Abstract
MicroRNAs are related to the development of hepatocellular carcinoma and can serve as potential therapeutic targets. Therapeutic strategies increasing tumor-suppressive microRNAs and reducing oncogenic microRNAs have been developed. Herein, the effects of simultaneously altering two microRNAs using MS2 virus-like particles were studied. The sequences of microRNA-21-sponge and pre-microRNA-122 were connected and cloned into a virus-like particle expression vector. Virus-like particles containing microRNA-21-sponge and pre-microRNA-122 sequences were prepared and crosslinked with a cell-specific peptide targeting hepatocellular carcinoma cells. Delivery effects were studied using RT-qPCR and functional assays to investigate the level of target mRNAs, cell toxicity, and the effects of proliferation, invasion, and migration. Virus-like particles delivered miR-21-sponge into cells, with the Ct value reaching 10 at most. The linked pre-miR-122 was processed into mature miR-122. The mRNA targets of miR-21 were derepressed as predicted and upregulated 1.2–2.8-fold, and the expression of proteins was elevated correspondingly. Proliferation, migration, and invasion of HCC cells were inhibited by miR-21-sponge. Simultaneous delivery of miR-21-sponge and miR-122 further decreased proliferation, migration, and invasion by up to 34%, 63%, and 65%, respectively. And the combination promoted the apoptosis of HCC cells. In conclusion, delivering miR-21-sponge and miR-122 using virus-like particles modified by cell-specific peptides is an effective and convenient strategy to correct microRNA dysregulation in hepatocellular carcinoma cells and is a promising therapeutic strategy for hepatocellular carcinoma.
Keywords: Delivery, virus-like particles, microRNA, competitive endogenous RNA, cancer therapy, hepatocellular carcinoma
Impact statement
Hepatocellular carcinoma patients at an advanced stage may not benefit from systemic therapy or may develop resistance to therapy. New therapeutic targets are widely studied, including microRNAs. MicroRNAs are involved in the pathogenesis of hepatocellular carcinoma. Altering microRNAs is a promising therapeutic strategy to suppress hepatocellular carcinoma and restore sensitivity to target drugs. Previous studies altered microRNAs by increasing tumor-suppressive microRNAs or decreasing oncomicroRNAs to therapeutically target hepatocellular carcinoma. In this study, a tumor-suppressive microRNA and an oncogenic microRNA were simultaneously altered in hepatocellular carcinoma cells, and enhanced suppressive effects were observed in these cells. The delivery strategy in this study provides a possible way to simultaneously target two dysregulated microRNAs in cells. This method can be extended to alter two or more microRNAs in cancer cells according to the microRNA profile of a subgroup of hepatocellular carcinoma cells.
Introduction
Hepatocellular carcinoma (HCC) is a common malignant carcinoma in the world with a high incidence and mortality rate. 1 For HCC patients without vascular invasion, tumor resection surgery or transplantation is performed to cure the disease. For cases of metastatic disease or extensive liver tumor burden, in the advanced stage of HCC, systemic therapy should usually be considered. 2 However, some patients may not benefit from systemic therapy, and some may develop resistance to it. 3
New therapeutic targets for HCC are widely studied to address the previously described therapy concerns. To date, many studies have focused on the potential therapeutic effects of targeting microRNAs for cancer.4,5 MicroRNAs are short noncoding RNAs that usually silence messenger RNAs and regulate physiological or pathological processes. 6 In HCC, miR-21 and miR-122 are two of the key microRNAs in the development of cancer. MiR-21 is presumed to be an oncogenic microRNA, and miR-122 is presumed to be a tumor suppression microRNA.7,8 Studies in vitro and in vivo have demonstrated that decreasing miR-21 and increasing miR-122 are potential approaches to suppress HCC.9,10 In addition, delivering miR-122 and anti-miR-21 is a promising approach to induce the secretion of cytokines and alter the immune microenvironment in HCC. 11
Increased expression of microRNAs can be achieved by delivering chemo-modified RNA using a nanocarrier, and siRNA or microRNA sponge technology is used to decrease microRNAs. 12 MS2 virus-like particles (VLPs) were developed as a vehicle to deliver therapeutic cargos, including the nucleic acids described above. 13 MS2 VLPs are nuclease resistant and can protect RNA cargos from degenerating in circulation. 12 MS2 VLPs are as small as 27.5 nm, compared with most nanoparticles, which are approximately 100–200 nm, 14 so VLPs leave circulation more easily and contact tumor cells independent of the apparatus. 15 The amino acid residues located outside the capsid make it possible to conjugate the VLPs with functional peptides, such as cell-specific peptides, penetrating peptides and lysosome escape peptides. 13 The conjugated peptides can improve the ability to target HCC cells and promote RNA cargo release in the cytoplasm. These effects can reduce the cytotoxicity of cargos and increase bioavailability.13,16 In this study, the dodecapeptide GE11, which is a ligand of EGFR, is used to conjugate with VLPs to target HCC cell lines, as previous studies have confirmed its delivery efficiency without off-target effects.17,18
Single-cell sequencing has revealed that some microRNAs increase and some decrease in a cell. The profile of microRNAs may facilitate the classification of liver cancer cells. 19 It can improve the effects of the microRNA-targeted therapeutic approach that altering microRNAs in each cell, which change in different directions. Recently, nucleic acid alteration relying on MS2 VLPs is a “one-way” method, and only upregulated or downregulated microRNAs are rescued. 13 Wrapping several microRNAs in VLPs might not ensure that a VLP contains all the desired microRNAs. 13 The clustered “Tough Decoy” inhibitors target more than one microRNA but only in one direction. 20
This study evaluated the therapeutic effects of simultaneously delivering miR-21-sponge and miR-122 to HCC cells by MS2 VLPs. It was also investigated whether this approach can suppress HCC cell lines better than miR-21-sponge alone in terms of molecular interactions and functions. Simultaneous alteration of two or more microRNAs may become a new strategy for HCC therapy.
Materials and methods
Preparation and identification of VLPs crosslinked with cell-specific peptides
The miR-21-sponge, pre-miR122-miR-21-sponge, and negative control (NC) sequences 21 were synthesized by Sangon Biotech (Sangon Biotech., China), and the sequences are listed in Table S1 in the Supplementary Material. The pre-miR122-miR-21-sponge hybrid was formed by connecting pre-miR-122 and miR-21-sponge sequences. Cassettes were added to both ends of the sponge sequence, which were presumed to make the sponge RNA more stable in the cytoplasm. 22 In addition, pac sites were added at both ends of the sequence to facilitate the assembly of MS2 VLPs and RNA. The sequences were cloned into the pACYC-MS2 expression vector, which coexpressed the RNA sequence and the capsid protein of the MS2 VLP. The plasmid containing pre-miR-122 was previously generated in our laboratory. 10 Then, VLPs containing the desired sequence of RNA were produced by the BL21 (DE3) prokaryotic expression system. 10 The GE11 cell-specific peptide which is modified with cysteine at N-terminal (Sangon Biotech., China) was crosslinked to the VLPs by sulfo-SMCC crosslinking reagent (Thermo Fisher, USA) according to the manufacturer’s instructions. To confirm that the desired RNA was packaged in the corresponding VLPs, GE11-VLP RNA was heated to release and tested by RT-qPCR. SDS-PAGE was performed to confirm that the cross-linker was connected to the corresponding VLPs and to identify the capsid protein and the number of cross-linkers linked to each MS2 monomer (see Supplementary Material, section 1 and section 2).
Cell culture
HepG2, Hep3B, and Huh7 liver cancer cell lines were cultured in our laboratory using either DMEM with high glucose (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, Israel), MEM (Gibco, USA) supplemented with 15% FBS, and DMEM with low glucose (Gibco, USA) supplemented with 10% FBS. SMMC7721 and SK-HEP1 cells were purchased from Shanghai Xin Yu Biotech and cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% FBS at 37°C and 5% CO2.
RT-qPCR
Cells were seeded in 24-well plates at 2.5–5 × 104 cells per well. Before dosing, cells were incubated with 40–100 nM chloroquine (Sigma, USA) to deprotonate the lysosomes for 4–24 h.13,23 GE11-VLPs were added to the cells and incubated for 48 h. There was an average of three independent experiments for each group. Total RNA was extracted using a QIAGEN cell RNA extraction kit according to the manufacturer’s instructions. Genome or exogenous DNA was digested using DNase I (Thermos Fisher, USA). cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Japan) with 1 μL of RNA template using hexa-random primers and oligo T7 primers. MiR-122 was reversed transcribed using stem-loop reverse transcription primers. Then, 1 μL of cDNA was tested by real-time PCR using TB Green Premix Ex Taq (Takara, Japan). The reverse transcription products were tested using the primers listed in Table S2 of the Supplementary Material. GAPDH was used as the internal control for mRNA and the sponge, and U6 was used as the internal control for microRNA.
Western blotting
We treated cells with 100 μg/mL GE11-VLPs carrying different RNA cargoes for 48 h. The cells in each group were lysed using RIPA lysis buffer supplemented with 1 mM PMSF (Beyotime Biotechnology, China). After centrifugation at 12,000 r/min for 5 min, the supernatant was collected, and protein concentrations were determined using a BCA assay kit (Thermo Fisher, USA). Twenty micrograms of each sample was separated by SDS-PAGE and transferred to PVDF membranes. 17 The membranes were then blocked with 5% fat-free milk/TBST buffer. PVDF membranes were cut according to the Prestained Protein Marker (Thermo Fisher, USA) and incubated with the corresponding antibody at 4°C overnight at a dilution of 1:1000 for RECK (Boster, China), 1:3000 for PDCD4 (Proteintech, USA), 1:10,000 for PTEN (Abcam, UK), 1:500 for IGF1R (CST, USA), and 1:5000 for β-actin (CWBIO, China). Then, membranes were washed using TBST five times for 4–5 min each time. Next, the membranes were incubated with secondary antibodies (CWBIO, China) at a dilution of 1:10,000 for 1 h at room temperature. TBST was used to wash the membranes five times, and ECL Western blotting substrate (Thermo Fisher, USA) was used to visualize the protein bands.
Cell toxicity and proliferation test
To observe the toxicity of GE11-VLPs, cells were seeded in 96-well plates and incubated with chloroquine as required. Then, GE11-VLPs were added to final concentrations of 10, 50, 100, and 500 μg/mL. Each concentration was tested in triplicate for each GE11-VLP. After dosing for 48 or 72 h, the culture medium was replaced with fresh medium, and 10 μL of CCK-8 reagent (Dojindo, Japan) was added to each well and incubated at 37°C for 1–2 h. Then, the ocular density (OD) value at 450 nm was measured by Multiskan FC Microplate Photometer (Thermo Fisher, USA).
To observe the effect of GE11-VLPs on the proliferation of liver cancer cells, cells were seeded in a 24-well plate and incubated with chloroquine as required, followed by the addition of GE11-VLPs to a final concentration of 100 μg/mL. After incubating for 0, 24, 48, and 72 h, the medium was replaced with fresh medium, and 50 μL of CCK-8 reagent was added to each well. After incubation at 37°C for 1 h, the OD value at 450 nm was measured.
Cell apoptosis assay
The cells were seeded in 24-well plates and incubated with chloroquine as required. Then, GE11-VLPs were added to a final concentration of 100 μg/mL and incubated for 48 h. The cell culture supernatant was transferred to a clean tube. Then, the cells were washed with PBS twice, and 100 μL of Accutase (BD Biosciences, USA) was added and incubated at 37°C for 5–10 min to detach the cells. The culture supernatant was then added to the corresponding well to neutralize Accutase. Collected cells were transferred to 1.5 mL tubes and centrifuged at 2000 r/min for 5 min. The cell pellets were carefully washed twice using PBS. Then, 300 μL of binding buffer was used to resuspend each cell pellet, and the cells were filtered using the falcon cell strainer (BD Biosciences, USA). Then, 3 μL of Annexin V-APC (Keygentech, China) and 3 μL of PI were added to each tube. The staining was processed in the dark at room temperature for 10–15 min, and cells were detected by an Accuri C6 plus flow cytometer system (BD Biosciences, USA). The results were analyzed using BD Accuri C6 Plus software.
Cell invasion and migration assay
Cells were seeded in 6-well plates and incubated with chloroquine as required and sequentially with GE11-VLPs for 24 h. Matrigel (Corning, USA) was diluted using DMEM without phenol red or FBS to a final concentration of 100 μg/mL. Transwell inserts (Corning, USA) for invasion assays were inserted into 24-well plates. Fifty microliters of Matrigel was added to the upper compartment of each Transwell and incubated for 5 h. Six hundred microliters of DMEM supplemented with 10% FBS was added to the lower compartment of each well and incubated for 1 h. Then, cells incubated with GE11-VLPs were detached by 0.5% trypsin EDTA (Gibco, USA) and counted. Next, 1–5 × 104 cells were seeded into the upper compartment in a volume of 100 μL. After 24 h, the cells on the surface of the upper compartment were removed by swab, and cells on the surface of the lower compartment were fixed with menthol for 30 min and stained with 1% crystal violet (Solarbio, China) for 1 h.
Statistical analysis
The relative expression of RNA tested by RT-qPCR was analyzed using the −2ΔCt method. T-test or one-way ANOVA was applied for statistical analysis to compare the relative expression of two or more groups. The OD values in cell proliferation assays and cell counts in cell migration and invasion assays were statistically analyzed by one-way ANOVA using SPSS (IBM, version 19.0) followed by the Bonferroni test to compare the values between two groups.
Results
Identification of the GE11-VLP
The packaged RNA was tested by RT-qPCR, sigmoidal shapes in amplification curves and sharp peaks in melt curves were observed (Supplementary Material, Figure S1), which indicated that the packaged RNA was specific as predicted. In addition, as shown in SDS-PAGE, the MS2 capsid monomer is approximately 14 kD. Each monomer was cross linked with 0–3 GE11 peptides after crosslinking (Supplementary Material, Figure S2).
The delivery and postprocessing of miR-21-sponge and pre-miR-122 in HCC cell lines
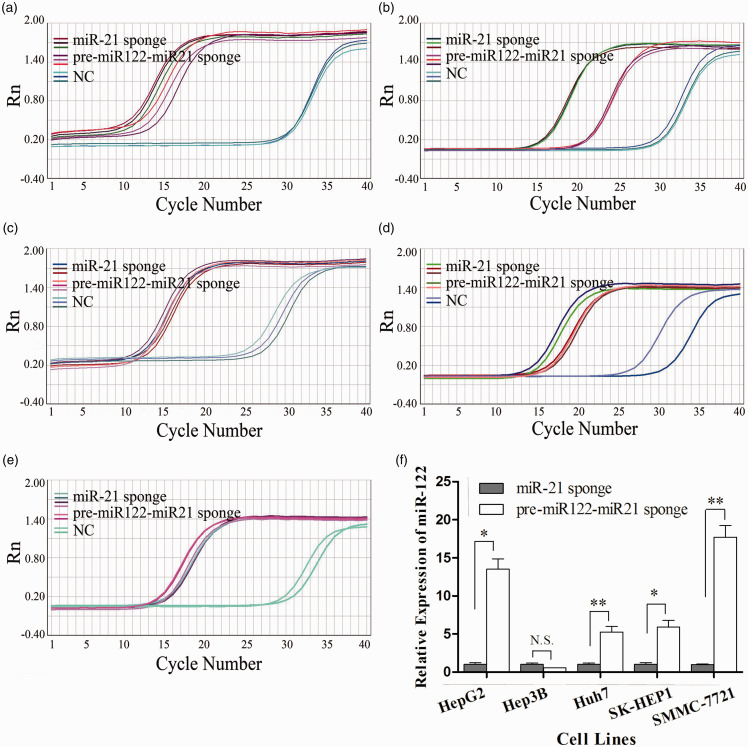
The miR-21-sponge RNA, which was extracted from cells in the miR-21-sponge-treated group or the pre-miR122-miR-21-sponge-treated group, was reverse transcribed and amplified specifically. Typical sigmoid amplification curves (Figure 1(a) to (e)) and specific melt curves were observed (data not shown). The average melting temperature was 80.4 and ranged from 79.6 to 81.1 in five different HCC cell lines of the treatment groups (see Supplementary Material, Table S3), which indicated that miR-21-sponge RNA was amplified specifically. The average Ct values of miR-21-sponge in the miR-21-sponge-treated group of HepG2, Hep3B, Huh7, SMMC-7721, and SK-HEP1 cells were 9.9, 12.3, 15.4, 14.5, and 14.1, respectively; the corresponding Ct values of these cells treated with pre-miR122-miR-21-sponge were 11.8, 20.8, 11.3, 13.6, and 12.9, respectively. As miR-21-sponge was generated, the sequence was not supposed to be amplified in the control group treated with NC RNA. However, sigmoid amplification curves were observed with an average Ct value of 28.5, which was higher than that of the treatment group by at least 10. Because the melt curves were bimodal and the melting temperature ranged from 72.3 to 77.6 in different cell lines, the amplification of the miR-21 RNA was supposed to be unspecific in the control group. The internal reference, GAPDH, was well amplified in all treatment and control groups (see Supplementary Material, Table S3), so the process of RNA determination was thought to be stable, and the results were reliable.
Figure 1.
Intracellular miR-21-sponge and miR-122 levels were detected by RT-qPCR. (a–e) The amplification curves of miR-21-sponge tested by RT-qPCR in the treatment and control groups of five HCC cell lines are shown. (a) HepG2, (b) Hep3B, (c) Huh7, (d) SMMC-7721, (e) SK-HEP1. (f) The relative expression of miR-122 in HCC cell lines is shown. (a) Each column represents an average of three independent experiments. *P < 0.05; **P < 0.01; N.S.: no significance. (A color version of this figure is available in the online journal.)
Compared with the miR-21-sponge-treated group, miR-122 was increased significantly by 13.5-, 5.3-, 17.7-, and 5.9-fold in the pre-miR122-miR-21-sponge-treated groups of HepG2, Huh7, SMMC-7721, and SK-HEP1 cells, respectively (Figure 1(f)). Because only mature miR-122 could be detected, the results indicated that pre-miR-122 was processed to mature miR-122 in cells. Exceptionally, in Hep3B cells, miR-122 expression showed no significant difference between the two groups (Figure 1(f), P = 0.133).
The alteration of miR-21 and miR122-targeted genes
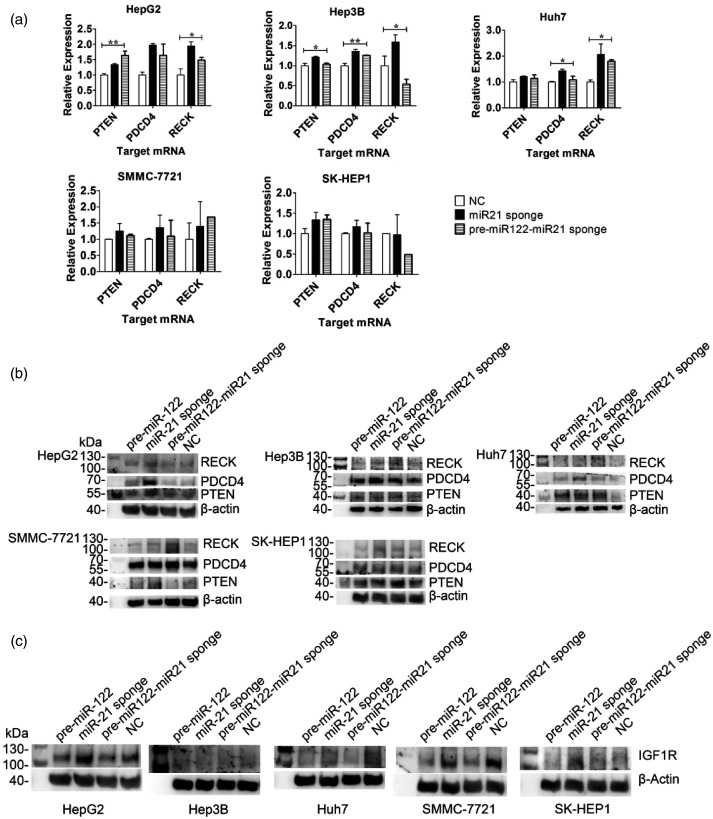
The target mRNA of miR-21 was detected by RT-qPCR and analyzed by relative quantitative methods (Figure 2(a)). The PTEN and PDCD4 gene expression levels were upregulated in all HCC cell lines in both treatment groups compared with the NC group. Among them, the levels of PTEN in HepG2 (P = 0.006) and Hep3B (P = 0.018) cells were significantly different. The RECK mRNA level of both treatment groups was upregulated in all HCC cell lines except for Hep3B and SK-HEP1. In the HepG2 and Huh7 cell lines, the RECK mRNA level of the treatment groups was increased to 1.5–2.1-fold (P = 0.013) and 1.8–2.9-fold (P = 0.05), respectively. Contrary to expectation, the level of RECK expression decreased in Hep3B and SK-HEP1 cells treated with pre-miR122-miR-21-sponge.
Figure 2.
(a) The expression levels of miR-21-targeted mRNAs, including PTEN, PDCD4, and RECK were determined in the treatment and control groups of five HCC cell lines. Each column represents an average of three independent experiments (*P < 0.05; **P < 0.01). (b) Western blot analysis of PTEN, PDCD4, and RECK protein in five HCC cell lines after treatment with GE11-VLPs carrying different RNA cargoes. β-actin protein was used as an internal control. (c) Western blot analysis of IGF1R protein in five HCC cell lines after treatment with GE11-VLPs carrying different RNA cargoes. (The bands of IGF1R protein in HepG2, and SK-HEP1 were incubated with the first antibody after stripping, so they share the same internal control with the bands of RECK protein.).
In Hep3B, Huh7, and SK-HEP1 cell lines, Western blot analysis revealed that PTEN, PDCD4, and RECK protein levels were upregulated by miR-21-sponge alone or in combination with pre-miR-122. In other groups that were treated with pre-miR-122 or the null-sequence, the expression of these proteins remained relatively low (Figure 2(b)). However, PDCD4 was upregulated only in the miR-21-sponge group but not in the pre-miR-122-miR-21-sponge group in HepG2 cells, so as to the PTEN in the SMMC-7721 cell line.
The IGF1R survival factor was detected by Western blot analysis, which revealed that in the five HCC cell lines, IGF1R expression was downregulated when treated with VLPs containing pre-miR-122 or pre-miR-122-miR-21-sponge (Figure 2(c)). Both VLPs elevated the cellular level of miR122, which is tumor suppressive microRNA.
Inhibitory effects on proliferation
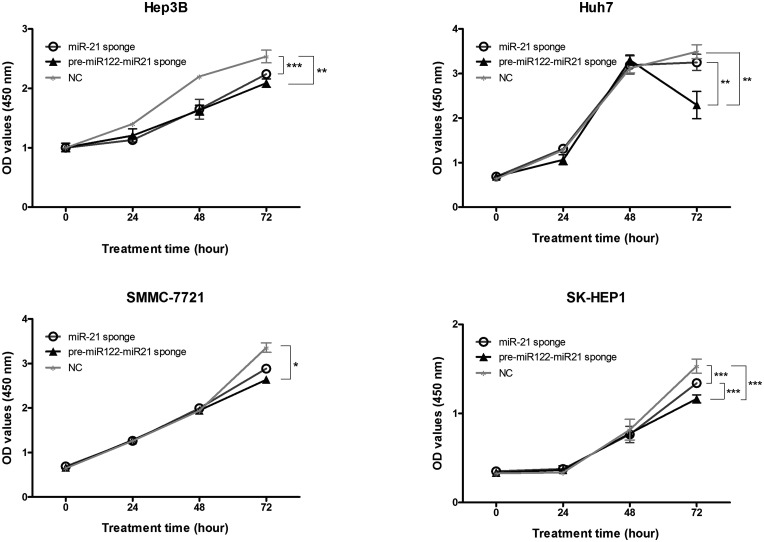
For HepG2 and Hep3B liver cancer cell lines, cell viability decreased significantly for both the miR-21-sponge group and the NC group at a concentration of 500 μg/mL (Figure S2 in the Supplementary Material). This result indicated that 500 μg/mL GE11-VLPs might have toxic effects on HCC cells instead of altering RNA function. Therefore, a final concentration of 100 μg/mL GE11-VLPs was incubated with liver cell lines, and the OD values at 450 nm differed on the third dosing day (Figure 3). Compared with the NC group treated with GE11-VLPs containing the NC sequence, the OD values at 450 nm of cells treated with GE11-VLPs containing the miR-21-sponge sequence decreased in Hep3B (P < 0.001), Huh7 (P = 0.715), SMMC-7721 (P = 0.215), and SK-HEP1 (P < 0.001) cell lines. The OD values further declined in the pre-miR122-miR-21-sponge group in the four cell lines, and a significant difference was observed in Huh7 (P = 0.006) and SK-HEP1 (P < 0.001) cells. The OD values in HepG2 cell lines showed no variation between the treatment and control groups (Supplementary Material, Figure S3).
Figure 3.
The OD values representing cell counts were detected by CCK-8 assay in the treatment and control groups of four HCC cell lines. Each dot represents three independent biological repeats of the results. *P < 0.05; **P < 0.01; ***P < 0.001.
The promotion of apoptosis
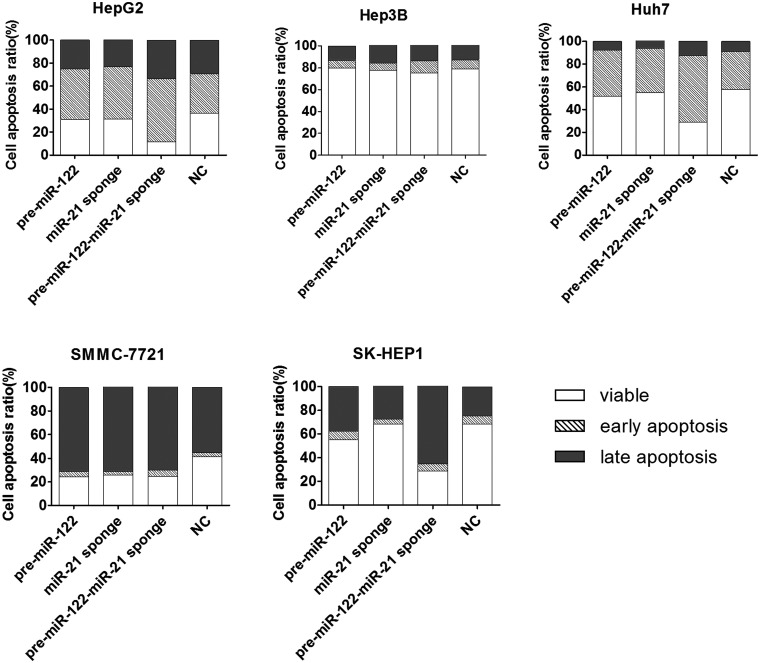
MiR-21-sponge increased the total apoptosis rate by 17.35% in comparison with the NC group for SMMC-7721 cells, and it increased the early apoptosis rate by 10.83% for HepG2 cells. For the other HCC cells treated with miR-21-sponge, no significantly higher rates of apoptosis were observed when compared with the negative control group, which indicated that miR-21 exhibits cytostatic effects rather than cytotoxic effects. 24 However, miR-122 increased the apoptosis rates of these HCC cells, and the activity was enhanced by miR-21-sponge, especially in the SK-HEP1 cell line. The total apoptosis rate of SK-HEP1 cells increased by 13.46% and 39.89% for the pre-miR-122-treated group and the pre-miR122-miR-21-sponge-treated group, respectively (Figure 4).
Figure 4.
The effects in inducing apoptosis of HCC cell lines when delivering the pre-miR122, miR-21-sponge, pre-miR122-miR-21-sponge, or NC sequence.
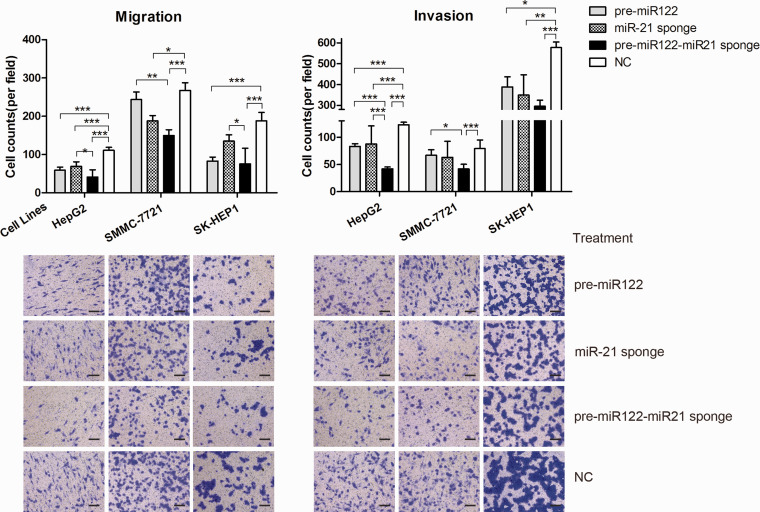
Inhibitory effects on cell invasion and migration
GE11-VLP-delivered pre-miR-122, miR-21-sponge and pre-miR122-miR-21-sponge inhibited migration and invasion ability (Figure 5). When compared with the NC group, the average number of pre-miR-122-treated HepG2 cells passing through the transwell membrane decreased to 53.25% (59/111) in the migration assay and decreased to 67.55% (83/123) in the invasion assay. And the average number of miR-21-treated HepG2 cells passing through the transwell membrane decreased to 62.51% (69/111) and 71.41% (88/123) in the migration assay and invasion assay, respectively. The cell number further decreased when miR-21-sponge was delivered combined with pre-miR-122, decreasing to 37.25% (41/111) and 34.18% (42/123) for migration and invasion assays, respectively. In migration assays for the SK-HEP1 cell line, the cell number decreased to 44.33% (83/187), 71.98% (135/187), and 40.22 (76/187) for the pre-miR-122 alone, miR-21-sponge, and pre-miR122-miR-21-sponge groups, respectively, and in invasion assays, the cell number decreased to 67.24% (388/578), 60.24% (349/578), and 51.21% (296/578), respectively. For the SMMC-7721 cell line, the combination group was also more effective than single treatments in migration and invasion assays, and cells passing through decreased to 55.95% (150/267) and 52.65% (41/79) when incubated with GE11-VLPs containing pre-miR122-miR-21-sponge, respectively.
Figure 5.
The average number of cells passing through the transwell membrane is shown for migration and invasion assays when delivering the pre-miR122, miR-21-sponge, pre-miR122-miR-21-sponge, or NC sequence to HCC cell lines. *P < 0.05; **P < 0.01; ***P < 0.001. The error bar represents SE, which is calculated from 10 different fields of view in two independent experiments. One representative field of view for each group is attached in the corresponding column (scale bar, 200 μm). (A color version of this figure is available in the online journal.)
Discussion
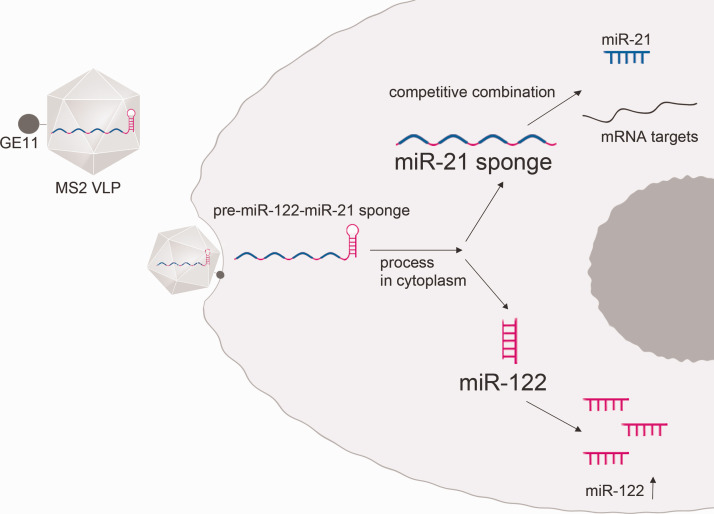
In this study, we demonstrated HCC suppression effects of simultaneous delivery of miR-122 and miR-21-sponge by MS2 VLPs (Figure 6). Herein, miR-21-sponge was linked with pre-miR122 to ensure that both RNAs entered cells. Five HCC cell lines were tested. At the molecular level, several target genes of miR-21 were found to be derepressed after treatment, including PTEN, PECD4, and RECK. For cell function assays, proliferation, cell migration, and invasion were reduced, and apoptosis was increased in miR-21-sponge-treated cells. Moreover, enhanced effects were observed when combined with miR-122.
Figure 6.
A diagram of the delivery strategy is shown. Pre-miR-122-miR-21-sponge is delivered into a cell by the GE11-MS2 VLP. It is further processed into miR-21-sponge and mature miR-122. The sponge is supposed to competitively combine with onco-microRNA miR-21, and tumor-suppressive microRNA miR-122 is supposed to increase. (A color version of this figure is available in the online journal.)
The competitive endogenous RNA hypothesis, which is presumed to be a main regulatory mechanism of microRNA-based therapeutic strategies, is that complementary RNA sequences competitively interact with microRNAs and repress the degeneration of downstream mRNA targets.12,25 MiR-21-sponge can serve as a competitive RNA to repress the functions of onco-microRNA miR-21.9,21 The PDCD4, PTEN, and RECK genes are common mRNA targets of miR-21 in HCC, and they are related to the proliferation, migration, invasion, and apoptosis of HCC cells.9,24,26 The results showed that most mRNA targets were derepressed. The expression of several mRNA targets significantly increased by 0.1–0.5 log10-fold, which is comparable to other knockdown technologies.9,27,28 Because the level of derepression relies on cell type and status, 29 although the ANOVA results did not indicate that relative mRNA expression was significantly different, it was confirmed by Western protein assays. In addition, the reduction in the RECK gene in Hep3B and SK-HEP1 cells was unexpected, which might result from the high level of variation between RECK Ct values (data not shown).
Cell function assays indicated that miR-21-sponge delivered by VLPs inhibited proliferation, cell migration, and invasion of more than one HCC cell line. Further repression was observed when combining miR-21-sponge together with miR-122. Further, the results might differ among cell lines. MiR-21-sponge and pre-miR122-miR-21-sponge were functionally active in SK-HEP1 cells. Treatment significantly influenced HepG2 cell migration and invasion but had no effects on cell proliferation. The diverse responses could be attributed to gene expression differences between cell lines,30,31 which might serve as proper models for animals and even patients in clinical trials in future applications.
The stem-loop primer ensures the specificity of detecting mature miR-122 instead of the pre-microRNA, which indicated that the GE11-VLP construct delivered pre-miR122-miR-21-sponge, which was processed to generate functional miR-122, so miR-122 increased in the targeted cell. It has been declared that the tumor-suppressing effects of delivering miR-122 to HCC cells in vitro and in vivo include repressing the expression of IGF1R at the protein level and inhibiting proliferation, cell migration, and invasion. 10 However, the expression of miR-122 was not significantly different when comparing between the two treatment groups of Hep3B cells, which may result from the relatively high baseline expression of miR-122. Correspondingly, no significant differences were observed between the two treatment groups in Hep3B cells in cell proliferation assays.
The limits of this study are that it remains at the cell level. Animal models are required to further declare the effects of this delivery strategy. GE11-VLPs delivered systemically were shown to be penetrable to HCC cell lines in a xenograft model in our laboratory.17,18 However, altering miR-21 may work in a disease-stage-dependent manner,32,33 and different methods of downregulating miR-21 may affect outcomes. So a proper experimental design is supposed to be conducted before further application of the therapy strategy. Besides, co-delivery of miR-122 and miR-21-sponge may enhance the HCC tumor-suppressive efficiency compared with altering miR-21 alone.
Recent studies have gradually revealed the microRNA profiles of HCC, and dysregulation of multiple microRNAs is common.19,34 Under the circumstance of the transcriptional background becoming clearer, targeting more than two microRNAs and altering them in different directions in each cell can be a new strategy to improve the antitumor effects of microRNA-based therapeutic methods. An extension of the VLP-based delivery system in this study may also be of use.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211035689 for Delivery of microRNA-21-sponge and pre-microRNA-122 by MS2 virus-like particles to therapeutically target hepatocellular carcinoma cells by Jiawei Zhang, Dandan Li, Rui Zhang, Rongxue Peng and Jinming Li in Experimental Biology and Medicine
Footnotes
AUTHORS’ CONTRIBUTIONS: J Z, DL, and RP designed the research study, performed the research, analyzed the data, and wrote and edited the manuscript; RZ and JL designed the research study, provided the funding, reviewed the manuscript, made a critical revision, and approved the submitted and final versions.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported by J. Li, National Natural Science Foundation of China (Grant 81772273).
ORCID iD: Jiawei Zhang https://orcid.org/0000-0001-8964-1848
SupplementAL MATERIAL: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 2021; 71:209–49 [DOI] [PubMed] [Google Scholar]
- 2.Benson AB, D'Angelica MI, Abbott DE, Abrams TA, Alberts SR, Anaya DA, Anders R, Are C, Brown D, Chang DT, Cloyd J, Covey AM, Hawkins W, Iyer R, Jacob R, Karachristos A, Kelley RK, Kim R, Palta M, Park JO, Sahai V, Schefter T, Sicklick JK, Singh G, Sohal D, Stein S, Tian GG, Vauthey JN, Venook AP, Hammond LJ, Darlow SD. Guidelines insights: hepatobiliary cancers, version 2.2019. J Natl Compr Canc Netw 2019; 17:302–10 [DOI] [PubMed] [Google Scholar]
- 3.Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, Xie M, Lei T, Zhang N, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer 2019; 18:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwok GT, Zhao JT, Weiss J, Mugridge N, Brahmbhatt H, MacDiarmid JA, Robinson BG, Sidhu SB. Translational applications of microRNAs in cancer, and therapeutic implications. Noncoding RNA Res 2017; 2:143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronte F, Bronte G, Fanale D, Caruso S, Bronte E, Bavetta MG, Fiorentino E, Rolfo C, Bazan V, Di Marco V, Russo A. HepatomiRNoma: the proposal of a new network of targets for diagnosis, prognosis and therapy in hepatocellular carcinoma. Crit Rev Oncol Hematol 2016; 97:312–21 [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466:835–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T, Yang Z, Kusumanchi P, Han S, Liangpunsakul S. Critical role of microRNA-21 in the pathogenesis of liver diseases. Front Med 2020; 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 2009; 284:32015–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, Huang SM, Jiang L, Kim D, Metz-Weidmann C, Pavlicek A, Pollard J, Reeves J, Rocnik JL, Scheidler S, Shi C, Sun F, Tolstykh T, Weber W, Winter C, Yu E, Yu Q, Zheng G, Wiederschain D. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res 2015; 13:1009–21 [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Jia T, Xu X, Chang L, Zhang R, Fu Y, Li Y, Yang X, Zhang K, Lin G, Han Y, Li J. Novel miR-122 delivery system based on MS2 virus like particle surface displaying cell-penetrating peptide TAT for hepatocellular carcinoma. Oncotarget 2016; 7:59402–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wischhusen JC, Chowdhury SM, Lee T, Wang H, Bachawal S, Devulapally R, Afjei R, Sukumar UK, Paulmurugan R. Ultrasound-mediated delivery of miRNA-122 and anti-miRNA-21 therapeutically immunomodulates murine hepatocellular carcinoma in vivo. J Control Release 2020; 321:272–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4:721–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, Brinker CJ, Caldeira Jdo C, Chackerian B, Wharton W, Peabody DS. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano 2011; 5:5729–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang D, Sun L, Huang L, Chen Y. Nanodrug delivery systems modulate tumor vessels to increase the enhanced permeability and retention effect. J Pers Med 2021; 11:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW. Analysis of nanoparticle delivery to tumours. Nat Rev Mater 2016; 1:16014 [Google Scholar]
- 16.Midoux P, Kichler A, Boutin V, Maurizot JC, Monsigny M. Membrane permeabilization and efficient gene transfer by a peptide containing several histidines. Bioconjug Chem 1998; 9:260–7 [DOI] [PubMed] [Google Scholar]
- 17.Chang L, Wang G, Jia T, Zhang L, Li Y, Han Y, Zhang K, Lin G, Zhang R, Li J, Wang L. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget 2016; 7:23988–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng L, Huang FZ, Cheng LF, Zhu YQ, Hu Q, Li L, Wei L, Chen DW. GE11-modified liposomes for non-small cell lung cancer targeting: preparation, ex vitro and in vivo evaluation. Int J Nanomed 2014; 9:921–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, Itzkovitz S. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab 2019; 1:899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollensen AK, Bak RO, Haslund D, Mikkelsen JG. Suppression of microRNAs by dual-targeting and clustered tough decoy inhibitors. RNA Biol 2013; 10:406–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollensen AK, Thomsen R, Bak RO, Petersen CC, Ermegaard ER, Aagaard L, Damgaard CK, Mikkelsen JG. Improved microRNA suppression by WPRE-linked tough decoy microRNA sponges. RNA 2017; 23:1247–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul CP, Good PD, Li SX, Kleihauer A, Rossi JJ, Engelke DR. Localized expression of small RNA inhibitors in human cells. Mol Ther 2003; 7:237–47 [DOI] [PubMed] [Google Scholar]
- 23.Roy S, Zhu D, Parak WJ, Feliu N. Lysosomal proton buffering of poly(ethylenimine) measured in situ by fluorescent pH-sensor microcapsules. ACS Nano 2020; 14:8012–23 [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu W, Yang K, He X, Chen S. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol 2010; 53:98–107 [DOI] [PubMed] [Google Scholar]
- 25.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 2011; 146:353–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan H, Wang S, Yu H, Zhu J, Chen C. Molecular pathways and functional analysis of miRNA expression associated with paclitaxel-induced apoptosis in hepatocellular carcinoma cells. Pharmacology 2013; 92:167–74 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Zha Y, Hu W, Huang Z, Gao Z, Zang Y, Chen J, Dong L, Zhang J. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (MiR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem 2013; 288:37082–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao X-J, Xu C-Z, Wang J-T, Li X-J, Wang M-M, Gu Y-H, Liang Z-G. miR-21 promotes proliferation and inhibits apoptosis of hepatic stellate cells through targeting PTEN/PI3K/AKT pathway. J Recept Signal Transduct Res 2018; 38:455–61 [DOI] [PubMed] [Google Scholar]
- 29.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods 2012; 9:840–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, Sekido Y, Kondo M, Hasegawa Y. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med 2015; 4:551–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caruso S, Calatayud AL, Pilet J, La Bella T, Rekik S, Imbeaud S, Letouzé E, Meunier L, Bayard Q, Rohr-Udilova N, Péneau C, Grasl-Kraupp B, de Koning L, Ouine B, Bioulac-Sage P, Couchy G, Calderaro J, Nault JC, Zucman-Rossi J, Rebouissou S. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology 2019; 157:760–76 [DOI] [PubMed] [Google Scholar]
- 32.Lai S, Iwakiri Y. Is miR-21 a potent target for liver fibrosis? Hepatology 2018; 67:2082–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caviglia JM, Yan J, Jang M-K, Gwak G-Y, Affo S, Yu L, Olinga P, Friedman RA, Chen X, Schwabe RF. MicroRNA-21 and dicer are dispensable for hepatic stellate cell activation and the development of liver fibrosis. Hepatology 2018; 67:2414–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan P, Pang P, Hu X, Wang A, Zhang H, Ma Y, Zhang K, Ye Y, Zhou B, Mao J. Specific MiRNAs in naïve T cells associated with hepatitis C virus-induced hepatocellular carcinoma. J Cancer 2021; 12:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211035689 for Delivery of microRNA-21-sponge and pre-microRNA-122 by MS2 virus-like particles to therapeutically target hepatocellular carcinoma cells by Jiawei Zhang, Dandan Li, Rui Zhang, Rongxue Peng and Jinming Li in Experimental Biology and Medicine