Abstract
Hepatitis E virus (HEV) causes 14 million infections and 60,000 deaths per year globally, with immunocompromised persons and pregnant women experiencing severe symptoms. Although ribavirin can be used to treat chronic hepatitis E, toxicity in pregnant patients and the emergence of resistant strains are major concerns. Therefore there is an imminent need for effective HEV antiviral agents. The aims of this study were to develop a drug screening platform and to discover novel approaches to targeting steps within the viral life cycle. We developed a screening platform for molecules inhibiting HEV replication and selected a candidate, isocotoin. Isocotoin inhibits HEV replication through interference with heat shock protein 90 (HSP90), a host factor not previously known to be involved in HEV replication. Additional work is required to understand the compound’s translational potential, however this suggests that HSP90-modulating molecules, which are in clinical development as anti-cancer agents, may be promising therapies against HEV.
Keywords: hepatitis E virus, hepatitis E, virus replication, antiviral drugs, host factor
Introduction
Hepatitis E virus (HEV) was first discovered in the late 1970’s, during an epidemic that spread through the Kashmir region of northern India(Khuroo and Khuroo, 2016). It is a positive-sense, single-stranded RNA virus of the Hepeviridae family with a genome measuring ~7.2kb in length(Tam et al., 1991). The virus contains three open reading frames (ORFs), of which ORF1 is the largest (~5100 bp) and encodes a number of functional domains, including a methyltransferase, putative cysteine protease (PCP) region, RNA helicase, and RNA-dependent RNA polymerase (RdRp)(LeDesma et al., 2019). ORF2 of HEV encodes the viral capsid protein, and ORF3 encodes a small protein essential for viral egress(Nimgaonkar et al., 2018). During viral genomic replication, an intermediate negative-sense RNA template is used to transcribe several fold greater amounts of positive-sense RNA strands(Varma et al., 2011).
The strains of HEV affecting humans fall under the genus Orthohepeviridae and are primarily transmitted through contaminated drinking water or the consumption of infected pork, venison, or wild boar meat(Meng, 2016). HEV infection most commonly manifests as self-limiting, acute hepatitis in healthy individuals, typically lasting for a month(Rein et al., 2012). However, pregnant women and immunocompromised individuals experience more severe symptoms. Pregnant women have up to a 30% mortality rate in the third trimester from HEV infection, particularly from genotype 1 strains of the virus(Pérez-Gracia et al., 2017). Immunocompromised individuals exposed to HEV can develop chronic infection, leading to the rapid progression of liver cirrhosis in as little as two years(Kumar et al., 2013). There are at least 20 million HEV infections across the world annually, and an estimated 60,000 deaths(Rein et al., 2012; Shukla et al., 2011).
Treatment options for HEV infection are limited. In patients who develop chronic hepatitis E while taking immunosuppressive drugs (e.g. after receiving organ transplantation), a reduction in the immunosuppressive regimen is first attempted. This results in clearance of the virus in one-third of patients(Kamar et al., 2010). When this is not successful, patients are typically treated with ribavirin, a nucleoside analog and broad-spectrum antiviral, and/or pegylated IFN-α (pegIFN-α)(Kamar et al., 2014). pegIFN-α is less commonly used, since it is associated with severe side effects and can lead to transplant rejection in organ transplant recipients(Haagsma et al., 2010). Ribavirin monotherapy is 78% effective in clearing chronic HEV infection, however it is highly teratogenic and cannot be used in pregnant patients(Kamar et al., 2014). Furthermore, HEV strains with “fitness-enhancing” mutations have been identified in patients showing clinical resistance to ribavirin treatment(Todt et al., 2018a). One clinical case study has suggested that sofosbuvir, a drug approved for hepatitis C virus treatment, may have a beneficial additive effect when used in combination with ribavirin, however other studies have observed no therapeutic benefit, and the use of this drug for HEV treatment remains controversial(van der Valk et al., 2017; van Wezel et al., 2019). There are currently no other clinically approved drugs for hepatitis E.
Here, we report a high-throughput screening assay for molecules exhibiting activity against host factors and HEV ORF1 proteins involved in viral replication. Using this assay, we found a compound with novel antiviral activity against HEV named isocotoin. We further report that isocotoin inhibits HEV replication by modulating heat shock protein 90 (HSP90). HSP90-modulating molecules, which are currently in clinical development as anti-cancer agents, may therefore be promising candidates for novel therapies against HEV.
Materials and Methods
Screening Assay.
The objective of this study was to identify small molecules with antiviral activity against proteins encoded by HEV ORF1. A high-throughput screening platform was developed to test a library of ~60,000 compounds from the Princeton University Small Molecule Screening Center against a fluorescent HEV replicon. The HEV replicon was derived from the pKernowC1p6 construct, kindly provided by Dr. Suzanne Emerson (NIAID). Images were acquired using the Operetta CLS High Content Screening System (PerkinElmer, Waltham, MA) and analyzed using a custom Python script (https://github.com/aploss/PLOCUS).
All follow-up in vitro experiments to characterize hits were conducted using Huh7 hepatoma cells, with each condition tested in at least three biological replicate wells. The only experiments done in a different cell line, HepG2/C3A, are displayed in Supplementary Figure 1 and were performed to validate the results in an alternative cell line. No data were excluded from analyses.
Dose titration assays.
Huh7 cells were seeded in a 96-well plate at a seeding density of 6,250 cells/well, or HepG2/C3A cells were seeded in a 96-well plate at a density of 8,000 cells/well. The next day, the cells were transfected with 50 ng/well p6/Gluc IVT RNA using TransIT-mRNA transfection reagent (Mirus Bio LLC, Madison, WI) according to the instructions. Five hours post-transfection, the cells were incubated in medium containing the compound of interest. Three wells were used per dose for triplicate data. Information for the compounds used in dose titration assays is shown in Supplementary Table 1. All compounds were dissolved in DMSO, and DMSO concentration was kept constant at 0.1% in all wells. Quantification of HEV RNA by RT-qPCR Assay.Huh7 cells were seeded 42,000 cells/well in 24-well plates. One day post-seeding, the cells were transfected with 10 ng/well KernowC1p6 or KernowC1p6-GAD (pol-) in vitro transcribed RNA. The next day, the medium on the cells transfected with KernowC1p6 was changed to 0.1% DMSO medium containing 25 μM, 12.5 μM, 6.25 μM, or 0 μM isocotoin. The negative control cells transfected with KernowC1p6-GAD were maintained in medium with 0.1% DMSO. The media on the cells was changed every 2 days to fresh drug-containing or DMSO-containing media. On day 6 cells were harvested and lysed in RLT buffer using 350 μL per well. Cell lysates were homogenized by pipetting 6–7 times with a needle syringe. Total RNA was extracted using BioBasic RNA MiniPreps SuperKit (Amherst, NY). RT-qPCR was performed on the samples using qScript XLT 1-Step RT-qPCR ToughMix (Quanta Bio) according to the manufacturer instructions. In vitro transcribed KernowC1p6 RNA of known concentration was serially diluted 1:10 and included as controls to generate a standard curve to calculate absolute RNA copies/μL in the samples.
Primers for HEV RNA quantification
Forward GGTGGTTTCTGGGGTGAC
Reverse AGGGGTTGGTTGGATGAA
FAM-BHQ Probe TGATTCTCAGCCCTTCGC
For a detailed description of the methods please see the Supplemental Information.
Results
Replicon-based high-throughput screening to identify inhibitors of HEV replication
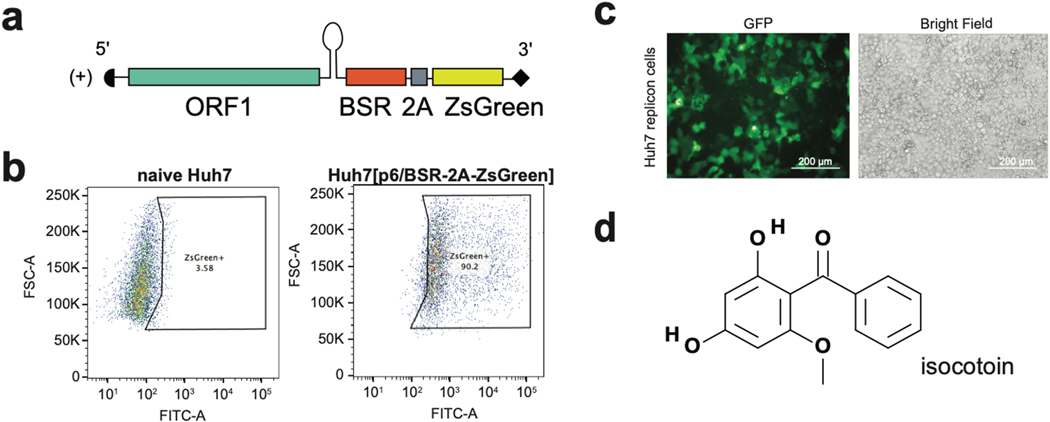
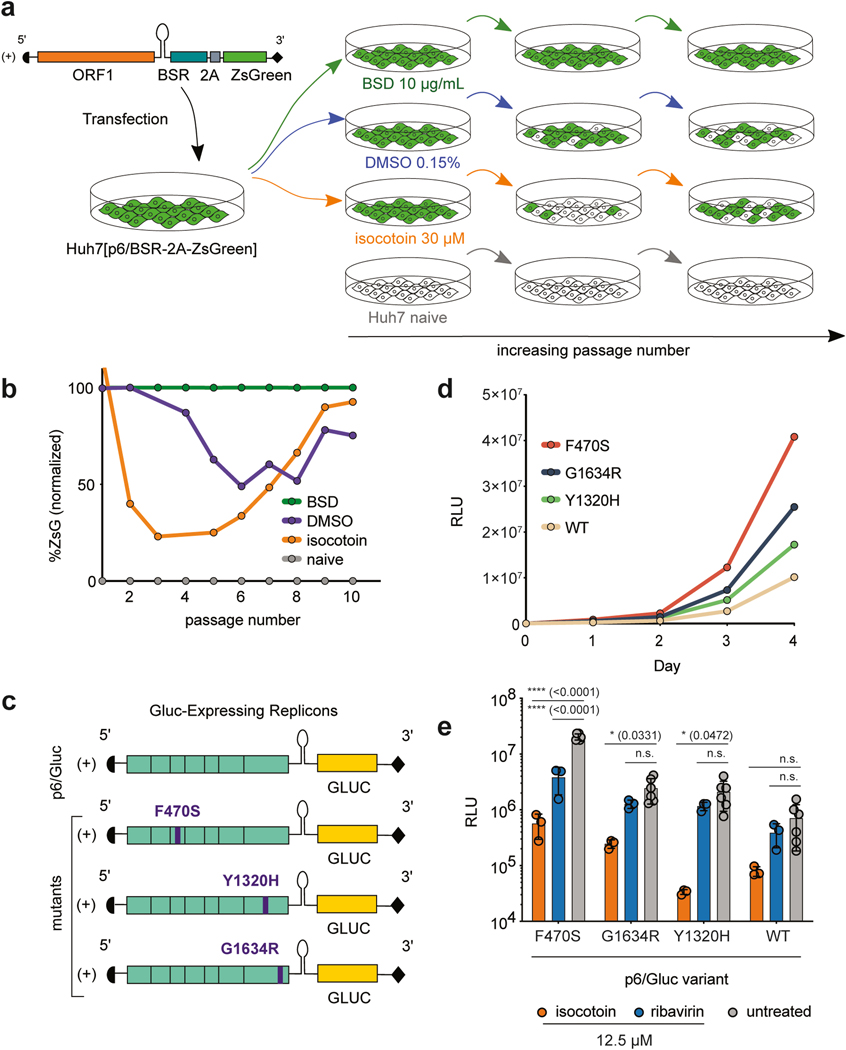
To identify small molecules with antiviral activity, functional domains within ORF1 were targeted since they (1) encompass the majority of domains encoded by the viral genome, (2) mediate numerous processes critical to the viral life cycle and interact with a number of cellular host factors, and (3) have roles in gene expression and genomic replication, which previously established treatments for other viral infections have targeted with great success(Benhamou et al., 2003; Lin et al., 2009; Malcolm et al., 2006; Stedman, 2014). We used a replicon-based screening assay in which ORFs 2 and 3 of HEV, which encode the capsid protein and an ion channel required for viral egress, respectively, were replaced in the viral genome with a blasticidin resistance gene (BSR) and a ZsGreen fluorescent reporter (Fig 1a). Huh7 cells were transfected with p6/BSR-2A-ZsGreen and placed under blasticidin selection to generate a population of cells highly expressing ZsGreen (Fig 1b). A collection of ~60,000 small molecules from the Princeton University Small Molecule Screening Center was tested against cells expressing the replicon genome, using decreased fluorescence as a readout to indicate inhibition of replication. Briefly, replicon-expressing cells were seeded in 384-well format and treated with a 50 μM dose of each compound for four days. A GFP channel image and bright-field image were taken of each well on day 4 using the Perkin Elmer Operetta High-Content Imaging System (Fig 1c). Fluorescence levels in the GFP channel images were quantified using a custom Python script (publicly sourced at: https://github.com/aploss/PLOCUS), and decreased fluorescence was used as a metric to select hits with putative antiviral activity. The brightfield images were used to remove false positive hits that showed decreased fluorescence due to cytotoxicity, resulting in 37 non-cytotoxic hits from the high-throughput assay (Appendix 1). Through dose titration assays on the hits, isocotoin was identified as a promising molecule to investigate mechanisms to inhibit HEV replication (Fig 1d).
Fig 1|. A replicon-based high-throughput screening assay identifies isocotoin as an inhibitor of HEV replication.
a, The p6/BSR-2A-ZsGreen replicon genome is derived from KernowC1p6, with open reading frames (ORFs) 2 and 3 replaced by a blasticidin resistance-conferring gene (BSR) and a ZsGreen fluorescence reporter, with a 2A self-cleaving peptide in between. b, Flow cytometric analysis of p6/BSR-2A-ZsGreen-transfected Huh7 cells (right; 90.2% ZsGreen+) versus naïve Huh7 cells (left; 3.58% ZsGreen+). c, GFP-channel (left) and brightfield (right) images taken of p6/BSR-2A-ZsGreen-expressing Huh7 cells. d, Structure of isocotoin.
In vitro functional validation of isocotoin
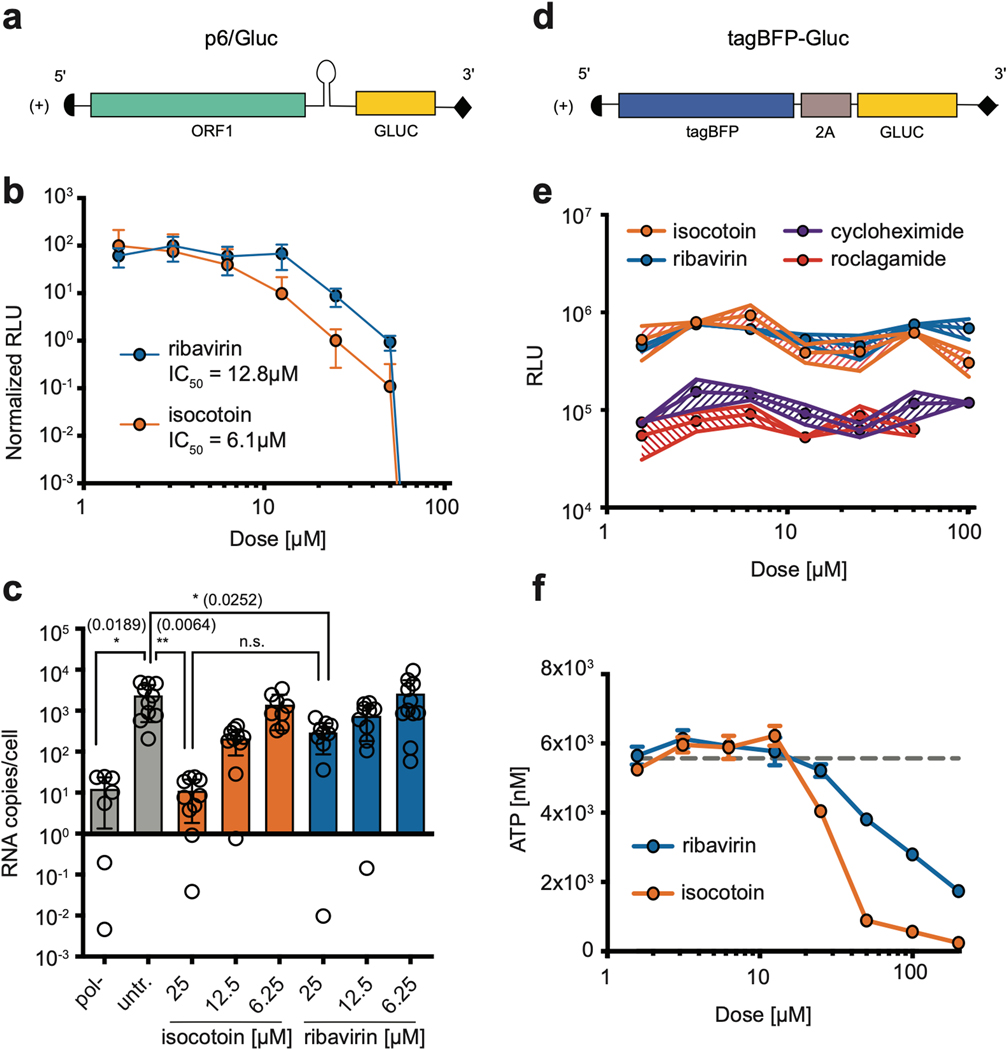
To further verify the antiviral effect of isocotoin against HEV replication, in vitro functional assays were performed using a secreted Gaussia luciferase (Gluc)-expressing HEV replicon derived from the KernowC1p6 HEV strain, abbreviated p6/Gluc (Fig 2a), and supernatant Gluc levels were measured as a proxy for viral replication(Shukla et al., 2011). Dose titration assays, in which Huh7 or HepG2/C3A hepatoma cells were transfected with in vitro-transcribed p6/Gluc RNA and then treated with ribavirin or isocotoin for 4 days, revealed that isocotoin effectively inhibits viral replication (Fig 2b, Supplementary Fig 1a). In Huh7 cells, isocotoin exhibits an IC50 value of 6.1 μM as compared to 12.8 μM for ribavirin (Fig 2b). Therefore based on our in vitro assays, isocotoin was more potent than ribavirin, which is currently used to treat chronic hepatitis E in solid organ transplant recipients in a clinical setting. RT-qPCR assays confirmed that isocotoin also inhibits replication of the full-length KernowC1p6 genome in a dose-dependent manner, both using transfection of full-length KernowC1p6 RNA in Huh7 cells (Fig 2c), and in an infection system in HepG2/C3A cells (Supplementary Fig 1b).
Fig 2|. In vitro functional validation of isocotoin.
a, Schematic of p6/Gluc replicon. b, Isocotoin inhibits p6/Gluc replication more efficiently than ribavirin in vitro. n=9 wells per data point, collected over three repeat experiments. c, Isocotoin inhibits replication of full-length HEV (KernowC1-p6 strain) more efficiently than ribavirin. n=3 wells per data point, measured in quadruplicate. p values calculated using ordinary one-way ANOVA followed by Tukey’s multiple comparisons test are indicated in parentheses. d, Schematic of T7-tagBFP-Gluc. e, Isocotoin and ribavirin do not inhibit translation of T7-tagBFP-Gluc. n=3 wells per data point. f, Isocotoin is non-cytotoxic to Huh7 cells up to 12.5 μM. n=3 wells per data point. GLUC=secreted Gaussia Luciferase; RLU=relative light units. Error bars indicate 1 SD from mean.
To demonstrate that isocotoin was not simply inhibiting general protein translation, we created a reporter construct, tagBFP-Gluc, containing Gluc and the fluorescent protein tagBFP but not any HEV-derived viral proteins, that could be in vitro-transcribed from a T7 promoter (Fig 2d). Capped tagBFP-Gluc RNA was transfected into cells which were then treated with varying doses of isocotoin for 4 days. Two known translation inhibitors cycloheximide and roclagamide (an analog of silvestrol), were included as controls along with ribavirin (a nucleoside analog that does not affect translation)(Sidwell et al., 1972; Todt et al., 2018b). Whereas cycloheximide and roclagamide suppressed Gluc levels ~10-fold, isocotoin did not lead to a significant decrease in Gluc levels as compared to ribavirin (Fig 2e).
To ensure that cytotoxicity did not account for the observed reductions in replication, an ATP-based cell viability assay was performed on Huh7 cells after treatment with isocotoin or ribavirin. Isocotoin displayed cytotoxicity at doses 25 μM and above, but not up to 12.5 μM (Fig 2f). Since isocotoin exhibits antiviral effects with an IC50 value of 6.1 μM, its antiviral effects cannot be attributed to cytotoxicity. Therefore, isocotoin specifically interferes with HEV replication, in contrast with silvestrol, which inhibits HEV primarily through general suppression of protein translation(Todt et al., 2018b).
Testing isocotoin against diverse HEV genotypes
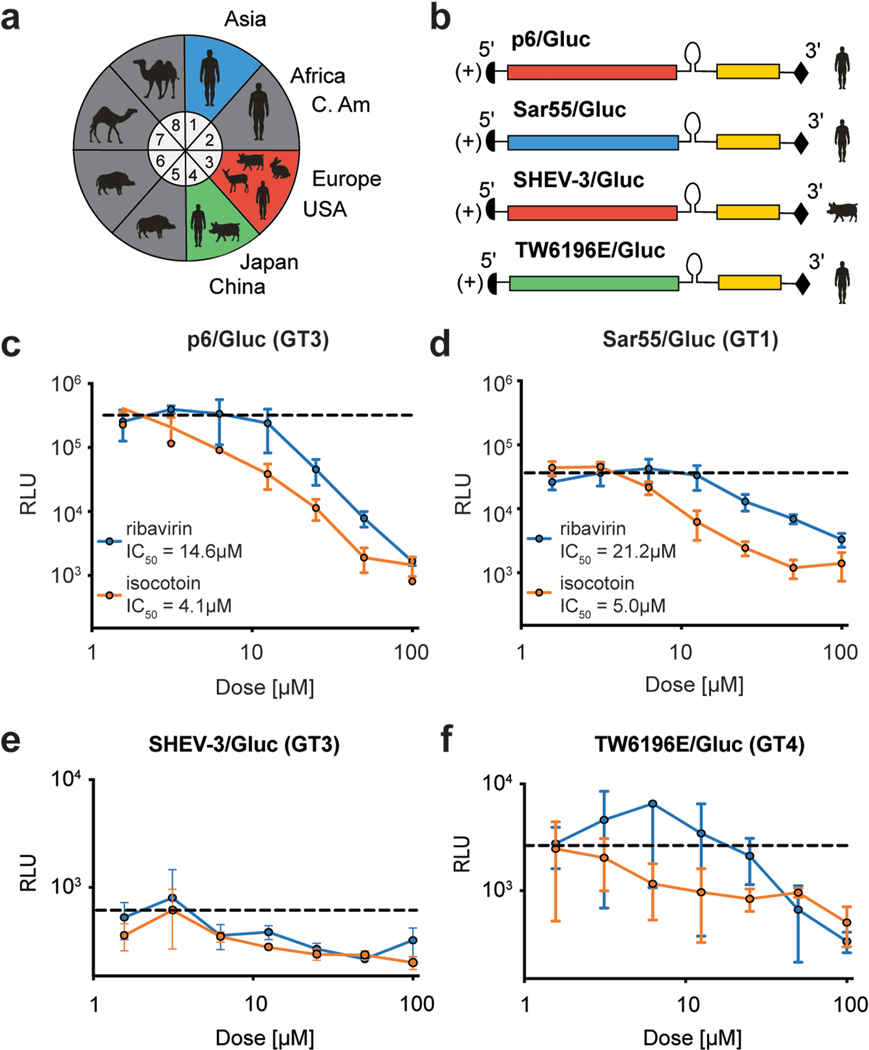
Next, we aimed to determine whether isocotoin exhibits antiviral activity against genetically diverse HEV genotypes. At least eight genotypes (GT) of HEV (GT 1–8) have been identified in mammals, with genotypes 1–4 accounting for the majority of reported infections in humans (Fig 3a)(Meng, 2016). Of these, the KernowC1p6 (GT 3) and Sar55 (GT 1) strains replicate robustly in cell culture systems and are frequently used for in vitro studies(Shukla et al., 2011; Tsarev et al., 1992). To evaluate its pan-genotypic antiviral efficacy, isocotoin was additionally tested against Gluc-expressing replicons from the Sar55 (GT 1), SHEV-3 (GT 3, swine-derived), and TW6196E (GT 4) HEV strains (named Sar55/Gluc, SHEV-3/Gluc, and TW6196E/Gluc, respectively) (Fig 3b)(Ding et al., 2018). Dose titration experiments against Sar55/Gluc demonstrated that isocotoin is effective in inhibiting replication and has a lower IC50 than ribavirin (Fig 3d). The replicative capacities of SHEV-3 and TW6196E in cell culture are much lower, but a modest inhibitory effect was still observed with isocotoin, albeit with large error bars (Figs 3e–f).
Fig 3|. Isocotoin inhibits replication of genetically diverse strains of HEV.
a, Of the 8 HEV genotypes categorized in Orthohepevirus A, genotypes 1–4 have most frequently been identified in human hosts. b, Gluc-expressing replicon genomes were generated for the Sar55 HEV strain from genotype (GT) 1, the SHEV-3 strain (GT 3), and the TW6196E strain (GT 4). The original Sar55 and TW6196E strains were derived from human patients whereas the SHEV-3 strain was derived from a swine host. c-d, Isocotoin inhibits replication of the p6/Gluc (GT3) and Sar55/Gluc (GT1) replicon strains. Dashed lines indicate Gluc levels for untreated cells. e-f, Isocotoin inhibits replication of the SHEV-3/Gluc (GT3) and TW6196E/Gluc (GT4) replicons albeit with large error bars due to poor replicative capacity of these strains in cell culture. RLU=relative light units. All data points represent the mean of triplicate wells (n=3). Error bars indicate 1 SD from mean.
Testing isocotoin against other positive-sense RNA viruses
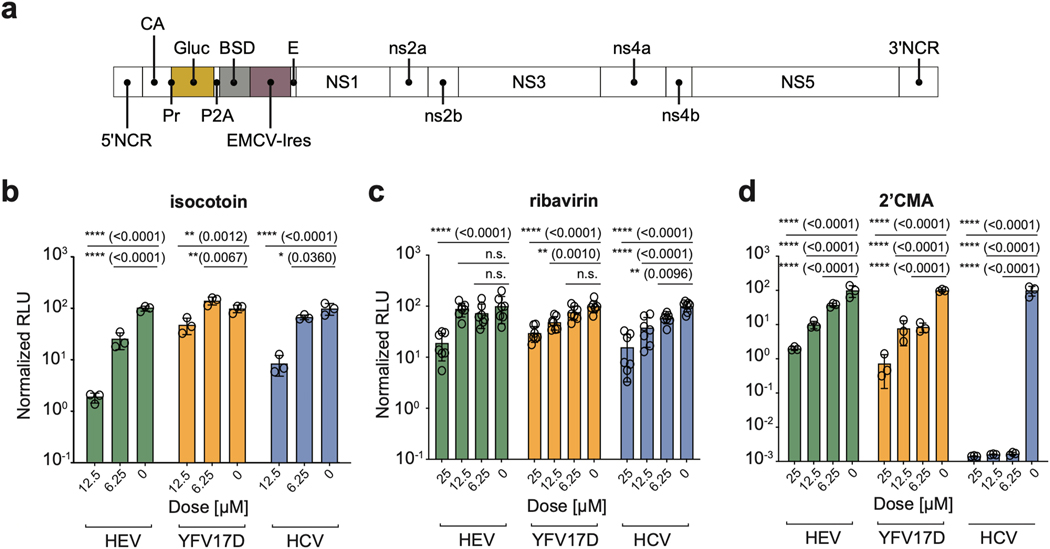
To examine whether the antiviral properties of isocotoin were specific to HEV or broadly effective against other positive-sense, single-stranded RNA viruses, we tested it against Gluc-expressing genomes from hepatitis C virus (Jc1(p7nsGluc2A), abbreviated ‘HCV’) and yellow fever virus 17-D vaccine strain (pACNR-FLYF-17D-Gluc-IRES-bsd, abbreviated ‘YFV17D’) (Figs 4a–b)(Marukian et al., 2008). Ribavirin, which used to be part of the standard care for treating HCV, and 2’-C-Methyladenosine, which is a specific inhibitor of the HCV NS5B protein, were used as controls (Figs 4c–d, Supplementary Fig 2). Although ribavirin had only minor inhibitory effects on any of the viruses under the experimental conditions used here, isocotoin notably showed dose-dependent inhibitory effects against all three viruses. Collectively, these data suggested that isocotoin operated through a common inhibitory mechanism against the three viruses tested.
Fig 4|. Isocotoin inhibits replication of other positive-sense RNA viruses.
a, Schematic of pACNR-FLYF-17D-Gluc-IRES-bsd construct, abbreviated YFV17D, which is derived from pACNR/FLYF-17D (GenBank ID: AY640589). The other reporter genomes have been described elsewhere. b, Isocotoin inhibits replication of Gluc-expressing HEV, YFV17D, and HCV genomes to a greater extent than ribavirin. c-d, Ribavirin and 2’CMA, a potent and specific inhibitor of HCV replication, were tested against the three viruses as controls. n=3–7 wells per data point. To determine p-values, ordinary two-way ANOVA test was performed followed by Dunnett’s multiple comparisons test with all means compared to that of the 0 μM dose. RLU values are normalized to untreated conditions. NCR=non-coding region; CA=capsid; Gluc=secreted Gaussia luciferase; BSD=blasticidin resistance gene; E=envelope; RLU=relative light units; 2’CMA=2’-C-Methyladenosine. Error bars indicate 1 SD from mean.
Suboptimal dosing experiments to identify adaptive mutations
To determine isocotoin’s mechanism of action, suboptimal dosing experiments were performed in which HEV was passaged in the presence of minimally cytotoxic doses of isocotoin for a prolonged time. The virus was expected to evolve resistance mutations that could be sequenced and used to identify the compound’s target (Fig 5a). Cells transfected with p6/BSR-2A-ZsGreen were maintained in 30 μM isocotoin, 10 μg/mL blasticidin, or 0.15% DMSO (concentration of vehicle for isocotoin), and fluorescence levels were analyzed at each passage with flow cytometry. Though some toxicity was observed at the 30 μM isocotoin dose, cells were still able to grow at a reduced rate and recover from repeated passaging. Naïve Huh7 hepatoma cells were included as a negative control for fluorescence.
Fig 5|. Isocotoin inhibits strains exhibiting higher replicative capacity in vitro.
a, Schematic of suboptimal dosing experiment. b, p6/BSR-2A-ZsGreen-expressing Huh7 cells serially passaged in medium containing 30 μM isocotoin showed a decrease in ZsGreen expression up to passage 5 and a subsequent increase in ZsGreen expression between passages 5–10. At passage 10, the F470S point mutation was found in 2/10 colonies sequenced. c, Schematic of the full-length viral genome and Gluc-expressing replicons. The p6/Gluc[F470S], p6/Gluc(Debing et al.), and p6/Gluc[G1634R] strains were derived from p6/Gluc and contain point mutations in the PCP, RdRp, and RdRp and regions respectively. d, Replication kinetics for p6/Gluc[F470S], p6/Gluc[G1634R], p6/Gluc[Y1320H], and p6/Gluc-WT over 4 days. e, Treatment of mutant strains with 12.5 μM of isocotoin or ribavirin. n=3 wells per data point for the treatment conditions, n=6 wells per data point for the untreated conditions. To determine p-values, ordinary two-way ANOVA test was performed followed by Dunnett’s multiple comparisons test with all means compared to that of the untreated conditions. BSR = blasticidin resistance gene; BSD = blasticidin; ZsG = ZsGreen; RLU = relative light units. Error bars indicate 1 SD from mean.
Isocotoin treatment led to a sharp decrease in ZsGreen levels during the first few passages, with a subsequent increase in ZsGreen levels starting at passage 6 (Fig 5b). By passage 9, ZsGreen signal approached the levels seen in the positive control (cells under selection with blasticidin) (Fig 5b). Intracellular RNA was extracted from both passage 1 and passage 10 cell lysates and reverse transcribed to produce cDNA. Overlapping segments covering ORF1 were PCR-amplified, sequenced, and aligned to identify any regions that had mutated from passage 1 to passage 10. Although no one dominant mutation was found, a highly conserved palindromic ‘FCCF’ peptide sequence, located at residues 470–473 in the putative cysteine protease region, was found to contain a single phenylalanine-to-serine point mutation in 30% of the clones (either at the 470 or the 473 position). These F470S and F473S single point mutations were engineered into the wild-type p6/Gluc replicon, and p6/Gluc[F470S] exhibited a higher replicative capacity in vitro than either the parental p6/Gluc strain or the p6/Gluc[F473S] strain (Fig 5d, Supplementary Figure 3), which was replication-defective. These data suggested that the emergence of the F470S point mutation in passage 10 cells may have been due to the enhanced replicative capacity of this strain and not isocotoin resistance.
Testing isocotoin against HEV strains harboring mutations associated with ribavirin resistance
Emergence of ribavirin-resistant strains poses a considerable problem for the treatment of hepatitis E. Previous studies characterizing clinical ribavirin-resistant strains in vitro found that G1634R and Y1320H point mutations in the RdRp region of ORF1 led to enhanced replicative capacity in vitro(Debing et al., 2014; Debing et al., 2016). We generated p6/Gluc[G1634R] and p6/Gluc[Y1320H] mutant replicons to compare their replicative capacities to that of the newly identified p6/Gluc[F470S] (Fig 5c). Measurement of supernatant Gluc over 4 days post-transfection in Huh7 cells revealed that p6/Gluc[F470S] exhibited a higher replicative capacity than both p6/Gluc[G1634R] and p6/Gluc[Y1320H] (Fig 5d). All three higher-replicating strains demonstrated greater sensitivity to isocotoin than to ribavirin (Fig 5e).
Isocotoin directly binds heat shock protein 90 (HSP90)
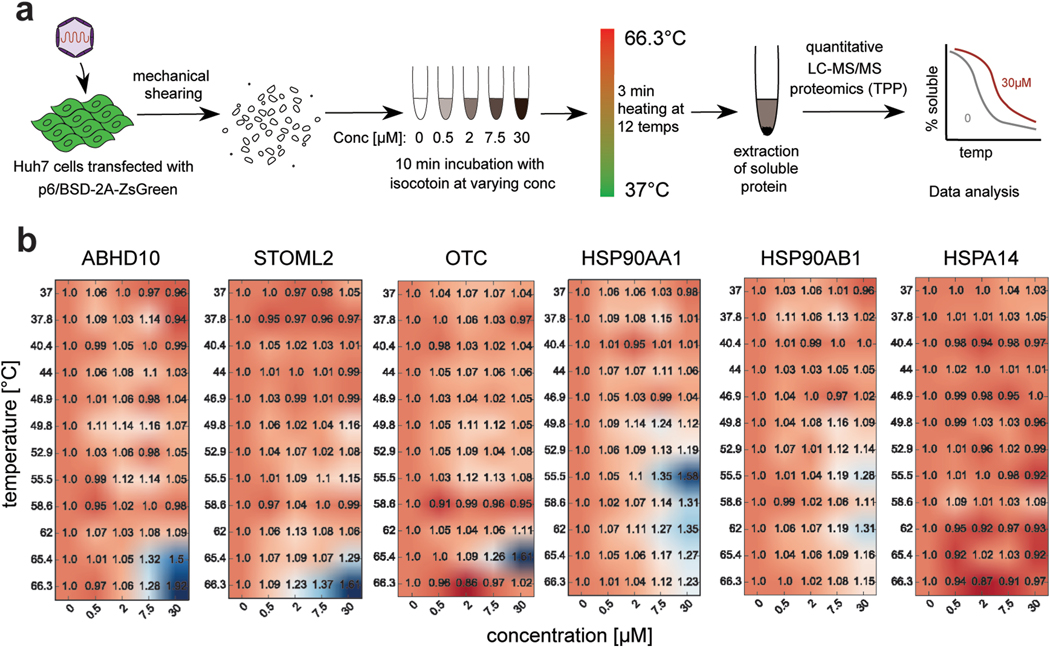
To further explore isocotoin’s mechanism of action, thermal proteome profiling was performed to identify direct binding targets of isocotoin (Fig 6a)(Becher et al., 2016; Savitski et al., 2014). Briefly, this assay identifies proteins whose stability changes in the presence of the drug due to binding interactions with the drug. Thirty-one targets were identified exhibiting thermal stabilization in the presence of isocotoin (Supplementary Table 2, PRIDE partner repository identifier PXD014485). Though no viral proteins were identified, the hits included Hsp90-α1 and Hsp90-β, two HSP90 isoforms known to play broad pro-viral roles (Fig 6b)(Geller et al., 2012). HSP90 are a family of conserved, abundant, and constitutively expressed molecular chaperones assisting in the maturation and localization of hundreds of cellular proteins(Schopf et al., 2017). A diverse panel of viruses, including herpes simplex viruses, simian virus 40, and HCV, rely on HSP90 for critical functions, including virus internalization, localization, and viral protein complex assembly(Geller et al., 2012). Furthermore, there is evidence that HSP90 directly binds to the macro domain of HEV ORF1, though this interaction has not previously been functionally validated(Ojha and Lole, 2016). Many viruses are hyper-dependent on HSP90 function, such that HSP90 inhibition disproportionately impairs the viral life cycle as compared to regular host functions(Geller et al., 2012). HSP90 is reported to bind the HEV capsid protein for intracellular trafficking of the virus during the early stages of infection (Zheng et al., 2010). However, since our assay employed a replicon lacking the capsid protein gene (ORF2), we hypothesized that HSP90 must play an additional, yet previously uncharacterized role specifically in HEV genomic replication by binding to HEV ORF1.
Fig 6|. Two-dimensional thermal proteome profiling identifies HSP90 as a binding target of isocotoin.
a, Schematic of two-dimensional thermal proteome profiling (2D-TPP) workflow. b, 2D-TPP data for proteins ABHD10, STOML2, OTC, HSP90AA1, HSP90AB1, and HSPA14. Numbers in heatmaps indicate fold-change in soluble protein at the indicated isocotoin concentration and heating temperature. ABHD10, STOML2, OTC, and HSP90AA1 were among hits showing the greatest increase in the soluble protein fraction upon addition of drug. HSP90AB1 showed a relatively modest but identifiable increase. HSPA14 is shown as an example of a protein that did not produce a thermal shift.
Hsp90 is an essential host factor for HEV replication.
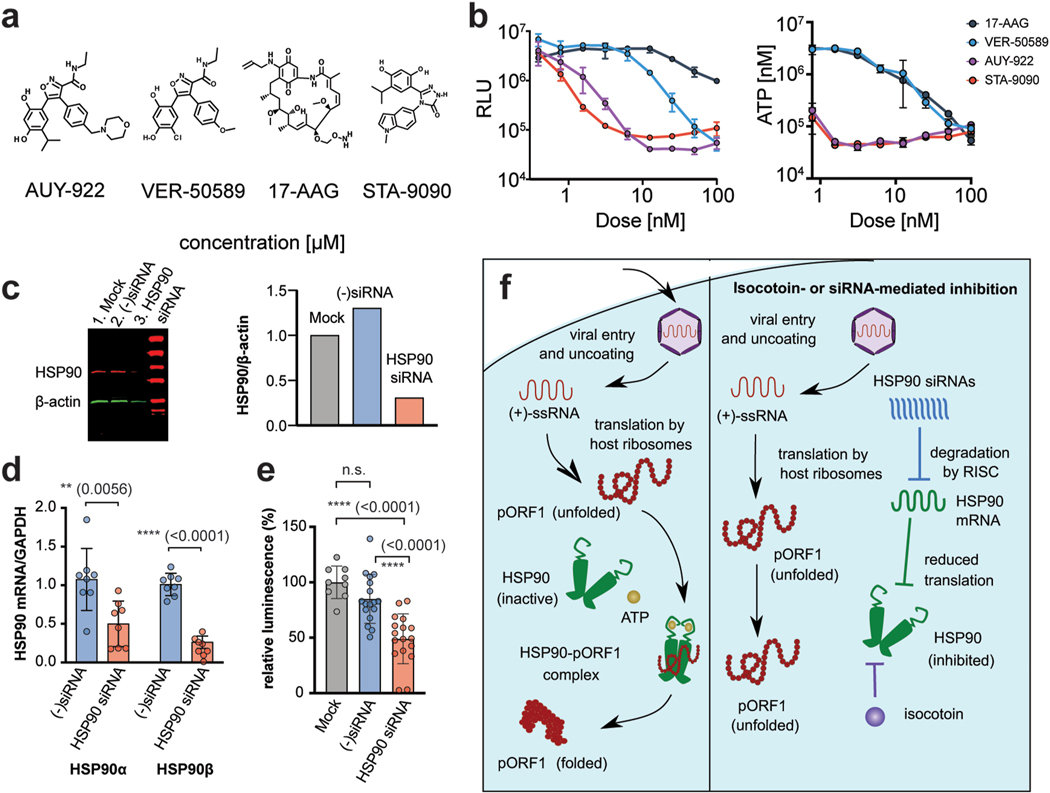
The HSP90 family was additionally of interest to us because isocotoin bears structural similarities to two potent HSP90 inhibitors, AUY-922 and VER-50589 (Fig 7a), which were identified through structure-activity relationship analysis (Supplementary Fig 4) (Eccles et al., 2008; Sharp et al., 2007). These compounds, along with the other known HSP90 inhibitors STA-9090 and 17-AAG, were tested against p6/Gluc replication (Fig 7b)(Lin et al., 2008; Schulte and Neckers, 1998). All four compounds inhibited HEV replication, confirming the significant role of HSP90 in viral replication. Unlike isocotoin, however, the four compounds exhibited cytostatic properties inhibiting cell growth(Fig 7b).
Fig 7|. Isocotoin inhibits HEV replication through interference with HSP90.
a, Chemical structures of known HSP90 inhibitors AUY-922, STA-9090, VER-50589, and 17-AAG. b, AUY-922, STA-9090, VER-50589, and 17-AAG are potent inhibitors of HEV (p6/Gluc strain) replication (left panel). ATP-based live cell assay showed that known HSP90 inhibitors 17-AAG, VER-50589, AUY-922, and STA-9090 are inhibitors of cell growth at concentrations required for HEV inhibition (right panel). c, Treatment with HSP90-specific siRNA results in a 69% reduction in HSP90 protein levels relative to mock-transfected cells at 72h post-transfection. HSP90 bands were first normalized to β-actin band intensity and then to the mock-transfected cells for each lane. d, Treatment with HSP90-specific siRNA resulted in a 54% reduction in HSP90AA1 and 74% reduction in HSP90AB1 mRNA levels. Data are combined from two repeat experiments and normalized to cellular GAPDH levels. P values were calculated using unpaired t-tests. e, Treatment with HSP90-specific siRNA resulted in reduced viral replication of p6/Gluc as measured via supernatant Gluc 3 days post-transfection. Gluclevels decreased 51% compared to mock-transfected cells and 36% compared to cells treated with negative control seed sequence-matched siRNA. Data are combined from two repeat experiments and normalized to viral replication levels in cells treated with transfection reagent only. p values calculated using ordinary one-way ANOVA followed by Tukey’s multiple comparisons test are indicated in parentheses. f, Hypothesized mechanism for isocotoin-mediated inhibition of pORF1 folding.
To further corroborate these data, we also performed an siRNA-mediated knockdown of HSP90AA1 and HSP90AB1 to examine the effect on viral replication. HSP90 protein levels were reduced 69%, and mRNA levels were reduced 54% for HSP90AA1 and 74% for HSP90AB1 post-siRNA transfection (Figs 7c–d, Supplementary Fig 5). Seed sequence-matched negative control siRNAs did not demonstrate knockdown of HSP90 protein or mRNA (Figs 7c–d). Knockdown of HSP90AA1 and HSP90AB1 decreased viral replication by 51% compared to cells treated with transfection reagent only (“mock”) and by 36% compared to cells treated with seed sequence-matched negative control siRNA (Fig 7e). The incomplete inhibition of viral replication is likely due to incomplete knockdown of the HSP90 as demonstrated by western blot (Fig 7c). In summary, our chemical and genetic data collectively demonstrate that HSP90 is essential for HEV replication, and we propose that isocotoin inhibits HEV replication by binding HSP90 (Fig 7f).
Discussion
This study provides an important step forward in developing therapeutics against HEV infection, where there is a dire need for new treatments, especially for vulnerable patient populations such as immunocompromised individuals and pregnant women. We present a robust high-throughput screening platform to assess small molecules for inhibitory activity against HEV replication, and report that isocotoin was identified as a candidate through this assay. We further show that isocotoin directly binds to two HSP90 proteins, and that targeted inhibition or depletion of HSP90 proteins severely impairs HEV replication. Collectively, our data provide the first evidence that HSP90 proteins play an essential role in HEV genomic replication and suggest that HSP90 inhibitors may provide a novel therapeutic approach for treating hepatitis E. Furthermore since HSP90 inhibitors employ a distinct mechanism of action from ribavirin, they may be effective in cases of ribavirin-resistance, or as combination therapy with ribavirin.
Isocotoin was identified using the p6/BSR-2A-zsGreen replicon, a reporter genome that can be used for future high-throughput studies. We also developed a publicly sourced custom script to rapidly quantify fluorescence in large image datasets and to select promising candidate wells (https://github.com/aploss/PLOCUS, Appendix 1). One limitation of our screening assay was that due to the large number of compounds in the library, only one dose (50 μM) per compound was tested. This dose was chosen based on the in vitro efficacy of ribavirin, but as a result, compounds that were effective at lower doses but cytotoxic at the 50 μM dose would not have been selected in the screening assay. However, the assay is adaptable to testing compounds at lower doses, or at multiple doses to calculate dose titrations. Therefore, we hope that this platform will be a tractable tool for future screening assays against HEV replication.
Two-dimensional thermal proteome profiling (2D-TPP) was used to identify binding targets of isocotoin, towards determining its mechanism of action(Becher et al., 2016). The hits identified did not include any HEV viral proteins, which was consistent with our data demonstrating that isocotoin is effective against several different viruses (HCV and YFV-17D) and therefore unlikely to be specifically targeting an HEV protein. Of the hits identified, we were most intrigued by HSP90 for three reasons: (1) isocotoin bears structural similarities to other known HSP90 inhibitors AUY-922 and VER-50589, (2) HSP90 has previously been shown to interact with HEV ORF1, and (3) HSP90 proteins are known to play pro-viral roles for a large number of other viruses(Ojha and Lole, 2016). Furthermore, though the 2D-TPP measured a relatively modest shift in stability for HSP90 proteins upon the addition of isocotoin, protein complexes such as HSP90 typically show smaller thermal shifts from binding interactions due to the initial presence of many pre-existing stabilizing interactions(Mateus et al., 2016). Our assays testing other HSP90 inhibitors against HEV and measuring HEV replication in the presence of HSP90 knockdown confirm the importance of HSP90 for the viral life cycle. However, the other hits identified through 2DTPP, e.g. proteins within the dynactin complex, septin proteins, and ACAT2, remain interesting areas for follow-up on whether they play roles during HEV replication (Supplementary Table 2).
Using the CellTiterGlo assay, which measures ATP levels as a proxy for viable cells, we found that isocotoin has a similar cytotoxicity profile in vitro to ribavirin. A caveat is that the cytotoxicity assays were performed in hepatoma cell lines, which are more susceptible to HSP90 inhibition than non-tumor derived cell lines(Miyata et al., 2013). Therefore cytotoxicity in non-tumor liver cells may actually be lower than our reported results. Indeed, when we treated mice for seven days with 50 mg/kg doses of isocotoin, we did not observe any obvious toxicity, though these observations remain preliminary since extensive follow-up work is required to understand the pharmacokinetic properties of the compound.
A suboptimal dosing experiment also resulted in the discovery of the F470S fitness-enhancing mutation in the putative cysteine protease (PCP) region of p6/Gluc. The F470S mutation was not detectable during the initial passages of the experiment based on convential Sanger sequencing, but was detected in a subset of clones by passage 10. It could have arisen either from de novo mutation, or through selection of a minor quasispecies present in the original inoculum that was not detectable by conventional Sanger sequencing. We found that the F470S mutant strain replicates more robustly in vitro than the wild-type p6/Gluc strain and two previously discovered mutant strains with fitness-enhancing mutations (G1634R and Y1320H, both in the RNA-dependent RNA polymerase region of ORF1)(Debing et al., 2014; Debing et al., 2016). The PCP region of HEV, so-named due to sequence similarity with the rubella virus protease, is poorly understood, and its protease activity remains highly controversial (Koonin et al., 1992). The discovery of a replication-enhancing mutation in this region provides a look into the important functional sites of this enigmatic domain. Due to its robust replicative capacity, the F470S strain may also prove useful for in vitro studies that require higher replication levels, such as viral production assays.
The precise role of HSP90 in HEV genomic replication remains unknown, however it is likely that as a molecular chaperone, it aids in the folding and assembly of proteins in HEV ORF1. This would be consistent with the pro-viral roles HSP90 is known to play for many other viruses and remains an important area for follow-up work(Geller et al., 2012).
Conclusion
We report a tractable screening assay for the identification of molecules inhibiting HEV replication, and we provide the first evidence that HSP90 proteins play an essential role in HEV genomic replication. From our screening platform, we report a compound named isocotoin showing superior activity in vitro to the currently available clinical treatment, ribavirin. This work increases our understanding of host-virus interactions during the HEV life cycle, and suggests that HSP90 may be a viable therapeutic target for treating hepatitis E.
Supplementary Material
Highlights.
We developed a high-throughput screening platform to identify molecules inhibiting hepatitis E virus (HEV) replication
Through this platform, we identified a new anti-HEV compound, isocotoin
Isocotoin inhibits HEV replication in in vitro studies more effectively than ribavirin, the only currently available treatment against HEV
Based on 2D thermal proteome profiling, isocotoin binds to the molecular chaperone HSP90
siRNA-mediated knockdown of HSP90 impairs HEV replication
Acknowledgements
Suzanne Emerson and Patricia Farci (NIAID) kindly provided the pSK Sar55, pKernowC1p6, pKernowC1p6/Gluc constructs. The plasmids pGEM-9zf-pSHEV3 and pGEM-7Zf(-)-TW6196E encoding the infectious pSHEV3(gt. 3) and TW6196 (gt. 4) clones, respectively were a kind gift from X.J. Meng (Virginia Tech). The pACNR/FLYF-17D plasmid, used to generate the YFV-17D Gluc-expressing replicon, was a kind gift of Charles M Rice, Rockefeller University, NY, USA. For the mouse infection experiments Chris Walker (Nationwide Children’s Hospital) sent stool filtrate from HEV-infected rhesus macaques. We thank Christina DeCoste and Katherine Rittenbach of the Molecular Biology Flow Cytometry Resource Facility, and Gary Laevsky of the Molecular Biology Confocal Microscopy Core Facility for outstanding technical help. We are grateful to the members of the Ploss lab for critical discussions and comments on the manuscript.
Funding: The work was supported in part by grants from Princeton University, an Investigator in Pathogenesis Award by the Burroughs Wellcome Fund (to AP) and support from the New Jersey Health Foundation. A.M. is supported by a fellowship from the EMBL Interdisciplinary Postdoc (EIPOD) program under Marie Curie Actions COFUND.
List of Abbreviations:
- HEV
Hepatitis E virus
- HSP90
heat shock protein 90
- ORF
open reading frame
- PCP
putative cysteine protease
- RdRp
RNA-dependent RNA polymerase
- DMSO
dimethyl sulfoxide
- BSR
blasticidin resistance gene
- Gluc
Gaussia luciferase
- 2D-TPP
Two-dimensional thermal proteome profiling
Footnotes
Competing interests
I.N., H.K. and A.P. have filed a patent application on the use of isocotoin and related compounds for the treatment of hepatitis E.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Becher I, Werner T, Doce C, Zaal EA, Tögel I, Khan CA, Rueger A, Muelbaier M, Salzer E, Berkers CR, Fitzpatrick PF, Bantscheff M, Savitski MM, 2016. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat Chem Biol 12, 908–910. [DOI] [PubMed] [Google Scholar]
- Benhamou Y, Tubiana R, Thibault V, 2003. Tenofovir disoproxil fumarate in patients with HIV and lamivudine-resistant hepatitis B virus. N Engl J Med 348, 177–178. [DOI] [PubMed] [Google Scholar]
- Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J, 2014. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147, 1008–1011 e1007; quiz e1015–1006. [DOI] [PubMed] [Google Scholar]
- Debing Y, Ramiere C, Dallmeier K, Piorkowski G, Trabaud MA, Lebosse F, Scholtes C, Roche M, Legras-Lachuer C, de Lamballerie X, Andre P, Neyts J, 2016. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol 65, 499–508. [DOI] [PubMed] [Google Scholar]
- Ding Q, Nimgaonkar I, Archer NF, Bram Y, Heller B, Schwartz RE, Ploss A, 2018. Identification of the Intragenomic Promoter Controlling Hepatitis E Virus Subgenomic RNA Transcription. mBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, Patterson L, de Haven Brandon A, Gowan S, Boxall F, Aherne W, Rowlands M, Hayes A, Martins V, Urban F, Boxall K, Prodromou C, Pearl L, James K, Matthews TP, Cheung KM, Kalusa A, Jones K, McDonald E, Barril X, Brough PA, Cansfield JE, Dymock B, Drysdale MJ, Finch H, Howes R, Hubbard RE, Surgenor A, Webb P, Wood M, Wright L, Workman P, 2008. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res 68, 2850–2860. [DOI] [PubMed] [Google Scholar]
- Geller R, Taguwa S, Frydman J, 2012. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta 1823, 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagsma EB, Riezebos-Brilman A, van den Berg AP, Porte RJ, Niesters HG, 2010. Treatment of chronic hepatitis E in liver transplant recipients with pegylated interferon alpha-2b. Liver Transpl 16, 474–477. [DOI] [PubMed] [Google Scholar]
- Kamar N, Abravanel F, Selves J, Garrouste C, Esposito L, Lavayssière L, Cointault O, Ribes D, Cardeau I, Nogier MB, Mansuy JM, Muscari F, Peron JM, Izopet J, Rostaing L, 2010. Influence of immunosuppressive therapy on the natural history of genotype 3 hepatitis-E virus infection after organ transplantation. Transplantation 89, 353–360. [DOI] [PubMed] [Google Scholar]
- Kamar N, Mallet V, Izopet J, 2014. Ribavirin for chronic hepatitis E virus infection. N Engl J Med 370, 2447–2448. [DOI] [PubMed] [Google Scholar]
- Khuroo MS, Khuroo NS, 2016. Hepatitis E: Discovery, global impact, control and cure. World J Gastroenterol 22, 7030–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW, 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subhadra S, Singh B, Panda BK, 2013. Hepatitis E virus: the current scenario. Int J Infect Dis 17, e228–233. [DOI] [PubMed] [Google Scholar]
- LeDesma R, Nimgaonkar I, Ploss A, 2019. Hepatitis E Virus Replication. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TI, Lenz O, Fanning G, Verbinnen T, Delouvroy F, Scholliers A, Vermeiren K, Rosenquist A, Edlund M, Samuelsson B, Vrang L, de Kock H, Wigerinck P, Raboisson P, Simmen K, 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob Agents Chemother 53, 1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Bear M, Du Z, Foley KP, Ying W, Barsoum J, London C, 2008. The novel HSP90 inhibitor STA-9090 exhibits activity against Kit-dependent and -independent malignant mast cell tumors. Exp Hematol 36, 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm BA, Liu R, Lahser F, Agrawal S, Belanger B, Butkiewicz N, Chase R, Gheyas F, Hart A, Hesk D, Ingravallo P, Jiang C, Kong R, Lu J, Pichardo J, Prongay A, Skelton A, Tong X, Venkatraman S, Xia E, Girijavallabhan V, Njoroge FG, 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob Agents Chemother 50, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB, 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48, 1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus A, Määttä TA, Savitski MM, 2016. Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Sci 15, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ, 2016. Expanding Host Range and Cross-Species Infection of Hepatitis E Virus. PLoS Pathog 12, e1005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Nakamoto H, Neckers L, 2013. The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des 19, 347–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimgaonkar I, Ding Q, Schwartz RE, Ploss A, 2018. Hepatitis E virus: advances and challenges. Nature reviews. Gastroenterology & hepatology 15, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha NK, Lole KS, 2016. Hepatitis E virus ORF1 encoded non structural protein-host protein interaction network. Virus Res 213, 195–204. [DOI] [PubMed] [Google Scholar]
- Pérez-Gracia MT, Suay-García B, Mateos-Lindemann ML, 2017. Hepatitis E and pregnancy: current state. Rev Med Virol 27, e1929. [DOI] [PubMed] [Google Scholar]
- Rein DB, Stevens GA, Theaker J, Wittenborn JS, Wiersma ST, 2012. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55, 988–997. [DOI] [PubMed] [Google Scholar]
- Savitski MM, Reinhard FB, Franken H, Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R, Dovega RB, Klaeger S, Kuster B, Nordlund P, Bantscheff M, Drewes G, 2014. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J, 2017. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol 18, 345–360. [DOI] [PubMed] [Google Scholar]
- Schulte TW, Neckers LM, 1998. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol 42, 273–279. [DOI] [PubMed] [Google Scholar]
- Sharp SY, Prodromou C, Boxall K, Powers MV, Holmes JL, Box G, Matthews TP, Cheung KM, Kalusa A, James K, Hayes A, Hardcastle A, Dymock B, Brough PA, Barril X, Cansfield JE, Wright L, Surgenor A, Foloppe N, Hubbard RE, Aherne W, Pearl L, Jones K, McDonald E, Raynaud F, Eccles S, Drysdale M, Workman P, 2007. Inhibition of the heat shock protein 90 molecular chaperone in vitro and in vivo by novel, synthetic, potent resorcinylic pyrazole/isoxazole amide analogues. Mol Cancer Ther 6, 1198–1211. [DOI] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU, 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108, 2438–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK, 1972. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177, 705–706. [DOI] [PubMed] [Google Scholar]
- Stedman C, 2014. Sofosbuvir, a NS5B polymerase inhibitor in the treatment of hepatitis C: a review of its clinical potential. Therap Adv Gastroenterol 7, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR, 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todt D, Meister TL, Steinmann E, 2018a. Hepatitis E virus treatment and ribavirin therapy: viral mechanisms of nonresponse. Curr Opin Virol 32, 80–87. [DOI] [PubMed] [Google Scholar]
- Todt D, Moeller N, Praditya D, Kinast V, Friesland M, Engelmann M, Verhoye L, Sayed IM, Behrendt P, Dao Thi VL, Meuleman P, Steinmann E, 2018b. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antiviral Res 157, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarev SA, Emerson SU, Reyes GR, Tsareva TS, Legters LJ, Malik IA, Iqbal M, Purcell RH, 1992. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci U S A 89, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk M, Zaaijer HL, Kater AP, Schinkel J, 2017. Sofosbuvir shows antiviral activity in a patient with chronic hepatitis E virus infection. J Hepatol 66, 242–243. [DOI] [PubMed] [Google Scholar]
- van Wezel EM, de Bruijne J, Damman K, Bijmolen M, van den Berg AP, Verschuuren EAM, Ruigrok GA, Riezebos-Brilman A, Knoester M, 2019. Sofosbuvir Add-on to Ribavirin Treatment for Chronic Hepatitis E Virus Infection in Solid Organ Transplant Recipients Does Not Result in Sustained Virological Response. Open Forum Infect Dis 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma SP, Kumar A, Kapur N, Durgapal H, Acharya SK, Panda SK, 2011. Hepatitis E virus replication involves alternating negative- and positive-sense RNA synthesis. J Gen Virol 92, 572–581. [DOI] [PubMed] [Google Scholar]
- Zheng ZZ, Miao J, Zhao M, Tang M, Yeo AE, Yu H, Zhang J, Xia NS, 2010. Role of heat-shock protein 90 in hepatitis E virus capsid trafficking. J Gen Virol 91, 1728–1736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.