Abstract
Phthalate, a plasticizer, endocrine disruptor, and potential carcinogen, is degraded by a variety of bacteria. This degradation is initiated by phthalate dioxygenase (PDO), a Rieske oxygenase (RO) that catalyzes the dihydroxylation of phthalate to a dihydrodiol. PDO has long served as a model for understanding ROs despite a lack of structural data. Here we purified PDOKF1 from Comamonas testosteroni KF1 and found that it had an apparent kcat/Km for phthalate of 0.58 ± 0.09 μM−1s−1, over 25-fold greater than for terephthalate. The crystal structure of the enzyme at 2.1 Å resolution revealed that it is a hexamer comprising two stacked α3 trimers, a configuration not previously observed in RO crystal structures. We show that within each trimer, the protomers adopt a head-to-tail configuration typical of ROs. The stacking of the trimers is stabilized by two extended helices, which make the catalytic domain of PDOKF1 larger than that of other characterized ROs. Complexes of PDOKF1 with phthalate and terephthalate revealed that Arg207 and Arg244, two residues on one face of the active site, position these substrates for regiospecific hydroxylation. Consistent with their roles as determinants of substrate specificity, substitution of either residue with alanine yielded variants that did not detectably turnover phthalate. Together, these results provide critical insights into a pollutant-degrading enzyme that has served as a paradigm for ROs and facilitate the engineering of this enzyme for bioremediation and biocatalytic applications.
Keywords: phthalate dioxygenase, Comamonas testosteroni KF1, Rieske oxygenase, mononuclear iron, isophthalate, terephthalate
Abbreviations: BPDO, biphenyl dioxygenase; DHP, cis-4,5-dihydrodiol phthalate; HPLC, high-pressure liquid chromatography; NDO, naphthalene dioxygenase; PDO, phthalate dioxygenase; RO, Rieske oxygenase; TCA, tricarboxylic acid
Phthalate is an endocrine-disrupting pollutant and potential carcinogen that is extensively used as a plasticizer in a wide range of consumer products, including plastics, medicines, and cosmetics (1, 2, 3). As phthalate is not covalently bound in these materials, it readily leaches into the environment, potentially exposing humans and other organisms to its detrimental health effects through inhalation, ingestion, and absorption (2). Phthalate is also a major catabolic intermediate in the biodegradation of phthalate esters and some polycyclic aromatic hydrocarbons, such as fluoranthene, fluorene, and phenanthrene (4).
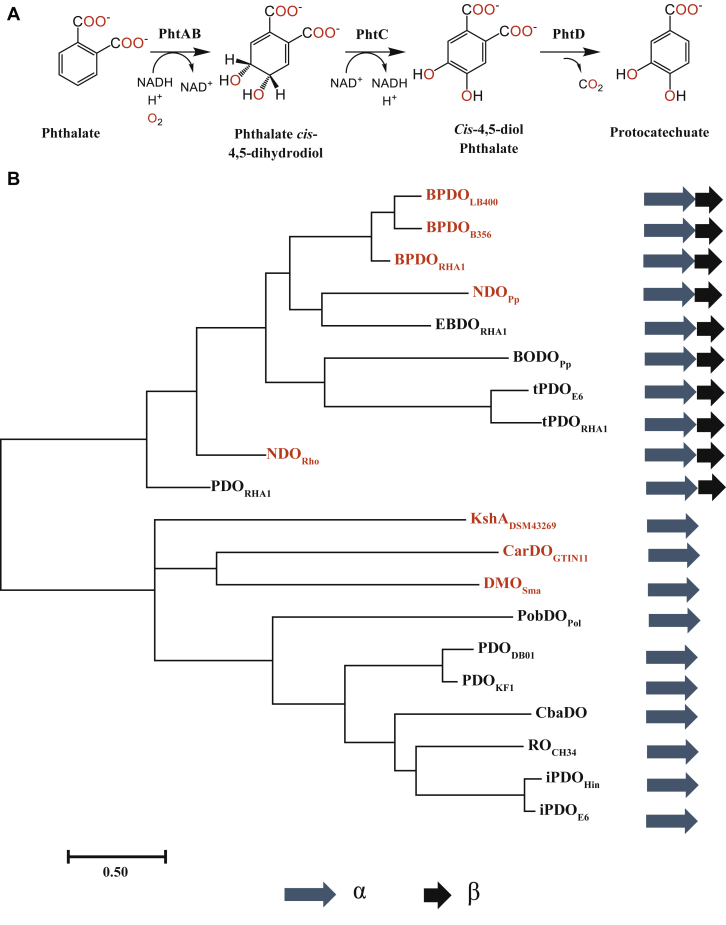
A wide range of bacterial strains are able to grow on phthalate, including proteobacteria such as Burkholderia cepacia DB01 (5, 6) and Comamonas testosteroni (7), and actinobacteria such as Rhodococcus jostii RHA1 (8) and Arthrobacter keyseri 12B (9). In these strains, the first step in phthalate catabolism is dihydroxylation of the aromatic diacid catalyzed by the phthalate dioxygenase (PDO). PDODB01 from B. cepacia DB01 and PDOKF1 of C. testosteroni KF1 catalyze the dihydroxylation of phthalate to cis-4,5-dihydrodiol phthalate (DHP) (6) (Fig. 1A). This differs from the reaction catalyzed by the actinobacterial PDOs, which is 3,4-dihydroxylation (9). Nevertheless, in both cases, the dihydrodiol is eventually converted to protocatechuate and ultimately leads to the formation of tricarboxylic acid (TCA) cycle intermediates for energy and biomass (6). Moreover, both classes of PDOs are Rieske oxygenases (RO).
Figure 1.
Phthalate catabolic pathway and phylogeny of select ROs.A, catabolism of phthalate to protocatechuate by the Pht enzymes of C. testosteroni KF1. PhtA and PhtB correspond to PDO and PDR, which catalyze the 4,5-dihydroxylation of phthalate. Protocatechuate is catabolized to TCA cycle intermediates. B, phylogenetic relationship of PDOs. Unrooted phylogenetic tree was calculated using a sequence alignment of 20 Rieske oxygenase α-subunits. The proteins are abbreviated using the protein name and strain as follows (GenPeptID): biphenyl dioxygenase from Comamonas testosteroni B356 (BPDOB356, AAC44526), Paraburkholderia xenovorans LB400 (BPDOLB400, AAB63425), and Rhodococcus jostii RHA1 (BPDORHA1, BAA06868); ethylbenzene dioxygenase from R. jostii RHA1 (EBDORHA1, BAC92718); naphthalene dioxygenase from Pseudomonas putida 9816-4 (NDOPp, P0A110), and Rhodococcus sp. NCIMB 12038 (NDORho, AAD28100); terephthalate dioxygenase from Comamonas testosteroni E6 (tPDOE6, AIJ48578), and R. jostii RHA1 (tPDORHA1, ABH00392); benzoate dioxygenase from Pseudomonas putida (BODOPp, WP_011600771); carbazole dioxygenase from Sphingomonas sp. GTIN11 (CarDOGTIN11, AAL37976); 3-ketosteroid 9α-hydroxylase from Rhodococcus rhodochrous DSM43269 (KshADSM43269, B6V6V5); dicamba monooxygenase from Stenotrophomonas maltophilia (DMOSma, AAV53699); phthalate dioxygenase from Comamonas testosteroni KF1 (PDOKF1, EED67076), Rhodococcus jostii RHA1 (PDORHA1, BAD36800) and Burkholderia cepacia DB01 (PDODB01, WP_011881604); 3-chlorobenzoate dioxygenase from Comamonas testosteroni BR60 (CbaDO, Q44256); phenoxybenzoate dioxygenase from Pseudomonas oleovorans (PobDOPol, Q52185); RO from Cupriavidus metallidurans CH34 (ROCH34, ABF12407); isophthalate dioxygenase from Comamonas testosteroni E6 (iPDOE6, BAH70269), and Hydrogenophaga intermedia (iPDOHin, CDN90362). Structurally characterized proteins are marked with red.
ROs are a family of enzymes best known for catalyzing the NAD(P)H-dependent dihydroxylation of aromatic compounds pollutants (10, 11). However, they catalyze a range of oxidation chemistries and have been the focus of extensive investigation, in part because of their potential use in bioremediation and biocatalysis (12, 13). RO systems comprise two or three components: an oxygenase and a reductase that transfers electrons from NAD(P)H to the oxygenase, either directly or via a ferredoxin (14). The oxygenase contains a mononuclear iron where catalysis occurs and a Rieske-type iron-sulfur cluster ([2Fe-2S]) that mediates electron transfer to the catalytic center. Structural studies on ROs such as naphthalene dioxygenase (NDO) (10, 15, 16, 17, 18, 19) have identified key features that are responsible for substrate specificity and have provided valuable insight into the catalytic mechanism of these enzymes (12). For example, a small N-terminal domain harbors the [2Fe-2S] cluster while the catalytic center occurs in a larger C-terminal domain (20). All ROs characterized to date are either α3 trimers or have an additional small subunit to form an α3β3 hexamer.
The PDO systems of B. cepacia DB01 and C. testosteroni KF1 comprise two components: an oxygenase and a reductase, PDR, encoded by phtA and phtB, respectively. PDR contains a flavin mononucleotide and a ferredoxin-type [2Fe-2S] center (21). Work on PDODB01 pioneered our understanding of RO function (11, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34). For example, these studies were the first to establish that ROs are iron-dependent (20). Similarly, ENDOR spectroscopy of the PDO Rieske center provided the first evidence that one of the cluster irons has bis-thiolate coordination while the other has bis-imidazole coordination (31). Additional spectroscopic studies of PDO revealed that substrate binding results in a decrease in the coordination number of the mononuclear iron center from six to five (26, 33).
Despite extensive efforts, the structure of PDO has been elusive. Based on gel filtration chromatography, the enzyme was initially proposed to be an α4 tetramer (20). Further analysis, including analytical centrifugation and mass spectrometry, indicated that PDODB01 is a hexamer comprising two stacked α3 trimers, a configuration that has not been shown in crystal structures of ROs to date (24). Moreover, PDOs share less than 20% amino acid sequence identity with other structurally characterized ROs. Thus, it has been difficult to identify key residues beyond the metal ligands and an acidic residue (28) proposed to mediate the interaction between the Rieske cluster and the mononuclear iron (35). An atomic-resolution structure of PDO would address a major gap in our understanding of these enzymes and provide insights into the molecular determinants of substrate specificity and regiospecificity.
Herein, we present a biochemical and structural characterization of PDOKF1. Steady-state kinetics were used to evaluate the enzyme’s apparent specificity for phthalate. Crystal structures of PDOKF1 were solved for the resting state enzyme as well as in complex with phthalate and terephthalate. We also report the structure of a homolog, ROCH34. The structures reveal that PDOKF1 has unique structural features that rationalize the enzyme’s function. The roles of key residues in substrate specificity and catalysis were evaluated using site-directed mutagenesis. Additionally, the structures were compared to those of related oxygenases to understand the dihydroxylation reaction catalyzed by other PDOs.
Results
Bioinformatic analysis and phylogenetic relationship of PDOKF1
Phylogenetic analysis of a variety of PDOs and characterized ROs revealed that PDOKF1 is closely related to PDODB01, with which it shares 81% amino acid sequence identity (Fig. 1B). These two enzymes form a larger clade with isophthalate dioxygenases iPDOE6 from C. testosteroni E6 (36) and iPDOHin from Hydrogenophaga intermedia, a 3-chlorobenzoate dioxygenase, CbaDO, from C. testosteroni BR60 (37) as well as ROCH34, an RO of unknown function with which PDOKF1 shares ∼35% amino acid sequence identity. All of these enzymes are α-type ROs (20, 36). By contrast, the actinobacterial phthalate 3,4-dioxygenases, such as PDORHA1, are αβ-type enzymes. Accordingly, these PDOs are more closely related to other αβ-type ROs such as NDO and biphenyl dioxygenase (BPDO) (38, 39, 40). The terephthalate dioxygenases tPDOE6 from C. testosteroni E6 (41) and tPDORHA1 from R. jostii RHA1 (42) are also αβ-type ROs and cluster accordingly. Finally, the sequence alignments indicate that in PDOKF1, the mononuclear iron is coordinated by His181, His186, and Asp343, the Rieske center is coordinated by Cys70, His72, Cys89, and His92 and that Asp178 bridges the metallocenters (Fig. S1). This is the same residue numbering as in PDODB01.
Purification and biochemical activity
PDOKF1 and ROCH34 were heterologously produced as His-tagged (Ht-) proteins for structural studies. In addition, PDOKF1 was produced as an untagged protein for kinetic studies as it had higher specific activity. Ht-PDOKF1 and PDOKF1 were purified to ∼99% and ∼95% apparent homogeneity, respectively (Fig. S2A), at yields of up to 20 mg protein per liter culture. Preparations of Ht-PDOKF1 and PDOKF1 contained 2.9 ± 0.2 and 2.7 ± 0.5 iron per monomer. Ht-PDOKF1 had a molecular weight of ∼300 kDa according to size exclusion chromatography (SEC), consistent with a homohexameric quaternary structure, while ROCH34 had a molecular weight of ∼150 kDa, consistent with a homotrimer (Fig. S2B). We also heterologously produced PDRKF1 and RO-RCH34, the cognate reductases of PDOKF1 and ROCH34 for activity assays.
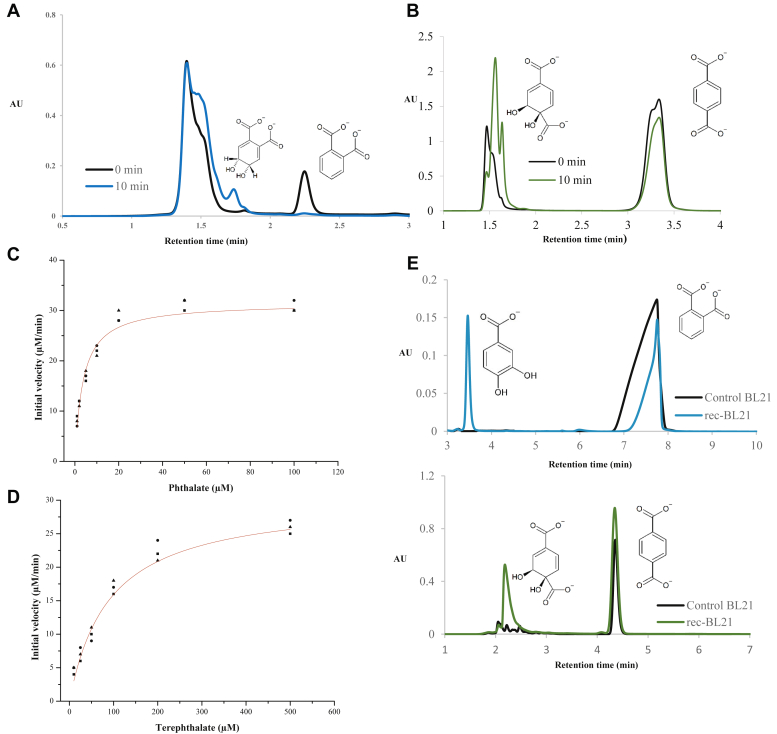
The ability of the enzymes to catalyze the dihydroxylation of phthalate was evaluated using an HPLC assay. When equimolar amounts of PDOKF1 and PDRKF1 were incubated with 100 μM phthalate (retention time, tR = 2.26 min) in the presence of 1 mM NADH (0.1 M Tris, pH 8), phthalate was consumed accompanied by the formation of a product with tR = 1.73 min (Fig. 2A) and an m/z value of 194.95 (Fig. S3A). The m/z value of phthalate was 158.92. Overall, the tR and an m/z values are consistent with the product being cis-4,5-dihydrodiol phthalate (20). The identity of the product was further confirmed by its transformation to protocatechuate by PhtCD as described below. Product formation was dependent on the presence of NADH. Moreover, the stoichiometry of NADH oxidized to phthalate hydroxylated was 1 ± 0.4:1 (Table 1). By contrast, ROCH34 did not detectably transform phthalate (data not shown).
Figure 2.
Biochemical analyses.A, conversion of phthalate by PDOKF1 and PDRKF1. Purified enzymes (10 μM each) were incubated with 100 μM phthalate in the presence of 100 μM NADH. The reaction product was analyzed by HPLC. B, conversion of terephthalate by purified PDOKF1 and PDRKF1. Purified enzymes (10 μM each) were incubated with 500 μM terephthalate in the presence of 200 μM NADH. C, dependence of initial velocity of oxygen consumption on the phthalate concentration in air-saturated buffer. Red lines represent fits of the Michaelis–Menten equation to the data. D, dependence of initial velocity of oxygen consumption on the terephthalate concentration in air-saturated buffer. Red lines represent fits of the Michaelis–Menten equation to the data. E, conversion of phthalate and terephthalate to protocatechuate and cis-dihydrodiol terephthalate, respectively with recombinant E. coli BL21, named R1 cells containing phthalate catabolic genes (phtABCD encoding PDO, PDR, phthalate cis-4,5-dihydrodiol dehydrogenase, and cis-4,5-diol phthalate decarboxylase, respectively).
Table 1.
Apparent steady-state kinetic parameters of PDOKF1 for different substratesa
| kcat (s−1) | Km (μM) | kcat/Km (mM−1s−1) | Couplingb | |
|---|---|---|---|---|
| Phthalate | 2.1 ± 0.1 | 3.6 ± 0.4 | 583 ± 87 | 1.0 ± 0.4 |
| Terephthalate | 2.0 ± 0.1 | 90 ± 12 | 22 ± 4 | 5.0 ± 0.2 |
Experiments were performed using air-saturated 50 mM Tris, pH 8.0, at 25 °C. The reported parameters are based on oxygen consumption.
Relative amount of NADH oxidized per mol of aromatic substrate hydroxylated.
In addition to phthalate, PDOKF1 catalyzed the NADH-dependent transformation of terephthalate. The product’s retention time (Fig. 2B) and m/z value (Fig. S3B) are consistent with the product being cis-1,2-dihydrodiol terephthalate (41). However, the dihydroxylation of terephthalate was not well coupled to NADH oxidation: the stoichiometry of NADH oxidized to terephthalate hydroxylated was ∼5:1. Finally, ROCH34 did not detectably transform terephthalate and neither enzyme transformed isophthalate, 3-chlorobenzoate, 2-chlorobenzoate, 3-phenoxybenzoate, or 4-phenoxybenzoate (data not shown). The ability of PDOKF1 to transform phthalate and terephthalate was confirmed using recombinant E. coli BL21 cells. Briefly, cells harboring the phthalate catabolic genes, phtABCD, converted phthalate to protocatechuate while control E. coli BL21 did not (Fig. 2E). The identity of protocatechuate was assigned with its retention time (Fig. S3C). Cells harboring PhtABCD also transformed terephthalate to the presumed dihydrodiol, indicating that PhtC is unable to catalyze the dehydrogenation of cis-1,2-dihydrodiol terephthalate.
We next used an oxygraph assay to evaluate the apparent steady-state kinetic parameters of PDOKF1 in air-saturated buffer. PDOKF1 catalyzed oxygen consumption in the presence of NADH and phthalate (50 mM Tris, pH 8.0, at 25 °C). The initial rate of consumption displayed Michaelis–Menten kinetics with respect to phthalate and terephthalate concentration (Fig. 2, C and D). However, the apparent specificity (kcat/Km) of PDOKF1 for phthalate was over 25-fold higher than for terephthalate and the apparent Km value was ∼20-fold lower (Table 1). Considering these experiments measured oxygen consumption, and terephthalate turnover was poorly coupled to NADH consumption, the true difference in apparent specificity is likely greater than 100-fold.
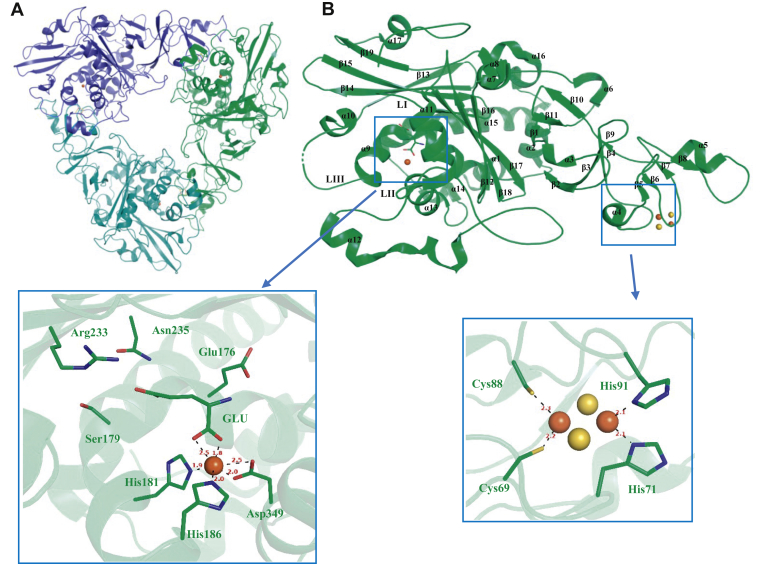
The crystal structure of ROCH34
Although we were unable to identify a substrate for ROCH34, solving its structure provided a means of obtaining structures of PDOKF1. ROCH34 was crystallized and solved to 1.8 Å resolution with single-wavelength anomalous dispersion (SAD) phasing using Fe as anomalous scatterer. The crystals belonged to the P63 space group and contained a single monomer in the asymmetric unit. The overall statistics of data collection, refinement procedures, and protein stereochemistry are within the standard values (Table 2). The crystallographic symmetry of ROCH34 suggests a trimeric architecture (α3 arrangement) with each α subunit related to another subunit by a noncrystallographic threefold axis (Fig. 3A). Each ROCH34 protomer has 437 residues and structurally can be divided into the Rieske domain and the catalytic domain (Fig. 3B). The Rieske domain in ROCH34 (residues 26–153) consists of eight α-helices and three β-sheet structures with a total of eight β-strands. In the Rieske center, Fe1 is coordinated with Cys69 and Cys88, at 2.2 Å and 2.3 Å distances, respectively, while Fe2 is coordinated with His71 and His91 at distances of 2.1 Å each. The two sulfide ions bridge the two iron ions and form a flat rhombic arrangement. The catalytic domain is dominated by an eight-stranded antiparallel β-sheet that extend over ten α-helices. The active site is formed by two helices (α9 and α11) and a β-sheet from the top. Three loops (LI, 246–253; LII, 291–330; LIII, 186–199) form the outer entrance of the cavity from three sides. The mononuclear iron-containing active site groove consists of residue Ser179, Glu176, His181, His186, Arg233, Asn235, and Asp349 (Fig. 3B). The mononuclear Fe atom is coordinated at 1.9 Å and 2.0 Å distances to His181 and His186, respectively, as well as bidentately to Asp349 with 2.0 Å and 2.5 Å distances (Fig. 3B). Additionally, a glutamate molecule, which originates from the mother liquor, binds the mononuclear iron in a bidentate manner (1.8 Å and 2.5 Å from each of the carboxylate oxygen atoms), completing its coordination sphere (Fig. 3B). However, the two coordination sites of the GLU are usually occupied by solvent molecules, as observed in other RO structures (16, 43).
Table 2.
Data processing and refinement statistics
| ROCH34 | PDOKF1-native | PDOKF1-phthalate | PDOKF1-terephthalate | |
|---|---|---|---|---|
| Resolution range | 38.8–1.84 | 88.87–2.11 | 53.29–2.74 | 53.06–3.07 |
| Wavelength (Å) | 1.74 | 0.96 | 1 | 1 |
| Space group | P 63 | C 1 2 1 | C 1 2 1 | C 1 2 1 |
| Unit cell dimensions a b c (Å) α, β, γ (°) |
103.4 103.4 77.6 90 90 120 |
178.2 121.4 161.5 90 100.9 90 |
179.0 122.2 162.9 90 101.1 90 |
179.2 122.7 163.5 90 101.1 90 |
| Completeness (%) | 99.9 | 98.84 | 97.88 | 97.35 |
| Rmerge (%)a | 3.7 | 10 | 6 | 8.8 |
| I/σ(I) | 9.4 | 3.9 | 12.6 | 8.6 |
| Refinement | ||||
| Reflections used in refinement | 40,967 | 191,509 | 88,624 | 63,340 |
| Reflections used for R-free | 2026 | 9784 | 4362 | 3147 |
| R-workb | 0.16 | 0.17 | 0.2 | 0.26 |
| R-freeb | 0.21 | 0.22 | 0.28 | 0.3 |
| Number of non-hydrogen atoms | 3875 | 22,171 | 20,545 | 19,601 |
| Macromolecules | 3508 | 19,389 | 19,274 | 19,311 |
| Ligands | 27 | 154 | 42 | 42 |
| Solvent | 340 | 2628 | 1229 | 248 |
| Protein residues | 437 | 2459 | 2446 | 2454 |
| RMS (bonds) (Å)c | 0.011 | 0.013 | 0.008 | 0.008 |
| RMS (angles) (°)c | 1.78 | 2.11 | 1.58 | 1.6 |
| Favored (%) | 97.46 | 94.99 | 91.54 | 88.39 |
| Allowed (%) | 2.31 | 4.52 | 7.5 | 9.64 |
| Outliers (%) | 0.23 | 0.49 | 0.95 | 1.98 |
| Average B-factor (Å) | 43.99 | 37.7 | 50.73 | 44.09 |
| Macromolecules (Å) | 43.31 | 36.29 | 51.03 | 44.36 |
| Phthalate/terephthalate (Å) | 77.25 | 78.5 | ||
| Ligands (Å) | 60.09 | 44.1 | 47.51 | 43.8 |
| Solvent (Å) | 49.72 | 47.69 | 46.16 | 22.59 |
Rmerge = Σ| I − ⟨I⟩|/ΣI.
R = Σ|Fobs| − |Fcalc|/Σ|Fobs|. The Rfree is the R calculated on the 5% reflections excluded for refinement.
RMS is root mean square.
Figure 3.
The crystal structure of ROCH34.A, cartoon representation of ROCH34 α3 trimer. The three subunits A, B, and C are colored in green, teal, and blue, respectively. B, cartoon representation of ROCH34 α protomer (green) with secondary structure elements; the active site entrance is covered with three loops (LI, LII, and LIII), the active site residues are depicted with sticks, and the coordination of the mononuclear iron with residues and glutamate ligand are shown. The Rieske center and the coordinating residues are depicted as sticks, distances are indicated (in Å).
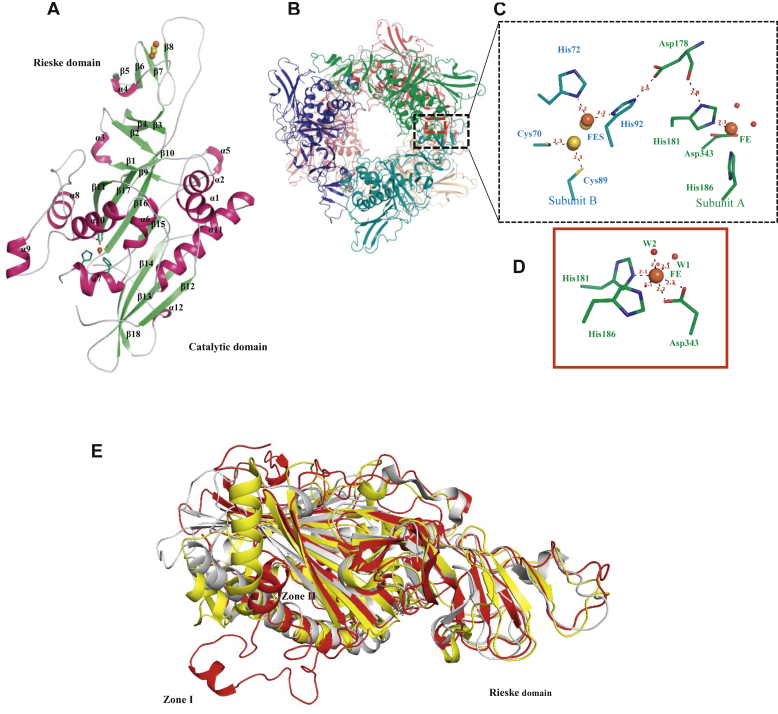
The structure of PDOKF1
PDOKF1 was crystallized and its structure was determined at 2.1 Å resolution using molecular replacement and the ROCH34 structure as a template. The overall data collection and refinement statistics are within the standard values (Table 2). The PDOKF1 crystal belongs to the C2 space group and contained six monomers in the asymmetric unit. The overall crystallographic symmetry operation shows that PDOKF1 is hexameric, consistent with the SEC measurement. The hexamer has a star-shaped arrangement in which one α3 is stacked on top of, and 60˚ offset from, the other (Fig. 4B). The modeled PDOKF1 protomer can be divided into the Rieske domain and the catalytic domain (Fig. 4A). Typical head-to-tail interaction interface between the subunits consists of many charged residues that form up to 28 hydrogen bonds and 16 salt bridges as calculated by PISA-CCP4i (44). The overall structure of PDOKF1 is very similar to that of ROCH34, with an RMSD of 0.98 Å over 274 Cα atoms. The Rieske domain of PDOKF1 is very similar to that of other α3 type ROs despite sharing less than 20% amino acid sequence identity with them over this domain. For example, the Rieske domains of PDOKF1 and DMO (45) have an RMSD of 1.0 Å over 90 Cα atoms (Fig. 4D). More specifically, the Rieske domain of PDOKF1 consists of five α-helices and three beta-sheet structures with a total of ten β-strands. The Fe1 of the Rieske cluster is coordinated with Cys70 and Cys89, at distances of 2.3 Å each. For its part, Fe2 is coordinated with His72 and His92 at distances of 2.2 Å each. The two sulfide ions bridge the two iron ions and form a flat rhombic arrangement typical of ROs (Fig. 4C). Within a single protomer, the mononuclear iron is located at 44 Å from the Rieske center (nearer iron). However, as typical of ROs, it is only at 12 Å (nearer iron) from the mononuclear iron of the adjacent α subunit within the trimer (39, 45, 46). The mononuclear iron is also seen to be very distant (44 Å) from the Rieske center of the stacked protomer (Fig. S4). Finally, an N-terminal helix extension present in CarDO, which mediates interactions with a ferredoxin, is absent in PDOKF1, consistent with the absence of this component in the PDOKF1 system.
Figure 4.
The crystal structure of PDOKF1.A, cartoon representation of PDOKF1 monomer with secondary structure elements (B) Cartoon representation of PDOKF1 α3α3 hexamer; six subunits A, B, C, D, E, and F are colored in green, teal, blue, red, salmon, and orange respectively. C, residue Asp178 found at the subunit–subunit interface bridge the Rieske center to the catalytic mononuclear iron center. D, the coordination of the mononuclear iron center with residues and two water molecules. Residues and bond distances (in Å) are labeled. E, structural superposition of a PDOKF1 protomer (red), DMO (gray), and CarDO (yellow). Extended region (residue 285–338) of the catalytic domain of PDOKF1 are involved in α3:α3 stacking interactions.
In contrast to the Rieske domain, the catalytic domain of PDOKF1 shows important differences with respect to that of other ROs (Fig. 4E). Most strikingly, this domain contains 286 residues (154–439), significantly more than any other structurally characterized RO. For example, the catalytic domains of DMO and CarDO comprise 220 and 240 residues, respectively. Much of the additional size is due to an extended region that contains protruded helices and loops (Zone I and II in Fig. 4E). Together, these protrusions form a bipod-like structure and enable the stacking of each α protomer with two protomers from the neighboring trimer (Fig. S5A). More specifically, this extended region comprising residues 285 to 338 forms two helices, each of which interacts with a separate protomer (α4 and α5). These interactions include a hydrogen bond and a salt bridge formation (Fig. S5B). The residues that mediate trimer stacking in PDOKF1 are largely conserved among proteobacterial PDOs, including PDODBO1 (Fig. S1), consistent with the latter being a homohexamer (24). ROCH34 harbors residues (292–300) that align with the extended region (Fig. S1); however, they adopt a different structure than that of PDOKF1 (Fig. S5C). Additionally, the residues constituting this helix are not conserved. Especially, ROCH34 lacks Glu293 and Thr294, which form critical intertrimer interactions in PDOKF1 (Fig. S1). Overall, these differences are consistent with ROCH34 being a trimer rather a homohexamer.
Other than the insertions that mediate trimer stacking, the catalytic domain is similar to that of other ROs. Briefly, the domain comprises V-shaped β-sheets formed from eight antiparallel β-strands. These sheets are surrounded by nine α-helices (Fig. 4A). The active site itself is formed by two α-helices and two β-sheets facing each other. A broad channel extends from the surface of the protein, providing substrates with access to the catalytic mononuclear iron. The mononuclear iron ion is coordinated to His181 and His186 at 2.1 Å each and is bidentate to Asp343 with 2.2 Å and 2.3 Å distances (Fig. 4D). Unlike ROCH34, the last two coordination sites of mononuclear iron are occupied by two water ligands, W1 and W2, at distances of 2.1 Å and 2.0 Å, respectively (Fig. 4D). As described previously, the coordination sphere of the mononuclear iron is conserved within ROs (38, 46). Asp178 bridges the mononuclear iron and the Rieske center of the neighboring subunit (Fig. 4C) as reported in PDODB01 (28). More specifically, Asp178 forms a hydrogen bond with His181, a mononuclear iron ligand, and His91 of the neighboring subunit where it coordinates the FeS cluster (Fig. 4C). The substrate-binding pocket is defined by Arg207, Arg231, Ser179, and Arg244 on one face and Phe278, Phe280, and Phe339 on the other face. These residues interact with phthalate as described below and are conserved in proteobacterial PDOs, including PDODB01 (Fig. S1).
The structure of PDOKF1 complexes with phthalate and terephthalate
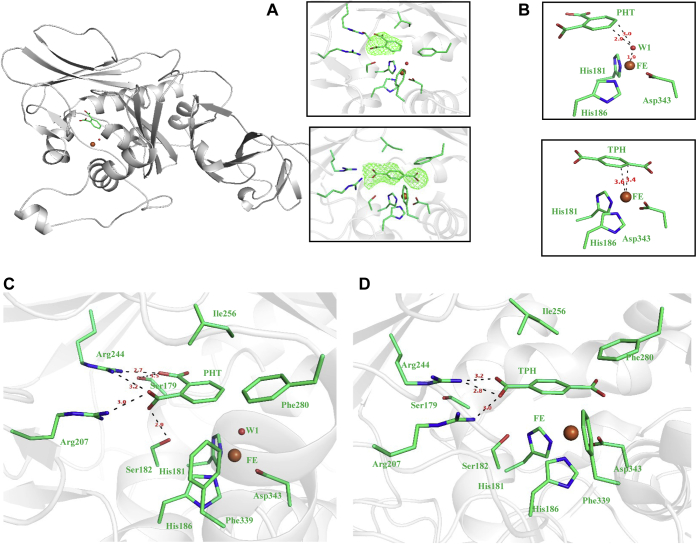
To identify residues responsible for binding phthalate, we soaked crystals of PDOKF1 with the substrate and determined a structure of the complex to 2.7 Å resolution. The main chain conformation in the PDOKF1:phthalate complex is nearly identical to that of substrate-free PDOKF1 (RMSD 0.24 Å over 366 Cα atoms). The phthalate molecule could be modeled in one chain of PDOKF1 structure that matched the observed electron density with a real space correlation coefficient (RSCC) of 0.84 (Fig. 5A). In contrast to the distorted octahedral geometry of mononuclear iron in the substrate-free enzyme, the mononuclear iron is pentacoordinate in the PDOKF1:phthalate complex (Fig. 5B), as observed in case of PDODB01 with magnetic circular dichroism (26, 33). Phthalate is positioned in the active site such that the carbon atoms that are dihydroxylated, C4 and C5, are 4.3 Å and 4.5 Å, respectively, away from mononuclear iron, consistent with earlier reported (32) distance (4.3 Å–6.5 Å) between phthalate and mononuclear iron. A solvent ligand is also present, between the substrate and the mononuclear iron, 2.9 Å and 3.0 Å from C4 and C5 carbon atoms, respectively (Fig. 5B). There is sufficient space between the metal ion and the substrate for O2 to displace the solvent ligand as suggested for NDO and other ROs (15, 47). Interestingly, Arg244 interacts with both carboxylate groups of the phthalate molecule, being positioned at 2.7 Å and 3.2 Å from C1 and C2 carboxylate groups, respectively. The C2 carboxylate also interacts with the side chain of Arg207, at a distance of 3.0 Å (Fig. 5C). These carboxylate groups are also hydrogen-bonded to the side chain hydroxyls of Ser179 and Ser182. These salt bridges and hydrogen-bond networks indicate that the carboxylate groups are critical binding determinants. Finally, Phe280 and Phe339, located on the other face of the active site, form π–π stacking interaction with the substrate’s aromatic ring (Fig. 5C).
Figure 5.
Structure of phthalate and terephthalate bound PDOKF1complexes to reveal the protein interactions that are important for correctly positioning substrate.A, electron density omit maps (contoured at 3σ) at 2.7 Å, 3.1 Å resolution for phthalate and terephthalate, respectively in the active site. B, the coordination state of the mononuclear iron in presence of phthalate and terephthalate. C, the phthalate molecule is anchored in the active site of PDOKF1via interactions that are highlighted here. D, similarly, the terephthalate molecule is anchored in the active site of PDOKF1via interactions that are highlighted here. The interactions are represented with black dotted lines. Residues and bond distances (in Å) are labeled.
The key phthalate-binding residues are conserved among PDOs from proteobacterial strains (Fig. S1). However, these residues are not all conserved in ROCH34. Thus, Ser182, Arg207, Phe280, and Phe339 correspond to Ala182, Trp201, Met271, and Leu346, respectively, in ROCH34 (Fig. S1). These substitutions are consistent with ROCH34’s inability to turnover phthalate. Finally, the sequence analysis reveals that Arg207 and Arg244 of PDOKF1 correspond to Arg218 and Met283, respectively, in PDORHA1 as discussed below. This highlights the different architectures of the PDOs that catalyze 4,5-dihydroxylation and 3,4-dihydroxylation, respectively.
To investigate the structural basis of why phthalate is a better substrate for PDOKF1 than terephthalate, we soaked PDOKF1 crystals with terephthalate and solved the structure of the resulting complex to 3.1 Å. In the PDOKF1:terephthalate structure, the ligand terephthalate also could be modeled in one chain of the PDOKF1 structure and has an RSCC value of 0.87. Like the PDOKF1:phthalate complex, the structure of the PDOKF1:terephthalate complex is very similar to that of the substrate-free enzyme (RMSD 0.26 Å over 367 Cα atoms). Interestingly, no density corresponding to a metal-bound solvent or O2 species was observed, perhaps due to the low resolution. The terephthalate molecule is positioned in the active site such that the carbon atoms that are dihydroxylated, C1 and C2, are at 3.4 Å and 3.6 Å, respectively from the mononuclear iron (Fig. 5B). Notably, the orientation of the terephthalate in the active site is consistent with that required for productive catalysis. Moreover, several of the interactions between the enzyme and the substrate observed in the PDOKF1:phthalate structure are conserved in the PDOKF1:terephthalate structure. The substrate’s C4 carboxylate interacts with the side chains of Arg244 and Arg207 at distances of 2.8 Å and 3.0 Å, respectively, analogous to the interactions between the C2 carboxylate of phthalate and these residues (Fig. 5D). Similarly, the aromatic ring of terephthalate forms a nonpolar interaction with Ile256 and π–π stacking interactions with Phe339 and Phe280 (Fig. 5D), as observed for phthalate. The striking difference between the bound substrates concerns the second carboxylate: in the case of terephthalate, the C1 carboxylate does not form hydrogen bonds or salt bridges with any residues. The markedly fewer interactions are consistent with the lower specificity of the enzyme for terephthalate.
Active site variants
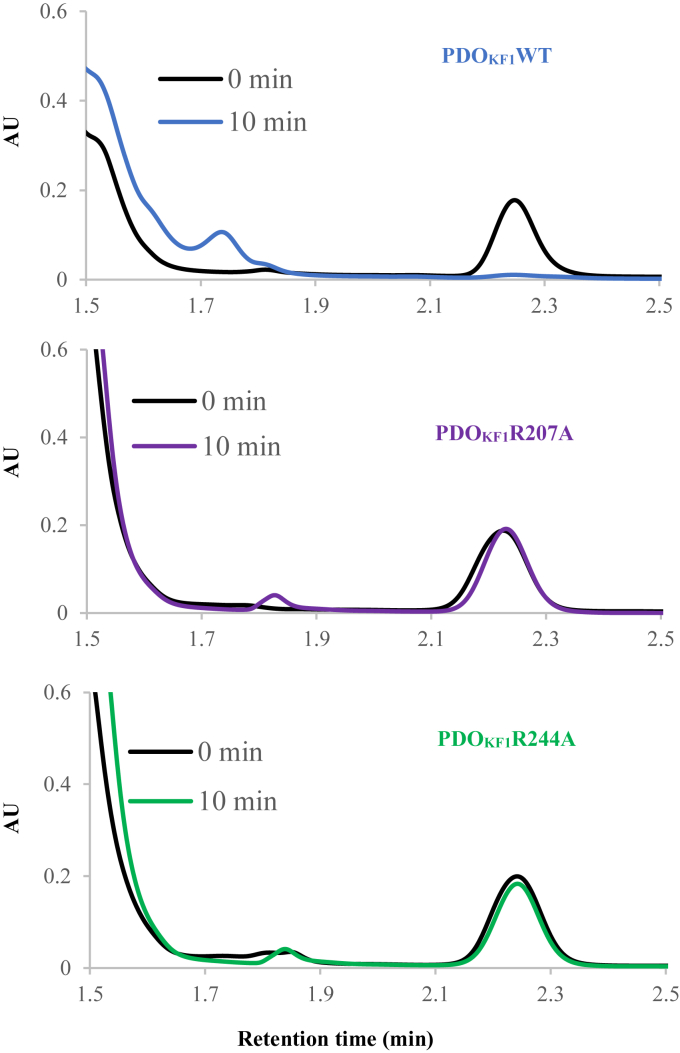
The roles of active site residues in determining the substrate specificity of PDOKF1 were probed using site-directed mutagenesis. More specifically, we generated the R207A and R244A variants of the enzyme and purified them as described for the wild-type enzyme. Preparations of both variants contained 2.9 ± 0.3 Fe per monomer. Using the HPLC-based assay, neither variant detectably transformed phthalate, indicating that these two residues are critical determinants of substrate specificity (Fig. 6).
Figure 6.
Site directed mutagenesis and additional activity of PDOKF1. Reactions of PDOKF1 wild-type and variants with phthalate. Purified enzymes (10 μM each) were incubated with 10 μM PDRKF1, 100 μM phthalate in the presence of 1 mM NADH. The reaction product was analyzed by HPLC.
Modeling of PDORHA1 and iPDOE6
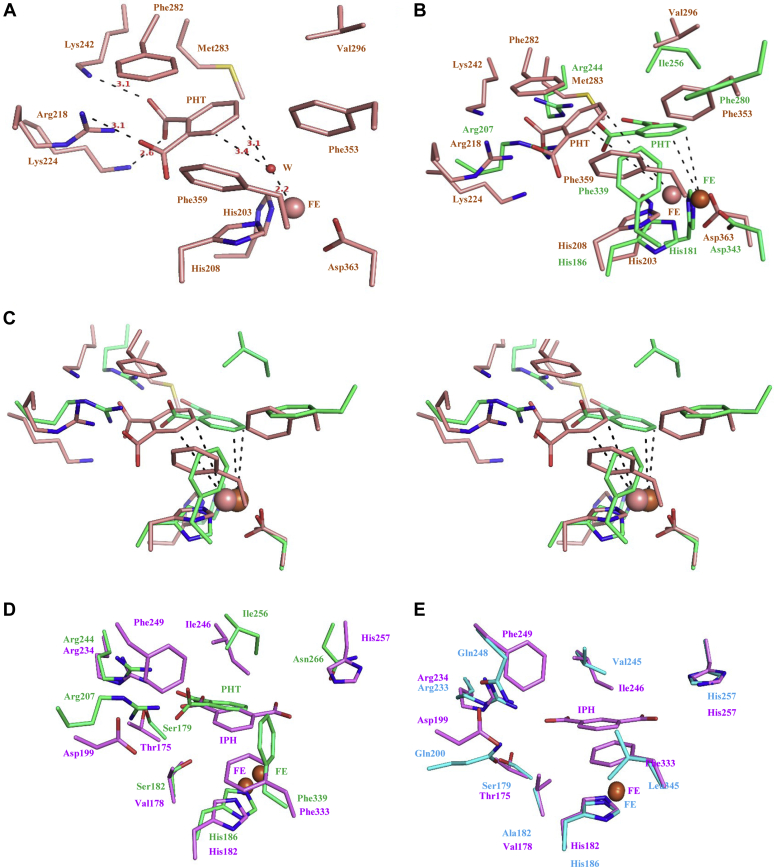
To better understand the structural features that determine the regiospecificity of PDOs, we modeled PDORHA1, an enzyme that catalyzes 3,4-dihydroxylation, and compared it with the structure of PDOKF1. A homology model of PDORHA1 generated using the structure of NDORho (19) has an RMSD of 0.15 Å over 325 aligned Cα atoms with the structure of NDORho. Despite significant architectural differences between actinobacterial PDOs and proteobacterial PDOs, the active site features are similar in terms of their constituent residues. As in PDOKF1, the PDORHA1 active site features basic residues on one face and nonpolar residues on the other, positioned to interact with the phthalate’s carboxyl moieties and aromatic ring, respectively (Fig. 7A). Consistent with enzyme’s regiospecificity, docking phthalate into the active site using AutoDock Vina yielded a pose with the atoms to be hydroxylated, C3 and C4, positioned 3.4 Å and 3.1 Å, respectively, from a water molecule, which is at 2.2 Å distance from mononuclear iron and occupies the presumed binding site for dioxygen (Fig. 7A). More specifically, Arg218, which corresponds to Arg207 in PDOKF1, interacts with the C2 carboxylate group of phthalate, while Lys224 and Lys242 residues interact with the C1 carboxylate. These interactions orient the phthalate molecule for its regiospecific 3,4-dihydroxylation (Fig. 7A). Comparison with the PDOKF1:phthalate complex revealed that Arg244 in PDOKF1 is replaced with Met283 in PDORHA1, and that this prevents the orientation of the substrate to enable 4,5-dihydroxylation (Fig. 7B).
Figure 7.
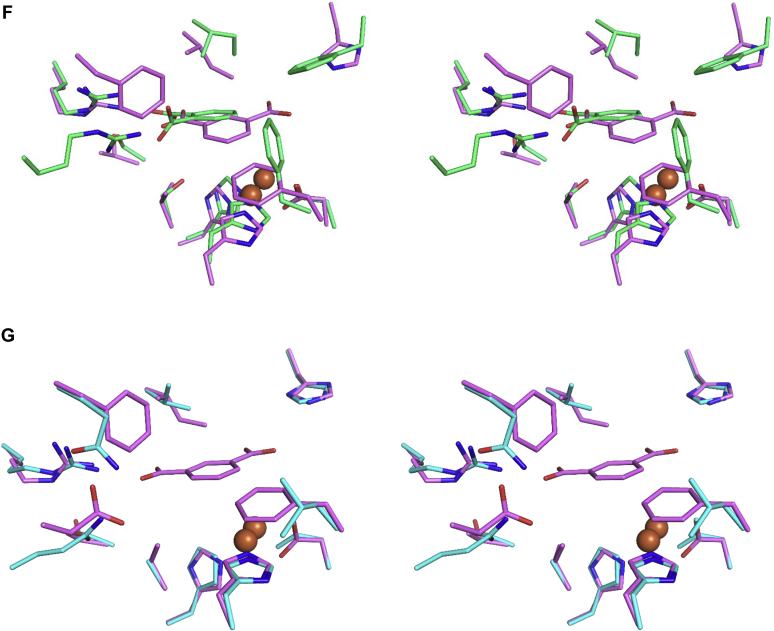
Structural comparison and active site analysis of PDORHA1and iPDOE6.A, active site of PDORHA1 with docked phthalate molecule, shown in salmon-colored stick. Residues and bond distances (in Å) are labeled. B, superposition of the structures of PDORHA1 and PDOKF1 (green). C, stereoview of superposition of the structures of PDORHA1 and PDOKF1. D, superposition of the structures of iPDOE6 and PDOKF1 (green). The isophthalate docked model of iPDOE6 is shown in violet-colored stick. E, superposition of the structures of iPDOE6 and ROCH34 (Cyan). The residues are labeled. F, stereoview of superposition of the structures of iPDOE6 and PDOKF1. G, stereoview of superposition of the structures of iPDOE6 and ROCH34.
To gain insight into the active site features of iPDOE6, we modeled the enzyme’s structure using the ROCH34 coordinates. The homology model of iPDOE6 has an RMSD of 0.19 Å over 309 aligned Cα atoms. Comparison with the structures of PDOKF1 and ROCH34 revealed significant similarities and differences in the active sites. First, Arg244, which interacts with a phthalate carboxylate in PDOKF1, is conserved in iPDOE6 (Arg234), and ROCH34 (Arg233) (Fig. 7, D and E). The iPDOE6 active site also harbors His257 that is positioned to interact with the meta-carboxylate group of the isophthalate. In the modeled structure, this residue corresponds to Asn266 of PDOKF1. Importantly, iPDOE6 does not have a second Arg in the active site to bind an ortho-carboxylate. Finally, Val178, Phe249, and Phe333 are positioned to stabilize the aromatic ring of isophthalate. These residues correspond to Ser182, Arg207, and Phe339 of PDOKF1, respectively (Fig. 7, D and E). This analysis indicates that Arg234 and His257 are critical determinants for the specificity of iPDOE6. Interestingly, ROCH34 also harbors His257. However, the residues predicted to interact with the substrate’s aromatic ring in iPDOE6, Val178, Val245, Phe249, and Phe333, correspond to Ala182, Ile246, Gln248, and Leu345 in ROCH34, suggesting this is a major reason why ROCH34 is unable to turnover isophthalate (Fig. 7E).
Discussion
The structural characterization of PDOKF1 validates and extends pioneering work done on PDODB01. Specific features of PDODB01 that are validated by the structure of PDOKF1 include the hexameric configuration of the former (24) and the bridging of the metallocenters by Asp178 (35). More particularly, because all functionally important residues are conserved between these two enzymes, the structure of PDOKF1 provides a framework for characterizing the molecular determinants of PDODB01’s properties. For example, PDODB01 contains the regions and specific residues that stabilize hexamer formation in PDOKF1, and these residues are unique to this clade of PDOs. Accordingly, we predict that the PDODB01 hexamer adopts the same star-shaped structure as the PDOKF1 hexamer. Similarly, all the substrate-binding residues of PDOKF1 are conserved in PDODB01, including Arg207 and Arg244, providing a basis for understanding the substrate specificity and regiospecificity of PDODB01.
The structural data together with the functional characterization of the variants establish that Arg244 and Arg207 are critical determinants of substrate specificity in the proteobacterial PDOs. The structure of the PDOKF1:phthalate complex revealed that these two residues on one face of the active site form salt bridges with carboxylate groups of phthalate. These residues are conserved among proteobacterial PDOs, highlighting their importance in correctly positioning phthalate for its regiospecific hydroxylation. Further, the positioning of these residues is such that they are unable to properly orientate either terephthalate or isophthalate in the active site, as indicated by PDOKF1’s poor turnover of terephthalate and inability to turn over isophthalate.
The data for the PDOKF1 and PDOKF1:phthalate structures corroborate the spectroscopic data showing the substrate-dependent change in the coordination geometry of PDODB01. Magnetic circular dichroism spectroscopic analyses indicated that in substrate-free PDODB01, the mononuclear iron is hexacoordinate and becomes pentacoordinate upon substrate binding (26, 33). Moreover, NMR evidence showed that the dissociating ligand is a water molecule (34). Consistent with these, the data presented here establish that the mononuclear iron exists in a distorted octahedral six-coordinate geometry in the resting state PDOKF1 (Fig. 3B) and is five-coordinate in the PDOKF1:phthalate complex (Fig. 5B). The mononuclear iron is six-coordinate in the resting state of NDO as well (48). The change in coordination state induced by substrate binding is thought to be mechanistically important as it enables the mononuclear iron to bind O2 in a side-on manner (13). Additionally, in PDODB01, mononuclear iron was reported (11) to be in the ferrous state at the end of the reaction during single turnover experiments. Based on this, it was proposed that two electrons for the reaction are provided by two Rieske centers and that the resulting stoichiometry is one product molecule produced per two Rieske centers oxidized (24). This phenomenon was proposed to be a consequence of a stacked trimer architecture placing two Rieske centers close to each mononuclear iron, facilitating electron transfer. The PDOKF1 structure demonstrates that the stacked trimer architecture does not position a second Rieske center sufficiently close to the mononuclear iron (Fig. S4) to enable transfer of second electron. However, with the current lack of biochemical and structural evidences, the “two Rieske centers for one mononuclear iron” cannot be proven.
The terephthalate-transforming activity of PDOKF1 is unlikely to be physiologically relevant. First, the dihydroxylation of terephthalate was not well coupled with NADH oxidation, indicating that the active site of PDOKF1 does not accommodate terephthalate well. This is confirmed by the structure of the PDOKF1:terephthalate complex, which reveals that one of the terephthalate carboxylates does not form any salt bridges with active site residues. Second, recombinant cells containing the phthalate catabolic pathway transformed terephthalate to the cis-dihydrodiol. While this confirms that PDOKF1 can dihydroxylate terephthalate, it also established that the subsequent phthalate catabolic enzymes are unable to transform the cis-diol product to protocatechuate. Finally, as C. testosteroni KF1 possesses a bonafide terephthalate catabolic pathway (41), terephthalate hydroxylation activity of PDOKF1 is expected to not be physiologically relevant.
The structural comparison of PDOKF1 with a homology model of PDORHA1 highlights possible determinants of the regiospecificity of PDORHA1. Specifically, while the overall structures of PDORHA1 and PDOKF1 are different, most of the active site residues are identical. Notably, Arg207, a key phthalate-binding residue in PDOKF1, corresponds to Arg218 in PDORHA1. An important difference in the active sites of these enzymes is Met283 in PDORHA1, which corresponds to Arg244 of PDOKF1. In PDORHA1, the phthalate binding pose equivalent of PDOKF1 would have a steric conflict between the phthalate and Met283 and thus prevent the phthalate molecule to get 4,5-dihydroxylated. Additionally, the interactions of Arg218, Lys224, and Lys242 in PDORHA1 with the C1 and C2 carboxylates of phthalate are critical to orientating the substrate for 3,4-dihydroxylation.
While the physiological role of ROCH34 is unclear, the functional and structural data are consistent with it not being either a PDO or an iPDO. The crystal structure of ROCH34 depicted common features of ROs related to mononuclear iron-binding residues and electron transfer residues. However, the structural analysis indicated that active site residues in ROCH34 differ from those of PDOKF1. More specifically, the lack of Arg207, Phe280, and Phe339 in ROCH34 likely explains this enzyme’s inability to transform phthalate. Comparison of the crystal structure of the PDOKF1 complex to the modeled iPDOE6 structure revealed structural features that explain the different reactivities of these two enzymes. Most strikingly, the occurrence of polar residue His257 in iPDOE6 at the position corresponding to Asn266 of PDOKF1 is consistent with the specificity of iPDOE6 for isophthalate. The ROCH34 structure lacks the equivalent of aromatic ring stabilizing residues Phe333, Phe249, Val178, Ile246 of iPDOE6, likely explaining its inability to bind isophthalate. Unfortunately, the genomic context of the gene encoding ROCH34 (protein_id, ABF12407) provides no insight into the function of the enzyme: the adjacent genes are predicted to encode an alcohol dehydrogenase (ABF12404) and an aldehyde dehydrogenase (ABF12403). Finally, although ROCH34 shares sequence similarity with phenoxybenzoate dioxygenase, ROCH34 did not transform either 3-phenoxybenzoate or 4-phenoxybenzoate.
In conclusion, this study provides important new insights into the dihydroxylation of phthalate by proteobacterial PDOs. The structures and mutagenesis data not only provide insights into the molecular details of PDOKF1, but also provide structural basis for better understanding many years of work on PDODB01. The findings also provide a structural basis for understanding the regiospecific 4,5-dihydroxylation by proteobacterial PDOs versus 3,4-dihydroxylation of actinobacterial PDOs.
Experimental procedures
Chemicals, reagents, and bacterial strains
All reagents were of analytical grade. Restriction enzymes, T4 DNA ligase used for cloning were from New England Biolabs. Phusion polymerase used was from Thermo scientific. Water for buffers was purified using a Merck Synergy Water Purification System to resistance of at least 18.2 MΩ. C. testosteroni KF1 was purchased from the German Collection of Microorganisms and Cell Cultures DSMZ. Cupriavidus metallidurans CH34 was purchased from the Microbial Type Culture Collection (MTCC).
Cloning and expression
DNA was purified, propagated, cloned, and amplified using standard protocols (49). Genes encoding RO components were amplified from genomic DNA of C. testosteroni KF1 and megaplasmid DNA of C. metallidurans CH34 using primers listed in Table S1. N- and C- terminal primers contained NdeI and XhoI sites, respectively. The PCR amplicons and vectors (either pET-28c or pET-41b) were digested with the appropriate restriction enzymes, gel purified, and ligated with T4 DNA ligase. The phthalate catabolic gene fragment (phtA encoding PDO, phtB encoding PDR, phtC encoding phthalate cis-4,5-dihydrodiol dehydrogenase, and phtD encoding cis-4,5-diol phthalate decarboxylase) was amplified from genomic DNA of C. testosteroni KF1 using primers listed in Table S1. The amplified fragment was digested with NcoI and HindIII enzymes and cloned into the pET-28c vector. The recombinant vector was transformed into competent E. coli DH5α cells and subsequently to E. coli BL21 λ(DE3) cells. The ligation products were transformed into chemically competent E. coli DH5α cells and spread onto lysogeny broth (LB) agar plates containing kanamycin (50 μg/ml). Multiple single colonies were picked for each construct and grown in 10 ml LB supplemented with kanamycin at 37 °C and 220 rpm overnight. Plasmid DNA was extracted and a positive clone was confirmed by restriction digestion and DNA sequencing. Further, the confirmed plasmid was transformed into chemically either competent E. coli BL21 λ(DE3) cells and spread onto LB agar plates containing kanamycin (50 μg/ml). A single transformed colony was inoculated into 10 ml of LB supplemented with kanamycin (50 μg/ml) and incubated at 37 °C and 220 rpm overnight. Cells were grown in LB supplemented with kanamycin (50 μg/ml) at 37 °C and 200 rpm until OD600 = 0.6, upon which culture was transferred to 16 °C and induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 16 h. Cells were pelleted (6000g, 10 min, 4 °C) and stored at −80 °C.
Purification of recombinant PDOKF1 and PDRKF1
For kinetic characterization, PDOKF1 and ROCH34 were produced in E. coli BL21 λ(DE3) with pET41PDOKF1 and pET41ROCH34 plasmid constructs, respectively, as the His-tagged form of these enzyme did not show any detectable activity. Cell culture collected from 1 l was resuspended in 10 ml purification buffer (20 mM Tris, pH 7.4) containing 5 mM PMSF, and cells were lysed using Constant Cell Disruptor at 20 psi. Cell debris was removed by centrifugation at 12,000 rpm for 1 h at 4 °C. The PDOKF1 was purified using a MonoQ 10/100 Gl column (GE Healthcare). The protein was eluted with a linear gradient from 0.2 to 0.6 M NaCl in 120 ml of 20 mM Tris, pH 8.0. Fractions containing PDOKF1 and ROCH34 were pooled, dialyzed into 20 mM Tris, pH 7.4, concentrated to ∼20 mg/ml, flash-frozen as beads in liquid N2, and stored at −80 °C until needed.
The ROCH34-Ht, RO-RCH34-Ht, PDOKF1-Ht, and PDRKF1-Ht were produced in E. coli BL21 λ(DE3) with pET28ROCH34, pET28RO-RCH34, pET28PDOKF1, and pET28PDRKF1 plasmid constructs, respectively. Cells were grown, cell extracts were produced as described above and resuspended in 10 ml purification buffer (20 mM Tris, pH 7.4, NaCl 300 mM) containing 5 mM PMSF, and cells were lysed using Constant Cell Disruptor operated at 20 psi. The supernatant solutions were incubated with the pre-equilibrated Ni-NTA affinity column. Bound proteins were eluted by an imidazole gradient from 100 to 500 mM with purification buffer and analyzed on an SDS-PAGE gel. Fractions containing pure protein bands were concentrated using an Amicon 30 kDa Millipore filter. N-terminal His tag was removed by incubation with TEV protease (1:10 M ratio) in dialysis buffer (100 mM sodium phosphate, pH 7.4; 50 mM NaCl). The cleaved protein mixture was purified by using a Reverse Ni-NTA column. The purity of the protein was checked on SDS-PAGE. Tag-free proteins were concentrated using an Amicon 30 kDa Millipore filter and dialyzed with 50 mM HEPES, pH 7.4, 50 mM NaCl.
Analytical methods
Protein concentration was determined using the Micro BCA protein assay kit (Pierce) using bovine serum albumin as a standard. UV–Visible spectra were recorded using a Cary 60 spectrophotometer. Iron concentrations were determined spectrophotometrically using the Ferene-S assay and FAS solution as a standard (50).
Phylogenetic analysis of RO sequences
Twenty sequences of ROs were selected for phylogenetic analysis based on biochemical characterization and preferred substrate. Sequences of α subunits were aligned using MUSCLE. Phylogenetic reconstruction and validation were performed using the neighbor-joining (NJ) method based on bootstrap analysis with 1000 replications using the Jukes–Cantor distance model in molecular evolutionary genetics analysis (MEGA) 7 software (51). Both maximum-likelihood and minimum-evolution methods were also employed to test the robustness of the tree.
Biochemical activity
The PDOKF1 activity was determined by measuring the consumption of phthalate by high-pressure liquid chromatography (HPLC). HPLC experiments were carried out on an X-Bridge (Waters) C18 column (5 μm, 150 × 4.6 mm) using a Waters HPLC system equipped with a binary pump (Waters; Binary pump 1525) and photodiode array (Waters; PDA 2998) detector. Chromatograms were analyzed on a Windows 10 platform with an EMPOWER software ver. 3 feature release 4 (Waters). The 1 ml assay mixture contained 100 mM Tris, pH 8.0 containing 100 μM Fe(NH4)2(SO4)2.6H2O, 200 μM substrate, 100 μM NADH, 10 μM each of PDOKF1 and PDRKF1. Portions (100 μl) of the reaction mixture were removed at various sampling times and quenched with 100 μl acetonitrile (100%) prior to analysis. The mobile phase used for HPLC analysis was a mixture of acidified water (68%) and acetonitrile (32%). The HPLC system was operated at a flow rate of 1 ml/min and the metabolites were detected at 240 nm. For substrate preference activity, PDOKF1 was incubated with 100 μM of phthalate, isophthalate, terephthalate, 3-chlorobenzoate, 2-chlorobenzoate, 3-phenoxybenzoate, or 4-phenoxybenzoate under the assay conditions described above, and the reaction mixtures were analyzed by HPLC.
Steady-state kinetic assays were performed by monitoring the consumption of O2 using a Clark-type polarographic O2 electrode OXYG1 (Hansatech) connected to a circulating water bath. Assays were performed in 1 ml of air-saturated 50 mM Tris, pH 8.0 at 25 °C, with 0.25 μM each of PDOKF1 and PDRKF1 and initiated by adding the substrate. The electrode was calibrated according to the manufacturer's instructions using air-saturated water and O2-depleted water via the addition of sodium dithionite. Reaction velocities were corrected for the background rate of O2 consumption, recorded prior to aromatic substrate addition. Steady-state kinetic parameters were evaluated by fitting the Michaelis–Menten equation to the initial velocities.
Site-directed mutagenesis
PCR amplification using mutagenic primers was used to introduce the desired substitution. In all, 25 μl reactions composed of 1x Phusion buffer (HF), 100 μM dNTPs, 2 ng/μl plasmid template, 1 μM primer (Table S1), 1 U Phusion DNA polymerase, and 1% DMSO. The parent template was digested using DpnI upon the visual confirmation of a successful amplification using agarose gel electrophoresis. The DpnI digest reaction was incubated at 37 °C for 1 h and 10 μl reaction volume was transformed into DH5α chemically competent cells. Mutations were verified by sequencing.
Whole cell biotransformation of phthalate and terephthalate
Cell-mediated substrate hydroxylation was detected and analyzed with HPLC. One mL of overnight recombinant BL21 λ(DE3) culture was added to two flasks containing 1 mM phthalate and terephthalate in 50 ml of MINERAL MEDIUM (BRUNNER), and IPTG (1 mM final concentration) was added. Cultures were grown for 4 h at 30 °C and centrifuged, and the supernatant was analyzed by HPLC and the cells were disrupted using Cell Disruptor (Constant Systems) and extracted using acetonitrile (GC-grade). The cell debris was removed with centrifugation. The extracted samples were concentrated to 500 μl and then filtered through 0.22 μm membrane filters. Filtered samples were analyzed with HPLC as described above. The HPLC experiment was carried out on a SunFire (Waters) C18 column (5 μm, 250 × 4.6 mm).
Crystallization of ROCH34 and PDOKF1
ROCH34 was concentrated up to 18 to 20 mg/ml and subjected to crystallization trials using the sitting drop vapor diffusion method at 20 °C. The initial crystals appeared in the Morpheus screen. The best diffracting crystals were obtained in an optimized condition, which contained 50 mM glycine, 25 mM lysine, 20 mM glutamate, and 14% polyethylene glycol 4000 reservoir solution, and protein-to-reservoir ratio of 1:1 was used. ROCH34 crystals were obtained at 20 °C. For data collection, ROCH34 crystals were cryoprotected in a reservoir solution containing 15% ethylene glycol and were flash-frozen in a nitrogen stream at 100 K. PDOKF1 was concentrated up to 12 to 15 mg/ml and subjected to crystallization trials using the sitting drop vapor diffusion method at 20 °C. The best diffracting crystals were obtained in an optimized condition that contained 100 mM HEPES buffer pH 7.5, 25 mM CaCl2, 25 mM MgCl2, 100 mM sodium potassium tartrate, and 12% polyethylene glycol 4000 as a reservoir solution with protein-to-reservoir ratio of 1:1. Within 2 days, deep red-colored PDOKF1 crystals were obtained at 20 °C. For data collection, crystals were cryoprotected in a reservoir solution containing 40% ethylene glycol and were flash-frozen in a nitrogen stream at 100 K. PDOKF1 crystals in complex with phthalate and terephthalate were obtained by soaking the crystals in cryoprotectant solutions containing 5 mM phthalate or 5 mM terephthalate for 5 min at 25 °C before freezing and data collection.
X-ray data collection and structure determination
Diffraction data were collected at the Home Source, Macromolecular crystallography Unit, IIC, IIT Roorkee, European Synchrotron Radiation Facility (ESRF), and Elettra Sincrotrone Trieste. The data quality was evaluated using PHENIX.XTRIAGE (52). The structure of ROCH34 was solved by a SAD phasing using Auto-Rickshaw (53) and was used as a search model for molecular replacement (MR) for PDOKF1 data, which was performed with Phaser-MR (54) of CCP4i. Iterative rounds of model-building in COOT (55) and refinement of atomic coordinates and B-factors in refmac5 (56) allowed for the correct placement of sidechains and loops. The NCS restraints were used throughout the refinement of PDOKF1 data. The data collection and refinement statistics are summarized in Table 2. The phthalate and terephthalate ligands could be modeled in only one chain of PDOKF1 structure. All figures of protein and ligand structures were prepared using PyMol (57) and UCSF Chimera (58).
Homology modeling and molecular docking
Homology modeling was performed with Modeller 9.21 (59). The ROCH34 structure served as a template for the homology modeling of iPDOE6, while the NDORho (PDB id: 2B1X) structure served for PDORHA1. The alignment (Fig. S1) was produced with the STAMP algorithm (60) implemented in Multiseq (61). Two passes were performed with the following parameters: similarity set to 3, comparison residues set to 10, and slow scan option as performed in Capyk & Eltis, 2012 (62). The program ESPript (63) was used for the visualization of multiple sequence alignments. Molecular docking of isophthalate and phthalate was carried out by docking using AutoDock Vina (64). The ligand molecules of iPDOE6 were prepared using ChemSketch (65). Autogrid4 was utilized for the grid selection, and the dimensions of the box were set to 6 Å × 7 Å × 7 Å for iPDOE6, and 6 Å × 6 Å × 6 Å for PDORHA1. The coordinates' center points were at x = 23.91, y = 7.65, and z = 45.48 for iPDOE6, and x = 25.12, y = 26.72, and z = 48 for PDORHA1. A total of ten docking poses were evaluated based on the proper distances of the substrates to the receptors, and the best pose with the binding energy of −5.9 kcal mol−1 for iPDOE6 and −7.2 kcal mol−1 for PDORHA1 was selected.
Data availability
The ROCH34 and PDOKF1 coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with accession numbers 7FHR, 7FJL, 7V25, 7V28. Other data are available from the corresponding author upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The Authors thank MCU, IIT Roorkee; DBT, India; ESRF (Grenoble), France; and Elettra Synchrotron Trieste, Italy for facilitating with X-ray data collection. P. K. thank IIT Roorkee for providing funds for the anaerobic chamber through SMILE grant.
Author contributions
P. K. conceptualization; E. K., L. D. E., A. K. S., S. T., D. S., N. N., B. W., M. S., J. K. M., and P. K. formal analysis; E. K., L. D. E., A. K. S., S. T., D. S., N. N., B. W., M. S., J. K. M., and P. K. investigation; P. K, L. D. E, J. K. M., and N. N. methodology; P. K. writing–original draft; J. K. M., L. D. E, N. N., E. K., and P. K. writing–review and editing.
Funding and additional information
This work was funded by National Bioscience Award by the Department of Biotechnology (DBT), India (Project No. BT/HRD/NBA/37/01/2015 (VIII)) and WTI 2019 grant by the Department of Science and Technology (DST), India (Project No. DST/TMD/EWO/WTI/2K19/EWFH/2019/8 (G)) to P. K. J. K. M. wants to thank DBT, Govt. of India, N. N. to CSIR, India and B. W and M. S. to MHRD, India for providing fellowship.
Edited by Ruma Banerjee
Supporting information
References
- 1.Lin H., Ge R.-S., Chen G.-R., Hu G.-X., Dong L., Lian Q.-Q., Hardy D.O., Sottas C.M., Li X.-K., Hardy M.P. Involvement of testicular growth factors in fetal Leydig cell aggregation after exposure to phthalate in utero. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7218–7222. doi: 10.1073/pnas.0709260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie M., Wu Y., Little J.C., Marr L.C. Phthalates and alternative plasticizers and potential for contact exposure from children’s backpacks and toys. J. Expo. Sci. Environ. Epidemiol. 2016;26:119–124. doi: 10.1038/jes.2015.71. [DOI] [PubMed] [Google Scholar]
- 3.Melnick R.L. Is peroxisome proliferation an obligatory precursor step in the carcinogenicity of di (2-ethylhexyl) phthalate (DEHP)? Environ. Health Perspect. 2001;109:437–442. doi: 10.1289/ehp.01109437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasai D., Iwasaki T., Nagai K., Araki N., Nishi T., Fukuda M. 2, 3-Dihydroxybenzoate meta-cleavage pathway is involved in o-phthalate utilization in Pseudomonas sp. strain PTH10. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-018-38077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keyser P., Pujar B.G., Eaton R.W., Ribbons D.W. Biodegradation of the phthalates and their esters by bacteria. Environ. Health Perspect. 1976;18:159. doi: 10.1289/ehp.7618159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang H.-K., Zylstra G.J. Novel organization of the genes for phthalate degradation from Burkholderia cepacia DBO1. J. Bacteriol. 1998;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakazawa T., Hayashi E. Phthalate metabolism in Pseudomonas testosteroni: Accumulation of 4, 5-dihydroxyphthalate by a mutant strain. J. Bacteriol. 1977;131:42–48. doi: 10.1128/jb.131.1.42-48.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrauchan M.A., Florizone C., Dosanjh M., Mohn W.W., Davies J., Eltis L.D. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: Redundancies and convergence. J. Bacteriol. 2005;187:4050–4063. doi: 10.1128/JB.187.12.4050-4063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton R.W. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 2001;183:3689–3703. doi: 10.1128/JB.183.12.3689-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson A., Parales J.V., Parales R.E., Gibson D.T., Eklund H., Ramaswamy S. Crystal structure of naphthalene dioxygenase: Side-on binding of dioxygen to iron. Science. 2003;299:1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 11.Tarasev M., Ballou D.P. Chemistry of the catalytic conversion of phthalate into its cis -dihydrodiol during the reaction of oxygen with the reduced form of phthalate dioxygenase. Biochemistry. 2005;44:6197–6207. doi: 10.1021/bi047724y. [DOI] [PubMed] [Google Scholar]
- 12.Lukowski A.L., Liu J., Bridwell-Rabb J., Narayan A.R.H. Structural basis for divergent C–H hydroxylation selectivity in two Rieske oxygenases. Nat. Commun. 2020;11:2991. doi: 10.1038/s41467-020-16729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovaleva E.G., Lipscomb J.D. Versatility of biological non-heme Fe (II) centers in oxygen activation reactions. Nat. Chem. Biol. 2008;4:186–193. doi: 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers M.S., Lipscomb J.D. Salicylate 5-hydroxylase: Intermediates in aromatic hydroxylation by a Rieske monooxygenase. Biochemistry. 2019;58:5305–5319. doi: 10.1021/acs.biochem.9b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauppi B., Lee K., Carredano E., Parales R.E., Gibson D.T., Eklund H., Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase–naphthalene 1, 2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 16.Carredano E., Karlsson A., Kauppi B., Choudhury D., Parales R.E., Parales J.V., Lee K., Gibson D.T., Eklund H., Ramaswamy S. Substrate binding site of naphthalene 1, 2-dioxygenase: Functional implications of indole binding1. J. Mol. Biol. 2000;296:701–712. doi: 10.1006/jmbi.1999.3462. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson A., Parales J.V., Parales R.E., Gibson D.T., Eklund H., Ramaswamy S. NO binding to naphthalene dioxygenase. J. Biol. Inorg. Chem. 2005;10:483–489. doi: 10.1007/s00775-005-0657-1. [DOI] [PubMed] [Google Scholar]
- 18.Ferraro D.J., Okerlund A.L., Mowers J.C., Ramaswamy S. Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase. J. Bacteriol. 2006;188:6986–6994. doi: 10.1128/JB.00707-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gakhar L., Malik Z.A., Allen C.C., Lipscomb D.A., Larkin M.J., Ramaswamy S. Structure and increased thermostability of Rhodococcus sp. naphthalene 1, 2-dioxygenase. J. Bacteriol. 2005;187:7222–7231. doi: 10.1128/JB.187.21.7222-7231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batie C.J., LaHaie E., Ballou D.P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J. Biol. Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 21.Correll C.C., Batie C.J., Ballou D.P., Ludwig M.L. Phthalate dioxygenase reductase: A modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1614. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 22.Tarasev M., Pullela S., Ballou D.P. Distal end of 105–125 loop – a putative reductase binding domain of phthalate dioxygenase. Arch. Biochem. Biophys. 2009;487:10–18. doi: 10.1016/j.abb.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coulter E.D., Moon N., Batie C.J., Dunham W.R., Ballou D.P. Electron paramagnetic resonance measurements of the ferrous mononuclear site of phthalate dioxygenase substituted with alternate divalent metal ions: Direct evidence for ligation of two histidines in the copper(II)-reconstituted protein. Biochemistry. 1999;38:11062–11072. doi: 10.1021/bi9904499. [DOI] [PubMed] [Google Scholar]
- 24.Tarasev M., Kaddis C.S., Yin S., Loo J.A., Burgner J., Ballou D.P. Similar enzymes, different structures: Phthalate dioxygenase is an α3α3 stacked hexamer, not an α3β3 trimer like “normal” Rieske oxygenases. Arch. Biochem. Biophys. 2007;466:31–39. doi: 10.1016/j.abb.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gassner G.T., Ballou D.P. Preparation and characterization of a truncated form of phthalate dioxygenase reductase that lacks an iron-sulfur domain. Biochemistry. 1995;34:13460–13471. doi: 10.1021/bi00041a025. [DOI] [PubMed] [Google Scholar]
- 26.Gassner G.T., Ballou D.P., Landrum G.A., Whittaker J.W. Magnetic circular dichroism studies on the mononuclear ferrous active site of phthalate dioxygenase from Pseudomonas cepacia show a change of ligation state on substrate binding. Biochemistry. 1993;32:4820–4825. doi: 10.1021/bi00069a017. [DOI] [PubMed] [Google Scholar]
- 27.Gassner G., Wang L., Batie C., Ballou D.P. Reaction of phthalate dioxygenase reductase with NADH and NAD: Kinetic and spectral characterization of intermediates. Biochemistry. 1994;33:12184–12193. doi: 10.1021/bi00206a022. [DOI] [PubMed] [Google Scholar]
- 28.Pinto A., Tarasev M., Ballou D.P. Substitutions of the “bridging” aspartate 178 result in profound changes in the reactivity of the Rieske center of phthalate dioxygenase. Biochemistry. 2006;45:9032–9041. doi: 10.1021/bi060216z. [DOI] [PubMed] [Google Scholar]
- 29.Batie C.J., Ballou D.P. Phthalate dioxygenase. Methods Enzymol. 1990;188:61. doi: 10.1016/0076-6879(90)88013-z. [DOI] [PubMed] [Google Scholar]
- 30.Tsang H.T., Batie C.J., Ballou D.P., Penner-Hahn J.E. X-ray absorption spectroscopy of the [2-iron-2-sulfur] Rieske cluster in Pseudomonas cepacia phthalate dioxygenase. Determination of core dimensions and iron ligation. Biochemistry. 1989;28:7233–7240. doi: 10.1021/bi00444a015. [DOI] [PubMed] [Google Scholar]
- 31.Gurbiel R.J., Batie C.J., Sivaraja M., True A.E., Fee J.A., Hoffman B.M., Ballou D.P. Electron-nuclear double resonance spectroscopy of nitrogen-15-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 32.Tierney D.L., Gassner G.T., Luchinat C., Bertini I., Ballou D.P., Penner-Hahn J.E. NMR characterization of substrate binding in the phthalate dioxygenase system. Biochemistry. 1999;38:11051–11061. doi: 10.1021/bi990431y. [DOI] [PubMed] [Google Scholar]
- 33.Pavel E.G., Martins L.J., Ellis W.R., Jr., Solomon E.I. Magnetic circular dichroism studies of exogenous ligand and substrate binding to the non-heme ferrous active site in phthalate dioxygenase. Chem. Biol. 1994;1:173–183. doi: 10.1016/1074-5521(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 34.Bertini I., Luchinat C., Mincione G., Parigi G., Gassner G.T., Ballou D.P. NMRD studies on phthalate dioxygenase: Evidence for displacement of water on binding substrate. J. Biol. Inorg. Chem. 1996;1:468–475. [Google Scholar]
- 35.Tarasev M., Pinto A., Kim D., Elliott S.J., Ballou D.P. The “bridging” aspartate 178 in phthalate dioxygenase facilitates interactions between the Rieske center and the iron (II)- mononuclear center. Biochemistry. 2006;45:10208–10216. doi: 10.1021/bi060219b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuhara Y., Inakazu K., Kodama N., Kamimura N., Kasai D., Katayama Y., Fukuda M., Masai E. Characterization of the isophthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 2010;76:519–527. doi: 10.1128/AEM.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatsu C.H., Straus N.A., Wyndham R.C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3, 4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 38.Kumar P., Mohammadi M., Viger J.-F., Barriault D., Gomez-Gil L., Eltis L.D., Bolin J.T., Sylvestre M. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J. Mol. Biol. 2011;405:531–547. doi: 10.1016/j.jmb.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colbert C.L., Agar N.Y., Kumar P., Chakko M.N., Sinha S.C., Powlowski J.B., Eltis L.D., Bolin J.T. Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P., Mohammadi M., Dhindwal S., Pham T.T.M., Bolin J.T., Sylvestre M. Structural insights into the metabolism of 2-chlorodibenzofuran by an evolved biphenyl dioxygenase. Biochem. Biophys. Res. Commun. 2012;421:757–762. doi: 10.1016/j.bbrc.2012.04.078. [DOI] [PubMed] [Google Scholar]
- 41.Fukuhara Y., Kasai D., Katayama Y., Fukuda M., Masai E. Enzymatic properties of terephthalate 1,2-dioxygenase of Comamonas sp. strain E6. Biosci. Biotechnol. Biochem. 2008;72:2335–2341. doi: 10.1271/bbb.80236. [DOI] [PubMed] [Google Scholar]
- 42.Hara H., Eltis L.D., Davies J.E., Mohn W.W. Transcriptomic analysis reveals a bifurcated terephthalate degradation pathway in Rhodococcus sp. strain RHA1. J. Bacteriol. 2007;189:1641–1647. doi: 10.1128/JB.01322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furusawa Y., Nagarajan V., Tanokura M., Masai E., Fukuda M., Senda T. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 2004;342:1041–1052. doi: 10.1016/j.jmb.2004.07.062. [DOI] [PubMed] [Google Scholar]
- 44.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Robert L.D., Rydel T.J., Storek M.J., Sturman E.J., Moshiri F., Bartlett R.K., Brown G.R., Eilers R.J., Dart C., Qi Y. Dicamba monooxygenase: Structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation. J. Mol. Biol. 2009;392:481–497. doi: 10.1016/j.jmb.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 46.Nojiri H., Ashikawa Y., Noguchi H., Nam J.-W., Urata M., Fujimoto Z., Uchimura H., Terada T., Nakamura S., Shimizu K. Structure of the terminal oxygenase component of angular dioxygenase, carbazole 1, 9a-dioxygenase. J. Mol. Biol. 2005;351:355–370. doi: 10.1016/j.jmb.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 47.Dumitru R., Jiang W.Z., Weeks D.P., Wilson M.A. Crystal structure of dicamba monooxygenase: A Rieske nonheme oxygenase that catalyzes oxidative demethylation. J. Mol. Biol. 2009;392:498–510. doi: 10.1016/j.jmb.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohta T., Chakrabarty S., Lipscomb J.D., Solomon E.I. Near-IR MCD of the nonheme ferrous active site in naphthalene 1, 2-dioxygenase: Correlation to crystallography and structural insight into the mechanism of Rieske dioxygenases. J. Am. Chem. Soc. 2008;130:1601–1610. doi: 10.1021/ja074769o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J., Fritsch E.F., Maniatis T. Cold spring harbor laboratory press; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 50.Zabinski R., Münck E., Champion P.M., Wood J.M. Kinetic and Mössbauer studies on the mechanism of protocatechuic acid 4, 5-oxygenase. Biochemistry. 1972;11:3212–3219. doi: 10.1021/bi00767a012. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. Phenix: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panjikar S., Parthasarathy V., Lamzin V.S., Weiss M.S., Tucker P.A. On the combination of molecular replacement and single-wavelength anomalous diffraction phasing for automated structure determination. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1089–1097. doi: 10.1107/S0907444909029643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLano W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 58.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell R.B., Barton G.J. Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 61.Roberts E., Eargle J., Wright D., Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:1–11. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Capyk J.K., Eltis L.D. Phylogenetic analysis reveals the surprising diversity of an oxygenase class. J. Biol. Inorg. Chem. 2012;17:425–436. doi: 10.1007/s00775-011-0865-9. [DOI] [PubMed] [Google Scholar]
- 63.Gouet P., Courcelle E., Stuart D., Metoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 64.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter A.D. ACS Publications; Washington, DC: 1997. ACD/ChemSketch 1.0 (Freeware); ACD/ChemSketch 2.0 and its Tautomers, Dictionary, and 3D Plug-Ins; ACD/HNMR 2.0; ACD/CNMR 2.0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ROCH34 and PDOKF1 coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with accession numbers 7FHR, 7FJL, 7V25, 7V28. Other data are available from the corresponding author upon reasonable request.