Abstract
Huntington's disease (HD) causes neurological impairments, as well as muscle dysfunction, including smaller neuromuscular junctions (NMJs). This study assessed the expression levels of the subunits of the nicotinic acetylcholine receptor (nAChR) in muscles of the R6/2 mouse model of HD. Based on our previous findings of reduced NMJ size in R6/2 mice, it was hypothesized that muscles from R6/2 mice would also show an altered expression pattern of nAChR subunits compared to wild-type (WT) mice. Therefore, the mRNA levels of nAChR subunits were quantified in R6/2 and WT mouse muscles using qRT-PCR. Denervated muscles from WT mice served as positive controls for alterations in nAChR expression. Although some changes in nAChR subunit expression occurred in R6/2 muscles, the expression levels closely resembled WT. However, the expression of nAChR subunit-ε (Chrne) was significantly decreased in R6/2 muscles relative to WT. This study demonstrates that only minor changes in nAChR subunit expression occurs in R6/2 mouse muscles and that reduction in Chrne expression may be related to a reduction in NMJ size in R6/mice.
Keywords: Acetylcholine receptor, Denervation, Huntington's disease, R6/2, Skeletal muscle
Highlights
-
•
Huntington's Disease(R6/2) mouse muscles showed only minor alterations in the expression of nAChR subunits.
-
•
The primary change being a reduction in the expression of nAChR-ε subunit.
-
•
In contrast and as expected, denervation caused major changes in nAChR subunits.
-
•
Lastly, R6/2 mouse skeletal muscle showed no changes in nAChR-γ subunit expression at any disease stage.
1. Introduction
Huntington's Disease (HD) is a heritable (autosomal dominant) neurodegenerative disease that results in progressive loss of both cognitive and neuropsychological functions [1,2]. While multiple studies have focused on the progressive debilitations in neurological function associated with HD [3], a primary hallmark of HD is a progressive loss of motor function including chorea (uncontrollable dance-like movements), rigidity (rigid muscular contractions), dystonia (abnormal repetitive movements or posture), and bradykinesia (slower than normal movements) [4]. The abnormalities in motor function are associated with a loss of body mass in human subjects and animal models [5] and a loss of muscle mass in animal models of the disease [[6], [7], [8]].
At the molecular and cellular levels, HD is caused by a trinucleotide (CAG) repeat expansion (TRE) in the first intron of the human IT15 gene (interesting transcript 15, also known as HTT) that encodes for a protein now known as Huntingtin [9]. Normally the HTT gene includes a tract of ∼19 CAG trinucleotide repeats in the first exon of the gene that upon translation encode a polyglutamine (polyQ) tract in the amino terminal region of the protein. In HD individuals the length of the CAG tract is expanded to 40 or more CAG repeats. The length of the CAG repeat is also related to the age of disease onset, with a longer CAG tract correlating with an earlier age of disease onset [10] and a greater deficit in body mass in humans and mouse models of the disease [6]. Multiple transgenic and knock-in animal models of HD have been generated to replicate the pathology of HD [11]. The most commonly used model being the R6/2 transgenic mouse which includes the first intron of the human HTT gene (including ∼144 CAG repeats) inserted into the mouse genome [12].
Although specific defects in muscle metabolism, RNA processing, and function have been observed in mouse models of HD [[13], [14], [15], [16]], the loss in motor function has been largely attributed to decrements in central nervous system control of movement and muscular symptoms described as “denervation-like” [17]. Recent studies have suggested that limb muscle fiber denervation and reinnervation, due to the possible loss or dysfunction of spinal motoneurons and subsequent reinnervation by functional motoneurons, may play a role in the muscle debilitations in a different animal model of HD, the BACHD mouse [18,19]. However, we recently demonstrated that neither muscle fiber denervation, as determined by both anatomical and functional analyses, nor expression of some key molecular markers of denervation were evident in late-stage R6/2 mouse muscle [20]. However, we did find that the NMJs in R6/2 mice were smaller than in WT mice. Therefore, we assessed the expression levels of all five of the nAChR subunits in multiple muscles of late stage R6/2 mice in order to determine if altered expression levels of any of the subunits might help to explain the reduction in NMJ size.
2. Materials and methods
All animal procedures were approved by the Animal Care and Use Committee of Wright State University and followed all guidelines and regulations set forth by the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH Publication No. 8023). Animals were obtained from a breeding colony of R6/2 mice at Wright State University, housed and maintained as previously described [20]. Animal genotypes were determined from tail DNA from young mice (7–14 days of age) performed by Laragen Inc. (Culver City, CA). Mice were euthanized by isoflurane inhalation (∼2 g/L), followed by cervical dislocation. For the majority of analyses, the R6/2 and wild-type mice used in this study were between 11 and 14 weeks of age and considered late-stage. To assess for potential changes in Chrng (γ-subunit) mRNA expression at earlier ages, the tibialis anterior (TA) muscle from mice ranging in age from 3 to 14 weeks were utilized. For these analyses, animals were grouped as follows: early-stage, ages 23–46 days of age; mid-stage, ages 47–69 days of age; and late-stage, ages 70–92 days of age [16]. No effort was made to select mice of a specific sex. Further, to our knowledge, sex has not been shown to impact the expression of the nAChR subunits in skeletal muscle. The sexes of the animals used for this study are as follows: Denervated (see methodology below) and control gastrocnemius from SWR/J mice (n = 7 male and 0 female for control; and 7 male and 0 female for denervated); late-stage R6/2 and WT gastrocnemius (n = 5 male and 1 female for WT; and 1 male and 5 female for R6/2), late-stage R6/2 and WT extensor digitorum longus (EDL) (n = 5 male and 1 female for WT; and 1 male and 5 female for R6/2), late-stage R6/2 and WT soleus (n = 6 male and 4 female for WT; and 6 male and 4 female for R6/2); early-stage R6/2 and WT TA (n = 9 male and 1 female for WT; and 6 male and 4 female for R6/2); mid-stage R6/2 and WT TA (n = 2 male and 7 female for WT; and 3 male and 7 female for R6/2); and late-stage R6/2 and WT TA (n = 5 male and 1 female for WT; and 1 male and 5 female for R6/2).
As a positive control for altered expression of nAChR subunits, the expression levels of the nAChR subunits were analyzed in denervated muscles of mice. For these procedures, SWR/J mice were anesthetized with isoflurane and the left hindlimb muscles denervated via complete transection (and removal of a 3 mm section) of the sciatic nerve as previously described [20]. Animals were denervated for a period of 7 days and euthanized as described above.
The gastrocnemius, EDL, soleus, and TA muscles were removed from R6/2 and WT mice. The muscles were rapidly weighed, snap frozen in liquid nitrogen, and stored at −80 °C until analyzed. Total RNA was isolated using the trizol technique and stored at −80 °C. One μg of total RNA was used for the synthesis of cDNA using SuperScript III reverse transcriptase (ThermoFisher). Quantitative real-time polymerase chain reaction (qRTPCR) methods were used for the quantification of the nAChR subunit mRNAs, and Scn5a using TaqMan® based assays (Applied Biosystems™, Foster City, CA) for each specific transcript along with β-2-microglobulin as a normalizing gene (Table 1). The ΔΔCT method was used for calculation of relative expression levels [21]. For analyses of age-related (early-, mid-, late-stages) changes in nAChR subunits expression, the Chrng (γ-subunit) mRNA was chosen, because our current data (as shown in Fig. 2A) suggest that it is the gene that is most highly responsive to denervation.
Table 1.
Reverse transcriptase polymerase chain reaction gene expression assays.
| Common Name | Gene nomenclature | TaqMan® Expression Assaya |
|---|---|---|
| nAChR, α subunit | Chrna1 | Mm00431629_m1 |
| nAChR, β subunit | Chrnb1 | Mm00680412_m1 |
| nAChR, δ subunit | Chrnd | Mm00445545_m1 |
| nAChR, ε subunit | Chrne | Mm00437411_m1 |
| nAChR, γ subunit | Chrng | Mm00437419_m1 |
| Voltage-gated Na+ channel, cardiac isoform | Scn5a | Mm01342518_m1 |
| β-2-microglobulin | B2m | Mm00437762_m1 |
All TaqMan® Expression Assays were obtained from Applied Biosystems™, Foster City, CA. Abbreviations: nAChR, nicotinic acetylcholine receptor.
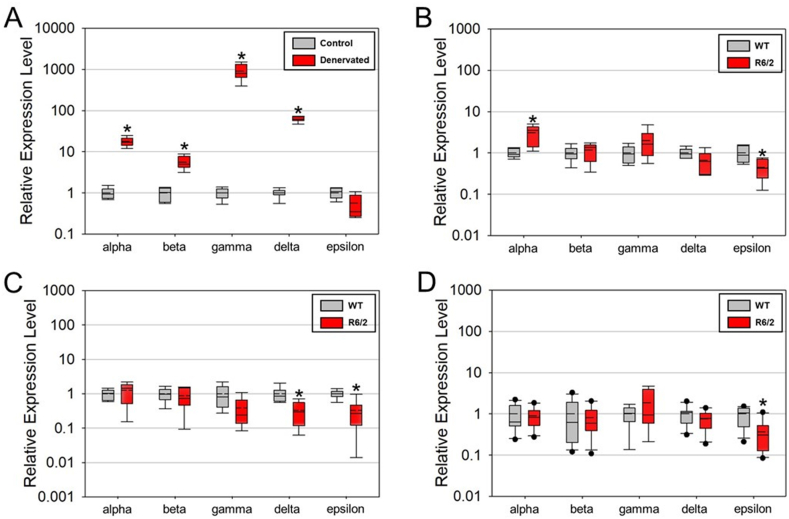
Fig. 2.
Relative expression levels as determined by real-time RT-PCR for the subunits of the nicotinic acetylcholine receptor: Chrna1 (alpha); Chrnb1 (beta); Chrng (gamma); Chrnd (delta); and Chrne (epsilon) in A) Denervated gastrocenemius muscle (n = 7 per group) relative to control; B) gastrocnemius muscle (n = 6 per group) from late-stage R6/2 and WT mice; C) EDL muscle (n = 6 per group) from late-stage R6/2 and WT mice; and D) soleus muscle (n = 11 WT and 10 R6/2) from late-stage R6/2 and WT mice. Denervation (A) resulted in major elevations in the expression levels of all subunits except Chrne which was reduced. In contrast R6/2 mice show no (or minor) changes in nicotinic acetylcholine receptor subunits, except for a consistent decrease in Chrne. The asterisks denote significantly different from control at p < 0.05.
The data are graphed as box and whisker plots with the mean (dashed line) and median (solid line) displayed in the middle of each box. The whiskers extend from the upper and lower quartiles to the upper and lower extremes, respectively and outliers are displayed as individual points. Two-tailed Student's t-tests were used for statistical comparisons between groups (WT vs R6/2 or control vs denervated) using SigmaPlot 13 software with a p-value ≤ 0.05 denoting statistical significance (Systat Software, San Jose, CA).
3. Results
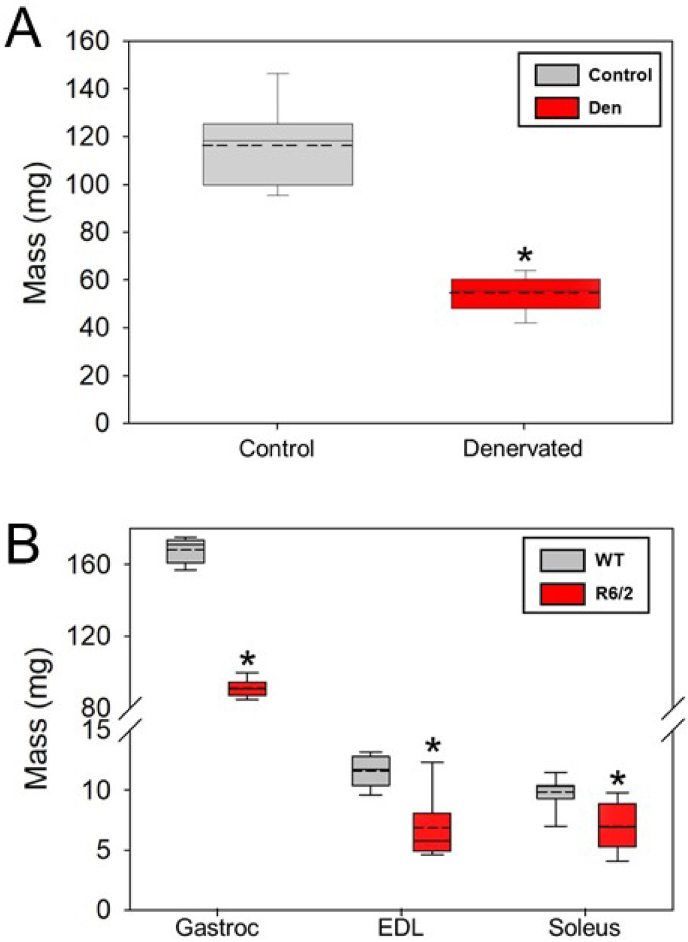
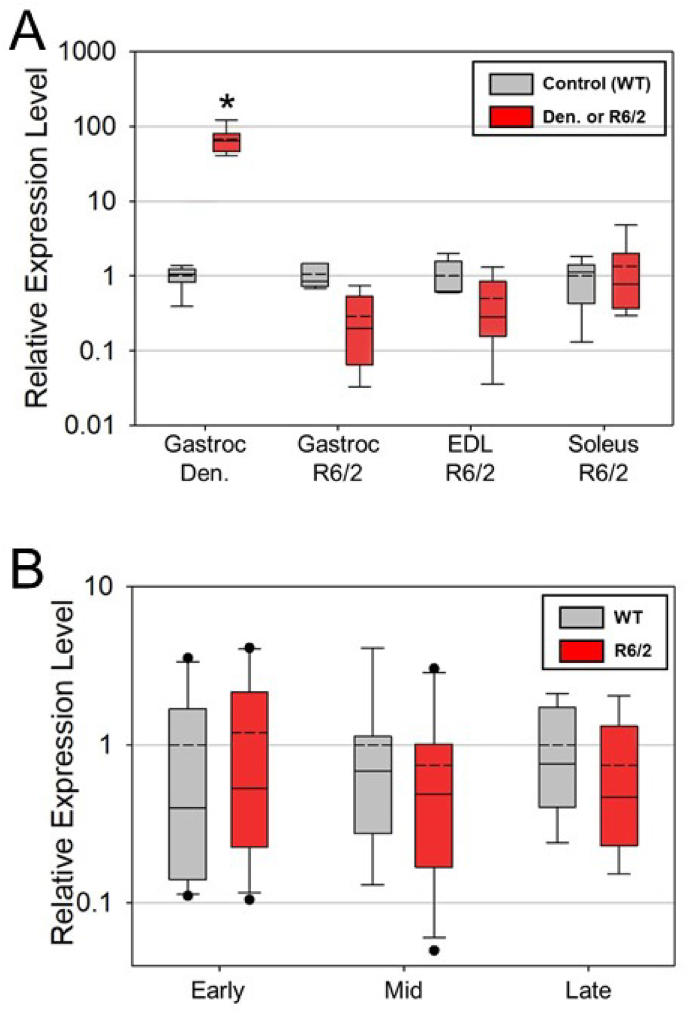
A reduction in muscle mass was observed for both HD and denervated muscles compared to wild-type controls (Fig. 1). As expected, the expression of all five nAChR subunits were significantly altered following denervation of mouse gastrocnemius muscle (Fig. 2A). After denervation, Chrna1 (α-subunit), Chrnb1 (β-subunit), Chrnd (δ-subunit), Chrng (γ-subunit) were significantly elevated by 18x, 5.5x, 61x, and 916x, respectively, whereas the Chrne (ε-subunit) was decreased to ∼57% of control (Fig. 2A). In contrast, in HD mice, the nAChR subunit Chrne (ε-subunit) was significantly decreased in all muscles of R6/2 mice relative to WT. The only nAChR subunit that was upregulated in R6/2 mice was the Chrna1 (α-subunit) in the gastrocnemius muscle, and it was only increased by 3x (Fig. 2B). Further, Chrnd (δ-subunit) was decreased in the EDL (Fig. 2B–D). There were no other instances of an elevated nAChR subunit in HD muscle (Fig. 2B–D). The cardiac isoform of the Na+ channel, Scn5a, was significantly elevated (67x) only in denervated, but not HD muscle (Fig. 3A).
Fig. 1.
A) Gastrocnemius (Gastroc) muscle masses from control (n = 7) and denervated (n = 7) mice. Denervation resulted in a near 50% loss in muscle mass. B) Gastroc (n = 6 per group), extensor digitorum longus (EDL, n = 6 per group), and soleus (n = 11 WT and 10 R6/2) muscle masses from late-stage R6/2 and wild type (WT) mice. Muscles in R6/2 mice show significant reductions in muscle mass. The asterisks denote significantly different from control at p < 0.05.
Fig. 3.
A. Relative expression levels as determined by real-time RT-PCR for the cardiac isoform of the sodium channel (Scn5a) in Denervated gastrocenemius (Gastroc Den.) muscle (n = 7 per group) relative to control; and gastrocnemius (Gast) muscle (n = 6 per group) from late-stage R6/2 and WT mice; EDL muscle (n = 6 per group) from late-stage R6/2 and WT mice; and soleus (Sol) muscle (n = 11 WT and 10 R6/2) from late-stage R6/2 and WT mice. The asterisks denote significantly different from control at p < 0.05. B. Relative expression levels as determined by real-time RT-PCR for Chrng in the tibialis anterior muscle at 3 disease stages (ages) according to Miranda et al. [16] as follows: Early-stage, ages 23–46 days of age (n = 10 per group); Mid-stage, ages 47–69 days of age (n = 9 WT and 10 R6/2); and Late-stage, ages 70–92 days of age (n = 6 per group). No significant differences were observed between R6/2 and WT at any time point.
To assess the potential for changes in nAChR subunit expression at younger ages in R6/2 mice relative to controls, we assessed the expression of Chrng (γ-subunit), because it was the nAChR subunit that was the most sensitive to denervation as shown in Fig. 2A. As shown in Fig. 3B, there were no significant differences in Chrng expression at any of the three stages in the tibialis anterior muscle.
4. Discussion
The only consistent change in expression of the nAChR subunits in HD muscles was a reduction in ε-subunit (Chrne) expression. This may relate to the reduction in miniature endplate currents, evoked endplate currents, and the size of the motor endplates in fast HD muscle fibers, as identified using voltage-clamp electrophysiology and α-bungarotoxin labeling [20]. It is possible that the reduced expression of one of the subunits (in this case ε) could result in a reduced number of fully formed receptors and junctional size, which could explain the smaller endplate size and currents in fast R6/2 muscle fibers. As α-bungarotoxin labels the motor endplate by binding to the fully formed nAChRs present at the endplate, a reduction in the number of fully formed nAChRs might result in a physically smaller endplate as assessed by α-bungarotoxin labeling [20]. However, it must be emphasized that, despite their smaller size, all neuromuscular junctions (NMJ) analyzed by Khedraki et al. [20] in HD muscle showed an anatomically and physiologically intact synapse. Thus, the molecular changes in R6/2 skeletal muscle, such as Clcn1 miss-splicing, more likely reflect a myopathy triggered by muscle autonomous expression of the mutant Htt gene rather than occurring consequential to denervation and are consistent with a less-mature muscle phenotype [16].
HD muscle does not respond like denervated muscle, as the profile of expression of the subunits of the nAChR in HD muscle does not mimic the changes that occur with denervation. Following denervation, the mouse gastrocnemius showed greatly elevated levels of the mRNAs for the Chrna1 (α-subunit, 18x), Chrnb1 (β-subunit, 5.5x), Chrnd (δ-subunit, 61x), and particularly Chrng (γ-subunit, 916x). The highly elevated expression of the nAChR γ-subunit after denervation is consistent with our previous data on the mouse gastrocnemius muscle [20] and several other studies. That the γ-subunit was not elevated in HD muscle, as shown by the present study (soleus, EDL, and gastrocnemius muscles), nor at any stage of development of the HD symptoms in R6/2 mice (Fig. 3), and our previous study on soleus and tibialis anterior muscles [20] implies that denervation is likely not present in late-stage HD muscle. Also, the expression of the cardiac isoform of the voltage gated sodium channel, Scn5a, which is a positive marker for denervation, was not increased in late-stage HD muscle. Thus, the muscles from HD mice do not appear to be responding to a denervation signal (or lack of innervation signal).
Regarding a potential mechanism for the reduction in Chrne expression, altered Chrne expression has been linked to alterations in growth factor expression (specifically neuregulin) in the central nervous system of schizophrenia patients [22]. In addition, altered splicing patterns of Chrne, as induced by genetic mutations that result in expression of a truncated Chrne, have been implicated in causing congenital myasthenic syndrome [23]. Further, single-nucleotide polymorphisms in the neuregulin-1 gene are associated with psychosis in Huntington's disease patients [24,25]. Finally, fibroblast growth factor 18 (FGF18), expressed in spinal motor neurons, appears to positively modulate the expression of Chrne at neuromuscular junctions during development via the FGF receptor 2, whereas absence of FGF18 results in smaller NMJs and reduced Chrne in NMJs [26]. Thus, future studies will address these relationships to skeletal muscle in Huntington's disease by assessing whether reductions in neural and non-neural growth factors (including neuregulin) may be modulating the expression and pre-mRNA splicing of Chrne in skeletal muscle and in turn limit the amount of available Chrne for formation of a normal sized and functional synapse at the neuromuscular junction, as well as possibly impacting growth and atrophy of skeletal muscle in R6/2 mice.
In summary, skeletal muscles from late-stage HD mice do not show major alterations in nAChR subunit expression, other than a consistent reduction in Chrne in fast muscles. This reduction in Chrne expression could partially explain the reduced size and functional alterations of R6/2 mouse NMJs that likely contribute to muscle hyperexcitability [20] and are consistent with a less-mature muscle phenotype [16]. In total, these data support the idea that HD is a multisystem disorder and that muscular changes are a consequence of muscle autonomous expression of the mutant Htt gene.
Authors contributions
Briana Simpson performed the majority of the PCR experiments and assisted in the writing of the manuscript and the generation of figures. Mark M. Rich performed denervation experiments, provided muscle samples for analyses, and read, edited, and approved the final manuscript. Andrew A. Voss provided R6/2 muscle samples, assisted in conceiving the study, and read, edited and approved the final manuscript. Robert J. Talmadge performed RNA isolations, purification, and cDNA synthesis, some PCR experiments, statistical analyses, conceived the study, and wrote, edited, and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by NIH/NIAMS grant NS099850 (AAV and RJT) and an NIH STEP-UP Program Summer Research Fellowship to BS.
References
- 1.Roos R.A. Huntington's disease: a clinical review. Orphanet J. Rare Dis. 2010;5:40. doi: 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoli R.A., Botturi A., Ciammola A., Silani V., Prunas C., Lucchiari C., Zugno E., Caletti E. Neuropsychiatric burden in Huntington's disease. Brain Sci. 2017;7:67. doi: 10.3390/brainsci7060067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barboza L.A., Ghisi N.C. Evaluating the current state of the art of Huntington disease research: a scientometric analysis. Braz. J. Med. Biol. Res. 2018;51 doi: 10.1590/1414-431x20176299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berardelli A., Noth J., Thompson P.D., Bollen E.L., Currà A., Deuschl G., van Dijk J.G., Töpper R., Schwarz M., Roos R.A. Pathophysiology of chorea and bradykinesia in Huntington's disease. Mov. Disord. 1999;14:398–403. doi: 10.1002/1531-8257(199905)14:3<398::aid-mds1003>3.0.co;2-f. http://www.ncbi.nlm.nih.gov/pubmed/10348461 [DOI] [PubMed] [Google Scholar]
- 5.Aziz N.A., van der Marck M.A., Pijl H., Olde Rikkert M.G.M., Bloem B.R., Roos R.A.C. Weight loss in neurodegenerative disorders. J. Neurol. 2008;255 doi: 10.1007/s00415-009-0062-8. 1872–80. [DOI] [PubMed] [Google Scholar]
- 6.Aziz N.A., van der Burg J.M.M., Landwehrmeyer G.B., Brundin P., Stijnen T., EHDI Study Group R.A.C., Roos R.A.C. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology. 2008;71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 7.She P., Zhang Z., Marchionini D., Diaz W.C., Jetton T.J., Kimball S.R., Vary T.C., Lang C.H., Lynch C.J. Molecular characterization of skeletal muscle atrophy in the R6/2 mouse model of Huntington's disease. Am. J. Physiol. Metab. 2011;301 doi: 10.1152/ajpendo.00630.2010. E49–E61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribchester R., Thomson D., N.W.-E.J. of, undefined . Wiley Online Libr; 2004. Progressive Abnormalities in Skeletal Muscle and Neuromuscular Junctions of Transgenic Mice Expressing the Huntington's Disease Mutation.https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1460-9568.2004.03783.x (n.d.) [DOI] [PubMed] [Google Scholar]
- 9.MacDonald M., Ambrose C., Duyao M., R.M.- Cell, undefined . Elsevier; 1993. A Novel Gene Containing a Trinucleotide Repeat that Is Expanded and Unstable on Huntington's Disease Chromosomes.https://www.sciencedirect.com/science/article/pii/009286749390585E (n.d.) [DOI] [PubMed] [Google Scholar]
- 10.Walker F.O. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 11.Ramaswamy S., McBride J.L., Kordower J.H. Animal models of Huntington's disease. ILAR J. 2007;48:356–373. doi: 10.1093/ilar.48.4.356. [DOI] [PubMed] [Google Scholar]
- 12.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 13.Lee J.E., Cooper T.A. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters C.W., Varuzhanyan G., Talmadge R.J., Voss A.A. Huntington disease skeletal muscle is hyperexcitable owing to chloride and potassium channel dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9160–9165. doi: 10.1073/pnas.1220068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda D.R., Wong M., Romer S.H., McKee C., Garza-Vasquez G., Medina A.C., Bahn V., Steele A.D., Talmadge R.J., Voss A.A. Progressive Cl- channel defects reveal disrupted skeletal muscle maturation in R6/2 Huntington's mice. J. Gen. Physiol. 2017;149:55–74. doi: 10.1085/jgp.201611603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribchester R.R., Thomson D., Wood N.I., Hinks T., Gillingwater T.H., Wishart T.M., Court F.A., Morton A.J. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington's disease mutation. Eur. J. Neurosci. 2004;20:3092–3114. doi: 10.1111/j.1460-9568.2004.03783.x. [DOI] [PubMed] [Google Scholar]
- 18.Valadão P.A.C., de Aragão B.C., Andrade J.N., Magalhães-Gomes M.P.S., Foureaux G., Joviano-Santos J.V., Nogueira J.C., Ribeiro F.M., Tapia J.C., Guatimosim C. Muscle atrophy is associated with cervical spinal motoneuron loss in BACHD mouse model for Huntington's disease. Eur. J. Neurosci. 2017 doi: 10.1111/ejn.13510. [DOI] [PubMed] [Google Scholar]
- 19.de Aragão B., Rodrigues H., P.V.-N., undefined . Elsevier; 2016. Changes in Structure and Function of Diaphragm Neuromuscular Junctions from BACHD Mouse Model for Huntington's Disease.https://www.sciencedirect.com/science/article/pii/S0197018615300826 (n.d.) [DOI] [PubMed] [Google Scholar]
- 20.Khedraki A., Reed E.J., Romer S.H., Wang Q., Romine W., Rich M.M., Talmadge R.J., Voss A.A. Depressed synaptic transmission and reduced vesicle release sites in Huntington's disease neuromuscular junctions. J. Neurosci. 2017;37:8077–8091. doi: 10.1523/JNEUROSCI.0313-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-deltadeltaCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. http://www.sciencedirect.com/science/article/B6WN5-457MF4S-3/2/46ddfa7ac4e6bd9a57d68a58f1b633a3%0A [DOI] [PubMed] [Google Scholar]
- 22.Moises H.W., T Z., Gottesman I.I. The glial growth factors deficiancy and synaptic destabilization hypothesis of schizophrenia. BMC Psychiatr. 2002;2:8. doi: 10.1186/1471-244X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieb J.P., Kraner S., Rauch M.S., Steinlein O.K. Immature end-plates and utrophin deficiancy in congenital myasthenic syndrome caused by epsilon-AChR subunit truncating mutations. Hum. Genet. 2000;107:160–164. doi: 10.1007/s004390000359. [DOI] [PubMed] [Google Scholar]
- 24.Ohno K., Tsujino A., Shen X.-M., Milone M. Spectrum of splicing errors caused by CHRNE mutations affecting introns and intron/exon boundaries. J. Med. Genet. 2005 doi: 10.1136/jmg.2004.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuang D.W., Greenwood T.A., Jayadav S., Davis M., Shutes-David A., Bird T.A. A genetic study of psychosis in Huntington's Disease: evidence for the involvement of glutamate signaling pathways. J. Huntingtons. Dis. 2018;7:51–59. doi: 10.3233/JHD-170277. [DOI] [PubMed] [Google Scholar]
- 26.K. Ito, B. Ohkawara, H. Yagi, H. Nakashima, M. Tsushima, K. Ota, H. Konishi, A. Masuda, S. Imagama, H. Kiyama, N. Ishiguro, K. Ohno, Lack of Fgf18 Causes Abnormal Clustering of Motor Nerve Terminals at the Neuromuscular Junction with Reduced Acetylcholine Receptor Clusters OPEN, (n.d.). 10.1038/s41598-017-18753-5. [DOI] [PMC free article] [PubMed]