Abstract
In 1997, a cluster of multiresistant invasive serogroup 19 pneumococcus infections, including two fatalities, was reported in Washington State. Further investigation identified other cases. Fourteen Washington Streptococcus pneumoniae isolates, four from Alaska, and eight isolates from eastern Canada with reduced penicillin susceptibility (MIC of ≥1 μg/ml) were included in the study. Pulsed-field gel electrophoresis (PFGE) with ApaI, SacII, and SmaI restriction enzymes and IS1167 and mef restriction fragment length polymorphism (RFLP) pattern analysis were performed. Twenty of the 26 isolates had identical or related PFGE patterns, with two or all three enzymes, and identical or related IS1167 RFLP patterns, indicating that they were genetically related. These 20 isolates contained the mef gene conferring erythromycin resistance and had identical mef RFLP patterns. The PFGE and RFLP patterns were distinct from those of six multiresistant clones previously described and suggest that a new multiresistant clone has appeared in Washington, Alaska, and eastern Canada. This newly characterized clone should be included in the Pneumococcal Molecular Epidemiology Network.
Streptococcus pneumoniae is the leading bacterial cause of community-acquired pneumonia, otitis media, bacteremia, and meningitis in the United States (14). In the past 20 years, a worldwide increase in the incidence of antibiotic-resistant S. pneumoniae has been observed (3, 13, 30). Although more than 90 serotypes of S. pneumoniae exist, resistance to two or more different classes of antibiotics (i.e., multiresistance) is currently limited to a few major serotypes (6B, 9V, 14, 19F, and 23F) (11, 12, 30). The first non-penicillin-susceptible multiresistant S. pneumoniae strain, described in the 1970s, contained a conjugative transposon, Tn1545, which carried four resistance genes: erm(B) (macrolides, lincosamides, and streptogramin B), tet(M) (tetracycline), aphA-3 (aminoglycosides), and cat (chloramphenicol). This family of transposons has since disseminated through the pneumococcal population (5, 8). Since the first description of an S. pneumoniae clone, Spain-23F-1, other clones have been identified (18, 30). A Pneumococcal Molecular Epidemiology Network has been newly established to collect, study, and assign systematic number designations to S. pneumoniae clones that meet the Network's criteria (K. Klugman, Letter, ASM News 64:371, 1998).
In February 1997, the Washington State Health Department was notified of three cases of pneumonia due to ceftriaxone-resistant S. pneumoniae, two of which were fatal. Further investigation found that all three invasive isolates were from patients in the same community. The isolates were serogroup 19, were nonsusceptible to penicillin, and were resistant to ceftriaxone, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole. Although serogroup 19 represents approximately 10% of pneumococci tested in Washington, ceftriaxone-resistant S. pneumoniae had only rarely been identified in Washington (2, 10; Centers for Disease Control and Prevention [CDC], unpublished data). To determine the magnitude of the problem and to further characterize the isolates, we selected 14 serogroup 19 multiresistant S. pneumoniae isolates collected from the hospitals in the community where the original cluster occurred and from other hospitals throughout Washington State. We also included four randomly chosen Alaskan isolates from a pool of multiresistant serotype 19F isolates. We also chose eight Canadian isolates, serogroup 19, which had antibiograms, including resistance to erythromycin, cephalosporins, and penicillin, similar to those of the Washington isolates for comparison. These 26 isolates were compared with previously characterized multiresistant S. pneumoniae clones using pulsed-field gel electrophoresis (PFGE) and insertion sequence (IS) restriction fragment length polymorphism (RFLP) pattern typing.
(This study was presented in part as abstract 10638 at the Eighth International Congress of Infectious Diseases in Boston, Mass., May 1998, where it won the North American Pasteur-Merieux Connaught Award in Epidemiology.)
MATERIALS AND METHODS
Bacteria.
We examined 26 S. pneumoniae serogroup 19 isolates with diminished susceptibility to penicillin (MIC of >1 μg/ml) and resistance to at least three other antibiotics (Table 1). Seven isolates, including the initial outbreak cluster, were from three hospitals around Tacoma, Wash., and were collected during February and March 1997 (WA1 to WA7). Seven isolates were from other hospitals in Washington and were collected between December 1995 and April 1996 in a prior survey (WA8 to WA14). The Washington isolates were from adults; most were from hospitals in the Puget Sound region, which includes Tacoma and represents the major portion of the state's population. The Arctic Investigations Program (AIP), National Center for Infectious Disease, CDC, provided four serogroup 19 isolates from adults treated at hospitals in Alaska (AK15 to AK18). The Alaskan isolates were randomly chosen from a pool of multiresistant serotype 19F isolates. Eight isolates (CN19 to CN26) from children in metropolitan Toronto and the neighboring urban Peel region of Canada were chosen because they were serogroup 19 and had antibiograms similar to those of the Washington isolates, being resistant to erythromycin, cephalosporins, and penicillin. Some clinical and antimicrobial susceptibility data from the Canadian isolates have been previously reported (15). The isolates outside of Washington allowed us to determine if genetically related S. pneumoniae strains were present in regions outside Washington State. In addition, six isolates representing six known, characterized clones (267-Spain-23F-1, 681-Spain-6B-2, 665-France-9V-3, 17219-South Africa-19A-7, 50803-South Africa-6B-8, and 51702-South Africa-19A-unnumbered) were provided by the Pneumococcal Diseases Research Unit at the South African Institute for Medical Research, University of the Witwatersrand, Johannesburg, South Africa (SP27 to SP32).
TABLE 1.
Characteristics of S. pneumoniae isolates
| Isolatea | Date isolated (mo/yr)b | Specimen source | Serotype |
|---|---|---|---|
| Related isolates | |||
| WA1 | 2/97 | Blood | 19F |
| WA2 | 2/97 | Blood | 19F |
| WA3 | 2/97 | Blood | 19F |
| WA4 | 3/97 | Nasopharynx | 19F |
| WA5 | 3/97 | Sinus | 19F |
| WA7 | 3/97 | Sputum | 19 |
| WA8 | 12/95 | Sputum | 19F |
| WA9 | 12/95 | Sputum | 19A |
| WA10 | 12/95 | Blood | 19F |
| WA11 | 1/96 | Sinus | 19F |
| WA12 | 4/96 | Wound | 19F |
| WA13 | 6/96 | Blood | 19F |
| WA14 | 6/96 | Unknown | 19F |
| AK15 | 4/94 | Nasopharynx | 19F |
| AK16 | 8/94 | Blood | 19F |
| AK17 | 2/96 | Blood | 19F |
| AK18 | ?/97 | Blood | 19F |
| CN19 | 4/95 | Nasopharynx | 19F |
| CN20 | 4/95 | Nasopharynx | 19F |
| CN24 | 1/96 | Nasopharynx | 19F |
| Unrelated isolates | |||
| WA6 | 2/97 | Sputum | 19 |
| CN21 | 5/95 | Nasopharynx | 19F |
| CN22 | 6/95 | Nasopharynx | 19F |
| CN23 | 11/95 | Nasopharynx | 19F |
| CN25 | 1/95 | Blood | 19F |
| CN26 | 4/95 | Blood | 19F |
| Multiresistant clones | |||
| SP27 (267-Spain-23F-1) | 23F | ||
| SP28 (681-Spain-6B-2) | 6B | ||
| SP29 (665-France-9V-3) | 9V | ||
| SP30 (17219-South Africa-19A-7) | 19A | ||
| SP31 (50803-South Africa-6B-8) | 6B | ||
| SP32 (51702-South Africa-19A-unnumbered) | 19A |
AK, Alaska; CN, Canada; WA, Washington State.
?, month unknown.
Serology.
Serogroups were determined by the Quellung reaction (25) by our laboratory and confirmed by the University of Washington Medical Center and/or the AIP. Subtyping was performed by counterimmunodiffusion electrophoresis at the AIP.
Antibiograms.
The MIC was determined using agar dilution following the National Committee for Clinical Laboratory Standards guidelines (9, 19). The Washington and Canadian isolates were tested with the following antibiotics: penicillin, cefotaxime, ceftriaxone, cefprozil, ceftazidime, loracarbef, erythromycin, azithromycin, clindamycin, ciprofloxacin, grepafloxacin, levofloxacin, sparfloxacin, trovafloxacin, linezolid, and HMR3647 (a new ketolide) (Table 2). Isolates from Alaska were not included in the full antibiogram determination.
TABLE 2.
Antibiogram of Washington and Canadian S. pneumoniae isolates
| Isolate | MIC (μg/ml) of druga:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pen | Ceftri | Ctax | Cefpro | Ceftaz | Lora | Eryth | Azith | Clin | Cipro | Grepa | Levo | Spar | Trova | Linez | HMR | |
| Related isolates | ||||||||||||||||
| WA1 | 2 | 2 | 2 | 16 | 16 | 128 | 4 | 2 | 0.03 | 16 | 0.25 | 1 | 0.5 | 0.125 | 1 | 0.002 |
| WA2 | 2 | 1 | 2 | 16 | 16 | 64 | 4 | 4 | 0.016 | 16 | 0.25 | 1 | 0.5 | 0.06 | 1 | 0.002 |
| WA3 | 2 | 2 | 2 | 16 | 16 | 128 | 8 | 2 | 0.03 | 16 | 0.25 | 1 | 0.5 | 0.06 | 1 | 0.016 |
| WA4 | 2 | 1 | 2 | 16 | 16 | 128 | 4 | 2 | 0.03 | 16 | 0.25 | 1 | 0.5 | 0.125 | 1 | 0.002 |
| WA5 | 2 | 1 | 2 | 16 | 16 | 64 | 4 | 2 | 0.03 | 16 | 0.25 | 1 | 0.25 | 0.125 | 1 | 0.002 |
| WA7 | 2 | 1 | 2 | 16 | 16 | 64 | 4 | 4 | 0.125 | 16 | 0.25 | 1 | 1 | 0.25 | 1 | 0.004 |
| WA8 | 1 | 1 | 1 | 16 | 32 | 64 | 4 | 2 | 0.03 | 8 | 0.25 | 1 | 0.5 | 0.125 | 1 | 0.002 |
| WA9 | 1 | 1 | 1 | 16 | 16 | 64 | 2b | 4b | 0.06b | 8 | 0.25 | 1 | 1 | 0.125 | 1 | 0.002 |
| WA10 | 2 | 1 | 1 | 16 | 16 | 64 | 4 | 4 | 0.03 | 8 | 0.125 | 1 | 0.5 | 0.25 | 0.25 | 0.016 |
| WA11 | 1 | 1 | 1 | 16 | 16 | 128 | 4 | 4 | 0.03 | 16 | 0.008 | 1 | 0.5 | 0.25 | 1 | 0.002 |
| WA12 | 1 | 1 | 1 | 16 | 16 | 64 | 2 | 2 | 0.03 | 8 | 0.125 | 1 | 0.5 | 0.25 | 1 | 0.016 |
| WA13 | 1 | 1 | 1 | 8 | 16 | 64 | 2 | 2 | 0.06 | 16 | 0.125 | 0.5 | 0.5 | 0.25 | 1 | 0.016 |
| WA14 | 1 | 0.5 | 1 | 16 | 16 | 64 | 4 | 2 | 0.03 | 8 | 0.125 | 1 | 0.5 | 0.25 | 1 | 0.002 |
| CN19 | 2 | 1 | 1 | 16 | 32 | 128 | 2 | 2 | 0.25 | 1 | 0.25 | 1 | 0.25 | 0.125 | 1 | 0.016 |
| CN20 | 2 | 1 | 1 | 8 | 32 | 64 | 1 | 1 | 0.25 | 0.5 | 0.25 | 0.5 | 0.125 | 0.125 | 1 | 0.016 |
| CN24 | 2 | 0.5 | 0.03 | 8 | 32 | 64 | 1 | 2 | 0.25 | 0.5 | 0.25 | 1 | 0.25 | 0.125 | 1 | 0.032 |
| Unrelated isolates | ||||||||||||||||
| WA6 | 1 | 1 | 1 | 0.25 | 0.25 | 0.5 | 1 | 0.25 | 0.016 | 0.125 | 0.062 | 0.5 | 0.125 | 4 | 0.25 | 0.002 |
| CN21 | 0.25 | 0.125 | 0.06 | 0.5 | 8 | 8 | 0.5 | 0.5 | 0.25 | 1 | 0.25 | 1 | 0.25 | 0.125 | 1 | 0.032 |
| CN22 | 0.5 | 0.25 | 0.12 | 1 | 16 | 128 | 1 | 2 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.125 | 1 | 0.016 |
| CN23 | 2 | 0.5 | 0.03 | 16 | 32 | 64 | 2 | 4 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.125 | 1 | 0.032 |
| CN25 | 0.12 | 0.25 | 0.03 | 0.5 | 16 | 32 | 0.5 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.125 | 1 | 0.032 |
| CN26 | 0.5 | 0.5 | 0.03 | 1 | 16 | 64 | 0.5 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | 0.125 | 0.06 | 1 | 0.016 |
Pen, penicillin; Ceftri, ceftriaxone; Ctax, cefotaxime; Cefpro, cefprozil; Ceftaz, ceftazidime; Lora, loracarbef; Eryth, erythromycin; Azith, azithromycin; Clin, clindamycin; Cipro, ciprofloxacin; Grepa, grepafloxacin; Levo, levofloxacin; Spar, sparfloxacin; Trova, trovafloxacin; Linez, linezolid; HMR, HMR3647. Breakpoints (in micrograms per milliliter) per National Committee for Clinical Laboratory Standards guidelines (19), unless otherwise noted, are as follows. Penicillin: susceptible, ≤0.06; intermediate, ≥0.1; resistant, ≥2; ceftriaxone: susceptible, ≤0.5, intermediate, 1; resistant, ≥2; cefotaxime: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; cefprozil: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; ceftazidime: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; erythromycin: susceptible, ≤0.25; intermediate, 0.5; resistant, ≥1; azithromycin: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; clindamycin: susceptible, ≤0.25; intermediate, 0.5; resistant, ≥1; ciprofloxacin: susceptible, ≤2; intermediate, 4, resistant, ≥8; grepafloxacin: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; levofloxacin: susceptible, ≤2; intermediate, 4; resistant, ≥8; sparfloxacin: susceptible, ≤0.5; intermediate, 1; resistant, ≥2; trovafloxacin: susceptible, ≤1; intermediate, 2; resistant, ≥4. Because loracarbef, linezolid, and HMR3647 do not have established breakpoints, they are not listed. The breakpoints for ceftazidime, ciprofloxacin, sparfloxacin, and trovafloxacin are based upon the breakpoints of similar antibiotics (ceftriaxone and levofloxacin). Antibiograms of the four Alaskan isolates (AK14 to AK18) were determined by alternate methods; however, the isolates were resistant to penicillin, the cephalosporins, the macrolides, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole (data not shown) and carried the mef gene.
Isolate WA9 contained both the mef and ermB genes. MICs were determined after the isolate was exposed to a low level of erythromycin (0.5 μg/ml), with corresponding MICs of erythromycin, azithromycin, and clindamycin being 128 μg/ml (17).
Ceftriaxone was obtained from Difco (Detroit, Mich.). Clindamycin, erythromycin, cefotaxime, and penicillin were purchased from Sigma Chemical Co. (St. Louis, Mo.). The other antibiotics were provided by manufacturers as follows: ciprofloxacin, Bayer Corp., West Haven, Conn.; cefprozil, Bristol-Myers Squibb, Princeton, N.J.; grepafloxacin and ceftazidime, Glaxo-Wellcome Co., Triangle Park, N.C.; levofloxacin and HMR3647, Hoechst Marion Roussel, Paris, France; trovafloxacin and azithromycin, Pfizer, Inc., Groton, Conn.; loracarbef, Eli Lilly and Company, Indianapolis, Ind.; and linezolid, Pharmacia Upjohn, Kalamazoo, Mich. The antibiotic concentrations tested ranged from 0.002 to 16 μg/ml for the quinolones and ketolides and from 0.031 to 128 μg/ml for the other antibiotics. The bacterial isolates were tested against erythromycin, azithromycin, and clindamycin before and after exposure to a low level of erythromycin (0.5 μg/ml) to identify inducibly resistant isolates (17). Two S. pneumoniae strains, ATCC 6305 and ATCC 40619, were used as controls. The MIC breakpoints were available from the National Committee for Clinical Laboratory Standards for all of the antibiotics except loracarbef, grepafloxacin, linezolid, and HMR3647 (19).
Media and growth conditions.
Bacteria were grown on brucella blood agar (Difco) supplemented with 5% sheep red blood cells and incubated for 18 to 24 h with 5% CO2 at 36.5°C. Mueller-Hinton agar (Difco) supplemented with 5% sheep red blood cells and appropriate concentrations of antibiotics was used for the agar dilutions (19). Bacterial stocks were maintained at −70°C in sterile skim milk. Aliquots were subcultured onto appropriate media from the frozen stocks as needed. Purity was maintained by stringent aseptic techniques and confirmed by biochemical methods as described previously (25).
PFGE analysis.
Bacteria were grown for 18 to 24 h on brucella blood agar plates at 36.5°C in CO2, harvested, and made into blocks with 1% low-melting-point agarose (Bio-Rad Laboratories, Richmond, Calif.) in a PFGE mold provided by the manufacturer (Bio-Rad) as previously described (16, 23). The blocks were digested with proteinase K (Sigma) (100 μg/ml), washed, and stored at 4°C as described previously (16, 23). Gel plugs (approximately 3 by 6 mm) were cut from the blocks and digested for 20 to 24 h with 35 U of ApaI (Promega, Madison, Wis.) or SacII (Promega) at 37°C or SmaI (Promega) at 25°C as described previously (16, 23).
The digested gel blocks were embedded in a 1% agarose gel (SeaKem; FMC Corporation, Rockland, Maine) prepared with 0.5× Tris-borate-EDTA (TBE) (pH 8.0) and run using a CHEF-DRII (contour-clamped horizontal electrophoresis) apparatus (Bio-Rad) at 175 V for 20 h for SmaI-digested DNA gel plugs and 22 h for SacII- and ApaI-digested gel plugs. DNA bands were visualized by ethidium bromide and UV light and photographed as previously described (23, 31).
Analysis of the PFGE patterns was performed by visual inspection of the photographs. ApaI and SacII produced PFGE patterns of 10 to 15 DNA bands between 45.5 and 291.5 kb, while SmaI digests produced PFGE patterns with 8 to 12 DNA bands between 45.5 and 291.5 kb. The most common PFGE patterns were designated with the first letter of the enzyme and an assigned number (A37 to A50 for ApaI and S26 to S37 for SmaI). We did not start with A1 because these PFGE patterns have previously been assigned to S. pneumoniae 6B isolates from other parts of the United States (23). To distinguish SacII from SmaI patterns, C was used for the designated name for SacII patterns (C8 to C21). Isolates with DNA patterns that differed by three or fewer bands from the main pattern were considered to be related and given a subscripted number starting with 1 (A371, A382, etc.), as the band difference could be explained by one genetic event (23, 31). Isolates which had a difference of more than three DNA bands were considered unrelated and were given consecutive numbers as they appeared (A38, A39, A40, etc.). Isolates that were identical or highly related (three or fewer bands) by two or three restriction enzyme PFGE patterns were considered to be genetically related. We have found this criterion valuable in other studies with S. pneumoniae as well as for other pathogens (23, 31). This classification is more stringent than the five-band difference previously suggested by Tenover et al. (29).
IS1167 RFLP typing.
Whole-cell DNA extracts were prepared from isolates grown in 100 ml of brain heart infusion broth supplemented with 0.6% d-glucose plus 0.03% dl-threonine and incubated at 36.5°C in 5% CO2, as previously described (1, 16, 17). DNA was digested with 80 U of HindIII restriction enzyme and run on a 0.7% agarose gel in 0.5× TBE buffer at 100 V for 3.0 h (26). Southern blots were prepared from the agarose gels and hybridized with a 32P-labeled oligonucleotide probe, DAMO13 (5′-TGG ATA TTA TGG AGC CT-3′) (22, 32). This probe is specific for an upstream region of insertion sequence IS1167 (22, 32). The bands were counted, and patterns that differed in band number or size of band by more than one band were considered unrelated. RFLP typing and analysis were performed multiple times.
mef RFLP typing.
Whole-cell DNA extracts were prepared from isolates grown in 100 ml of brain heart infusion as described for the IS1167 RFLP typing. DNA was digested with 80 U of HindIII restriction enzyme and run on a 0.7% agarose gel in 0.5× TBE buffer at 100 V for 3.0 h (26). Southern blots were prepared from the agarose gels and hybridized with a 32P-labeled oligonucleotide probe, MF5 (5′-GGT GCT GTG ATT GCA TCT ATT AC-3′), that is specific for the mef gene (17, 28). The bands were counted and analyzed as described for the IS1167 RFLP analysis. RFLP typing and analysis were performed multiple times. This is the first time that the mef gene has been used for RFLP typing.
RESULTS
Antibiogram analysis.
Nonsusceptibility to penicillin was a criterion for inclusion in the study. The 22 Washington and Canadian isolates were also generally resistant to the cephalosporins. Cefprozil, ceftazidime, and loracarbef showed the highest MICs (MIC ranges, 8 to 16, 16 to 32, and 64 to 128 μg/ml, respectively) (Table 2). Of the 26 isolates, 20 were resistant to erythromycin and azithromycin (MIC, ≥1 μg/ml). One isolate (WA9) was inducibly resistant to the macrolides and clindamycin (MIC, ≥128 μg/ml for both) and carried both the ermB and mef genes (17, 27). The other 19 isolates carried the mef gene and were resistant to macrolides (MIC range, 1 to 8 μg/ml) but susceptible to clindamycin (MIC, ≤0.25 μg/ml). The quinolones, except for ciprofloxacin, had very low MICs. The MICs of linezolid were comparable to those of the quinolones. The new compound HMR3647 had the lowest MICs of any of the antibiotics tested (MIC range, 0.002 to 0.016 μg/ml).
PFGE analysis of the 26 clinical isolates.
Among the 26 S. pneumoniae isolates, 20 were considered to be genetically related because they had identical or highly related PFGE patterns with two or three enzymes (Table 3). Of these 20 isolates, 6 Washington isolates (WA7, WA8, WA9, WA11, WA12, and WA14) and 2 Alaska isolates (AK15 and AK18) of S. pneumoniae had identical PFGE patterns with all three enzymes. Three Washington isolates (WA1, WA3, and WA5) had identical patterns with two enzymes and had highly related PFGE patterns with one enzyme (ApaI). Two Washington isolates (WA2 and WA4) had highly related PFGE patterns with SacII and identical patterns with the other two enzymes. Two Washington isolates (WA10 and WA13) and one Alaska isolate (AK17) had identical PFGE patterns with one enzyme (SacII) and highly related patterns with the other two enzymes. One Alaskan (AK16) and three Canadian (CN19, CN20, and CN24) isolates had related PFGE patterns for each of the three enzymes.
TABLE 3.
Restriction enzyme PFGE patternsa
| Isolate | Isolation date (mo/yr)b | Origin | Pattern with enzyme:
|
||
|---|---|---|---|---|---|
| SmaI | SacII | ApaI | |||
| Related isolates | |||||
| WA8 | 12/95 | WA | S26 | C8 | A37 |
| WA9 | 12/95 | WA | S26 | C8 | A37 |
| WA11 | 1/96 | WA | S26 | C8 | A37 |
| WA12 | 4/96 | WA | S26 | C8 | A37 |
| WA14 | 6/96 | WA | S26 | C8 | A37 |
| WA7 | 3/97 | WA | S26 | C8 | A37 |
| AK15 | 4/94 | AK | S26 | C8 | A37 |
| AK18 | ?/97 | AK | S26 | C8 | A37 |
| WA3 | 2/97 | WA | S26 | C8 | A372 |
| WA5 | 3/97 | WA | S26 | C8 | A372 |
| WA1 | 2/97 | WA | S27 | C8 | A372 |
| WA2 | 2/97 | WA | S26 | C82 | A37 |
| WA4 | 3/97 | WA | S26 | C82 | A37 |
| WA10 | 12/95 | WA | S26 | C81 | A371 |
| WA13 | 6/96 | WA | S26 | C81 | A371 |
| AK17 | 2/96 | AK | S261 | C81 | A37 |
| AK16 | 8/94 | AK | S263 | C83 | A371 |
| CN19 | 4/95 | Canada | S263 | C83 | A372 |
| CN20 | 4/95 | Canada | S263 | C83 | A372 |
| CN24 | 1/96 | Canada | S263 | C83 | A373 |
| Unrelated isolates | |||||
| CN22 | 6/95 | Canada | S262 | C18 | A47 |
| CN23 | 11/95 | Canada | S262 | C19 | A49 |
| WA6 | 2/97 | WA | S28 | C9 | A38 |
| CN21 | 5/95 | Canada | S35 | C17 | A46 |
| CN25 | 1/95 | Canada | S36 | C20 | A48 |
| CN26 | 4/95 | Canada | S37 | C21 | A50 |
| Multiresistant clones | |||||
| SP27 (267-Spain-23F-1) | Spain | S264 | C10 | A39 | |
| SP28 (681-Spain-6B-2) | Spain | S29 | C11 | A40 | |
| SP29 (665-France-9V-3) | France | S30 | C12 | A41 | |
| SP30 (17219-S. Africa-19A-7) | S. Africa | S31 | C13 | A42 | |
| SP31 (50803-S. Africa-6B-8) | S. Africa | S32 | C14 | A43 | |
| SP32 (51702-S. Africa-19A) | S. Africa | S33 | C15 | A44 | |
Each ApaI, SacII, and SmaI PFGE pattern was assigned a letter and number designation (A 26 to A 50, C 8 to C 21, and S 26 to S 37). PFGE patterns have previously been assigned to isolates from other areas in the United States (reference 23 and unpublished data). Isolates having patterns which differed from these patterns by one to three bands were considered closely related and grouped into subtypes defined by subscripts (A261, A262, C81, etc.). Subscript 1 means one band added to the pattern, subscript 2 means one band deleted from the pattern, subscript 3 means two bands added or deleted, and subscript 4 means a three-band difference (23, 31). WA, Washington; AK, Alaska; S. Africa, South Africa.
?, month unknown.
The six remaining S. pneumoniae isolates, one Washington isolate (WA6) and five Canadian isolates (CN21, CN23, CN24, CN25, and CN26), had distinct PFGE patterns with each of the three enzymes and were not considered to be genetically related to the 20 isolates described above (Table 3).
PFGE comparison of related isolates with multiresistant clones.
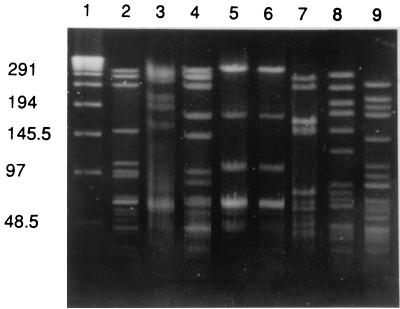
Isolate WA8, with the most common PFGE pattern for all three enzymes (A37, C8, and S26), was selected as a representative of the related isolates and was compared to the six previously known, multiresistant clones (Fig. 1). Five of the six clone isolates had unique PFGE patterns with all three enzymes from the PFGE patterns of isolate WA8. Isolate SP27 (267-Spain-23F-1) had distinct PFGE patterns with ApaI and SacII enzymes (Fig. 1) and a three-band difference for the SmaI PFGE pattern compared to the SmaI PFGE pattern of isolate WA8.
FIG. 1.
PFGE after ApaI restriction digestion. Lane 1, lambda ladder; lane 2, Washington isolate WA8; lane 3, 19F isolate from CDC; lane 4, 23F isolate from Spain; lanes 5 and 6, 19A isolates from South Africa; lane 7, 6B isolate from Spain; lane 8, 6B isolate from South Africa; lane 9, 9V isolate from France. The numbers on the side represent molecular weight standards in kilobases.
IS1167 RFLP analysis.
Of the 20 isolates with the same or related PFGE patterns, restriction fragment length patterns were either identical or had a one-band difference in the IS1167 RFLP patterns. The six clone isolates (SP27 to SP32) had unique IS1167 RFLP patterns distinctly different from that of the Washington clone (data not shown). The remaining Washington isolate (WA6) and the Canadian isolates (CN21, CN22, CN23, CN25, and CN26) had six- to nine-band differences in patterns from the Washington clone (data not shown).
mef RFLP analysis.
All 20 isolates with the same or related PFGE patterns had identical mef RFLP patterns and were distinct from unrelated S. pneumoniae isolates that carried the mef gene (Fig. 2). The unrelated Washington isolate (WA6) and the unrelated Canadian isolates (CN21, CN22, CN23, CN25, and CN26) had distinct RFLP patterns having one to five bands with more than one band different from the Washington clone (data not shown). In contrast, none of the clone isolates (SP27 to SP32) carried the mef gene, and RFLP analysis could not be performed.
FIG. 2.
Hybridization of HindIII-digested whole DNA with mef probe. Whole DNA was digested with HindIII and run on a 0.7% agarose gel for 3 h. The mef probe is oligonucleotide probe MF5. Lane 1, 02J1048; lane 2, WA4; lane 3, WA8; lane 4, WA12; lane 5, n011; lane 6, 915. Isolates 02J1048 (mef gene positive), n011 (mef gene positive), and 915 (mef gene negative) are unrelated S. pneumoniae isolates.
DISCUSSION
PFGE analysis using three different enzymes and IS1167 and mef RFLP typing identified a unique, multiresistant, serogroup 19 S. pneumoniae group of 20 isolates. This clone was characterized by reduced susceptibility to penicillin and resistance to extended-spectrum cephalosporins, erythromycin, and ciprofloxacin. Recent reports have indicated increases in ceftriaxone-resistant pneumococcus-caused illness in Washington (D. B. Jernigan, I. Kargacin, A. Poole, and J. Kobayashi, Program Abstr. 36th Annu. Meet. Infect. Dis. Soc. Am., abstr. 565-Sa, 1998). This newly identified clone most likely contributed to these increases. The Washington isolates were genetically related to the multiresistant isolates obtained from Alaska and eastern Canada but unrelated to five previously characterized, multiresistant clones (30). The Spanish isolate (SP27) representing the Spanish clone (Spain-23F-1) differed from the Washington clone in PFGE patterns with two enzymes, ApaI (Fig. 1) and SacII, and in IS1167 RFLP typing, indicating that it was completely different from the Washington clone. In previous work with S. pneumoniae and Neisseria gonorrhoeae, if any two isolates had an identical or related PFGE pattern with only one of three enzymes used, we did not consider the isolates to be related (23, 31).
The initial three isolates that prompted our investigation were fully resistant to penicillin, cefotaxime, erythromycin, and other antibiotics but were susceptible to vancomycin, the newer quinolones (grepafloxacin, levofloxacin, and trovafloxacin), linezolid, and the investigational antibiotic HMR3647 (Table 2). Reports of these multiresistant isolates in Tacoma, with their associated mortality, presented a challenge to clinicians choosing empiric treatment for severe community-acquired pneumonia. In regions where multiresistant isolates have been identified, clinicians may choose to request that vancomycin, newer quinolones, or linezolid be added to routine laboratory susceptibility testing of invasive S. pneumoniae isolates. In addition, the new compound HMR3647 may offer another therapeutic choice in the future.
The Washington clone was identified in both serotype 19F and serotype 19A isolates. Previous reports have identified capsular transformation of the Spanish clone (267-Spain-23F-1) from serotype 23F to 19F (6, 7, 20). Therefore, the presence of both serotype 19F and serotype 19A may reflect capsular transformation within serogroup 19. The 23-valent pneumococcal polysaccharide vaccine contains both serotype 19F and serotype 19A and is an underutilized prevention tool for multiresistant S. pneumoniae illness (4, 21). All three of the initial patients, including both fatal cases, were eligible to receive the vaccine because of underlying conditions (i.e., cancer and cardiovascular disease). However, there was no documentation that any of the patients had been vaccinated.
Most of the previous work by others to characterize S. pneumoniae clones by PFGE has used only one enzyme and has allowed up to five-band differences in banding patterns (18). Our laboratory used three enzymes, a three-band difference limit, and the requirement that two enzymes give identical or related PFGE patterns. Our laboratory used a one-band difference limit for both the IS1167 and the mef RFLP patterns (22, 31). Klugman and others have proposed that at least two molecular methods be used in identifying clones, as the PFGE with only one enzyme was not sufficient (27; K. Klugman, Letter). In this study, the PFGE and IS1167 and mef RFLP methods correlated well. This is the first demonstration that mef can be used for RFLP analysis of S. pneumoniae.
A recent report from Spain has described a serogroup 19 isolate that is nonsusceptible to penicillin and resistant to cefotaxime (24). Comparison of the Washington clone to this isolate would be of interest. Surveillance of invasive pneumococcal illness in Washington is continuing, which will allow us to monitor trends in antibiotic resistance and to detect any further increase in multiresistant pneumococcal infections. Based on our results, we suggest that this newly characterized clone has all of the characteristics required to be included in the recently organized Pneumococcal Molecular Epidemiology Network as Washington 19-14 (K. Klugman, Letter).
ACKNOWLEDGMENTS
We thank T. Fritsche at the University of Washington Medical Center Clinical Microbiology Laboratory for some of the S. pneumoniae isolates and A. Parkinson at the AIP of the CDC in Anchorage, Alaska, for the four Alaskan isolates and for subtyping the S. pneumoniae isolates. We also thank Renee Kanan of the University of Washington School of Public Health and Community Medicine Preventive Medicine Residency Program for the epidemiology background. We thank K. Klugman from the South African Institute for Medical Research in Johannesburg, South Africa, for the representative members of the multiresistant clones.
This study was supported in part by Glaxo-Wellcome.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler J C, Breiman R F, Campbell J F, Lipman H B, Broom C V, Faklam R R. Pneumococcal polysaccharide vaccine efficacy. JAMA. 1993;270:1820–1831. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Defining the public health impact of drug-resistant Streptococcus pneumoniae: report of a working group. Morbid Mortal Weekly Rep. 1996;45(RR-1 Suppl.):1–20. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Prevention of pneumococcal disease. Recommendations of the advisory committee on immunization practices (ACIP) Morbid Mortal Weekly Rep. 1997;46(RR-8 Suppl.):1–24. [PubMed] [Google Scholar]
- 5.Clewell D B, Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in Streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berron S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 8.Courvalin P, Carlier C. Tn1545: a conjugative shuttle transposon. Mol Gene Genet. 1987;206:259–264. doi: 10.1007/BF00333582. [DOI] [PubMed] [Google Scholar]
- 9.Doern G V. Susceptibility tests of fastidious bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 1342–1349. [Google Scholar]
- 10.Frick P, Black D, Duchin J, Deliganis S, McKee W M, Fritsche T R. Prevalence of antimicrobial drug-resistant Streptococcus pneumoniae in Washington State. West J Med. 1998;169:364–369. [PMC free article] [PubMed] [Google Scholar]
- 11.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann J, Cetron M S, Farley M M, Baughman W S, Facklam R R, Elliott J A, Deaver K A, Breiman R F. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med. 1995;333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs M R, Koornhof J J, Robins-Browne R M. Emergence of multiply resistant pneumococci. N Engl J Med. 1978;299:735–740. doi: 10.1056/NEJM197810052991402. [DOI] [PubMed] [Google Scholar]
- 14.Jernigan D B, Cetron M S, Breiman R F the Drug-Resistant Streptococcus pneumoniae Working Group. Minimizing the impact of drug-resistant Streptococcus pneumoniae (DRSP). A strategy from the DRSP working group. JAMA. 1996;275:206–209. [PubMed] [Google Scholar]
- 15.Kellner J D, McGeer A, Cetron M S, Low D E, Butler J C, Matlow A, Talbot J, Ford-Jones E L. The use of Streptococcus pneumoniae nasopharyngeal isolates from healthy children to predict features of invasive disease. Pediatr Infect Dis J. 1998;17:279–286. doi: 10.1097/00006454-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Luna V A, Roberts M C. The presence of the tetO gene in a variety of tetracycline resistant Streptococcus pneumoniae serotypes from Washington State. J Antimicrob Chemother. 1998;42:613–619. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- 17.Luna V A, Coates P, Eady E A, Cove J H, Nguyen T T H, Roberts M C. A variety of gram-positive bacteria carry mobile mef genes. J Antimicrob Chemother. 1999;44:19–25. doi: 10.1093/jac/44.1.19. [DOI] [PubMed] [Google Scholar]
- 18.Munoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin resistant clones of penicillin-binding protein profile, surface protein A typing and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. 1998. Approved standard M7-A4, M100-S8. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 20.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 21.Nuorti J P, Butler J C, Crutcher J M, Guevara R, Welch D, Holder P, Elliott J A. An outbreak of multidrug-resistant pneumococcal pneumonia and bacteremia among unvaccinated nursing home residents. N Engl J Med. 1998;338:1861–1868. doi: 10.1056/NEJM199806253382601. [DOI] [PubMed] [Google Scholar]
- 22.Robinson D A, Hollingshead S K, Musser J M, Parkinson A J, Briles D E, Crain M J. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J Mol Evol. 1998;47:222–229. doi: 10.1007/pl00006379. [DOI] [PubMed] [Google Scholar]
- 23.Rudolf K R, Parkinson A, Roberts M C. Molecular analysis by pulsed-field gel electrophoresis and antibiogram of Streptococcus pneumoniae serotype 6B isolates from selected areas within the United States. J Clin Microbiol. 1998;36:2703–2707. doi: 10.1128/jcm.36.9.2703-2707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz J, Sempere M, Simarro E, Fenoll A. Description of two new isolates of Streptococcus pneumoniae in Spain that are highly resistant to cefotaxime. Antimicrob Agents Chemother. 1998;42:2768–2769. doi: 10.1128/aac.42.10.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruoff K L. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 299–303. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 5.31–5.32. [Google Scholar]
- 27.Smith A M, Klugman K P. Pneumococcal diseases. In: Woodford N, Johnson A P, editors. Molecular bacteriology: protocols and clinical applications. Totowa, N.J: Humana Press, Inc.; 1998. pp. 139–156. [Google Scholar]
- 28.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasz A. Antibiotic resistance in Streptococcus pneumoniae. Clin Infect Dis. 1997;24(Suppl. 1):585–588. doi: 10.1093/clinids/24.supplement_1.s85. [DOI] [PubMed] [Google Scholar]
- 31.Xia M, Whittington W L, Holmes K K, Plummer F A, Roberts M C. Pulsed-field gel electrophoresis for genomic analysis of Neisseria gonorrhoeae. J Infect Dis. 1995;171:455–458. doi: 10.1093/infdis/171.2.455. [DOI] [PubMed] [Google Scholar]
- 32.Zhou L, Hui F M, Morrison D A. Characterization of IS1167, a new insertion sequence of Streptococcus pneumoniae. Plasmid. 1995;33:127–138. doi: 10.1006/plas.1995.1014. [DOI] [PubMed] [Google Scholar]