inflammation under metabolic syndrome, are involved in insulin resistance and have a long-term impact on systemic metabolism [25]. Therefore, the initiation and progression of NASH inflammation may affect a fraction of ADSCs, MSC lineages, and possibly immune-mediating inflammatory cells; however, gene expression profiles in NASH mouse-derived u-ADSCs were not significantly affected, except for a decrease in collagen matrix organization, enhanced cell cycle, and increased B cell proliferation; also, we found only in NASH (12w) u-ADSCs, an upregulation of genes related to metabolic processes, molecular modifications and intracellular transport of sterol and cholesterol. Furthermore, the therapeutic effects of NASH (4w) u-ADSCs and NASH (12w) u-ADSCs on mice with NASH-related cirrhosis were similar to the effect of wild-type u-ADSCs. In particular, the infiltration of inflammatory cells was reduced and the NAS and fibrosis improved; regarding the degree of improvement in individual items of the NAS, we confirmed that wild-type u-ADSCs and NASH (4w) u-ADSCs had a stronger effect on reducing lobular inflammation, conversely, NASH (12w) u-ADSCs reduced steatosis more efficiently; no major difference was observed in hepatocyte ballooning between treatments. M1/M2 macrophage ratio was relatively high in NASH (12w) u-ADSCs, which would cause a slight decrease in immunosuppression efficacy, moreover, this might explain why the therapeutic effect of NASH (12w) u-ADSCs was milder, as hepatic fibrosis development is recognized to be strongly associated with hepatic inflammation. In NASH-cirrhotic mice treated with NASH (12w) u-ADSCs, when compared to other treatments, a more profound decrease in steatosis was observed, which might be explained by a possible better capability of NASH (12w) u-ADSCs to affect the fatty acids composition by functions like metabolic processes, molecular modification and intracellular transport of sterol and cholesterol, as confirmed by gene expression results; furthermore, the higher relative frequency of the endothelial cell subfraction might confer an overall therapeutic effect [26,27]. Finally, the restorative effect on impaired liver tissues was confirmed in all groups, suggesting that the general properties of freshly isolated u-ADSCs were not significantly affected by NASH. We have previously reported that freshly isolated murine u-ADSCs without cell culture suppress inflammation in a murine model of acute hepatitis. In addition, we analyzed in detail cell fractions of subcutaneous adipose tissue-derived stromal cells of wild-type mice and found that this cell population contained an immunosuppressive subfraction similar to M2 type macrophages [6]. Based on these findings, u-ADSCs are not solely dependent on MSCs for reparative/regenerative properties, and other cells contribute to the therapeutic effect on hepatitis. Thus, the detailed characterization of u-ADSCs would be intriguing and provide insight into the mechanisms underlying functions of freshly isolated stromal cells of adipose tissue, particularly their reparative/restorative effects on injured organs. Clinical studies of freshly isolated adipose tissue-derived stromal cells are ongoing in various fields [28]. We have also conducted a clinical study [29] and an investigator-initiated trial [8] using freshly isolated autologous adipose tissue-derived stromal cells without culture for the treatment of liver cirrhosis. Moreover, cultured adipose tissue-derived stem cells have been investigated extensively for repair and regenerative therapy [30,31], and such studies are expected to provide important information for the development of novel cell therapies.
5. Conclusion
The pathogenesis of NASH is yet to be fully resolved, furthermore, continuous non-clinical and clinical studies of the use of u-ADSCs for the repair and restorative therapy of liver cirrhosis need to be performed, in order to successfully proceed with the application of somatic cell therapy, which does not require processing steps from isolation to treatment. The current non-clinical investigation provides a promising novel autologous cell therapy for NASH-related liver disease.
Data availability statement
All data regarding this study can be requested from the corresponding author.
Funding
This work was supported in part by MEXT/JSPS KAKENHI Grant#: JP17K09413 and JP20K08327.
Declaration of competing interest
None declared.
Acknowledgments
We sincerely thank Ms. Hatsune Mochida, Ms. Keiko Yoshida, and Ms. Maki Wakabayashi for their excellent assistance.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2021.11.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
Fig. S6.
References
- 1.Hernandez-Gea V., Friedman S.L. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 2.Yki-Jarvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2014;2:901–910. doi: 10.1016/S2213-8587(14)70032-4. [DOI] [PubMed] [Google Scholar]
- 3.Tilg H., Moschen A.R. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 4.Zuk P.A., Zhu M., Ashjian P., De Ugarte D.A., Huang J.I., Mizuno H., et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banas A., Teratani T., Yamamoto Y., Tokuhara M., Takeshita F., Quinn G., et al. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46:219–228. doi: 10.1002/hep.21704. [DOI] [PubMed] [Google Scholar]
- 6.Nasti A., Sakai Y., Seki A., Buffa G.B., Komura T., Mochida H., et al. The CD45(+) fraction in murine adipose tissue derived stromal cells harbors immune-inhibitory inflammatory cells. Eur J Immunol. 2017;47:2163–2174. doi: 10.1002/eji.201646835. [DOI] [PubMed] [Google Scholar]
- 7.Ishida K., Seki A., Kawaguchi K., Nasti A., Yamato M., Inui H., et al. Restorative effect of adipose tissue-derived stem cells on impaired hepatocytes through Notch signaling in non-alcoholic steatohepatitis mice. Stem Cell Res. 2021;54:102425. doi: 10.1016/j.scr.2021.102425. [DOI] [PubMed] [Google Scholar]
- 8.Sakai Y., Fukunishi S., Takamura M., Inoue O., Takashima S., Usui S., et al. Regenerative therapy for liver cirrhosis based on intrahepatic arterial infusion of autologous subcutaneous adipose tissue-derived regenerative (stem) cells: protocol for a confirmatory multicenter uncontrolled clinical trial. JMIR Res Protoc. 2020;9 doi: 10.2196/17904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamato M., Sakai Y., Mochida H., Kawaguchi K., Takamura M., Usui S., et al. Adipose tissue-derived stem cells prevent fibrosis in murine steatohepatitis by suppressing IL-17-mediated inflammation. J Gastroenterol Hepatol. 2019;34:1432–1440. doi: 10.1111/jgh.14647. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzawa N., Takamura T., Kurita S., Misu H., Ota T., Ando H., et al. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 2007;46:1392–1403. doi: 10.1002/hep.21874. [DOI] [PubMed] [Google Scholar]
- 11.Higashimoto M., Sakai Y., Takamura M., Usui S., Nasti A., Yoshida K., et al. Adipose tissue derived stromal stem cell therapy in murine ConA-derived hepatitis is dependent on myeloid-lineage and CD4+ T-cell suppression. Eur J Immunol. 2013;43:2956–2968. doi: 10.1002/eji.201343531. [DOI] [PubMed] [Google Scholar]
- 12.Jablonski K.A., Amici S.A., Webb L.M., Ruiz-Rosado Jde D., Popovich P.G., Partida-Sanchez S., et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veremeyko T., Yung A.W.Y., Anthony D.C., Strekalova T., Ponomarev E.D. Early growth response gene-2 is essential for M1 and M2 macrophage activation and plasticity by modulation of the transcription factor CEBPbeta. Front Immunol. 2018;9:2515. doi: 10.3389/fimmu.2018.02515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seki A., Sakai Y., Komura T., Nasti A., Yoshida K., Higashimoto M., et al. Adipose tissue-derived stem cells as a regenerative therapy for a mouse steatohepatitis-induced cirrhosis model. Hepatology. 2013;58:1133–1142. doi: 10.1002/hep.26470. [DOI] [PubMed] [Google Scholar]
- 15.Melief S.M., Zwaginga J.J., Fibbe W.E., Roelofs H. Adipose tissue-derived multipotent stromal cells have a higher immunomodulatory capacity than their bone marrow-derived counterparts. Stem Cells Transl Med. 2013;2:455–463. doi: 10.5966/sctm.2012-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcellin P., Heathcote E.J., Buti M., Gane E., de Man R.A., Krastev Z., et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 17.Feld J.J., Jacobson I.M., Hezode C., Asselah T., Ruane P.J., Gruener N., et al. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med. 2015;373:2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 18.Curry M.P., O'Leary J.G., Bzowej N., Muir A.J., Korenblat K.M., Fenkel J.M., et al. Sofosbuvir and velpatasvir for HCV in patients with decompensated cirrhosis. N Engl J Med. 2015;373:2618–2628. doi: 10.1056/NEJMoa1512614. [DOI] [PubMed] [Google Scholar]
- 19.Wong R.J., Aguilar M., Cheung R., Perumpail R.B., Harrison S.A., Younossi Z.M., et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 20.De Ugarte D.A., Morizono K., Elbarbary A., Alfonso Z., Zuk P.A., Zhu M., et al. Comparison of multi-lineage cells from human adipose tissue and bone marrow. Cells Tissues Organs. 2003;174:101–109. doi: 10.1159/000071150. [DOI] [PubMed] [Google Scholar]
- 21.Koh Y.J., Koh B.I., Kim H., Joo H.J., Jin H.K., Jeon J., et al. Stromal vascular fraction from adipose tissue forms profound vascular network through the dynamic reassembly of blood endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:1141–1150. doi: 10.1161/ATVBAHA.110.218206. [DOI] [PubMed] [Google Scholar]
- 22.Lafosse A., Desmet C., Aouassar N., Andre W., Hanet M.S., Beauloye C., et al. Autologous adipose stromal cells seeded onto a human collagen matrix for dermal regeneration in chronic wounds: clinical proof of concept. Plast Reconstr Surg. 2015;136:279–295. doi: 10.1097/PRS.0000000000001437. [DOI] [PubMed] [Google Scholar]
- 23.Prins H.J., Schulten E.A., Ten Bruggenkate C.M., Klein-Nulend J., Helder M.N. Bone regeneration using the freshly isolated autologous stromal vascular fraction of adipose tissue in combination with calcium phosphate ceramics. Stem Cells Transl Med. 2016;5:1362–1374. doi: 10.5966/sctm.2015-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai Y., Fukunishi S., Takamura M., Kawaguchi K., Inoue O., Usui S., et al. Clinical trial of autologous adipose tissue-derived regenerative (stem) cells therapy for exploration of its safety and efficacy. Regen Ther. 2021;18:97–101. doi: 10.1016/j.reth.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun S., Ji Y., Kersten S., Qi L. Mechanisms of inflammatory responses in obese adipose tissue. Annu Rev Nutr. 2012;32:261–286. doi: 10.1146/annurev-nutr-071811-150623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T., Torimura T., Iwamoto H., Masuda H., Naitou M., Koga H., et al. Prevention of liver fibrosis and liver reconstitution of DMN-treated rat liver by transplanted EPCs. Eur J Clin Invest. 2012;42:717–728. doi: 10.1111/j.1365-2362.2011.02637.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T., Tsutsumi V., Torimura T., Naitou M., Iwamoto H., Masuda H., et al. Human peripheral blood CD34-positive cells enhance therapeutic regeneration of chronically injured liver in nude rats. J Cell Physiol. 2012;227:1538–1552. doi: 10.1002/jcp.22873. [DOI] [PubMed] [Google Scholar]
- 28.Bora P., Majumdar A.S. Adipose tissue-derived stromal vascular fraction in regenerative medicine: a brief review on biology and translation. Stem Cell Res Ther. 2017;8:145. doi: 10.1186/s13287-017-0598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai Y., Takamura M., Seki A., Sunagozaka H., Terashima T., Komura T., et al. Phase I clinical study of liver regenerative therapy for cirrhosis by intrahepatic arterial infusion of freshly isolated autologous adipose tissue-derived stromal/stem (regenerative) cell. Regen Ther. 2017;6:52–64. doi: 10.1016/j.reth.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolle S.T., Duscher D., Taudorf M., Fischer-Nielsen A., Svalgaard J.D., Munthe-Fog L., et al. Ex vivo-expanded autologous adipose tissue-derived stromal cells ensure enhanced fat graft retention in breast augmentation: a randomized controlled clinical trial. Stem Cells Transl Med. 2020;9:1277–1286. doi: 10.1002/sctm.20-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geburek F., Roggel F., van Schie H.T.M., Beineke A., Estrada R., Weber K., et al. Effect of single intralesional treatment of surgically induced equine superficial digital flexor tendon core lesions with adipose-derived mesenchymal stromal cells: a controlled experimental trial. Stem Cell Res Ther. 2017;8:129. doi: 10.1186/s13287-017-0564-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data regarding this study can be requested from the corresponding author.