Abstract
Background: Non-small cell lung cancer (NSCLC) is the most common type of lung cancer affecting humans. However, appropriate biomarkers for diagnosis and prognosis have not yet been established. Here, we evaluated the gene expression profiles of patients with NSCLC to identify novel biomarkers. Methods: Three datasets were downloaded from the Gene Expression Omnibus (GEO) database, and differentially expressed genes were analyzed. Venn diagram software was applied to screen differentially expressed genes, and gene ontology functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed. Cytoscape was used to analyze protein-protein interactions (PPI) and Kaplan–Meier Plotter was used to evaluate the survival rates. Oncomine database, Gene Expression Profiling Interactive Analysis (GEPIA), and The Human Protein Atlas (THPA) were used to analyze protein expression. Quantitative real-time polymerase (qPCR) chain reaction was used to verify gene expression. Results: We identified 595 differentially expressed genes shared by the three datasets. The PPI network of these differentially expressed genes had 202 nodes and 743 edges. Survival analysis identified 10 hub genes with the highest connectivity, 9 of which (CDC20, CCNB2, BUB1, CCNB1, CCNA2, KIF11, TOP2A, NDC80, and ASPM) were related to poor overall survival in patients with NSCLC. In cell experiments, CCNB1, CCNB2, CCNA2, and TOP2A expression levels were upregulated, and among different types of NSCLC, these four genes showed highest expression in large cell lung cancer. The highest prognostic value was detected for patients who had successfully undergone surgery and for those who had not received chemotherapy. Notably, CCNB1 and CCNA2 showed good prognostic value for patients who had not received radiotherapy. Conclusion: CCNB1, CCNB2, CCNA2, and TOP2A expression levels were upregulated in patients with NSCLC. These genes may be meaningful diagnostic biomarkers and could facilitate the development of targeted therapies.
Keywords: non-small cell lung cancer, gene expression omnibus database, bioinformatics analysis, prognosis, diagnosis, biomarkers
Introduction
Lung cancer is highly invasive and metastatic and is a major cause of cancer-related deaths. Non-small cell lung cancer (NSCLC), the most common of lung cancer, accounts for 85% of all lung cancers. 1 Despite major advances in the diagnosis and treatment of NSCLC with the development of medical technology in recent years, the 5-year survival rate of patients with NSCLC is only 17%. 2 Changes in social lifestyles and environments have led to an increase in the incidence of NSCLC, and approximately 234 000 new cases of NSCLC are reported in the United States of America each year.3,4 Regardless of which therapeutic option is chosen, chemotherapy is still an essential adjuvant treatment for patients with lung cancer. However, serious adverse reactions can occur following administration of chemotherapy drugs, and the efficacy of these therapies is often not satisfactory.5,6 Therefore, there is an urgent need for new treatment strategies to complement traditional chemotherapy. Moreover, with commencement of the genomic era and advancements in molecular biology research, molecular mechanisms of life phenomena and disease have attracted much attention, and recent research on NSCLC has focused on the identification of novel targets to facilitate the development of new molecular targeted drugs.
With the development of sequencing technology and large-scale sequencing research, large amounts of sequencing data have been collected in many databases. 7 To discover biomarkers and potential targets related to cancer, researchers can resequence the tumor and then re-analyze the tumor from gene to protein expression.8,9 Furthermore, to avoid inaccurate experimental results owing to the use of multiple platforms or small sample sizes, comprehensive bioinformatics methods can be used to obtain valuable biological information in cancer research.10,11
Many novel targets and biomarkers of NSCLC have been reported in recent studies, providing substantial contributions to NSCLC research. For example, in a study by Hu et al. miR-210 was found to serve as a potential biomarker for NSCLC detection, and they found that the use of a set of multiple biomarkers may be a more comprehensive indicator than the analysis of miR-210 alone. 12 Additionally, Wang provided a dataset of NSCLC biomarkers with potential applications in prognosis and found that TOP2A may be a valuable biomarker for survival and prognosis in patients with NSCLC. 13 Saigusa and colleagues also revealed that new metabolites related to NRF2 activity may be diagnostic biomarkers for NRF2 activation, providing important insights into the selective toxicity of new metabolic nodes in NRF2-activated NSCLC. 14 Despite these findings, NSCLC remains a complex and diverse disease, and more genetic data and biological information are needed to improve diagnosis and treatment strategies.
In the present study, we selected three datasets, ie, GSE18842, 15 GSE44077, 16 and GSE19804, 17 from the Gene Expression Omnibus (GEO) database and conducted various bioinformatics analyses to evaluate gene expression and protein interactions in NSCLC tumor tissues, with the aim of elucidating the underlying molecular mechanisms in NSCLC and for establishing novel biomarkers for its diagnosis and treatment.
Materials and Methods
The gene expression profile data (GSE18842, GSE44077, and GSE19804) were downloaded from GEO (Supplementary Tables S1-S3). 18 The inclusion criteria for gene expression data were as follows: (1) the samples used for analysis were tissues, (2) all tissues were categorized as NSCLC or normal tissues, (3) samples were collected from the same species group, (4) probes could be converted, (5) complete information was available for analysis, and (6) the sample size of each study was larger than 10 samples. The array data for GSE18842 included 46 NSCLC tumor and 45 normal tissues as the control group. GSE19804 included 60 NSCLC tumors and 60 adjacent normal tissues. GSE44077 was composed of 66 tumor tissues and 55 normal tissues.
GEO2R was used to analyze differentially expressed genes (DEGs) between NSCLC samples and normal samples (http://www.ncbi.nlm.nih.gov/geo/geo2r). GEO2R is a web tool that can be used to compare and analyze DEGs in NSCLC samples and normal lung tissue samples through the Limma and GEOquery R packages of the Bioconductor project. Adjusted P values and |log2 fold change| (|log2FC|) values were used to assess the significance of DEGs, and the cut-off criteria were set as |log2FC| > 1 and adjusted P value < .01.
Venn diagram software in the Bioinformatics & Evolutionary Genomics platform (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to perform intersection analysis on the DEGs of our three independent samples. We screened out genes that were differentially expressed in the three independent samples and determined whether the selected genes were up- or downregulated based on log2FC values of DEGs between NSCLC and normal tissues.
Gene Ontology (GO), a commonly used bioinformatics tool for comprehensive evaluation of gene function; Kyoto Encyclopedia of Genes and Genomes (KEGG), a database for annotation of the advanced functions of biological systems at the molecular level; and the Database for Annotation, Visualization, and Integrated Discovery (DAVID; https://david.ncifcrf.gov/), an online tool for enriching and analyzing bioinformatics resources. 19 , were used to clarify gene functions and functional enrichment. Upregulated DEGs (uDEGs) and downregulated DEGs (dDEGs) were identified using DAVID for the three gene expression profile datasets. The cut-off criterion was set as a P value less than 0.05.
The STRING (http://string-db.org/) database, a search tool for retrieving interacting genes and providing important information regarding protein-protein interactions (PPIs), 20 was used to construct a PPI network of DEGs, and Cytoscape (version 3.7.2) was used to process and analyze the PPI network. 21 The cut-off criterion was a combined score greater than or equal to 0.9. Subsequently, we used a Cytoscape plug-in Molecular Complex Detection (MCODE) to detect important modules in the PPI network, with the following parameters: cut-off degree = 2, cut-off node score = 0:2, K-core = 2, and maximum depth = 100. Functional annotation of DEGs in the identified module was investigated with DAVID. The cut-off criterion was set to a P value less than 0.05.
The Kaplan–Meier Plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=lung), which provides information on the relationships of more than 54 000 genes (mRNAs, microRNAs, and proteins) with survival rates in 21 cancer types, 22 was used to investigate whether the top 10 hub genes were related to overall survival in patients with NSCLC. We also assessed relationships according to histological type, including adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC). Subsequently, we analyzed the association of overall survival rate with hub genes according to treatment strategies in order to compare the prognostic significance of hub genes under different treatment regimens. The selection criteria were as follows: hazard ratio (HR) within the 95% confidence interval (CI) and log-rank P value less than .05.
Next, we used the Cytoscape plug-in CytoHubba to identify hub genes, subnets of complex networks, and central elements in the network. The Biological Networks Gene Oncology Tool (BiNGO; version 3.0.3) plug-in in Cytoscape was used to analyze and visualize the biological processes (BPs) associated with the hub genes. The Oncomine database (http://www.oncomine.org) was used to compare the mRNA expression levels of hub genes between lung cancer tissues and normal control tissues and between different types of lung cancer. Data were collected from all related datasets.
Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) is a new web-based interactive analysis and visualization tool based on The Cancer Genome Atlas (TCGA) database and genotype tissue expression. 23 To further verify the 10 hub genes identified from the PPI network, differences in gene expression between LUSC, LUAD, and adjacent lung tissues were mapped in TCGA database and GTEx database using the GEPIA web tool Box Plots. The patient data were grouped according to the results per million reads (TPM). Log2(TPM + 1) was used as the logarithmic scale, and the criteria were as follows: |log2FC| > 1 and p < .01.
The Human Protein Atlas (THPA), which maps all human proteins in cells, tissues and organs by integrating various omics technologies (including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems). 24 , was used to assess protein-based differences between NSCLC and normal tissues. First, we downloaded the histological section images and corresponding information on overexpressed hub genes from normal bronchial respiratory tract epithelial tissues and lung cancer tissues obtained by immunohistochemistry from THPA. Because some antibody staining may be inconsistent, the detection results were reported as low, medium, or high according to the staining intensity and score of the stained cells. We then performed Mann–Whitney U tests using SPSS 23.0 software to compare the antibody staining levels of hub genes between normal lung tissues and bronchial epithelial tissues. The cut-off P value was set to .05.
All cell lines were obtained from Professor Haidan Liu (Changsha). A549 human NSCLC cells (epidermal growth factor receptor wild type) and HBE normal immortalized lung epithelial cells were purchased from American Type Culture Collection (ATCC). The cells were cultured in a humidified incubator at 37°C with 5% CO2 according to ATCC protocols. A549 cells were subjected to mycoplasma analysis and were cytogenetically tested and authenticated before being frozen. HBE cells were maintained in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Biological Industries) and 1% antibiotics. A549 cells were maintained in RPMI1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS (Biological Industries) and 1% antibiotics.
For quantitative real-time polymerase chain reaction (qPCR), HBE and A549 cells were seeded in 100-mm Petri dishes and grown for 1 day at 37 °C in an atmosphere containing 5% CO2. After 48 h, total RNA was extracted using a GeneJET RNA Purification Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Additionally, a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) was used to synthesize cDNA based on the manufacturer's recommendations. qPCR was then performed in a Thermo 9700 Fast Real-time PCR system using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). The qPCR reaction conditions were: UDG enzyme activation, 50℃ for 2 min, a hold; pre-denaturation, 95℃ for 2 min, a hold; denaturation, 95℃ for 3 s and annealing and extension, 60℃ for 30 s, 40 cycles. The melting curve selects the default setting. The reaction system used a 10ul reaction system, and the sample amount of cdna is 10 ng per reaction. The primers used were as follows: ARF5-F, 5′-ATCTGTTTCACAGTCTGGGACG-3′; ARF5-R, 5′-CCTGCTTGTTGGCAAATACC-3′; CDC20-F, 5′-AGACATTCACCCAGCATCAAG-3′; CDC20-R, 5′-GAGATGAGCTCCTTGTAATGGG-3′; CCNB1-F, 5′-GGCTTTCTCTGATGTAATTCTTGC-3′; CCNB1-R, 5′-GTATTTTGGTCTGACTGCTTGC-3′; TOP2A-F, 5′-ATCTAAACCTCTTGCAGCCC-3′; TOP2A-R, 5′-GCACCATTTATCAGCACCATG-3′; CCNB2-F, 5′-ACCTACTGCTTCTGTCAAACC-3′; CCNB2-R, 5′-TGTCCTCGATTTTGCAGAGC-3′; CCNA2-F, 5′-CCTTTCATTTAGCACTCTACACAG-3′; CCNA2-R, 5′-CCAGGGTATATCCAGTCTTTCG-3′; KIF11-F, 5′-ACCTCATGTTCCTTATCGAGAATC-3′; KIF11-R, 5′-GCATATTCCAATGTACTCAGAGTTTC-3′. ARF5 was used as an internal control, 25 and fold changes in mRNA levels were determined using the 2−ΔΔCT method.
All data were presented as means ± standard deviations. GraphPad Prism 8.0 (GraphPad Software) and SPSS software were used for all statistical analyses. Two-tailed Student's t-tests were used to assess the significance of differences between the two groups. Unless otherwise stated, results with P values less than .05 were considered statistically significant.
Results
Identification of Differentially Expressed Cells in Non-Small Cell Lung Cancer
According to GEO2R analysis, after standardizing the chip data, DEGs (4642 in GSE18842, 1960 in GSE19804, and 1235 in GSE44077) were identified. Among these DEGs, the three datasets together contained 594 genes, as demonstrated by Venn diagram analysis in the Bioinformatics & Evolutionary Genomics platform (Figure 1A and Supplementary Table S4). We matched the log2FC values of three independent datasets to DEGs and found that 177 uDEGs and 417 dDEGs were identified between NSCLC and normal tissues in all three independent datasets.
Figure 1.
Venn diagram, protein-protein interaction (PPI) network, the most significant module, and the top 10 highest scoring nodes of differentially expressed genes (DEGs). (A) DEG identification in three gene expression profile datasets (GSE18842, GSE44077, and GSE19804). In total, 594 DEGs were identified. (B) A PPI network was generated using Cytoscape (combined score ≥ 0.9). (C) The significant module obtained from the PPI network contained 24 nodes and 256 edges. (D) The interaction network of the 10 nodes with the highest screening scores. Upregulated genes are marked in dark red; downregulated genes are marked in dark blue. The red, orange, and yellow nodes represented the top 10 hub genes in the network.
Gene Ontology Functional Pathways and Analysis of Differentially Expressed Genes
GO analyses showed that uDEGS were mainly enriched in mitotic nuclear division, whereas dDEGs were mainly enriched in cell adhesion (Supplementary Table S5). Furthermore, cell component (CC) analysis indicated that most of the uDEGs were located in the midbody, whereas the dDEGs were mainly distributed in the proteinaceous extracellular matrix. Additionally, according to molecular function (MF) analysis, uDEGs were significantly associated with metalloendopeptidase activity, whereas dDEGs were associated with heparin binding (Table 1). KEGG pathway analysis suggested that most uDEGs were mainly involved in the BPs of the cell cycle, whereas most dDEGs were involved in cell adhesion (Table 2).
Table 1.
GO Function Annotation of uDEGs and dDEGs Associated With NSCLC (top five).
| Category | Term | Count | % | P-value |
|---|---|---|---|---|
| uDEGs | ||||
| GOTERM_BP_DIRECT | GO:0007067∼mitotic nuclear division | 22 | 0.07753304 | 7.26E-14 |
| GOTERM_BP_DIRECT | GO:0051301∼cell division | 23 | 0.081057269 | 7.34E-12 |
| GOTERM_BP_DIRECT | GO:0030574∼collagen catabolic process | 10 | 0.035242291 | 1.47E-08 |
| GOTERM_BP_DIRECT | GO:0007062∼sister chromatid cohesion | 11 | 0.03876652 | 8.61E-08 |
| GOTERM_BP_DIRECT | GO:0008283∼cell proliferation | 17 | 0.059911894 | 9.22E-07 |
| GOTERM_CC_DIRECT | GO:0030496∼midbody | 12 | 0.042290749 | 3.90E-08 |
| GOTERM_CC_DIRECT | GO:0000777∼condensed chromosome kinetochore | 10 | 0.035242291 | 1.31E-07 |
| GOTERM_CC_DIRECT | GO:0005819∼spindle | 10 | 0.035242291 | 2.20E-06 |
| GOTERM_CC_DIRECT | GO:0000922∼spindle pole | 9 | 0.031718062 | 9.04E-06 |
| GOTERM_CC_DIRECT | GO:0000776∼kinetochore | 8 | 0.028193833 | 1.14E-05 |
| GOTERM_MF_DIRECT | GO:0004222∼metalloendopeptidase activity | 12 | 0.042290749 | 1.37E-08 |
| GOTERM_MF_DIRECT | GO:0004252∼serine-type endopeptidase activity | 15 | 0.052863436 | 2.15E-07 |
| GOTERM_MF_DIRECT | GO:0019901∼protein kinase binding | 14 | 0.049339207 | 8.48E-05 |
| GOTERM_MF_DIRECT | GO:0003777∼microtubule motor activity | 7 | 0.024669604 | 1.32E-04 |
| GOTERM_MF_DIRECT | GO:0042802∼identical protein binding | 19 | 0.066960352 | 3.92E-04 |
| dDEGs | ||||
| GOTERM_BP_DIRECT | GO:0007155∼cell adhesion | 38 | 0.059311981 | 9.75E-12 |
| GOTERM_BP_DIRECT | GO:0001525∼angiogenesis | 23 | 0.035899357 | 4.70E-09 |
| GOTERM_BP_DIRECT | GO:0007166∼cell surface receptor signaling pathway | 25 | 0.03902104 | 9.87E-09 |
| GOTERM_BP_DIRECT | GO:0001570∼vasculogenesis | 10 | 0.015608416 | 3.37E-06 |
| GOTERM_BP_DIRECT | GO:0045766∼positive regulation of angiogenesis | 13 | 0.020290941 | 8.80E-06 |
| GOTERM_CC_DIRECT | GO:0005578∼proteinaceous extracellular matrix | 30 | 0.046825248 | 1.58E-12 |
| GOTERM_CC_DIRECT | GO:0005886∼plasma membrane | 146 | 0.227882874 | 3.56E-10 |
| GOTERM_CC_DIRECT | GO:0005887∼integral component of plasma membrane | 68 | 0.106137229 | 1.70E-09 |
| GOTERM_CC_DIRECT | GO:0016021∼integral component of membrane | 161 | 0.251295499 | 3.09E-07 |
| GOTERM_CC_DIRECT | GO:0045121∼membrane raft | 19 | 0.029655991 | 7.41E-07 |
| GOTERM_MF_DIRECT | GO:0008201∼heparin binding | 17 | 0.026534307 | 2.62E-07 |
| GOTERM_MF_DIRECT | GO:0005509∼calcium ion binding | 38 | 0.059311981 | 4.32E-07 |
| GOTERM_MF_DIRECT | GO:0030246∼carbohydrate binding | 16 | 0.024973466 | 1.72E-05 |
| GOTERM_MF_DIRECT | GO:0005178∼integrin binding | 11 | 0.017169258 | 7.15E-05 |
| GOTERM_MF_DIRECT | GO:0003779∼actin binding | 18 | 0.028095149 | 8.51E-05 |
Abbreviations: dDEGS: downregulated differentially expressed gene; GO: Gene Ontology; NSCLC: non-small cell lung cancer; uDEGs: upregulated differentially expressed genes.
Table 2.
KEGG Pathway Analysis of DEGs Associated With NSCLC.
| Category | Term | Count | % | P-value |
|---|---|---|---|---|
| uDEGs | ||||
| KEGG_PATHWAY | hsa04110:Cell cycle | 15 | 0.052863436 | 1.42E-10 |
| KEGG_PATHWAY | hsa04115:p53 signaling pathway | 8 | 0.028193833 | 1.29E-05 |
| KEGG_PATHWAY | hsa04114:Oocyte meiosis | 9 | 0.031718062 | 4.72E-05 |
| KEGG_PATHWAY | hsa04914:Progesterone-mediated oocyte maturation | 6 | 0.021145374 | .003663974 |
| KEGG_PATHWAY | hsa04512:ECM-receptor interaction | 6 | 0.021145374 | .003663974 |
| dDEGs | ||||
| KEGG_PATHWAY | hsa04514:Cell adhesion molecules (CAMs) | 12 | 0.018730099 | .001027649 |
| KEGG_PATHWAY | hsa05144:Malaria | 7 | 0.010925891 | .001487862 |
| KEGG_PATHWAY | hsa04270:Vascular smooth muscle contraction | 10 | 0.015608416 | .003069086 |
| KEGG_PATHWAY | hsa03320:PPAR signaling pathway | 7 | 0.010925891 | .007259593 |
| KEGG_PATHWAY | hsa04610:Complement and coagulation cascades | 7 | 0.010925891 | .00836134 |
Abbreviations: DEGs: differentially expressed genes; dDEGs: downregulated differentially expressed genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; NSCLC: non-small cell lung cancer; uDEGs: upregulated differentially expressed genes.
Protein-Protein Interaction Network Construction and Analysis of Modules
Next, we constructed a PPI network of DEGs. The PPI network contained 202 nodes and 743 edges, including 75 uDEGs and 124 DEGs (Figure 1B). According to the selection conditions, 19 sets of datasets were obtained, and the one with the highest cluster score was selected for subsequent in-depth analyses. The PPI network with the highest cluster score calculated using the MCODE plug-in generated 24 nodes and 256 edge-related modules, with a cluster score of 22.261 (Figure 1C and Supplementary Table S6). The top 10 highest scoring nodes, including CDC20, CCNB2, BUB1, CCNB1, CCNA2, KIF11, TOP2A, NDC80, ASPM, CDK1, were then evaluated using the Cytohubba plug-in (Table 3) and were used to build an interaction network in MCODE (Figure 1D). In addition to the 10 genes described above, other nodes in the module included KIF4A, PRC1, RRM2, PTTG1, MELK, TPX2, AURKA, CEP55, MAD2L1, NEK2, NUF2, DLGAP5, KIF2C, and PBK. The expression levels of all genes in the module were upregulated.
Table 3.
Top 10 in Network Ranked by Degree Method.
| Rank | Name | Type | Score |
|---|---|---|---|
| 1 | CDK1 | up | 42 |
| 2 | CDC20 | up | 40 |
| 3 | CCNB2 | up | 35 |
| 4 | BUB1 | up | 34 |
| 4 | CCNB1 | up | 34 |
| 6 | CCNA2 | up | 33 |
| 7 | KIF11 | up | 32 |
| 8 | TOP2A | up | 31 |
| 9 | NDC80 | up | 30 |
| 10 | ASPM | up | 29 |
Hub Gene Selection and Survival Analysis
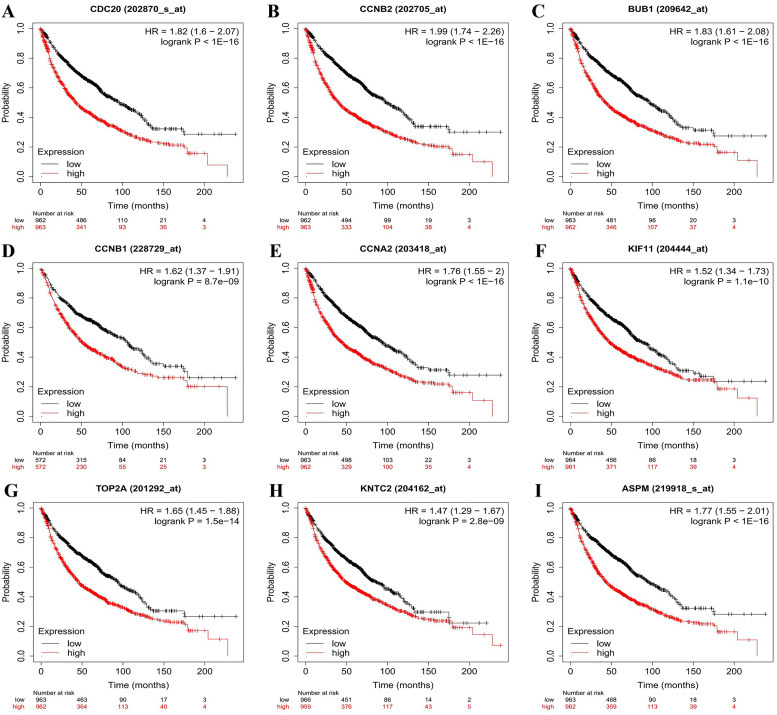
Using the Kaplan–Meier Plotter online database, we evaluated the relationships between nine of the 10 hub genes (excluding CDK1) and overall survival rates (Figure 2). The results indicated that the overexpressed hub genes were related to unfavorable overall survival in patients with NSCLC, as follows: CDC20 (HR = 1.82 [95% CI: 1.6-2.07], log-rank P < 1e − 16), CCNB2 (HR = 1.99 [95% CI: 1.74-2.26], log-rank P < 1e − 16), BUB1 (HR = 1.77 [95% CI: 1.55-2.01], log-rank P = 1e-16], CCNB1 [HR = 1.62 [95% CI: 1.37-1.91], log-rank P = 8.7e − 09], CCNA2 [HR = 1.76 [95% CI: 1.55-2], log-rank P < 1e − 16], KIF11 [HR = 1.52 [95% CI: 1.34-1.73], log-rank P = 1.1e − 10], TOP2A [HR = 1.65 [95% CI: 1.45-1.88], log-rank P = 1.5e − 14], NDC80 [HR = 1.47 [95% CI: 1.29-1.67], log-rank P = 2.8e − 09], ASPM [HR = 1.77 [95% CI: 1.55-2.01], log-rank P < 1e − 16]. Analysis of the BPs enriched for the nine hub genes is shown in Figure 3. Furthermore, using the Oncomine database, we confirmed that the nine hub genes were upregulated in NSCLC tissues compared with control tissues and cells [all P < 1e-4; Figure 4].
Figure 2.
Association of hub genes with overall survival rates. Kaplan–Meier Plotter was used to evaluate overall survival rates based high or low expression of (A) CDC20, (B) CCNB2, (C) BUB1, (D) CCNB1, (E) CCNA2, (F) KIF11, (G) TOP2A, (H) NDC80, and (I) ASPM in patients with non-small cell lung cancer (NSCLC).
Figure 3.
Analysis of biological processes (BPs) enriched in hub genes. BiNGO was used to assess the BPs enriched in hub genes. The color depth of the node reflects the corrected P value.
Abbreviation: BiNGO: Biological Networks Gene Oncology Tool.
Figure 4.
Evaluation of gene expression in cancer tissues and normal tissues using Oncomine. Oncomine was used to evaluate (A) CDC20, (B) CCNB2, (C) BUB1, (D) CCNB1, (E) CCNA2, (F) KIF11, (G) TOP2A, (H) NDC80, and (I) ASPM expression in cancer tissues versus normal tissues.
Verification of hub Genes Using Gene Expression Profiling Interactive Analysis
To determine the reliability of DEGs identified from GSE18842, GSE19804, and GSE44077, we used GEPIA for the assessment of hub gene expression levels in LUAD, LUSC, and normal lung tissues reported in the TCGA database. Consistent with the results of bioinformatics analysis of GEO data, the nine hub genes were identified, and their expression levels were found to be significantly increased (Figure 5).
Figure 5.
Expression levels of the nine hub genes. Heat map showing the expression levels of nine hub genes (CDC20, CCNB2, BUB1, CCNB1, CCNA2, KIF11, TOP2A, NDC80, and ASPM) in LUAD, LUSC, and normal lung tissues based on TCGA data analyzed using the GEPIA web tool. T represents LUAD or LUSC tumor tissues, and N represents normal lung tissues.
Abbreviations: GEPIA: Gene Expression Profiling Interactive Analysis; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; TCGA: The Cancer Genome Atlas.
Verification of hub Genes Based on Proteomics Analysis
Next, we used immunohistochemical images of proteins from THPA database to further verify the relevance of six hub genes in lung cancer tissues (Figure 6A). CDC20 (18/21), CCNB2 (17/26), CCNB1 (29/35), CCNA2 (10/11), KIF11 (31/33), and TOP2A (34/34) had high positive staining rates; however, only CDC20 (P = .000014), CCNB1 (P = .000021), and TOP2A (P = .000013) were compared by Mann–Whitney U test using normal lung cells, bronchial epithelial cells, and lung cancer tissues. The results showed statistically significant differences in the protein levels of these hub genes (Figure 6B), suggesting potential applications as biomarkers in the diagnosis of NSCLC.
Figure 6.
Verification of the protein expression levels of hub genes. (A) Immunohistochemistry images of six hub genes were obtained from https://www.proteinatlas.org/ENSG00000117399-CDC20/pathology/lung + cancer#ihc (CDC20), https://www.proteinatlas.org/ENSG00000157456-CCNB2/pathology/lung + cancer#ihc (CCNB2), https://www.proteinatlas.org/ENSG00000134057-CCNB1/pathology/lung + cancer#ihc (CCNB1), https://www.proteinatlas.org/ENSG00000145386-CCNA2/pathology/lung + cancer#ihc (CCNA2), https://www.proteinatlas.org/ENSG00000138160-KIF11/pathology/lung + cancer#ihc (KIF11), and https://www.proteinatlas.org/ENSG00000131747-TOP2A/pathology/lung + cancer#ihc (TOP2A) in THPA. (B) The antibodies used targeted CDC20 (HPA055288 and CAB004525), CCNB2 (CAB009575 and HPA008873), CCNB1 (CAB000115, CAB003804, and HPA061448), CCNA2 (CAB000114), KIF11 (HPA006916, HPA010568, and CAB017617), and TOP2A (HPA006458, HPA026773, and CAB002448). Mann–Whitney U tests were used for assessing significance, with a cut-off P value of .05.
Abbreviation: THPA: The Human Protein Atlas.
Verification of hub Gene Upregulation by Quantitative Polymerase Chain Reaction
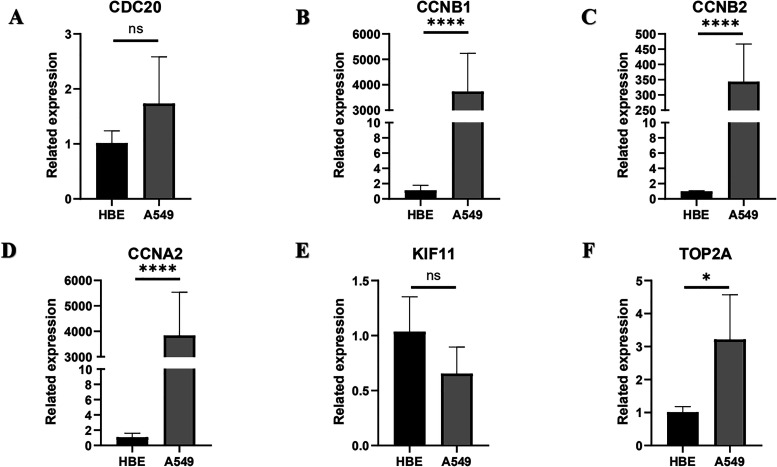
We then used qPCR to analyze the expression levels of the six hub genes (including three screened out and three unverified) in A549 and HBE cells. Consistent with bioinformatics analysis data, CCNB1, CCNB2, CCNA2, and TOP2A were significantly upregulated in A549 cells compared with that in HBE cells (Figure 7 and Supplementary Tables S7-S9).
Figure 7.
Expression levels of six hub genes in A549 and HBE cells. Expression levels were verified using RT-qPCR. (A) CDC20, (B) CCNB1, (C) CCNB2, (D) CCNA2, (E) KIF11, TOP2A. *P < .05, **P < .01, ***P < .001, ****P < .0001, ns: not significant.
Analysis of hub Gene Expression Levels in Different Types of Non-Small Cell Lung Cancer
The Oncomine database was used to analyze differences in the expression level of four hub genes in different types of NSCLCs reported in Hou's dataset. 26 The analytical results showed that compared with normal control tissues, gene expression levels were elevated in all types of NSCLCs, consistent with the verification results. Moreover, in a comparison between large cell lung carcinoma, LUAD, and LUSC, the expression levels of all four hub genes were highest in large cell lung carcinoma, followed by LUSC and then LUAD (Figure 8).
Figure 8.
Predictive ability of hub genes in distinguishing among different types of non-small cell lung cancer (NSCLC) tissues. Data were evaluated using Oncomine. (A) CCNB1, (B) CCNB2, (C) CCNA2, and (D) TOP2A.
Analysis of the Relationships Between Overall Survival and hub Gene Expression Based on Treatment Strategies in Patients With Non-Small Cell Lung Cancer
Finally, we used Kaplan–Meier Plotter online database to assess the relationships between overall survival rates and hub gene expression according to treatment strategies. As shown in Figure 9A, F, K, and P, the four hub genes significantly predicted overall survival for patients who underwent surgical resection only. Moreover, as shown in Figure 9B, G, L, and Q, among the four hub genes, only CCNB1 could predict overall survival in patients who underwent chemotherapy; however, the median survival time was lower for patients categorized as having high expression of the four hub genes than for patients categorized as having low expression. We also found that the four hub genes significantly predicted overall survival in patients who were not treated with chemotherapy (Figure 9C, H, M, and R), whereas no significant prediction ability was observed in patients who underwent radiotherapy (Figure 9D, I, N, and S), although the median survival time in patients with high expression was still shorter than that in patients with low hub gene expression. Additionally, in patients who were not treated with radiotherapy, CCNB1 and CCNA2 significantly predicted overall survival, and patients with high expression of the other two hub genes also had lower median survival times (Figure 9E, J, O, and T).
Figure 9.
Predictive ability of hub genes for overall survival in patients treated using different approaches. Kaplan–Meier Plotter was used to evaluate overall survival rates according to high and low expression of (A–E) CCNB1, (F–J) CCNB2, (K–O) CCNA2, and (P–T) TOP2A in patients with NSCLC. (A, F, K, and P) Surgical margin-negative patients; (B, G, L, and Q) chemotherapy; (C, H, M, and R) no chemotherapy; (D, I, N, and S) radiotherapy; (E, J, O, and T) no radiotherapy.
Discussion
Lung cancer is a major threat to human health, and although targeted immunotherapies have improved the quality of life of many patients, clinical outcomes in patients with NSCLC remain poor. To date, the pathogenic mechanisms of NSCLC have not been fully elucidated; however, multiple genes and pathways are known to be involved, resulting in complex biological behaviors. Accordingly, improving our understanding of the molecular mechanisms of NSCLC, particularly using genomics, transcriptomics, proteomics, and metabolomics analyses, is expected to lead to the development of better diagnostic and therapeutic strategies based on identification of novel biomarkers. 27
In this study, we selected 594 DEGs, including 177 uDEGs and 417 dDEGs, and found that the uDEGs were primarily involved in mitotic nuclear division, cell cycle, p53 signaling, oocyte meiosis, progesterone-mediated oocyte maturation, and extracellular matrix-receptor interactions, whereas the dDEGs were mainly enriched in cell adhesion, malaria, vascular smooth muscle contraction, peroxisome proliferator-activated receptor signaling, complement molecules, and coagulation cascade. Thus, this functional enrichment analysis provided insights into the signaling pathways involved in the occurrence and development of NSCLC. In normal and tumor cells, mitotic nuclear division, cell cycle, and meiotic division of oocytes are known to be important cellular processes, 28 and key tumor-related genes usually modulate tumor progression by regulating these cellular processes. 29 Furthermore, some studies have shown that cell adhesion molecules play crucial roles in tumor development, 30 consistent with our current results. Thus, although the specific mechanisms of disease progression are still not clearly defined, the signaling pathways identified above were confirmed to be related to NSCLC.
Next, we generated a PPI network of the DEGs. The top 10 genes and one relevant module extracted from the PPI network were all upregulated and survival analysis showed that nine of these genes (CDC20, CCNB2, BUB1, CCNB1, CCNA2, KIF11, TOP2A, NDC80, and ASPM) were significantly correlated with overall survival in patients with NSCLC. We further confirmed that CCNB1, CCNB2, CCNA2, and TOP2A were upregulated in NSCLC cells compared with normal lung cells and demonstrated that these four hub genes showed higher prognostic value in patients who underwent surgery and in patients who had not received chemotherapy. Furthermore, CCNB1 and CCNA2 had good prognostic value in patients who had not received radiotherapy. Overall, these findings suggested that the four hub genes may have applications as promising biomarkers in NSCLC.
CCNB1 encodes the regulatory protein cyclin B1, which is involved in mitosis. The gene product can interact with p34 (CDC2) to form a maturation promoting factor and has been shown to modulate the G2/M transition phase of the cell cycle. Moreover, CCNB1 has been shown to act as a potential diagnostic marker for rhabdomyosarcoma and estrogen receptor (ER)-positive breast cancer. 31 CCNB1 exerts carcinogenic effects in colorectal cancer cells and may be an effective target in the development of new treatments for colorectal cancer. 32 Researchers have also shown that overexpression of G2 and S phase-expressed-1 promotes cell proliferation, migration, and invasion by regulating CCNB1 and other pathways and predicts poor outcomes in patients with bladder cancer. 33 In a study of NSCLC, Arora et al. discovered that a network composed of miR-20b-5p, CCNB1, HMGA2, and E2F7 plays important roles in the development and progression of NSCLC, 34 suggesting that these targets may be effective prognostic biomarkers and may facilitate the development of novel treatments for NSCLC. Indeed, the cell cycle and immune surveillance mechanisms can be targeted through HMGA2 and E2F7, and the CCNB1 gene may be a crucial component in various cancers.
Cyclin B2, encoded by the CCNB2 gene, is a type B cyclin that has been shown to be upregulated in human cancer. CCNB2 is vital for controlling the cell cycle during the G2/M transition (mitosis) via modulation of p34 (CDC2). The subcellular localization of cyclins B1 and B2 differs; CCNB2 is mainly localized to the Golgi apparatus, whereas cyclin B1 is expressed near microtubules. Because CCNB2 also binds to transforming growth factor (TGF) βRII, CCNB2/CDC2 may have essential roles in TGF-β-mediated cell cycle transformation. 35 In several studies, the relative expression level of CCNB2 was found to be significantly higher in patients with cancer than in normal controls and individuals with benign disease. Another study showed that overexpression of CCNB2 protein was related to the clinical progression and poor prognosis in patients with NSCLC, suggesting that CCNB2 may be a biomarker of NSCLC. 36 Finally, overexpression of CCNB2 protein has been shown to be related to clinical progression and poor prognosis in hepatocellular carcinoma and nasopharyngeal carcinoma.37,38
Cyclin A2, encoded by CCNA2, is another cyclin family member and cell cycle regulator. The protein binds to and activates cyclin-dependent kinase 2, thereby facilitating the G1/S and G2/M transitions. 39 CCNA2 is also a prognostic biomarker for ER-positive breast cancer, 40 and overexpression of CCNA2 in tumor tissue predicts poor survival in patients with pancreatic ductal adenocarcinoma. 41
DNA topoisomerase II alpha, encoded by the TOP2A gene, modulates DNA topology during transcription by catalyzing the breakage and recombination of double-stranded and is also involved in DNA transcription and replication, chromatid separation, and chromosome condensation.42,43 The TOP2A gene has been used as a target for several anticancer drugs; however, mutations in TOP2A are associated with acquired resistance. Moreover, decreased enzyme activity may be involved in ataxia-telangiectasia. In previous studies, TOP2A was shown to be a therapeutic target for adrenocortical carcinoma and neuroblastoma tumors,44,45 and TOP2A gene amplification has been used to predict the response to anthracycline therapy in breast cancer. 46
There were some limitations to our study. First, although we repeated our experiments independently three times, some target genes did not show significant differences, likely owing to the influence of random errors. Additionally, we only compared the expression levels of the target genes in one cancer cell line (A549 cells) and one normal lung cell line (HBE cells); therefore, the results should be interpreted with caution. In the future, additional experiments using more clinical specimens and different cell lines are required to verify our findings.
Conclusion
In summary, in this study, we identified DEGs involved in the progression of NSCLC using integration of microarray data from multiple GEO datasets comprising large sample cohorts. We established a PPI network and determined the relationships of hub genes with diagnosis and prognosis in NSCLC. The diagnostic value and robustness of NSCLC predicted by the hub genes were evaluated using the GEPIA web tool based on TCGA datasets. We finally verified the hub genes at the protein level using THPA. The upregulation of CCNB1, CCNB2, CCNA2, and TOP2A was verified in cell experiments. However, the functions of these hub genes in NSCLC must be verified in vitro and in vivo in future studies. In addition, we found that the predictive ability of the identified hub genes differed according to the treatment strategy used in the patients as well as the histological type of NSCLC. Overall, our findings provide important insights into the treatment of NSCLC based on genomics.
.
Supplemental Material
Supplemental material, sj-xls-1-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-2-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-3-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-4-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-5-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-6-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-7-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-8-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-9-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Acknowledgments
The authors would like to thank Editage (www.editage.com) for English language editing.
Abbreviations
- BiNGO
Biological Networks Gene Oncology Tool
- BP
biological process
- CC
cellular component
- CI
confidence interval
- DAVID
The Database for Annotation, Visualization, and Integrated Discovery
- dDEG
downregulated differentially expressed gene
- DEG
differentially expressed gene
- FBS
fetal bovine serum
- GO
Gene Ontology
- GEO
Gene Expression Omnibus
- GEPIA
Gene Expression Profiling Interactive Analysis
- HR
hazard ratio
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MF
molecular function
- MCODE
Molecular Complex Detection
- NSCLC
non-small cell lung cancer
- PPI
protein-protein interaction
- qPCR
quantitative real-time polymerase chain reaction
- TCGA
The Cancer Genome Atlas
- THPA
The Human Protein Atlas
- uDEG
upregulated differentially expressed gene.
Footnotes
Ethical Statement: Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the the Natural Science Foundation for Young Scientists of Hunan Province (grant number 2016JJ4099, 8150020951).
Supplemental Material: Supplemental material for this article is available online.
ORCID iD: Li Xie https://orcid.org/0000-0002-2634-6187
References
- 1.Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709-719. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018(68):284-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. . Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [DOI] [PubMed] [Google Scholar]
- 5.Ostheimer C, Evers C, Palm F, et al. Mortality after radiotherapy or surgery in the treatment of early stage non-small-cell lung cancer: a population-based study on recent developments. J Cancer Res Clin Oncol. 2019;145:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. 2019;322:764-774. [DOI] [PubMed] [Google Scholar]
- 7.Prasad NB, Somervell H, Tufano RP, et al. Identification of genes differentially expressed in benign versus malignant thyroid tumors. Clin Cancer Res. 2008;14:3327-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lusito E, Felice B, D'Ario G, et al. Unraveling the role of low-frequency mutated genes in breast cancer. Bioinformatics. 2019;35:36-46. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Yang Y, Cheng L, et al. Identification of common genes refers to colorectal carcinogenesis with paired cancer and noncancer samples. Dis Markers. 2018;2018:3452739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of hippo-YAP pathway in colorectal cancer. J Hematol Oncol. 2017;10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Wang W, Xia P, et al. Identification of a five-lncRNA signature for predicting the risk of tumor recurrence in patients with breast cancer. Int J Cancer. 2018;143:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Peng Q, Zhu J, et al. Identification of miR-210 and combination biomarkers as useful agents in early screening non-small cell lung cancer. Gene. 2020;729:144225. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Chen R, Feng Z, et al. Identification of differentially expressed genes in non-small cell lung cancer. Aging. 2019;11:11170-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saigusa D, Motoike IN, Saito S, et al. Impacts of NRF2 activation in non-small-cell lung cancer cell lines on extracellular metabolites. Cancer Sci. 2020;111:667-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355-364. [DOI] [PubMed] [Google Scholar]
- 16.Kadara H, Fujimoto J, Yoo SY, et al. Transcriptomic architecture of the adjacent airway field cancerization in non-small cell lung cancer. J Natl Cancer Inst. 2014;106, dju004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19:2590-2597. [DOI] [PubMed] [Google Scholar]
- 18.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991-D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57. [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561-D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197-208. [DOI] [PubMed] [Google Scholar]
- 23.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L, Wu M, Lu Y, et al. MicroRNA-424 regulates cisplatin resistance of gastric cancer by targeting SMURF1 based on GEO database and primary validation in human gastric cancer tissues. Onco Targets Ther. 2019;12:7623-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu QF, Liu Y, Fan Y, et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3 K/AKT pathway. J Hematol Oncol. 2015;8(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623-1640. [DOI] [PubMed] [Google Scholar]
- 28.Hyun SY, Rosen EM, Jang YJ. Novel DNA damage checkpoint in mitosis: mitotic DNA damage induces re-replication without cell division in various cancer cells. Biochem Biophys Res Commun. 2012;423:593-599. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Yu Q, Zhang Y, et al. Tectonic 1 accelerates gastric cancer cell proliferation and cell cycle progression in vitro. Mol Med Rep. 2015;12:5897-5902. [DOI] [PubMed] [Google Scholar]
- 30.Falero-Perez J, Sorenson CM, Sheibani N. Retinal astrocytes transcriptome reveals Cyp1b1 regulates the expression of genes involved in cell adhesion and migration. PLoS One. 2020;15:e0231752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding K, Li W, Zou Z, et al. CCNB1 Is a prognostic biomarker for ER + breast cancer. Med Hypotheses. 2014;83:359-364. [DOI] [PubMed] [Google Scholar]
- 32.El-Huneidi W, Shehab NG, Bajbouj K, et al. Micromeria fruticosa induces cell cycle arrest and apoptosis in breast and colorectal cancer cells. Pharmaceuticals. 2020;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu A, Zeng S, Lu X, et al. Overexpression of G2 and S phase-expressed-1 contributes to cell proliferation, migration, and invasion via regulating p53/FoxM1/CCNB1 pathway and predicts poor prognosis in bladder cancer. Int J Biol Macromol. 2019;123:322-334. [DOI] [PubMed] [Google Scholar]
- 34.Arora S, Singh P, Rahmani AH, et al. Unravelling the role of miR-20b-5p, CCNB1, HMGA2 and E2F7 in development and progression of non-small cell lung cancer (NSCLC). Biology (Basel). 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daldello EM, Luong XG, Yang CR, et al. Cyclin B2 is required for progression through meiosis in mouse oocytes. Development. 2019;146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian X, Song X, He Y, et al. CCNB2 Overexpression is a poor prognostic biomarker in Chinese NSCLC patients. Biomed Pharmacother. 2015;74:222-227. [DOI] [PubMed] [Google Scholar]
- 37.Li R, Jiang X, Zhang Y, et al. Cyclin B2 overexpression in human hepatocellular carcinoma is associated with poor prognosis. Arch Med Res. 2019;50:10-17. [DOI] [PubMed] [Google Scholar]
- 38.Qian D, Zheng W, Chen C, et al. Roles of CCNB2 and NKX3-1 in nasopharyngeal carcinoma. Cancer Biother Radiopharm. 2020;35:208-213. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Ma XL, Tian FJ, et al. Downregulation of CCNA2 disturbs trophoblast migration, proliferation, and apoptosis during the pathogenesis of recurrent miscarriage. Am J Reprod Immunol. 2019;82:e13144. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, Lv Q, Guo Z. Transcriptomic signature predicts the distant relapse in patients with ER + breast cancer treated with tamoxifen for five years. Mol Med Rep. 2018;17:3152-3157. [DOI] [PubMed] [Google Scholar]
- 41.Dong S, Huang F, Zhang H, et al. Overexpression of BUB1B, CCNA2, CDC20, and CDK1 in tumor tissues predicts poor survival in pancreatic ductal adenocarcinoma. Biosci Rep. 2019;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu T, Zhang H, Yi S, et al. Mutual regulation of MDM4 and TOP2A in cancer cell proliferation. Mol Oncol. 2019;13:1047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Yu X, Davenport JW, Urtishak KA, et al. Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci. Genome Res. 2017;27:1238-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Z, Man X, Li Z, et al. Expression profiles analysis identifies the values of carcinogenesis and the prognostic prediction of three genes in adrenocortical carcinoma. Oncol Rep. 2019;41:2440-2452. [DOI] [PubMed] [Google Scholar]
- 45.Hou JY, Baptiste C, Hombalegowda RB, et al. Vulvar and vaginal melanoma: a unique subclass of mucosal melanoma based on a comprehensive molecular analysis of 51 cases compared with 2253 cases of nongynecologic melanoma. Cancer. 2017;123:1333-1344. [DOI] [PubMed] [Google Scholar]
- 46.Eltohamy MI, Badawy OM, El kinaai N, et al. Topoisomerase II alpha gene alteration in triple negative breast cancer and its predictive role for anthracycline-based chemotherapy (Egyptian NCI patients). Asian Pac J Cancer Prev. 2018;19:3581-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xls-1-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-2-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-3-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-4-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-5-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-6-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-7-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-8-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment
Supplemental material, sj-xls-9-tct-10.1177_15330338211060202 for Identification and Integrate Analysis of Key Biomarkers for Diagnosis and Prognosis of Non-Small Cell Lung Cancer Based on Bioinformatics Analysis by Ke Gong, Huiling Zhou, Haidan Liu, Ting Xie, Yong Luo, Hui Guo, Jinlan Chen, Zhiping Tan, Yifeng Yang and Li Xie in Technology in Cancer Research & Treatment