Abstract
Background
Atherosclerosis is a complex pathology in which dysfunctional endothelium, activated leucocytes, macrophages, and lipid‐laden foam cells are implicated, and in which plaque disruption is driven by many putative actors. This study aimed to identify accurate targetable biomarkers using new in vivo approaches to propose tools for improved diagnosis and treatment.
Methods and Results
Human scFv (single‐chain fragment variable) selected by in vivo phage display in a rabbit model of atherosclerosis was reformatted as scFv fused to the scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G format) antibodies. Their reactivity was tested using flow cytometry and immunoassays, and aorta sections from animal models and human carotid and coronary artery specimens. A pool of atherosclerotic proteins from human endarterectomies was co‐immunoprecipitated with the selected scFv‐Fc followed by mass spectrometry for target identification. Near‐infrared fluorescence imaging was performed in Apoe −/− mice after injection of an Alexa Fluor 647–labeled scFv‐Fc‐2c antibody produced in a baculovirus system with 2 additional cysteine residues (ie, 2c) for future coupling to nano‐objects for theranostic applications. One scFv‐Fc clone (P3) displayed the highest cross‐reactivity against atherosclerotic lesion sections (rabbit, mouse, and human) and was chosen for translational development. Mass spectrometry identified galectin‐3, a β‐galactoside‐binding lectin, as the leader target. ELISA and immunofluorescence assays with a commercial anti‐galectin‐3 antibody confirmed this specificity. P3 scFv‐Fc‐2c specifically targeted atherosclerotic plaques in the Apoe −/− mouse model.
Conclusions
These results provide evidence that the P3 antibody holds great promise for molecular imaging of atherosclerosis and other inflammatory pathologies involving macrophages. Recently, galectin‐3 was proposed as a high‐value biomarker for the assessment of coronary and carotid atherosclerosis.
Keywords: biomarkers, flow cytometry, human antibodies, imaging, in vivo phage display
Subject Categories: Animal Models of Human Disease, Proteomics, Biomarkers, Atherosclerosis, Basic Science Research
Nonstandard Abbreviations and Acronyms
- LOX1
lectin‐type oxidized low‐density lipoprotein receptor 1
- scFv
single‐chain fragment variable
- scFv‐Fc
single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G format
Clinical Perspective
What Is New?
This study highlights the potential of in vivo phage display for the discovery of new targetable biomarkers of atheroma plaque progression in the pathological microenvironment. Our findings show that galectin‐3 is an ideal molecular biomarker that can be targeted for diagnostic purposes.
P3 is the first human recombinant antibody that recognizes galectin‐3 both in animal models of atherosclerosis and in human specimens of atherosclerotic arteries and that can be easily used for translational development.
Manipulating the genes of the variable regions allowed the development of a format suitable for in vivo studies after coupling to theranostic nano‐objects.
What Are the Clinical Implications?
It is crucial to identify vulnerable patients at a higher risk of developing coronary and carotid atherosclerosis.
The human antibody against galectin‐3, which plays a key role in atherosclerosis pathogenesis, discovered in this study by combining in vivo phage display and proteomics could be a useful tool for developing a novel molecular imaging modality.
However, more studies are needed to better understand galectin‐3’s role (beneficial or detrimental) in atheroma plaque formation to determine its therapeutic potential, in addition to its confirmed potential as diagnostic biomarker for atherosclerosis or for inflammatory pathologies that involve macrophages.
Atherosclerosis, the main cause of death in Western countries, is a chronic and inflammatory disease characterized by the buildup of lipid‐rich plaques that clog the arteries. Atherosclerotic lesions result from the local accumulation of lipids, immune and nonimmune cells, and cellular debris. 1 Endothelial cells, activated by lipoproteins, express chemokines and adhesion molecules that contribute to the recruitment of monocytes. Monocytes then differentiate into macrophages that express scavenger receptors and CD36 molecules and uptake oxidized low‐density lipoprotein (LDL), leading to the formation of foam cells, the precursors of plaque instability and vulnerability. 2 , 3
To improve atherosclerosis diagnosis and treatment, the identification of new targetable biomarkers is crucial. Currently, the diagnosis of atheroma plaques is invasive and is often done only at advanced disease stages. Recently, we developed contrast agents functionalized with antibody fragments to target platelets and loaded with iron oxide for noninvasive magnetic resonance molecular imaging. These targeting objects can specifically recognize the atheroma plaque in Apoe− /− mice ex vivo and in vivo. 4
To identify new targetable proteins that are overexpressed in the atheroma plaque, we recently set up an in vivo phage display method to select antibody fragments from a human scFv (single‐chain fragment variable) library (MG‐Umab 5 ) in an hypercholesterolemic rabbit model. 6 The in vivo discovery of biomarkers by this approach should allow better understanding atherosclerosis pathogenesis and development of new tools for imaging modalities. Moreover, human antibodies show limited immunogenicity when used in the clinic. After high‐throughput flow cytometry analysis, this method led to the selection and retrieval of 142 scFv‐phages with complete and in‐frame VH and VL germline genes (Sanger sequencing of the whole scFv fragment). Immunohistochemistry experiments confirmed the ex vivo reactivity of 60% of these scFv‐phages in sections of rabbit aorta with atheroma. 6
Here, we characterized some of these selected in vivo human antibodies produced in HEK293 cells (scFv‐Fc [scFv fused to the crystallizable fragment of immunoglobulin G format]) to confirm their reactivity by flow cytometry assays with rabbit atheroma protein extracts and by immunohistochemistry using rabbit and mouse aorta sections and human endarterectomy specimens. Importantly, we selected only the antibodies that recognized atheroma in arterial tissue sections from 2 preclinical models and in human endarterectomy biopsies.
We then focused on P3 scFv‐Fc because of its high cross‐reactivity against protein extracts from atherosclerotic lesions in all tested species, and because it was highly represented during the in vivo phage‐display selection, as reported in our previous work using third‐generation next‐generation sequencing. 7 By mass spectrometry and ELISA assays, we identified galectin‐3 as the P3 scFv‐Fc target. Galectin‐3, a member of β‐galactoside‐binding proteins, plays key roles in several physiological and pathophysiological processes. 8 Besides its expression in endothelial cells, galectin‐3 is also overexpressed by macrophages, the main inflammatory cells in the atheromatous plaque, 9 , 10 and is involved in monocyte attraction and macrophage activation. 11 In agreement, we found that P3 scFv‐Fc and an antibody against the macrophage receptor LOX1 (lectin‐type oxidized LDL receptor 1) colocalized in human endarterectomy sections.
Finally, we showed the ex vivo binding of P3 scFv‐Fc to its target in atherosclerotic aorta of Apoe− /− mice by fluorescence. Because it has been suggested that the expression of galectin‐3 fluctuates during plaque progression, 9 our findings could lead to the development of novel specific contrast agents functionalized with the P3 human antibody for noninvasive preclinical imaging of atherosclerosis and other inflammatory diseases.
Methods
The data and analytical methods will be made available to other researchers for purposes of reproducing the results or replicating the procedure. However, because of a patent, study materials will not be made available to other researchers.
Animal Models
All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85‐23, revised 1996) and were approved by the Bordeaux Ethics Committee (CEEA50). All preclinical experiments described in this publication were approved by the Animal Care and Use Committee of Bordeaux, France (no. 50120192‐A).
Adult male New Zealand White rabbits, weighing between 2.5 and 3.0 kg, were obtained from Charles Rivers Laboratories (St. Germain sur l'Arbresle, France). For 6 to 8 months, rabbits were fed a fat atherogenic diet that included 0.3% (w/w) cholesterol. To promote the development of complicated plaques, rabbits were subject to a surgical inflammatory injury 1 to 2 months after the beginning of the diet. De‐endothelialization of thoracic and abdominal aortic areas was mechanically induced by 3 inflations and retractions of a 4‐F Fogarty balloon catheter (Edwards Lifesciences, Maurepas, France). Surgery was performed under anesthesia induced by intramuscular injection of 50 mg/kg ketamine (Merial, France) and 5 mg/kg xylazine (Bayer Healthcare, France) and maintained by mask inhalation of 1.5% to 2% of isoflurane. Preventive antithrombotic treatment was heparin sodium solution (1000 IU) (Heparin Choay; Sanofi Synthelabo, Paris, France). Preoperative and postoperative analgesia were performed by administration of 100 mg of aspirin (Injectable Aspegic; Sanofi Synthelabo), and tolfedine (4 mg/kg; 2 subcutaneous injections 48 hours apart), respectively. 12 Rabbits were euthanized by a single intravenous injection of pentobarbital in the marginal ear vein (120 mg/kg) (CEVA Santé Animale, France).
Female, 6‐week‐old Apoe− /− mice (17–18 g in weight) were purchased from Charles River Laboratories (Saint Germain Nuelles, France) and housed under a 12‐hour light/dark cycle with food and water provided ad libitum. Mice were fed a high cholesterol diet (0.15% cholesterol) for 24 weeks to promote the development of atherosclerotic lesions. Animals were cared for in accordance with the institutional guidelines, and they were familiarized with their environment for at least 7 days before initiation of any experiments. For ex vivo studies and aorta isolation, animals were euthanized by intraperitoneal injection of Exagon (Axience, France) (300 mg/kg), diluted with 0.9% NaCl (1:4) under general anesthesia with 5% isoflurane.
Human Specimen Collection
Human tissue specimens were provided by Dr Ducasse, vascular surgeon at CHU Pellegrin Hospital (Bordeaux, France). Human samples were from patients who underwent endarterectomy after an acute vascular event. Human coronary artery samples were harvested from patients with end‐stage heart failure during heart transplantation. All clinical interventions took place at CHU Pellegrin (Bordeaux, France) and at Haut‐Lévèque Hospital (Pessac, France). All work with human tissues was approved by the Bordeaux Ethics Committee (Committee for the Protection of Persons Southwest and Overseas) and by the Research Ministry in France (authorization no. DC −2016‐2724). The Bordeaux ethics committee waived the need for the patient written consent because surgical waste no longer attached to the person is considered res nullius. Patients were informed by the clinicians; if they did not express their opposition to research, the deidentified samples were immediately processed (paraffin embedding or protein extraction). All study procedures complied with the ethical standards of the Declaration of Helsinki.
In Vivo Selection of scFv‐Phages and Screening by Flow Cytometry of Individual Clones
In Vivo Selection
ScFv‐phages from the optimized (in‐frame scFv selection) MG‐Umab library (fully human scFv) 5 , 13 were selected by in vivo phage display in the rabbit model of atherosclerosis. The scFv‐phages were isolated from atherosclerotic lesions, located from the aortic arch to the iliac bifurcations. Briefly, they were recovered from the endothelial layer and then from the intratissular (ie, subendothelial layer in the intima) and intracellular (ie, cells in the plaque, such as macrophages, foam cells, T cells) fractions after extensive washes between each fraction. This process was repeated for each round of biopanning. Three cycles of selection were performed to enrich for specific antibodies, as described in our previous studies. 6 , 7 Then, the in vivo–selected scFv‐phages were screened by flow cytometry for the identification of clones of interest. 6
Flow Cytometry Screening
Rabbit Protein Extraction and Coupling to Magnetic Beads
Protein extraction from rabbit atherosclerotic lesions was performed as previously described. 12 Briefly, proteins were solubilized with the T‐PER lysis buffer (Thermo Fisher Scientific, France) complemented with a protease inhibitor cocktail (Thermo Fischer Scientific) and a Polytron TP‐20 Homogenizer 8 (Kinematica, Lucerne, Switzerland). After 2 centrifugation steps at 13 000g at 4°C for 45 minutes to discard the insoluble material in the supernatant, the protein concentration of every soluble extract was determined using the Bradford Assay Kit, according to the manufacturer’s instructions (Thermo Fisher Scientific).
Fifty micrograms of protein extracts were covalently coupled to 300 nm of magnetic Carboxyl Adembeads according to the manufacturer’s instructions (Ademtech, France). Three batches of protein‐coupled beads per sample were used.
Phage Antibody Preparation
Individual phage‐infected XL1 blue bacteria were grown in 96‐well plates (Greiner Bio One, France) in 500 µL triptone/yeast extract (2TY)/ampicillin/5% glucose supplemented with 10 µg/mL tetracycline. After overnight incubation, 25 µL of bacterial culture were inoculated in 500 µL 2xTYGAT medium and incubated for 3 hours. Phage production was induced by adding 25 µL of 2TY containing 3.10 8 M13KO7 helper phage particles (Stratagene, France). After infection at 37°C for 1 hour, bacteria were pelleted, resuspended in 500 µL 2TY/ampicillin/5% glucose medium supplemented with 40 µg/mL kanamycin, and grown at 26°C under rotation (New Brunswick Scientific, Edison, NJ) overnight. Bacteria were spun down at 10 000g for 10 minutes, and supernatants were used immediately for flow cytometry assay.
scFv‐Phage Screening by Flow Cytometry
Binding of scFv‐phages or scFv‐Fc fusion antibodies to atherosclerotic protein extracts was determined by flow cytometry. Forty microliters of atherosclerotic rabbit protein‐coated beads (5 µg/mL) were added to 100 µL of scFv‐phages and incubated at 4°C under rotation for 3 hours. This was followed by incubation with mouse anti‐pVIII (Abcam, France) or rabbit anti‐human Fcγ (Jackson Immunoresearch, USA) primary antibodies (1:1000) at 4°C under rotation overnight, and by incubation with Alexa Fluor 488– labeled anti‐mouse antibodies (Life Technologies, France) (1:40) for scFv‐phage detection.
Validation of the scFv Fragment VH‐VL Sequences by Sequencing
Sanger sequencing was performed using primers for the scFv flanking regions in the phagemid (5′‐TGCAAATTCTATTTCAAGGAGAC‐3′ and 5′‐AGAATCATCAGATAAAGTAATCC‐3′). Antibody gene fragments were analyzed using the IMGT/V‐QUEST database (www.imgt.org/IMGT_vquest/vquest) for V germline determination and Complementarity‐Determining Region analyses. 14
Production of Recombinant scFv‐Fc Antibodies in HEK‐293 FreeStyle and Baculovirus Expression Systems
Antibody Production and Purification From HEK‐293 Cells
ScFv‐Fc was produced by transient transfection using the FreeStyle 293‐F expression system (Invitrogen, France). Expression vectors were prepared by producing (using Invitrogen GeneArt) the scFv‐encoding cDNA fragments as linear fragments with optimized codons for Homo sapiens. HEK‐293 FreeStyle cells were transfected with the purified expression vectors, according to the supplier instructions, with PEI 250 kDa (Sigma‐Aldrich) at 1:2 ratio (DNA:transfection reagent). After 7 days of production at 37°C and 8% CO2 in F17 medium supplemented with 8 mmol/L glutamine, scFv‐Fc antibodies were purified by 1‐step affinity chromatography with HiTrap FF protein A (GE Healthcare, France) on an ÄKTA avant 80 chromatography system. Molecules were eluted with 25 mmol/L citrate, pH3.0, and dialyzed against PBS, filtered (0.2 µm), and stored at 4°C until use.
Antibody Production and Purification From Insect Cells
The fusion ScFv‐Fc P3 with 2 cysteine residues (scFv‐Fc‐2c) was produced using the baculovirus expression system. 15 Briefly, the cDNA encoding the P3 scFv was polymerase chain reaction–amplified with the following forward and reverse primers: 5′‐GCTACTTAAGGGTGTCCAGTGCCAGGTGCAGCTGCAGCAGTCTGGACCCGG‐3′ and 5′‐GCTACGTACGCTTGATTTCCAGCTTGGTGCCGCCT‐ 3′. The polymerase chain reaction fragment was inserted into a specific transfer vector in frame with the sequence encoding the immunoglobulin (Ig) G1 signal peptide at the 5′ end and with a cDNA encoding the human IgG1 fragment variable domain with 2 extra cysteine residues at the C‐terminus. Sf9 cells were cotransfected by lipofection with the transfer vector and purified viral DNA in the presence of 40 µL of DOTAP liposomal transfection reagent (Roche). Recombinant viruses were isolated by plaque assay. ScFv‐Fc‐2c was produced by infecting Sf9 cells, adapted to grow in serum‐free medium, with the selected recombinant virus at a multiplicity of infection of 3 plaque‐forming units per cell. At day 3 after infection, cell culture supernatant was harvested, and scFv‐Fc‐2c was purified using HiTrap FF, protein A, as recommended by the manufacturer (GE Healthcare), and an ÄKTA purifier system. ScFv‐Fc‐2c was filter‐sterilized through a 0.22 µm filter (Millex GP; Millipore) and stored at 4°C until use.
Reactivity Analysis of scFv‐Fc Clones by Flow Cytometry and Immunohistochemistry
Flow Cytometry Analyses
Maintenance of reactivity after scFv‐Fc reformatting was first assessed by flow cytometry using the same protocol described for scFv‐phages. Briefly, scFv‐Fc was diluted at 10 µg/mL in PBS buffer and incubated with beads coated with protein extracts from rabbit atherosclerotic lesions (5 µg/mL) at 4°C under rotation for 3 hours. This was followed by incubation with rabbit anti‐human Fcγ primary (1:65 dilution; Jackson Immunoresearch) and Alexa Fluor 488–labeled anti‐rabbit (1:30 dilution, Life Technologies) secondary antibodies.
Immunohistochemistry Analysis of Rabbit and Mouse Aorta Tissue Sections and of Human Endarterectomy Specimens
The immunoreactivity of scFv‐Fc antibodies was evaluated using tissue sections prepared from paraffin‐embedded human, mouse, and rabbit atheromatous plaque specimens. Tissue sections were deparaffinized and rehydrated. Blocking steps (H2O2 blocking and unspecific site blocking with PBS/1% BSA/0.2% Triton X‐100) and a retrieval step (10 mmol/L Tris, 1 mmol/L EDTA, 0.05% Tween 20, pH9) were performed. For human tissue sections, another blocking step was performed by incubation with 5% goat serum and the F(ab′)2 fragment goat anti‐human IgG (Heavy +Light chains) (Jackson Immunoresearch) and goat anti‐human IgG (Fcγ specific) (Jackson Immunoresearch), diluted at 100 µg/mL. After washes with PBS/1% BSA, sections were incubated with the scFv‐Fc antibodies at 10 µg/mL at 4°C overnight. After washes with PBS/1% BSA/0.025% Triton X‐100, sections were incubated with the secondary horseradish peroxidase–conjugated goat anti‐human (Fcγ specific) antibody (1:1000) (Jackson Immunoresearch). Negative control samples were incubated with only the secondary antibody. Antibody binding was revealed using the 3,3’‐diaminobenzidine system (DAKO, France).
Target Identification by Immunoprecipitation and Mass Spectrometry Analysis
Coimmunoprecipitations
The Invitrogen Dynabeads co‐immunoprecipitation kit (14321D; Thermo Fisher Scientific) was used following the manufacturer’s instructions. Briefly, 20 µg of P3 scFv‐Fc or C4 scFv‐Fc (control antibody) or 10 µg of a commercial Gal3 Ab antibody (anti‐galectin‐3) (Abcam) was added to 1 mg of Dynabeads in coupling buffer and incubated at 37°C under rotation overnight. Proteins from human endarterectomy specimens were extracted and solubilized following the same protocol used for proteins from rabbit atherosclerotic lesions. After extraction, proteins were pooled and stored at −80°C. Dynabeads/antibody (P3 or C4) complexes were equilibrated in extraction buffer and then incubated with 1 mg of pooled human protein homogenates at 4°C under rotation for 1 hour. The Dynabeads/commercial Gal3 Ab complexes were incubated with 3 µg of Gal3R (recombinant human galectin‐3). Then, beads were washed, and the co‐immunoprecipitated proteins were eluted in the elution buffer (2xSDS Laemmli buffer supplied with the kit). One aliquot of each eluate was directly analyzed by SDS‐PAGE polyacrylamide gel (4%–10%) and stained with silver nitrate. The rest of each sample was sent to a mass spectrometry facility for further processing.
Mass Spectrometry Analysis
Sample Preparation and Protein Digestion
Samples were solubilized in Laemmli buffer and were concentrated and cleaned on SDS‐PAGE gels. Separation was stopped when proteins entered the resolving gel. After colloidal blue staining, all of the bands present in the relevant lane were extracted from the SDS‐PAGE gel and cut in 1×1‐mm pieces. Gel pieces were destained in 25 mmol/L ammonium bicarbonate 50% acetonitrile (ACN), rinsed twice in ultrapure water, and shrunk in ACN for 10 minutes. After ACN removal, gel pieces were dried at room temperature, covered with trypsin solution (10 ng/µL in 50 mmol/L NH4HCO3), rehydrated at 4°C for 10 minutes, and incubated at 37°C overnight. They were then incubated in 50 mmol/L NH4HCO3 at room temperature with rotary shaking for 15 minutes. Supernatants were collected, and the H2O/ACN/HCOOH (47.5:47.5:5) extraction solution was added to the gel slices for 15 minutes. The extraction step was repeated twice. Supernatants were pooled and dried in a vacuum centrifuge. Digests were solubilized in 0.1% HCOOH. 16 , 17
Nano‐Scale Liquid Chromatographic Tandem Mass Spectrometry Analysis and Label‐Free Quantitative Data Analysis
Peptide mixtures were analyzed with an UltiMate 3000 nano–liquid chromatography system (Dionex, Amsterdam, the Netherlands) coupled to an Electrospray Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific). Ten microliters of peptide digests were loaded onto a 300‐µm‐ID×5‐mm C18 PepMap trap column (LC Packings) at a flow rate of 10 µL/min. Peptides were eluted from the trap column onto an analytical 75‐mm‐ID×50‐cm C18 Pep‐Map column (LC Packings) with a 4% to 40% linear gradient of solvent B in 48 minutes (solvent A was 0.1% formic acid and solvent B was 0.1% formic acid in 80% ACN). The separation flow rate was set at 300 nL/min. The mass spectrometer operated in positive ion mode at 1.8‐kV needle voltage. Data were acquired using the Xcalibur 4.1 software in data‐dependent mode. Mass spectrometry (MS) scans (m/z 375–1500) were recorded at a resolution of R=120 000 (at m/z 200) and an automatic gain control target of 4×105 ions collected within 50 milliseconds. Dynamic exclusion was set to 60 seconds, and top speed fragmentation in higher‐energy C‐trap dissociation (HCD) mode was performed over a 3‐second cycle. Tandem Mass Spectrometry (MS/MS) scans with a target value of 3×103 ions were collected in the ion trap with a maximum fill time of 300 milliseconds. Additionally, only +2 to +7 charged ions were selected for fragmentation. Other settings were as follows: no sheath, no auxiliary gas flow, heated capillary temperature=275°C, normalized HCD collision energy=30%, and isolation width=1.6 m/z. The monoisotopic precursor selection was set to peptide and the intensity threshold to 5×103.
Database Search and Result Processing
The obtained data were analyzed with SEQUEST and Proteome Discoverer 2.3 (Thermo Fisher Scientific) using the Homo sapiens Reference Proteome Set (from Uniprot 2019‐05; 73 645 entries). Spectra from peptides higher than 5000 Da or lower than 350 Da were rejected. The search parameters were as follows: mass accuracy of the monoisotopic peptide precursor and of peptide fragments was set to 10 ppm and 0.6 Da, respectively. Only b‐ and y‐ions were considered for mass calculation. Methionine oxidation (+16 Da) and N‐terminal acetylation (+42 Da) were considered as variable modifications and cysteine carbamidomethylation (+57 Da) as fixed modification. Two missed trypsin cleavages were allowed. Peptide validation was performed with the Percolator algorithm, 18 and only high‐confidence peptides were retained (ie, false positive rate=1% at the peptide level). Peaks were detected and integrated using the Minora algorithm embedded in Proteome Discoverer.
ELISA Assay
Three independent ELISA assays were performed in triplicate. ELISA plates were coated (at 4°C overnight) with recombinant human galectin‐3 (Abcam), galectin‐3BP (Abcam), galectin‐1 (Abcam), GPαIIbβ3 integrin (Enzyme Research Laboratory, UK), or BSA (all at 5 µg/mL diluted in carbonate buffer). Then, each well was blocked with PBS/5% milk at 37°C for 1 hour. A mouse antibody against galectin‐1 (Abcam) and rabbit antibodies against human galectin‐3 and galectin‐3BP (Abcam) and the mouse AP2 antibody (a gift from Dr Nurden) against the glycoprotein αIIbβ3 (GPαIIbβ3) integrin were used as positive controls. The human P3 scFv‐Fc and control negative (CN) scFv‐Fc (irrelevant antibody, a gift from Laboratoire Français du Fractionnement et des Biotechnologies antibodies (diluted to 50 µg/mL), and the commercial antibodies (diluted to 10 µg/mL) were added to the respective wells at room temperature for 2 hours, followed by horseradish peroxidase–conjugated secondary antibodies. Each step was followed by extensive washes in PBS/0.1% Tween 20. The final wash was with PSB alone, and the antibody reaction was evaluated with the o‐phenylenediamine dihydrochloride system for ELISA (Sigma‐Aldrich). Color absorbance was immediately read at 405 nm in a plate reader (Chameleon; Thermo Fisher Scientific ).
Co‐staining Experiments
Human Tissue Sections
Paraffin‐embedded atherosclerotic lesions from human endarterectomy and coronary artery specimens were used in co‐staining experiments. All of the steps including the retrieval process and classical blocking steps were as described for immunohistochemistry. The following antibody combinations were used: (1) P3 scFv‐Fc diluted to 50 µg/mL in PBS/1% BSA with the anti‐human galectin‐3 antibody (10 µg/mL) (Abcam), and (2) P3 scFv‐Fc diluted to 50 µg/mL in PBS/1% BSA with the rabbit anti‐human LOX1 antibody (5 µg/mL). After incubation with these antibody combinations at 4°C overnight, sections were washed and then incubated (room temperature for 1 hour) with the following secondary fluorescent antibodies: Alexa Fluor 568–labeled anti‐human (1:200) for P3 scFv‐Fc, Alexa Fluor 488–labeled anti‐rabbit (1:1000) for the anti‐LOX1 antibody, or Alexa Fluor 488–labeled anti‐mouse (1:200) for the anti‐galectin‐3 antibody. After washes, sections were mounted with Vectashield (VWR, France).
For all experiments, adjacent sections incubated only with secondary antibodies were used as a negative control.
Images were acquired with a Nanozoomer 2.0 HT slide scanner and the fluorescence imaging module (Hamamatsu Photonics, France) using an UPS APO 20X NA 0.75 objective and an additional 1.75× lens, leading to a final magnification of 35×. Virtual slides were acquired with a TDI‐3CCD camera. Fluorescent images were acquired with a mercury lamp (LX2000 200W; Hamamatsu Photonics, Massy, France), and the filter was set for DAPI and/or GFP (green fluorescent protein)/Alexa Fluor 488, and/or Alexa Fluor 568, and/or Alexa FLuor 647/Cy5 fluorescence.
Mouse Tissue Sections
For immunofluorescence co‐staining of atherosclerotic lesions from Apoe−/− mouse aorta samples, the following antibodies were tested (at 4°C overnight): P3 scFv‐Fc (diluted to 50 µg/mL in PBS/1% BSA) and rabbit anti‐LOX1 antibody (5 µg/mL). After washes and incubation (room temperature for 1 hour) with the secondary fluorescent antibodies Alexa Fluor 647–labeled anti‐human (1:100) for scFv‐Fc and Alexa Fluor 488 anti‐rabbit (1:1000) for the anti‐LOX1 antibody, sections were washed, stained with DAPI (Thermo Fisher Scientific), and mounted with ProLong Gold (Life Technologies).
For all experiments, adjacent sections incubated only with secondary antibodies were used as negative controls.
Ex Vivo Fluorescence Imaging of P3 scFv‐Fc‐2c
The chests of 3 mice (n=2 Apoe−/− and n=1 control) were opened by thoracotomy, the heart exposed, and the right atrium cut. A 30‐gauge needle was inserted in the left ventricle. PBS/heparin (50 IU/mL per 2.5 mL; Sanofi Aventis, France) was inoculated, followed by 10 mL of PBS. Perfusion was continued with 2 mL of PBS containing the human P3 scFv‐Fc‐2c or the control human IgG antibody (100 µg for both) coupled to Alexa Fluor 647 according to the manufacturer’s instructions and using the Alexa Fluor 647 Antibody Labeling Kit (Thermo Fisher Scientific). After 20 minutes, mice were perfused with 5 mL of 4% v/v paraformaldehyde. The aorta was removed and embedded in an 80‐mm Petri dish containing 0.8% p/v high‐grade, 245 low‐melting‐point agarose.
Two sets of images of the same aortas were acquired. The first set of images was taken with a fluorescent ultramicroscope (light sheet imaging macroscopy; LaVision Biotech, France), equipped with a light cube for Alexa Fluor 647 acquisition. For the second set of images, aortas were imaged with 2 different confocal microscopes. The first was a Leica TCS SP8 mounted on an upright stand DM6 FS (Leica Microsystems, Mannheim, Germany) equipped with 405, 488, 552, and 638 lasers. The scanning was done using a conventional scanner (10–1800 Hz). The microscope was equipped with a galvanometric stage to do fast z acquisition and a motorized xy stage.
Then, a more resolutive confocal microscope was used (a Leica TCS SP5 on an upright stand DM6000 (Leica Microsystems), controlled by the software LAS AF and using objective HC PL FLUOTAR 10× dry NA 0.30). The fluorescent molecule was excited with a laser Helium‐Neon 633 nm, and the emission of fluorescence was collected on a conventional Photomultiplier Tube (PMT) from 650 to 720 nm. The transmitted light image was done at the same time on a PMT on transmission pathway. The 3‐dimensional mosaic was done automatically on 4×2 positions by using the Tile Scan module included in the software controlling the motorized stage (Märzhäuser, Wetzlar, Germany). Finally, a maximum intensity projection was applied to represent the localization of the fluorescence signal through the thickness of the aorta.
The microscopy experiments were performed at the Bordeaux Imaging Center of the Neurosciences Institute of the University of Bordeaux, France.
Image Quantification
Immunofluorescence Image Quantification
Image processing and analysis were done automatically with a Fiji‐ImageJ macro. 19 After opening the images from the ndpis source file with the Bio‐formats plugin, a median filter was applied to remove noise. Then, for each tissue section, a different automatic threshold was applied for the red (P3 scFv‐Fc) and green (mouse anti‐galectin‐3 and rabbit anti‐LOX1 antibodies) channels to measure the area of positive pixels. Then, a logical AND was applied to create the colocalization image from the 2 separated channels and to determine the percentage of colocalization.
Ex Vivo Fluorescence Image Quantification
Image processing and analysis were done automatically with a Fiji‐ImageJ macro. 19 After applying a median filter to remove noise, the tissue contour was automatically drawn using an automatic threshold in a brightfield image. This contour was then transferred to the fluorescence image (P3 scFv‐Fc‐2c or IgG antibodies) to measure the intensity of all the pixels inside the contour and the intensity of the pixels above a fixed threshold.
Statistical Analysis
All ELISA measurements were performed in triplicate and repeated in 3 independent experiments. Triplicate values were averaged, and differential analyses were then conducted using the Kruskal‐Wallis methodology. The Dunn test method was then used to identify significant difference between proteins. Analyses were performed using the R environment (R Foundation for Statistical Computing, Vienna, Austria).
Results
In Vivo Selection and Individual Screening by Flow Cytometry of scFv‐Phages and Reformatting Into scFv‐Fc Fragments
After the third round of in vivo biopanning of the MG‐Umab library, >800 different scFv‐phages were screened using protein extracts from rabbit atherosclerotic lesions. Overall, ≈200 scFv‐phages (24%) recognized the atherosclerotic protein extracts (binding at least 2‐fold above the background noise of wild‐type phages) in flow cytometry experiments. Figure 1A and 1B show the results of a typical experiment with 4 positive scFv‐phages (P3, D4, C8, and C4). The E9 clone was chosen as a negative control for all of the experiments. The gating strategy to analyze the binding of scFv‐phages and scFv‐Fc clones on rabbit atheromatous proteins coupled to beads is illustrated in Figure 1A.
Figure 1. Flow cytometry analyses. A, Example of gating strategy to analyze the binding of scFv (single‐chain fragment variable)‐phages on rabbit atheromatous proteins coupled to beads.
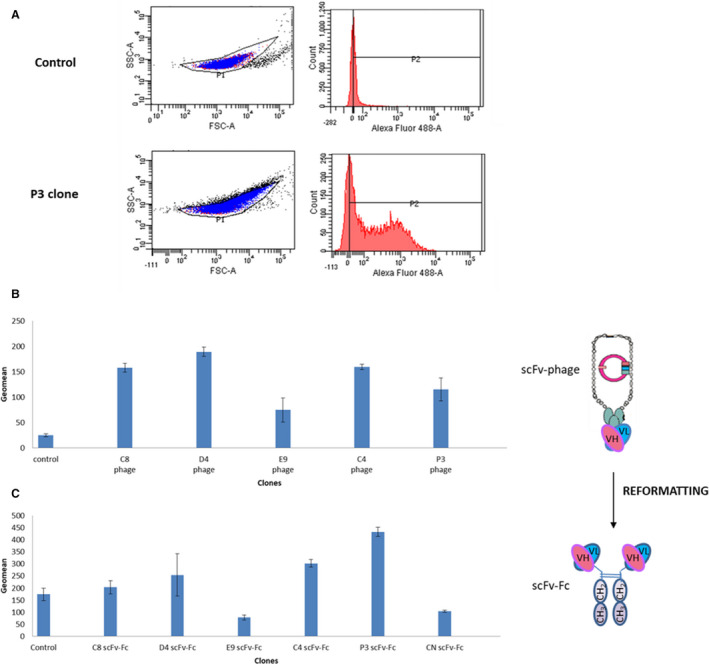
The control (mouse anti‐pVIII and Alexa Fluor 488–conjugated anti‐mouse antibodies) and the P3 clone are illustrated. Binding of scFv‐phages (B) and soluble scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) (C) on rabbit atheromatous proteins coupled to beads. Protein binding was detected using a mouse anti‐pVIII antibody and an Alexa Fluor 488‐conjugated anti‐mouse antibody for scFv‐phages, and using a rabbit anti‐human Fc antibody and an Alexa Fluor 488–conjugated anti‐rabbit antibody for scFv‐Fc. Mean values±SD were calculated using the geometric mean of fluorescence (P2 geomean) obtained with 3 different antibody batches. The error bars represent the mean values±SD. CN indicates control negative; FSC‐A,forward scatter area; VH, variable heavy; VL, variable light domains.
On the basis of the immunohistochemistry results in rabbit aorta sections with atheromatous lesions and the antibody sequence integrity, 142 scFv‐phages were finally considered for further investigation. Fifty of them were successfully produced in HEK‐293 FreeStyle cells as soluble scFv‐Fc fragments with a concentration >5 µg/mL (ie, threshold value for screening tests in supernatants). After scFv‐Fc engineering, analysis of the antibody reactivity in supernatants by flow cytometry using protein extracts from rabbit atherosclerotic lesions 6 (Figure 1C) showed that 60% of the selected scFv‐Fc maintained their reactivity.
In Vitro Validation of the Bioreactivity of Reformatted Clones by Immunohistochemistry Assays
ScFv‐Fc capacity to recognize their targets in artery sections with atheromatous lesions from different mammalian species was assessed, as done for scFv‐phages. 6 Figure 2 shows the results obtained with the P3, C4, D4, and C8 scFv‐Fc antibodies. In sections of human endarterectomy specimens, the P3 scFv‐Fc signal was strong in clusters of foam cells (arrowheads) and in areas with a necrotic core. In aorta sections from hypercholesterolemic rabbits, P3 staining was observed in the adventitia (A), in the intima (I) subendothelial area, and in some spindle‐shaped cells in the disorganized media layer (M). In sections from Apoe−/− mouse aorta with atheroma plaques, the necrotic area (arrowhead) and adventitia (A) were clearly stained. In sections of the human endarterectomy sample, the C4 scFv‐Fc antibody detected foam cells in necrotic cores of the subendothelial area and near to the tunica media (M) layer (arrowhead). Similarly, in atherosclerotic rabbit sections, specific C4 labeling was observed near the tunica media/intima (M/I) interface and in the necrotic core (arrowhead). In Apoe−/− mouse aorta sections, C4 scFv‐Fc recognized the tunica intima (I) (arrowhead), although some unspecific background signal was observed because of degradation and detachment of the adventitia tissue on the slide (asterisks). D4 scFv‐Fc only labeled a few groups of cells (arrowhead) in the atheroma plaque of the human carotid specimen. However, this signal might not be specific when compared with the background signal obtained by incubation with only the secondary antibody (upper panel, horseradish peroxidase‐goat anti‐human Fcγ). This is because of the presence of human antibodies in the plaque, despite the different specific blocking steps with goat serum, F(ab′)2 fragment goat anti‐human IgG (H+L), and goat anti‐human IgG (Fcγ specific). Conversely, in rabbit aorta sections, D4 scFv‐Fc clearly labeled the intima (I) and the endothelium (arrowhead). In the Apoe−/− mouse aorta section, D4 scFv‐Fc only labeled the adventitia (A) (arrowhead). C8 scFv‐Fc did not show any significant labeling in all section types, with the exception of an area close to the tunica media (M, arrowhead) in rabbit aorta sections with atheromatous lesions, and the adventitia (A) in Apoe−/− mouse atheroma sections (arrowhead). Because the final aim of the study was to use in vivo–selected human antibodies as targeting moieties for the detection of atheromatous plaques in patients, the best translatable scFv‐Fc antibodies were clearly P3 and C4, which displayed strong labeling of atheroma plaques in both animal models and also in human carotid sections. Moreover, the sequence of P3 was chosen as the reference for a recently published third‐generation sequencing project to study clone enrichment during the in vivo phage display selection. 7 Specifically, it was among the best enriched sequences, and clones belonging to the same P3 clonotype with well‐characterized somatic mutations in the VL gene were identified. On the basis of these previous results and because P3 reformatted as scFv‐Fc showed the highest binding by flow cytometry and immunohistochemistry experiments, the rest of the study focused on the identification of the P3 scFv‐Fc target, and C4 scFv‐Fc was used as a comparison in LC‐MS/MS analyses.
Figure 2. Immunohistochemical analysis of arterial tissue sections from atheromatous rabbits and Apoe−/− mice and human endarterectomy specimens using the indicated scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G).
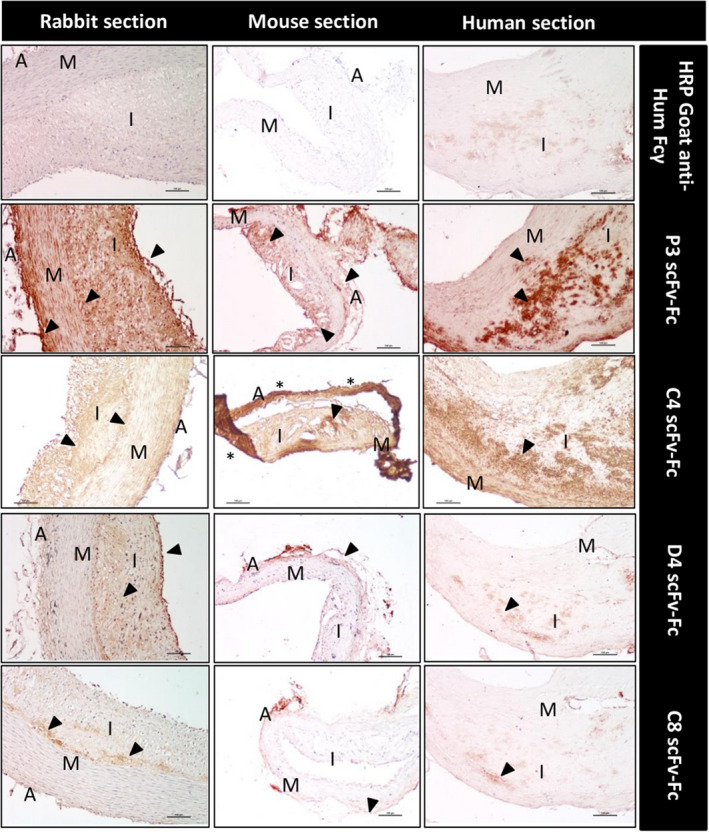
The different areas in transversal sections are identified: adventitia (A), media (M), and intima (I). Sections were incubated with the indicated scFv‐Fc antibodies (P3, C4, D4, and C8), followed by the HRP‐conjugated goat anti‐human Fcγ antibody, and the DAB substrate kit reagent. The yellow‐brown staining indicates the presence of the antigen recognized by the scFv‐Fc. No staining was observed in mouse and rabbit sections incubated with the secondary antibody alone, and there is low background noise in the human sections (upper panels). Scale bars, 100 µm. Nuclei were counterstained with hematoxylin. DAB indicates 3,3′‐diaminobenzidine; and HRP, horseradish peroxidase.
Antigen Identification by Immunoprecipitation and Proteomic Analyses
To identify the antigen recognized by P3 scFv‐Fc among the proteins extracted from endarterectomy specimens, after immunoprecipitation of human endarterectomy proteins with the P3 and C4 scFv‐Fc clones (P3‐PH [proteins from human biopsies], C4‐PH), samples were separated by SDS‐PAGE followed by silver staining to visualize the major bands. In samples immunoprecipitated with P3 scFv‐Fc, a single band with a relative molecular mass of 60 kDa was observed on the SDS‐PAGE gel (Figure 3, SDS‐PAGE 1). Conversely, after immunoprecipitation with C4 scFv‐Fc, multiple bands were detected (Figure 3, SDS‐PAGE 2). Extraction and analysis by LC‐MS/MS of the different bands (Table S1) showed that in both cases, the most represented proteins in the MS/MS spectra were keratin, filamin, vimentin, desmoglein, desmoplakin, and junction plakoglobin. However, galectin‐3, a new biomarker of atherosclerosis, 20 , 21 was the highest represented protein in the P3‐PH sample compared with C4‐PH extract (abundance=35 326 405 in P3‐PH and 1 266 731 in C4‐PH; C4‐PH/P3‐PH abundance ratio=0.037 (Table S1). Because monomeric galectin‐3 is subject to modifications, such as self‐association (dimerization or oligomerization), which increase its range of biological activity, 22 this might explain the band detected at 60 kDa (theoretical molecular weight of galectin‐3=30 kDa). To test this hypothesis, Gal3R was immunoprecipitated with a commercial anti‐Gal3 Ab, and the immunoprecipitate (Gal3 Ab‐Gal3R) was separated by SDS‐PAGE. As observed with P3 scFv‐Fc, a single band was detected (Figure 3, SDS‐PAGE 3). Moreover, the Gal3R protein was identified by LC‐MS/MS analysis for the Gal3 Ab‐Gal3R immunoprecipitate, with an abundance of 30 552 241. Comparison of galectin‐3 abundance in Gal3Ab‐Gal3R and P3‐PH gave a ratio close to 1 (Gal3Ab‐Gal3R/P3‐PH ratio=0.739), indicating a comparable galectin‐3 amount in the 2 immunoprecipitates (Table S1).
Figure 3. SDS‐PAGE analysis of immune complexes.
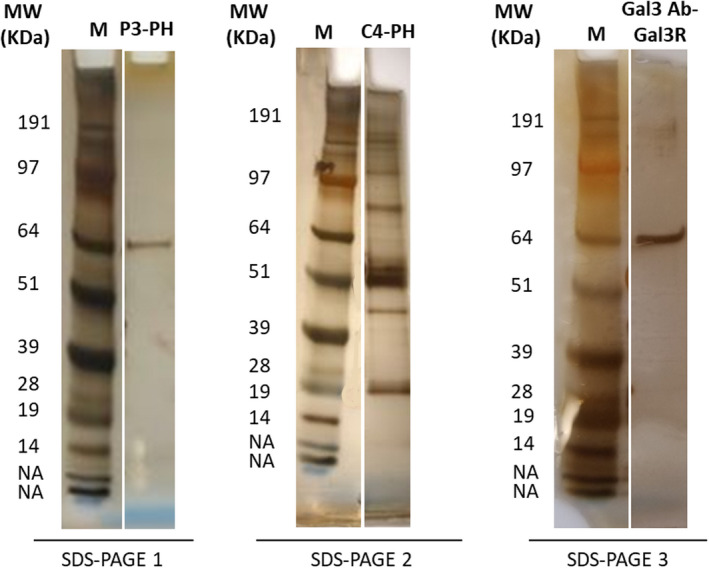
After immunoprecipitate elution in 2×SDS Laemmli buffer, immune complexes were separated on polyacrylamide gels (4%–10%) and stained with silver nitrate. SDS‐PAGE 1 and 2: proteins extracted from human endarterectomy specimens were immunoprecipitated with the P3 and C4 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) (from mammalian cells), respectively. SDS‐PAGE 3: Gal3R (recombinant galectin‐3 protein) was immunoprecipitated with a commercial anti‐GAL3 Ab (anti‐galectin‐3 antibody). MW indicates molecular weight; M, marker; NA, non attributed; PH, proteins from human biopsies.
Then, to assess the specificity of P3 scFv‐Fc binding to galectin‐3, an ELISA assay was performed using recombinant galectin‐3 and galectin‐3BP proteins, which is the main galectin‐3 ligand, 23 and also recombinant galectin‐1, another β‐galactoside lectin family member, and other irrelevant proteins such as BSA and the GPαIIbβ3 integrin (Figure 4A). Globally, average OD values tended to significantly differ (P Kruskas‐Wallis=0.0503) between studied proteins. Deeper analyses revealed that OD was significantly higher in galectin‐3 than in galectin‐3BP (P=0.022), BSA (P=0.018), and galectin‐1 (P=0.002) (Figure 4A). These results highlight that P3 scFv‐Fc bound specifically to galectin‐3, and did not recognize galectin‐3BP, galectin‐1 protein, and BSA. Some cross‐reactivity was observed with GPαIIbβ3, with a P value=0.1087 for galectin‐3 versus GPαIIbβ3. This could be explained by the fact that integrins, such as GPα3β1 24 and GPαvβ3, 25 are among the galectin‐3 ligands. Commercial antibodies against all these recombinant proteins and the AP2 (AP2 clone) antibody were used as positive controls, and the irrelevant CN antibody (scFv‐Fc format) as a negative control (Figure 4B).
Figure 4. Binding of P3 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) and control antibodies to recombinant proteins by ELISA assay.
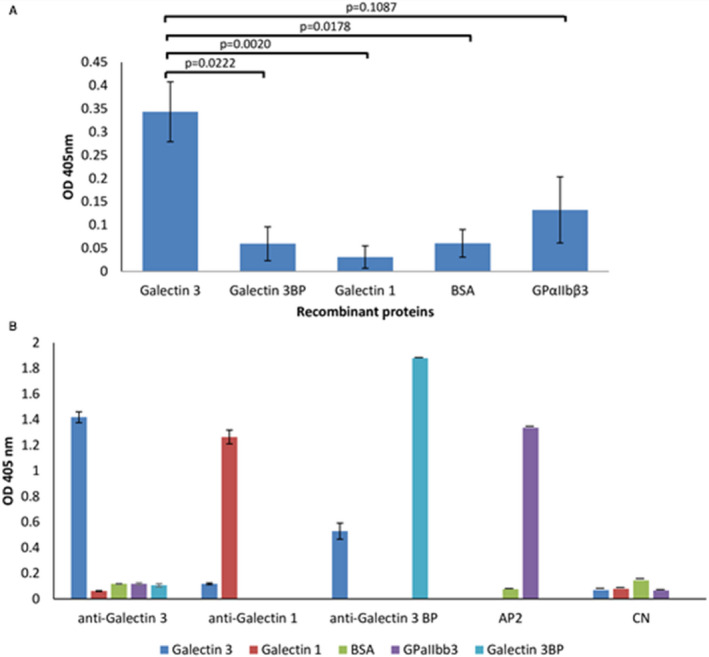
A, P3 scFv‐Fc showed stronger binding to recombinant galectin‐3 than to the galectin‐3BP (galectin‐3 binding protein) ligand. Galectin‐1 and Glycoprotein αIIbβ3 (GPαIIbβ3) were used to assess the specificity of galectin‐3 binding. An irrelevant antigen (BSA) served as a negative control. P3 scFv‐Fc binding was detected with a horseradish peroxidase (HRP)‐conjugated anti‐human Fcγ antibody, and absorbance was measured at 405 nm. Data are the means of 3 independent experiments repeated in triplicate. According to the Kruskal‐Wallis test followed by the Dunn test, differential analyses provided the following P values: P=0.0222 for galectin‐3 vs galectin‐3BP, P=0.0020 for galectin‐3 vs galectin‐1, P=0.0178 for galectin‐3 vs BSA, and P=0.1087 for galectin‐3 vs GPαIIbβ3. B, The specificity of the commercial antibodies against galectin‐3, galectin‐1, and galectin‐3BP, of the mouse AP2 (AP2 clone) antibody (from Dr Nurden) against GPαIIbβ3 integrin, and of CN (control negative) scFv‐Fc (negative control; from Laboratoire Français de Fractionnement et de Biotechnologies) was assesses by ELISA with recombinant galectin‐3, galectin‐1, galectin‐3BP, BSA, and GPαIIbβ3. The anti‐galectin‐3 and anti‐galectin‐3BP antibodies were detected with an HRP‐conjugated anti‐rabbit antibody. The galectin‐1 and AP2 antibodies were detected with an HRP‐conjugated anti‐mouse antibody. CN was detected with a peroxidase‐conjugated anti‐human FCγ antibody. The error bars reprement the mean values±SD.
To further validate the identified target in human endarterectomy samples, immunofluorescence experiments were performed with P3 scFv‐Fc and a commercial murine antibody against human galectin‐3. Image analysis (Figure 5A through 5D) showed that P3 scFv‐Fc labeled tunica intima areas that were recognized also by the commercial anti‐galectin‐3 antibody as indicated by the enlarged merge image (64.62% of colocalization with the commercial anti‐galectin‐3 antibody) (Figure 5C).
Figure 5. Colocalization of P3 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) with an anti‐galectin‐3 antibody and an anti‐LOX1 (lectin‐type oxidized LDL receptor 1) antibody in the intima of human endarterectomy specimens and coronary sections.
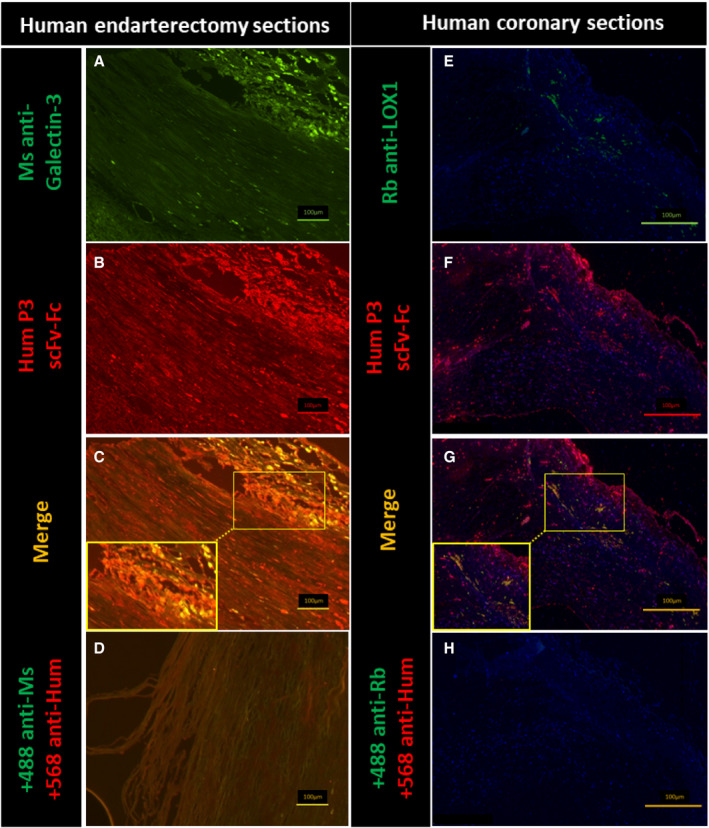
A through D, Co‐staining of P3 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) and an anti‐galectin‐3 antibody in human endarterectomy sections by immunofluorescence. An Alexa Fluor 488 anti‐mouse antibody (+488 anti‐Ms [mouse]) was used to reveal the specific binding of the commercial anti‐human galectin‐3 antibody (Ms anti‐galectin‐3) (A and C). The Alexa Fluor 568 anti‐human antibody (+568 anti‐Hum) was used to reveal the specific binding of P3 scFv‐Fc (Hum P3 scFv‐Fc) (B and C). Before image merging, the red and green fluorescence signals were adjusted to comparable levels. The yellow color indicates colocalization of the antigens recognized by P3 scFv‐Fc and the anti‐human galectin‐3 antibody (C). Secondary antibodies alone were used as negative controls (D). Size bars: 100 µm. E through H, The Alexa Fluor 488 anti‐rabbit antibody (+488 anti‐Rb [rabbit]) was used to reveal the specific binding of the commercial anti‐LOX1 antibody (Rb anti‐LOX1) (E and G). The Alexa Fluor 647 anti‐human antibody (+568 anti‐Hum) was used to reveal the specific binding of P3‐scFv‐Fc (Hum P3 scFv‐Fc) (F and G). Before image merging, the red and green fluorescence signals were adjusted to comparable levels. The yellow color indicates colocalization of the antigens labeled by P3 scFV‐Fc and anti‐LOX1 antibody (G). As galectin‐3 is also expressed also by other cell types (ie, mast cells, eosinophils, neutrophils, endothelial cells, and activated T and B cells 8 , 27 ), areas outside the macrophage location also are stained by P3 scFv‐Fc (G). Size bars: 100 μm.
Because galectin‐3 is overexpressed by macrophages, 26 human coronary biopsies were analyzed using P3 scFv‐Fc and a rabbit antibody against LOX1 (the macrophage receptor for oxidized LDL) (Figure 5E through 5H). The enlarged merge image shows the colocation between P3 scFv‐Fc and LOX1, especially in the subendothelial and lipid core areas (79.31%) (Figure 5G). Because galectin‐3 is expressed also by other cell types (eg, mast cells, eosinophils, neutrophils, endothelial cells, and activated T and B cells), 8 , 27 areas outside the macrophage location were also stained by P3 scFv‐Fc (Figure 5F and 5G).
P3 ScFv‐Fc Labels Atheroma Plaques in Apoe−/− Mice
To assess whether P3 scFv‐Fc could be used for atheroma diagnostic imaging, first we costained aorta sections from Apoe−/− mice (a model of atherosclerosis) with P3 scFv‐Fc and the anti‐LOX1 antibody (Figure 6). The colocalization (in yellow) of the P3 scFv‐Fc and anti‐LOX1 antibodies (84.24%) indicated the presence of galectin‐3 in the area of foam cells. Next, the P3 scFv‐Fc binding profile was characterized ex vivo in the aorta of Apoe−/− and wild‐type mice (Figure 7 and Figure S1). For this experiment, the P3 antibody with 2 extra cysteines at the C‐terminus (P3 scFv‐Fc‐2c) was used. This modification is crucial for coupling the antibody to nanoparticles in future magnetic resonance imaging experiments. P3 scFv‐Fc‐2c specificity was assessed and validated by flow cytometry (data not shown). Compared with the signal obtained from control human IgG in Apoe −/− mouse aorta, macroscale fluorescence imaging revealed a significant signal of P3 scFv‐Fc‐2c in the aortic root (83.33%) (yellow arrowheads in Figure 7). Conversely, P3 scFv‐Fc‐2c and the human IgG control did not give any significant signal in the aorta of wild‐type mice.
Figure 6. In vitro immunofluorescence analysis of Apoe−/− aorta sections with P3 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) and an anti‐LOX1 (lectin‐type oxidized LDL receptor 1) antibody.
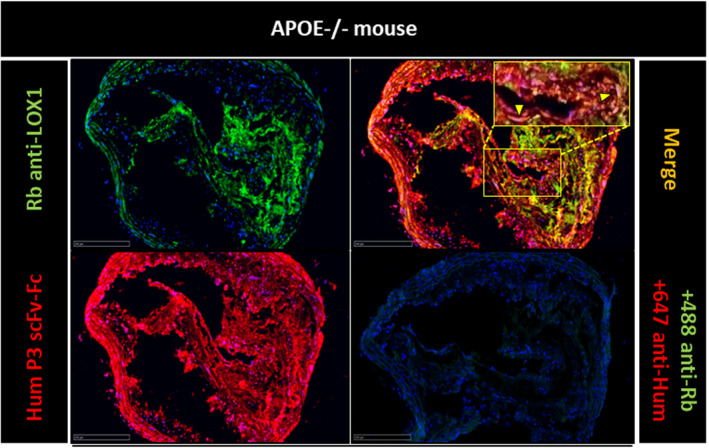
Colocalization analysis by fluorescence microscopy in Apoe−/− mouse aorta sections of the antigens recognized by P3 scFv‐Fc and the commercial anti‐LOX1 antibody. The Alexa Fluor 488 anti‐rabbit antibody (+488 anti‐Rb) was used to reveal the specific binding of the anti‐LOX1 antibody (Rb anti‐LOX1). The Alexa Fluor 647 anti‐human antibody (+647 anti‐Hum) was used to reveal the specific binding of P3 scFv‐Fc (Hum P3 scFv‐Fc). Before image merging, the red and green fluorescence signals were adjusted to comparable levels. The yellow color and yellow arrowheads indicate colocalization of the antigens recognized by P3 scFv‐Fc and the anti‐LOX1 antibody. Size bars: 250 μm.
Figure 7. Ex vivo imaging of P3 scFv‐Fc (single‐chain fragment variable fused to the crystallizable fragment of immunoglobulin G) in Apoe−/− and wild‐type (WT) mice using a fluorescent ultramicroscope.
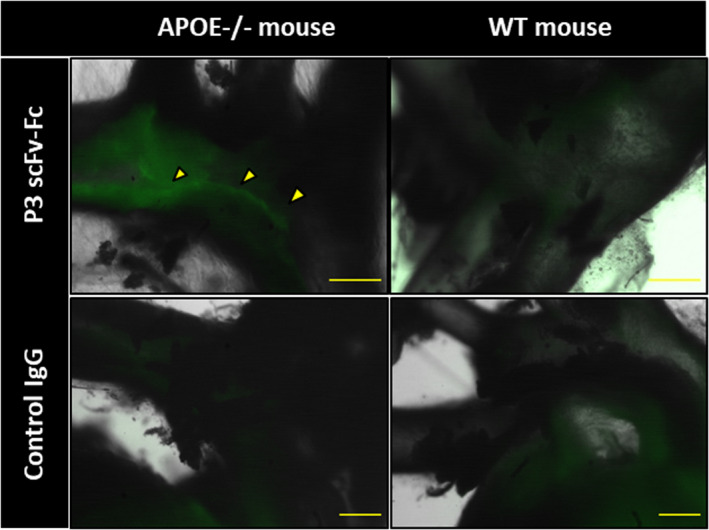
Fluorescence macroscopy analysis of P3 scFv‐Fc coupled to Alexa Fluor 568 dye after ex vivo injection in Apoe−/− and WT mice. A human immunoglobulin (Ig)G coupled to Alexa Fluor 568 was used as negative control antibody. P3 scFv‐Fc shows specific labeling of the atheroma in the Apoe− /− mouse (yellow arrowheads). Size bars: 500 μm.
In addition to this first set of images, the specific binding of P3 scFv‐Fc‐2c on atheromatous plaques was confirmed by the confocal microcopy images shown in Figure S1 (yellow arrowheads, Figure S1B and S1F).
Discussion
Nowadays, there is a rising interest in accessing and characterizing the molecular and cellular components of atheroma plaques to find strategies for reducing the associated risk of stroke and myocardial infarction. It is acknowledged that the plaque composition more than the narrowing of arteries defines the condition of plaque rupture. Imaging technologies are frequently used to determine the molecular composition of atheroma plaques, and specific contrast agents are urgently needed. This specificity can be offered by human antibodies that can target atherosclerosis biomarkers for diagnostic purposes.
Phage display is an efficient tool for biomarker identification for several tissues and cell types. Keller et al developed the direct cell phage display approach to select human scFv that targets blood and lymphatic cells in cancer. 28 In atherosclerosis, the combination of this method and fresh human tissues allowed the selection of peptides against CD100. 29
In 1998, Arap et al set up the first in vivo phage display method to map endothelial cells in cancer. 30 Later on, Arap et al, 31 Staquicini et al, 32 and Krag et al 33 initiated a human vascular mapping project and identified peptides that recognize tumor cells in patients with cancer. In atherosclerosis, in vivo phage selection in animal models led to the discovery of peptides 34 and antibodies 35 , 36 that bind to a large panel of targets in their microenvironment, thus allowing studying the atheroma plaque composition. One important limitation of in vivo phage display using antibody fragments is the identification of the target among all of the overexpressed proteins extracted from tissues. Unlike peptides that can be compared with protein databases, 32 in vivo antibody discovery requires more complex methodologies for target identification to better understand the pathogenesis and for biotherapeutic innovation. However, the myriad of unknown targets, which is a drawback of in vivo phage display using antibodies, also represents a large panel of antibody targets in their microenvironment. Here, immunoprecipitation and mass spectrometry analyses were used to show that galectin‐3, a protein of 25 to 30 kDa, is the target of P3 scFv‐Fc in proteins extracted from human endarterectomy samples.
Galectin‐3 is a member of a family of 15 β‐galactoside‐binding proteins, named galectins, characterized by the presence of conserved carbohydrate recognition domains. Galectin‐3 is the only chimera‐type galectin in vertebrates, with a single carbohydrate recognition domain and a nonlectin N‐terminal domain. Galectins are synthesized in the cytoplasm and secreted through a nonclassical exocytic pathway 37 to interact with cell surface glycans. They regulate the immune system by modulating monocyte/macrophage functions. 38 Galectin‐3 is implicated in different biological processes, including cell activation, anti‐apoptotic activity, cytokine secretion, and cell migration. 22 , 39 , 40 , 41 , 42 Galectin‐3 also strongly induces P‐selectin, which interacts with PSGL1 (P‐selectin glycoprotein ligand 1) on leukocyte to form platelet‐leukocyte aggregates. 43 The interaction between platelets and leukocytes promotes their activation, which is crucial for triggering inflammation, vascular remodeling, and thrombosis. Galectin‐3 implication in different processes in cancer 44 , 45 , 46 , 47 and inflammation led to the development of molecules to block the underlying mechanisms. 48
Currently, much interest is focused on the role of galectin‐3 in cardiovascular diseases, particularly in atherosclerosis. 20 , 24 , 42 , 49 , 50 , 51 Galectin‐3 protein was first detected in carotid samples from endarterectomies. 26 Galectin‐3 has been considered as an inflammation amplifier during atherosclerotic plaque progression 10 because of its close interrelation with macrophages and foam cells. Zhu et al showed that 125I‐oxidized LDL and 125I‐acetylated LDL are actively endocytosed by galectin‐3‐expressing chinese hamster ovary (CHO) cells. Moreover, incubation with acetylated‐LDL led to intracellular accumulation of cholesteryl esters, highlighting galectin‐3 role in endocytosis of advanced glycation end‐proteins and modified LDLs. 52 More recently, Madrigal‐Matute et al reported that galectin‐3 can modulate oxidative stress by stimulating superoxide production in monocytes and regulates the adhesion of monocytes/macrophages to endothelial cells. 50 Because of its implication in various diseases, galectin‐3 therapeutic potential has been evaluated using inhibitors and animal knock‐out models. 53 In atherosclerosis, inactivation of Lgals3 (the gene encoding galectin‐3) in Apoe−/− mice reduces the atheroma thickness. 54 , 55 Moreover, in patients on chronic statin treatment, galectin‐3 level was elevated, and the macrophage number was reduced within plaques, suggesting that this protein is a biomarker of plaque inflammation severity. 56 Dysfunction of the endothelial barrier is the starting point of atherosclerosis; however, the exact role of galectin‐3 in endothelial cells is unclear. A recent study showed that the interaction between galectin‐3 and integrin β1 promotes different inflammatory factors, leading to endothelial cell stress and apoptosis. 57 Galectin‐3 might have both pro‐ and antiatherosclerotic roles. For instance, increased accumulation of galectin‐3–negative macrophages has been observed in advanced human, rabbit, and mouse plaques compared with early lesions. 58 Although many studies reported high galectin‐3 expression in plaque macrophages, the functional heterogeneity of the macrophage population needs to be taken into account when assessing galectin‐3 expression. Single‐cell technologies, such as single‐cell RNA sequencing and cytometry by time‐of‐flight, allowed identifying different macrophage clusters with different gene expression profiles and phenotypes. Besides the resident‐like and proinflammatory subsets, the newly described anti‐inflammatory TREM2hi macrophages display enrichment in lipid metabolism, cholesterol efflux, oxidative phosphorylation, and catabolism, and also strongly express galectin‐3. 59 , 60 , 61 The TREM2hi subset of macrophages has been linked to lipid uptake and foam cell formation, 62 which are mainly associated with plaque progression. However, in agreement with the recent publication by Di Gregoli et al 58 emphasizing that galectin‐3 identifies a subset of macrophages with a beneficial role in plaque regression, the gene expression profile of foamy macrophages was also associated with plaque‐resolving parameters, such as efferocytosis and tissue repair. 62
Moreover, during atherosclerosis progression, foamy macrophage apoptosis may contribute to disease worsening. Although in the present study we did not compare plaque composition and phenotype in early and advanced atherosclerotic lesions in animal models and in human samples, we did observe significant differences in galectin‐3 expression in human samples (data not shown). Additional analyses of macrophages from plaques at different disease stages would be useful to characterize galectin‐3 expression level in nonfoamy, foamy, and apoptotic macrophages and to determine whether our anti‐galectin‐3 antibody P3 scFv‐Fc could be used as a marker of plaque stability or progression.
All of these studies indicate that galectin‐3 is an ideal target for imaging modalities and for developing biotherapeutics to regulate its role in atherosclerosis by directly targeting this protein or one of its ligands (eg, galectin‐3 binding protein, 63 integrins GPα3β1, 24 and GPαvβ3 25 ), which are all overexpressed in atherosclerosis. Because P3 scFv‐Fc can bind to galectin‐3 ex vivo, future studies should determine whether this antibody can act as an antagonist or agonist on one of the mechanisms implicated in the inflammation process that characterizes atherosclerosis. If necessary, random mutations could be introduced in P3 scFv‐Fc using human polymerases 5 , 64 to increase its affinity for galectin‐3. Moreover, our recent study performed by third‐generation sequencing of scFv clones issued from the in vivo selection 7 highlighted sequences related to the P3 scFv clone with point mutations.
In conclusion, our study shows that the combination of in vivo antibody selection and in vitro characterization of the target by considering the pathological microenvironment is a good starting point for developing diagnostic and biotherapeutic molecules that can easily be transferred to the clinic.
Sources of Funding
This work was supported by grant ANR‐13‐BSV5‐0018 from the French National Research Agency Program named ATHERANOS and a public grant from the French National Research Agency in the context of the Investments for the Future Program, reference ANR‐10‐LABX‐57 named TRAIL and ANR‐10‐LABX‐53 named MabImprove.
Disclosures
None.
Supporting information
Table S1
Figure S1
Acknowledgments
The microscopy work was done at Bordeaux Imaging Center, a service unit of CNRS‐INSERM and Bordeaux University, a member of the national France BioImaging infrastructure supported by the French National Research Agency (ANR‐10‐INBS‐04). The help of C. Poujol and S. Marais is acknowledged. The statistical analysis was done by D.‐A. Tregouet, Centre Bordeaux Population Research, Inserm U1219.
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.120.016287
For Sources of Funding and Disclosures, see page 17.
Contributor Information
Audrey Hemadou, Email: audrey.hemadou@orange.fr.
Marie‐Josée Jacobin‐Valat, Email: marie-josee.jacobin-valat@rmsb.u-bordeaux.fr.
References
- 1. Hansson GK, Libby P. The immune response in atherosclerosis: a double‐edged sword. Nat Rev Immunol. 2006;6:508–519. [DOI] [PubMed] [Google Scholar]
- 2. Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2000;20:1262–1275. DOI: 10.1161/01.ATV.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 3. Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HF, Freeman MW. Scavenger receptors class A‐I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277:49982–49988. DOI: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- 4. Lariviere M, Lorenzato CS, Adumeau L, Bonnet S, Hemadou A, Jacobin‐Valat MJ, Noubhani A, Santarelli X, Minder L, Di Primo C, et al. Multimodal molecular imaging of atherosclerosis: nanoparticles functionalized with scFv fragments of an anti‐alphaIIbbeta3 antibody. Nanomed Nanotechnol Biol Med. 2019;22: 102082. [DOI] [PubMed] [Google Scholar]
- 5. Mondon P, Souyris N, Douchy L, Crozet F, Bouayadi K, Kharrat H. Method for generation of human hyperdiversified antibody fragment library. Biotechnol J. 2007;2:76–82. DOI: 10.1002/biot.200600205. [DOI] [PubMed] [Google Scholar]
- 6. Hemadou A, Laroche‐Traineau J, Antoine S, Mondon P, Fontayne A, Le Priol Y, Claverol S, Sanchez S, Cerutti M, Ottones F, et al. An innovative flow cytometry method to screen human scFv‐phages selected by in vivo phage‐display in an animal model of atherosclerosis. Sci Rep. 2018;8:15016. DOI: 10.1038/s41598-018-33382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hemadou A, Giudicelli V, Smith ML, Lefranc MP, Duroux P, Kossida S, Heiner C, Hepler NL, Kuijpers J, Groppi A, et al. Pacific biosciences sequencing and IMGT/HighV‐QUEST analysis of full‐length single chain fragment variable from an in vivo selected phage‐display combinatorial library. Front Immunol. 2017;8:1796. DOI: 10.3389/fimmu.2017.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sciacchitano S, Lavra L, Morgante A, Ulivieri A, Magi F, De Francesco GP, Bellotti C, Salehi LB, Ricci A. Galectin‐3: one molecule for an alphabet of diseases, from a to z. Int J Mol Sci. 2018;19. DOI: 10.3390/ijms19020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papaspyridonos M, McNeill E, de Bono JP , Smith A, Burnand KG, Channon KM, Greaves DR. Galectin‐3 is an amplifier of inflammation in atherosclerotic plaque progression through macrophage activation and monocyte chemoattraction. Arterioscler Thromb Vasc Biol. 2008;28:433–440. DOI: 10.1161/ATVBAHA.107.159160. [DOI] [PubMed] [Google Scholar]
- 10. Novak R, Dabelic S, Dumic J. Galectin‐1 and galectin‐3 expression profiles in classically and alternatively activated human macrophages. Biochim Biophys Acta. 2012;1820:1383–1390. DOI: 10.1016/j.bbagen.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 11. Sano H, Hsu DK, Yu L, Apgar JR, Kuwabara I, Yamanaka T, Hirashima M, Liu FT. Human galectin‐3 is a novel chemoattractant for monocytes and macrophages. J Immunol. 2000;165:2156–2164. DOI: 10.4049/jimmunol.165.4.2156. [DOI] [PubMed] [Google Scholar]
- 12. Deramchia K, Jacobin‐Valat MJ, Laroche‐Traineau J, Bonetto S, Sanchez S, Dos Santos P, Massot P, Franconi JM, Martineau P, Clofent‐Sanchez G. By‐passing large screening experiments using sequencing as a tool to identify scFv fragments targeting atherosclerotic lesions in a novel in vivo phage display selection. Int J Mol Sci. 2012;13:6902–6923. DOI: 10.3390/ijms13066902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Renaut L, Monnet C, Dubreuil O, Zaki O, Crozet F, Bouayadi K, Kharrat H, Mondon P. Affinity maturation of antibodies: optimized methods to generate high‐quality ScFv libraries and isolate IgG candidates by high‐throughput screening. Methods Mol Biol (Clifton. NJ). 2012;907:451–461. [DOI] [PubMed] [Google Scholar]
- 14. Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT((r)) tools for the nucleotide analysis of immunoglobulin (IG) and t cell receptor (TR) V‐(D)‐J repertoires, polymorphisms, and IG mutations: IMGT/V‐QUEST and IMGT/HighV‐QUEST for NGS. Methods Mol Biol. 2012;882:569–604. [DOI] [PubMed] [Google Scholar]
- 15. Juliant S, Leveque M, Cerutti P, Ozil A, Choblet S, Violet ML, Slomianny MC, Harduin‐Lepers A, Cerutti M. Engineering the baculovirus genome to produce galactosylated antibodies in lepidopteran cells. Methods Mol Biol. 2013;988:59–77. [DOI] [PubMed] [Google Scholar]
- 16. Fessart D, Martin‐Negrier ML, Claverol S, Thiolat ML, Crevel H, Toussaint C, Bonneu M, Muller B, Savineau JP, Delom F. Proteomic remodeling of proteasome in right heart failure. J Mol Cell Cardiol. 2014;66:41–52. DOI: 10.1016/j.yjmcc.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 17. Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi‐supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. DOI: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 18. Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8:3737–3745. DOI: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9:676–682. DOI: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aksan G, Gedikli O, Keskin K, Nar G, Inci S, Yildiz SS, Kaplan O, Soylu K, Kilickesmez KO, Sahin M. Is galectin‐3 a biomarker, a player‐or both‐in the presence of coronary atherosclerosis? J Investig Med. 2016;64:764–770. DOI: 10.1136/jim-2015-000041. [DOI] [PubMed] [Google Scholar]
- 21. Falcone C, Lucibello S, Mazzucchelli I, Bozzini S, D'Angelo A, Schirinzi S, Totaro R, Falcone R, Bondesan M, Pelissero G. Galectin‐3 plasma levels and coronary artery disease: a new possible biomarker of acute coronary syndrome. Int J Immunopathol Pharmacol. 2011;24:905–913. DOI: 10.1177/039463201102400409. [DOI] [PubMed] [Google Scholar]
- 22. Suthahar N, Meijers WC, Sillje HHW, Ho JE, Liu FT, de Boer RA . Galectin‐3 activation and inhibition in heart failure and cardiovascular disease: an update. Theranostics. 2018;8:593–609. DOI: 10.7150/thno.22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeRoo EP, Wrobleski SK, Shea EM, Al‐Khalil RK, Hawley AE, Henke PK, Myers DD Jr, Wakefield TW, Diaz JA. The role of galectin‐3 and galectin‐3‐binding protein in venous thrombosis. Blood. 2015;125:1813–1821. DOI: 10.1182/blood-2014-04-569939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van der Hoeven NW , Hollander MR, Yildirim C, Jansen MF, Teunissen PF, Horrevoets AJ, van der Pouw Kraan TC , van Royen N . The emerging role of galectins in cardiovascular disease. Vascul Pharmacol. 2016;81:31–41. DOI: 10.1016/j.vph.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 25. Markowska AI, Liu FT, Panjwani N. Galectin‐3 is an important mediator of VEGF‐ and bFGF‐mediated angiogenic response. J Exp Med. 2010;207:1981–1993. DOI: 10.1084/jem.20090121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nachtigal M, Al‐Assaad Z, Mayer EP, Kim K, Monsigny M. Galectin‐3 expression in human atherosclerotic lesions. Am J Pathol. 1998;152:1199–1208. [PMC free article] [PubMed] [Google Scholar]
- 27. Liu FT. Regulatory roles of galectins in the immune response. Int Arch Allergy Immunol. 2005;136:385–400. DOI: 10.1159/000084545. [DOI] [PubMed] [Google Scholar]
- 28. Keller T, Kalt R, Raab I, Schachner H, Mayrhofer C, Kerjaschki D, Hantusch B. Selection of scFv antibody fragments binding to human blood versus lymphatic endothelial surface antigens by direct cell phage display. PLoS One. 2015;10:e0127169. DOI: 10.1371/journal.pone.0127169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luque MC, Gutierrez PS, Debbas V, Martins WK, Puech‐Leao P, Porto G, Coelho V, Boumsell L, Kalil J, Stolf B. Phage display identification of CD100 in human atherosclerotic plaque macrophages and foam cells. PLoS One. 2013;8:e75772. DOI: 10.1371/journal.pone.0075772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science (New York, N.Y.). 1998;279:377–380. [DOI] [PubMed] [Google Scholar]
- 31. Arap W, Pasqualini R. The human vascular mapping project. Selection and utilization of molecules for tumor endothelial targeting. Haemostasis. 2001;31:30–31. [PubMed] [Google Scholar]
- 32. Staquicini FI, Cardo‐Vila M, Kolonin MG, Trepel M, Edwards JK, Nunes DN, Sergeeva A, Efstathiou E, Sun J, Almeida NF, et al. Vascular ligand‐receptor mapping by direct combinatorial selection in cancer patients. Proc Natl Acad Sci USA. 2011;108:18637–18642. DOI: 10.1073/pnas.1114503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette‐Radoux S, Thomas C. Selection of tumor‐binding ligands in cancer patients with phage display libraries. Cancer Res. 2006;66:7724–7733. DOI: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 34. Chung J, Shim H, Kim K, Lee D, Kim WJ, Kang DH, Kang SW, Jo H, Kwon K. Discovery of novel peptides targeting pro‐atherogenic endothelium in disturbed flow regions ‐Targeted siRNA delivery to pro‐atherogenic endothelium in vivo. Sci Rep. 2016;6:25636. DOI: 10.1038/srep25636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robert R, Jacobin‐Valat MJ, Daret D, Miraux S, Nurden AT, Franconi JM, Clofent‐Sanchez G. Identification of human scFvs targeting atherosclerotic lesions: selection by single round in vivo phage display. J Biol Chem. 2006;281:40135–40143. DOI: 10.1074/jbc.M609344200. [DOI] [PubMed] [Google Scholar]
- 36. Deramchia K, Jacobin‐Valat MJ, Vallet A, Bazin H, Santarelli X, Sanchez S, Dos Santos P, Franconi JM, Claverol S, Bonetto S, et al. In vivo phage display to identify new human antibody fragments homing to atherosclerotic endothelial and subendothelial tissues. Am J Pathol. 2012;180:2576–2589. DOI: 10.1016/j.ajpath.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 37. Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem. 2003;270:2109–2119. DOI: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 38. Paclik D, Werner L, Guckelberger O, Wiedenmann B, Sturm A. Galectins distinctively regulate central monocyte and macrophage function. Cell Immunol. 2011;271:97–103. DOI: 10.1016/j.cellimm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 39. Wang L, Guo XL. Molecular regulation of galectin‐3 expression and therapeutic implication in cancer progression. Biomed Pharmacother. 2016;78:165–171. DOI: 10.1016/j.biopha.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 40. Menini S, Iacobini C, Blasetti Fantauzzi C, Pesce CM, Pugliese G. Role of galectin‐3 in obesity and impaired glucose homeostasis. Oxid Med Cell Longev. 2016;2016:9618092. DOI: 10.1155/2016/9618092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Venkatraman A, Hardas S, Patel N, Singh Bajaj N, Arora G, Arora P. Galectin‐3: an emerging biomarker in stroke and cerebrovascular diseases. Eur J Neurol. 2018;25:238–246. DOI: 10.1111/ene.13496. [DOI] [PubMed] [Google Scholar]
- 42. Fort‐Gallifa I, Hernandez‐Aguilera A, Garcia‐Heredia A, Cabre N, Luciano‐Mateo F, Simo JM, Martin‐Paredero V, Camps J, Joven J. Galectin‐3 in peripheral artery disease. Relationships with markers of oxidative stress and inflammation. Int J Mol Sci. 2017;18:973. DOI: 10.3390/ijms18050973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schattner M. Platelets and galectins. Ann Transl Med. 2014;2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ahmed H, AlSadek DM. Galectin‐3 as a potential target to prevent cancer metastasis. Clin Med Insights Oncol. 2015;9:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Song L, Tang JW, Owusu L, Sun MZ, Wu J, Zhang J. Galectin‐3 in cancer. Clinica Chimica Acta. 2014;431:185–191. DOI: 10.1016/j.cca.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 46. Eliaz I. The role of galectin‐3 as a marker of cancer and inflammation in a stage IV ovarian cancer patient with underlying pro‐inflammatory comorbidities. Case reports in oncology. 2013;6:343–349. DOI: 10.1159/000353574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruvolo PP. Galectin 3 as a guardian of the tumor microenvironment. Biochim Biophys Acta. 1863;2016:427–437. DOI: 10.1016/j.bbamcr.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 48. Demotte N, Wieers G, Van Der Smissen P, Moser M, Schmidt C, Thielemans K, Squifflet JL, Weynand B, Carrasco J, Lurquin C, et al. A galectin‐3 ligand corrects the impaired function of human CD4 and CD8 tumor‐infiltrating lymphocytes and favors tumor rejection in mice. Can Res. 2010;70:7476–7488. DOI: 10.1158/0008-5472.CAN-10-0761. [DOI] [PubMed] [Google Scholar]
- 49. Hogas S, Bilha SC, Branisteanu D, Hogas M, Gaipov A, Kanbay M, Covic A. Potential novel biomarkers of cardiovascular dysfunction and disease: cardiotrophin‐1, adipokines and galectin‐3. Arch Med Sci. 2017;13:897–913. DOI: 10.5114/aoms.2016.58664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Madrigal‐Matute J, Lindholt JS, Fernandez‐Garcia CE, Benito‐Martin A, Burillo E, Zalba G, Beloqui O, Llamas‐Granda P, Ortiz A, Egido J, et al. Galectin‐3, a biomarker linking oxidative stress and inflammation with the clinical outcomes of patients with atherothrombosis. J Am Heart Assoc. 2014;3. DOI: 10.1161/JAHA.114.000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He XW, Li WL, Li C, Liu P, Shen YG, Zhu M, Jin XP. Serum levels of galectin‐1, galectin‐3, and galectin‐9 are associated with large artery atherosclerotic stroke. Sci Rep. 2017;7:40994. DOI: 10.1038/srep40994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu W, Sano H, Nagai R, Fukuhara K, Miyazaki A, Horiuchi S. The role of galectin‐3 in endocytosis of advanced glycation end products and modified low density lipoproteins. Biochem Biophys Res Commun. 2001;280:1183–1188. DOI: 10.1006/bbrc.2001.4256. [DOI] [PubMed] [Google Scholar]
- 53. Dong R, Zhang M, Hu Q, Zheng S, Soh A, Zheng Y, Yuan H. Galectin‐3 as a novel biomarker for disease diagnosis and a target for therapy (review). Int J Mol Med. 2018;41:599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. MacKinnon AC, Liu X, Hadoke PW, Miller MR, Newby DE, Sethi T. Inhibition of galectin‐3 reduces atherosclerosis in apolipoprotein E‐deficient mice. Glycobiology. 2013;23:654–663. DOI: 10.1093/glycob/cwt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nachtigal M, Ghaffar A, Mayer EP. Galectin‐3 gene inactivation reduces atherosclerotic lesions and adventitial inflammation in ApoE‐deficient mice. Am J Pathol. 2008;172:247–255. DOI: 10.2353/ajpath.2008.070348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kadoglou NP, Sfyroeras GS, Spathis A, Gkekas C, Gastounioti A, Mantas G, Nikita KS, Karakitsos P, Liapis CD. Galectin‐3, carotid plaque vulnerability, and potential effects of statin therapy. Eur J Vasc Endovasc Surg. 2015;49:4–9. DOI: 10.1016/j.ejvs.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 57. Chen X, Lin J, Hu T, Ren Z, Li L, Hameed I, Zhang X, Men C, Guo Y, Xu D, et al. Galectin‐3 exacerbates ox‐LDL‐mediated endothelial injury by inducing inflammation via integrin beta1‐RhoA‐JNK signaling activation. J Cell Physiol. 2019;234:10990–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Di Gregoli K, Somerville M, Bianco R, Thomas AC, Frankow A, Newby AC, George SJ, Jackson CL, Johnson JL. Galectin‐3 identifies a subset of macrophages with a potential beneficial role in atherosclerosis. Arterioscler Thromb Vasc Biol. 2020;40:1491–1509. DOI: 10.1161/ATVBAHA.120.314252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Willemsen L, de Winther MP . Macrophage subsets in atherosclerosis as defined by single‐cell technologies. J Pathol. 2020;250:705–714. DOI: 10.1002/path.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lin JD, Nishi H, Poles J, Niu X, McCauley C, Rahman K, Brown EJ, Yeung ST, Vozhilla N, Weinstock A, et al. Single‐cell analysis of fate‐mapped macrophages reveals heterogeneity, including stem‐like properties, during atherosclerosis progression and regression. JCI insight. 2019;4. DOI: 10.1172/jci.insight.124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A. Single‐cell RNA‐Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–1674. DOI: 10.1161/CIRCRESAHA.117.312509. [DOI] [PubMed] [Google Scholar]
- 62. Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim KW, Jang MY, Seok Jang H, Yun TJ, Lee SH, et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ Res. 2018;123:1127–1142. DOI: 10.1161/CIRCRESAHA.118.312804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Inohara H, Akahani S, Koths K, Raz A. Interactions between galectin‐3 and Mac‐2‐binding protein mediate cell‐cell adhesion. Cancer Res. 1996;56:4530–4534. [PubMed] [Google Scholar]
- 64. Mondon P, Grand D, Souyris N, Emond S, Bouayadi K, Mutagen KH. A random mutagenesis method providing a complementary diversity generated by human error‐prone DNA polymerases. Methods Mol Biol (Clifton. NJ). 2010;634:373–386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figure S1


