Abstract
We present a genome assembly from an individual male Canis lupus orion (the grey wolf, subspecies: Greenland wolf; Chordata; Mammalia; Carnivora; Canidae). The genome sequence is 2,447 megabases in span. The majority of the assembly (98.91%) is scaffolded into 40 chromosomal pseudomolecules, with the X and Y sex chromosomes assembled.
Keywords: Canis lupus, Canis lupus orion, grey wolf, Polar wolf, Greenland wolf, genome sequence, chromosomal
Species taxonomy
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Laurasiatheria; Carnivora; Caniformia; Canidae; Canis; Canis lupus Linnaeus 1758 (NCBI:txid9612).
Background
The grey wolf, Canis lupus, is the largest species within the group wolf-like canids (Subtribe: Canina) and the member with the largest geographic distribution. Originally wolves were found throughout Eurasia, with the exception of tropical Southeast Asia, and all of North America. This vast distribution contains numerous habitats, encompassing wolf ecotypes adapted to the diverse environments throughout their distribution. The wolf is locally extinct in several places, such as the UK, Ireland and Brittany, yet it still holds much of its original distribution; the global population is estimated to be in the order of 200–250 thousand individuals ( Jhala et al., 2018).
Once numerous, wolves were eradicated from the islands of Great Britain in the 15th century and Ireland in the 18th century. There have been proposals to reintroduce populations of wolves to the Scottish Highlands to manage populations of red deer, which have a negative effect on biodiversity through overgrazing ( Nilsen et al., 2007). The Scottish Highlands are considered to be the only location in Great Britain that could support a healthy population of wolves; however, objections of livestock owners are likely to prevent their reintroduction in the near future ( Wilson, 2004). The reintroduction of wolves elsewhere has led not only to the reestablishment of this apex predator, but also to marked improvements in biodiversity in the ecosystem as a whole ( Ripple et al., 2014). Wolves reintroduced into the Yellowstone National Park, Wyoming, USA, in 1995 predated grazing animals such as wapiti ( Cervus canadensis) that preserved grasslands. The subsequent changes in prey behaviour led to trophic cascades that resulted in the reestablishment of tree species and an associated increase in populations of species that rely directly and indirectly on this habitat ( Ripple & Beschta, 2012).
Wolves have historically been found in Northwest, Northeast and East Greenland ( Dawes et al., 1986). Wolves were extirpated from East Greenland through hunting by 1939 and were absent from this area for the next 40 years ( Marquard-Petersen, 2012). In around 1979, a pair of wolves travelled from the north of the island and began a recolonisation of East Greenland, establishing a population of around 23 wolves ( Marquard-Petersen, 2011). A recent assessment found no trace of wolves for a number of years in East Greenland, while a population of up to 32 animals is still found in the northernmost parts of Greenland. Since the population in East Greenland was located entirely within the Northeast Greenland National Park, affording the wolves legal protection, it is unlikely that this extinction event was driven by hunting ( Marquard-Petersen, 2021).
Domestic dogs share a common ancestor with Eurasian wolves around 33,000 years ago ( Skoglund et al., 2015; Wang et al., 2016). In this regard, the Greenland wolf or Polar wolf reference genome described herein is highly relevant for dog and/or Eurasian wolf genomics. The Polar wolf is a North American wolf, an outgroup to dogs and Eurasian wolves ( Gopalakrishnan et al., 2019; Sinding et al., 2018), which will aid in making a minimally reference-biased representation of diversity in re-sequenced genomes ( Gopalakrishnan et al., 2017). The Polar wolf is also the North American wolf type with the least coyote-like ancestry ( Sinding et al., 2018); thus, it is probably the closest possible outgroup to dogs and Eurasian wolves with the least amount of exotic admixture that other North American wolves carry. Finally, this reference genome permits detailed genomic investigations of Polar wolves themselves, as a precise reference, to identify rare genomic variation. The genome is therefore an overall useful resource for research in the Polar wolf itself, a small, isolated and understudied population, but also canids, wolves and dogs overall.
Genome sequence report
The genome was sequenced from a single male C. lupus subspecies orion collected from Siorapaluk, Greenland (latitude 77.785278, longitude -70.631389) in 2016. A total of 28-fold coverage in Pacific Biosciences single-molecule long reads and 74-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 135 missing/misjoins and removed 9 haplotypic duplications, reducing the assembly length by 0.2% and the scaffold number by 42.1%, and increasing the scaffold N50 by 15.9%.
The final assembly has a total length of 2,447 Mb in 82 sequence scaffolds with a scaffold N50 of 66 Mb ( Table 1). Of the assembly sequence, 98.91% was assigned to 40 chromosomal-level scaffolds (named by synteny to an assembly for C. lupus familiaris, breed labrador: GCF_014441545.1), including 38 autosomes and the X and Y chromosomes ( Figure 1– Figure 4; Table 2). The assembly has a BUSCO ( Simão et al., 2015) completeness of 95.5% (single 93.0%, duplicated 2.4%) using the carnivora_odb10 reference set. While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
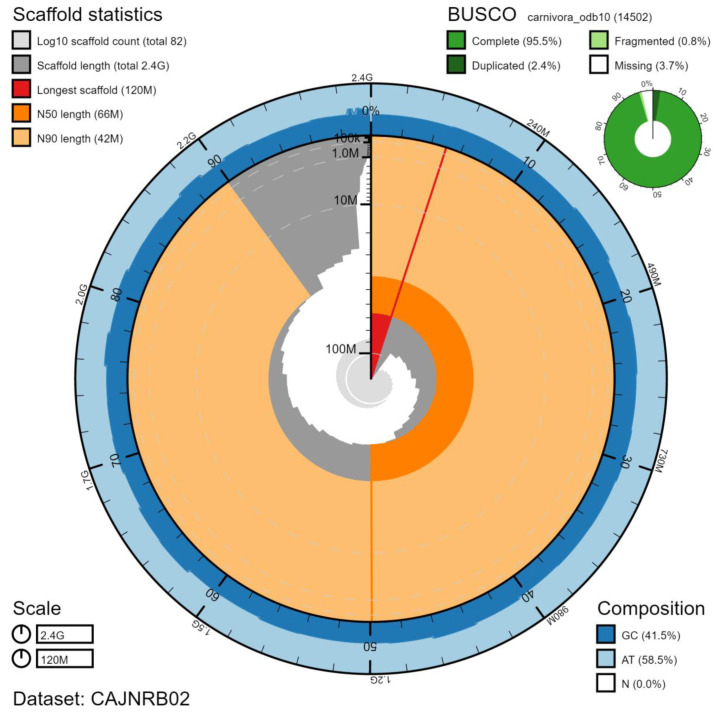
Figure 1. Genome assembly of Canis lupus, mCanLor1.2: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 2,447,463,909 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (124,665,963 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (65,778,685 and 41,774,919 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the carnivora_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/mCanLor1.2/dataset/CAJNRB02/snail.
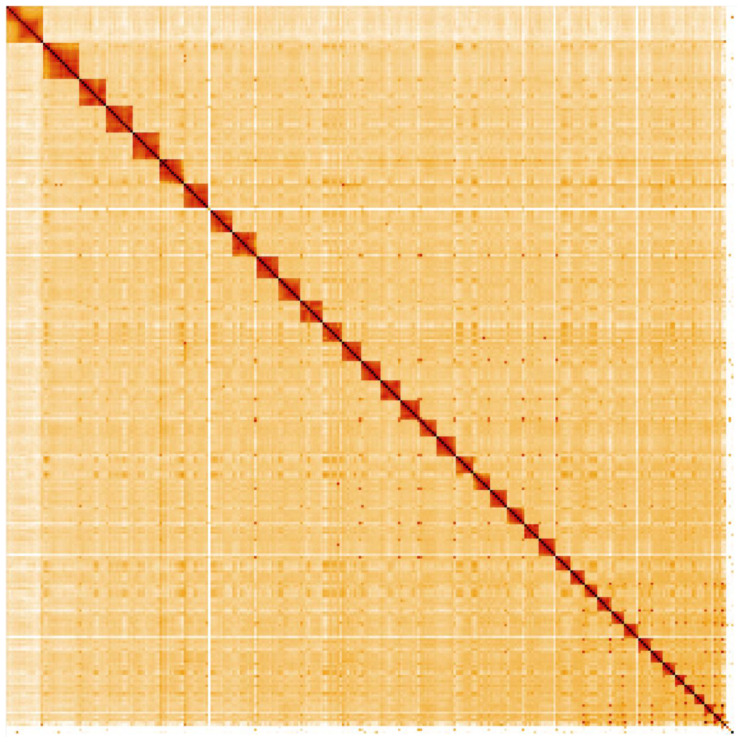
Figure 4. Genome assembly of Canis lupus, mCanLor1.2: Hi-C contact map.
Hi-C contact map of the mCanLor1.2 assembly, visualised in HiGlass. Chromosomes are shown in size order from left to right and top to bottom.
Table 1. Genome data for Canis lupus, mCanLor1.2.
| Project accession data | |
|---|---|
| Assembly identifier | mCanLor1.2 |
| Species | Canis lupus |
| Specimen | mCanLor1 |
| NCBI taxonomy ID | NCBI:txid9612 |
| BioProject | PRJEB43200 |
| BioSample ID | SAMEA7532739 |
| Isolate information | Male, muscle |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR6406204, ERR6406205,
ERR6412029, ERR6412030, ERR6412359, ERR6412360 |
| 10X Genomics Illumina | ERR6054484-ERR6054491 |
| Hi-C Illumina | ERR6511153 |
| Illumina RNA-Seq | ERR6054492 |
| Genome assembly | |
| Assembly accession | GCA_905319855.2 |
| Accession of alternate
haplotype |
GCA_905319845.1 |
| Span (Mb) | 2,447 |
| Number of contigs | 248 |
| Contig N50 length (Mb) | 34 |
| Number of scaffolds | 82 |
| Scaffold N50 length (Mb) | 66 |
| Longest scaffold (Mb) | 123 |
| BUSCO * genome score | C:95.8%[S:94.6%,D:1.2%],
F:2.0%,M:2.2%,n:4104 |
*BUSCO scores based on the carnivora_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/Canis%20lupus/dataset/CAJNRB02/busco.
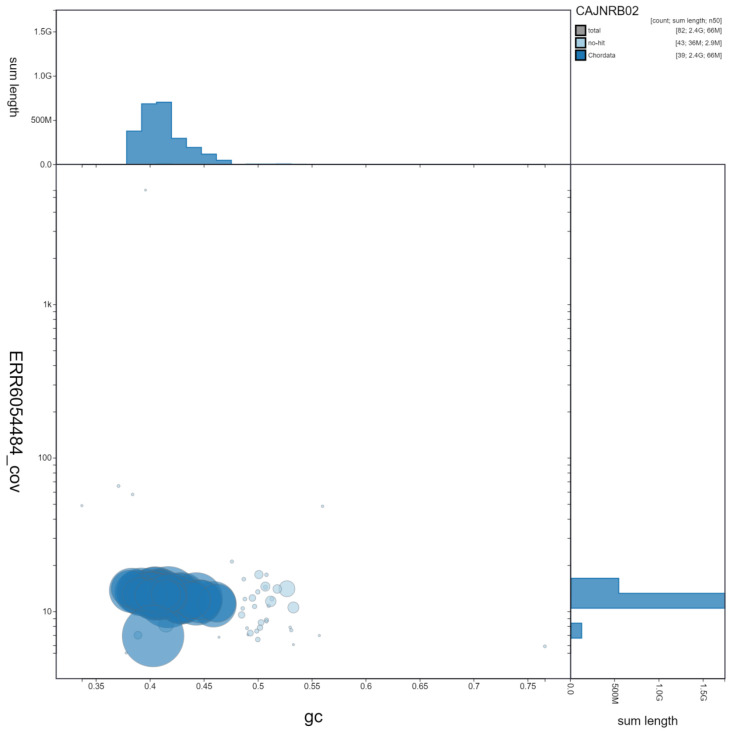
Figure 2. Genome assembly of Canis lupus, mCanLor1.2: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/mCanLor1.2/dataset/CAJNRB02/blob.
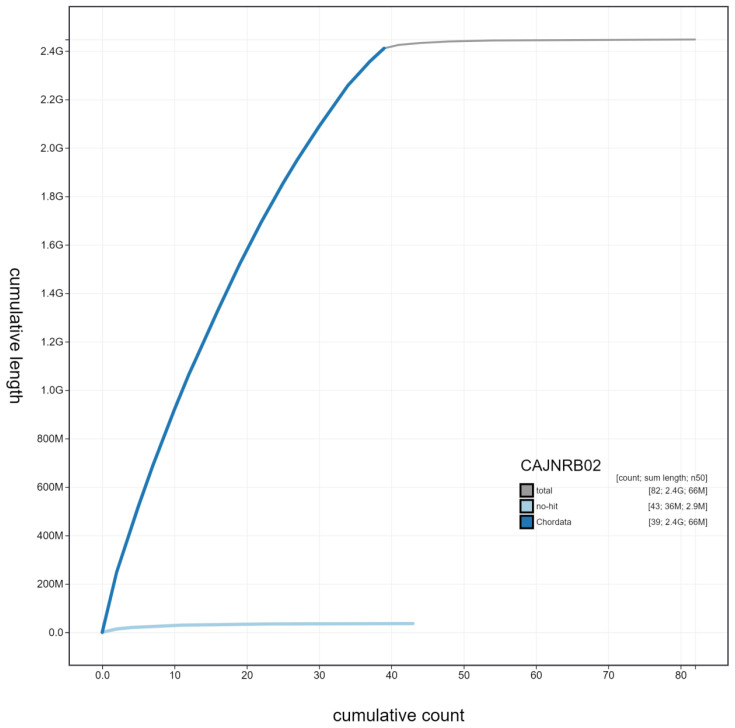
Figure 3. Genome assembly of Canis lupus, mCanLor1.2: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/mCanLor1.2/dataset/CAJNRB02/cumulative.
Table 2. Chromosomal pseudomolecules in the genome assembly of Canis lupus, mCanLor1.2.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| HG994383.1 | 1 | 122.96 | 41.7 |
| HG994387.1 | 2 | 86.40 | 42.9 |
| HG994384.1 | 3 | 93.48 | 40.5 |
| HG994386.1 | 4 | 88.63 | 40.4 |
| HG994385.1 | 5 | 89.78 | 44.3 |
| HG994389.1 | 6 | 78.39 | 42.8 |
| HG994388.1 | 7 | 82.29 | 41.1 |
| HG994390.1 | 8 | 77.59 | 40.8 |
| HG994394.1 | 9 | 66.79 | 45.9 |
| HG994393.1 | 10 | 71.93 | 42.9 |
| HG994391.1 | 11 | 75.75 | 40.4 |
| HG994392.1 | 12 | 73.73 | 39.2 |
| HG994397.1 | 13 | 65.44 | 40.3 |
| HG994400.1 | 14 | 62.79 | 39 |
| HG994395.1 | 15 | 65.78 | 40.5 |
| HG994398.1 | 16 | 63.67 | 41.5 |
| HG994396.1 | 17 | 65.96 | 41.9 |
| HG994402.1 | 18 | 57.59 | 43 |
| HG994403.1 | 19 | 56.75 | 38.7 |
| HG994401.1 | 20 | 59.77 | 44.6 |
| HG994405.1 | 21 | 53.11 | 40.4 |
| HG994399.1 | 22 | 63.45 | 38.3 |
| HG994406.1 | 23 | 52.96 | 40 |
| HG994407.1 | 24 | 49.88 | 44.7 |
| HG994404.1 | 25 | 53.62 | 41.6 |
| HG994409.1 | 26 | 46.11 | 46.2 |
| HG994408.1 | 27 | 48.75 | 40.8 |
| HG994413.1 | 28 | 42.48 | 43.9 |
| HG994412.1 | 29 | 44.09 | 38.9 |
| HG994415.1 | 30 | 41.62 | 41.6 |
| HG994411.1 | 31 | 44.76 | 41 |
| HG994414.1 | 32 | 41.77 | 38.1 |
| HG994417.1 | 33 | 32.66 | 39.4 |
| HG994410.1 | 34 | 45.90 | 41.6 |
| HG994419.1 | 35 | 28.53 | 42 |
| HG994416.1 | 36 | 33.43 | 39 |
| HG994418.1 | 37 | 31.50 | 40.1 |
| HG994420.1 | 38 | 26.44 | 41.5 |
| HG994381.1 | X | 124.67 | 40.3 |
| HG994382.1 | Y | 6.54 | 41.5 |
| HG998573.1 | MT | 0.02 | 39.6 |
| - | Unplaced | 29.74 | 50.3 |
Methods
A single 4-year-old male C. lupus orion (mCanLor1) was collected from Siorapaluk, Greenland (latitude 77.785278, longitude -70.631389) by The Ministry of Fisheries, Hunting and Agriculture, Government of Greenland. The animal was put down by the local municipal bailiff in Siorapaluk on 13 January 2016. The wolf had little fear of humans, persistently entered the village and could not be chased away. It was therefore decided that the wolf should be killed to protect villagers and dogs in Siorapaluk. After termination, the skull of the specimen was confiscated by the authorities and made available for the purposes of research to the Greenland Institute of Natural Resources.
DNA was extracted from the muscle tissue of mCanLor1 at the Wellcome Sanger Institute (WSI) Scientific Operations core from the whole organism using the Qiagen MagAttract HMW DNA kit, according to the manufacturer’s instructions. RNA (from the same muscle tissue) was extracted in the Tree of Life Laboratory at the WSI using TRIzol, according to the manufacturer’s instructions. RNA was then eluted in 50 μl RNAse-free water and its concentration RNA assessed using a Nanodrop spectrophotometer and Qubit Fluorometer using the Qubit RNA Broad-Range (BR) Assay kit. Analysis of the integrity of the RNA was done using Agilent RNA 6000 Pico Kit and Eukaryotic Total RNA assay.
Pacific Biosciences HiFi circular consensus and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Poly(A) RNA-Seq libraries were constructed using the NEB Ultra II RNA Library Prep kit. DNA sequencing was performed by the Scientific Operations core at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II and Illumina HiSeq X instruments. RNA sequencing was performed using an Illumina MiSeq instrument. Further 10X sequencing was performed at SciLifeLab, Stockholm, Sweden. DNA was extracted using the automatic KingFisher™ Duo Prime Purification System (Thermo Fisher Scientific, Bremen, Germany) following the manufacturer's protocol. Following this, Illumina TruSeq PCR-free libraries were constructed and sequencing performed on HiSeq X. Hi-C data were generated at SciLifeLab, Stockholm, Sweden using the Dovetail Hi-C kit and sequenced on HiSeq X.
Assembly was carried out with Hifiasm ( Cheng et al., 2021). Haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020). Scaffolding with Hi-C data ( Rao et al., 2014) was carried out with SALSA2 ( Ghurye et al., 2019). The Hi-C scaffolded assembly was polished with the 10X Genomics Illumina data by aligning to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012). One round of the Illumina polishing was applied. The mitochondrial genome was assembled with MitoHiFi ( Uliano-Silva et al., 2021). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation ( Howe et al., 2021) was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext. Regions of concern were identified and resolved using 10X longranger and genetic mapping data. The genome was analysed within the BlobToolKit environment ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Hifiasm | 0.12 | Cheng et al., 2021 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| SALSA2 | 2.2 | Ghurye et al., 2019 |
| longranger align | 2.2.2 |
https://support.10xgenomics.com/
genome-exome/software/pipelines/latest/ advanced/other-pipelines |
| freebayes | 1.3.1-17-gaa2ace8 | Garrison & Marth, 2012 |
| MitoHiFi | 1 | Uliano-Silva et al., 2021 |
| gEVAL | N/A | Chow et al., 2016 |
| PretextView | 0.1.x | https://github.com/wtsi-hpag/PretextView |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| BlobToolKit | 2.6.2 | Challis et al., 2020 |
Data availability
European Nucleotide Archive: Canis lupus (Greenland wolf). Accession number PRJEB43200; https://identifiers.org/ena.embl/PRJEB43200.
The genome sequence is released openly for reuse. The C. lupus genome sequencing initiative is part of the Darwin Tree of Life (DToL) project and the Vertebrate Genomes Project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated using the RNA-Seq data and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328). The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
Author information
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.4893704.
Members of the Wellcome Sanger Institute Tree of Life programme are listed here: https://doi.org/10.5281/zenodo.5377053.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.4790456.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.5013542.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783559.
References
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, et al. : Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat Methods. 2021;18(2):170–75. 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL - a web-based browser for evaluating genome assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes PR, Elander M, Ericson M: The Wolf ( Canis Lupus) in Greenland: A Historical Review and Present Status. Arctic. 1986;39(2):119–32. 10.14430/arctic2059 [DOI] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing.arXiv: 1207.3907.2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Castruita JAS, Sinding MS, et al. : The Wolf Reference Genome Sequence ( Canis Lupus Lupus) and Its Implications for Canis Spp. Population Genomics. BMC Genomics. 2017;18(1):495. 10.1186/s12864-017-3883-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sinding MS, Ramos-Madrigal J, et al. : Interspecific Gene Flow Shaped the Evolution of the Genus Canis. Curr Biol. 2019;29(23):4152. 10.1016/j.cub.2019.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhala Y, Boitani L, Phillips M: IUCN Red List of Threatened Species: Canis Lupus. IUCN Red List of Threatened Species. 2018. 10.2305/IUCN.UK.2018-2.RLTS.T3746A163508960.en [DOI] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquard-Petersen U: Invasion of Eastern Greenland by the High Arctic Wolf Canis Lupus Arctos. Wildl Biol. 2011;17(4):383–88. 10.2981/11-032 [DOI] [Google Scholar]
- Marquard-Petersen U: Decline and Extermination of an Arctic Wolf Population in East Greenland, 1899-1939. Arctic. 2012;65(2):121–243. 10.14430/arctic4197 [DOI] [Google Scholar]
- Marquard-Petersen U: Sudden Death of an Arctic Wolf Population in Greenland. Polar Res. 2021;40. 10.33265/polar.v40.5493 [DOI] [Google Scholar]
- Nilsen EB, Milner-Gulland EJ, Schofield L, et al. : Wolf Reintroduction to Scotland: Public Attitudes and Consequences for Red Deer Management. Proc Biol Sci. 2007;274(1612):995–1002. 10.1098/rspb.2006.0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripple WJ, Beschta RL: Trophic cascades in Yellowstone: The first 15 years after wolf reintroduction. Biol Conserv. 2012;145(1):205–13. 10.1016/j.biocon.2011.11.005 [DOI] [Google Scholar]
- Ripple WJ, Estes JA, Beschta RL, et al. : Status and ecological effects of the world's largest carnivores. Science. 2014;343(6167):1241484. 10.1126/science.1241484 [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Sinding MS, Gopalakrishan S, Vieira FG, et al. : Population Genomics of Grey Wolves and Wolf-like Canids in North America. PLoS Genet. 2018;14(11):e1007745. 10.1371/journal.pgen.1007745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Ersmark E, Palkopoulou E, et al. : Ancient Wolf Genome Reveals an Early Divergence of Domestic Dog Ancestors and Admixture into High-Latitude Breeds. Curr Biol. 2015;25(11):1515–19. 10.1016/j.cub.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Uliano-Silva M, Nunes JGF, Krasheninnikova K, et al. : marcelauliano/MitoHiFi: mitohifi_v2.0.2021. 10.5281/zenodo.5205678 [DOI] [Google Scholar]
- Wang GD, Zhai W, Yang HC, et al. : Out of Southern East Asia: The Natural History of Domestic Dogs across the World. Cell Res. 2016;26(1):21–33. 10.1038/cr.2015.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ: Could We Live with Reintroduced Large Carnivores in the UK? Mamm Rev. 2004;34(3):211–32. 10.1111/j.1365-2907.2004.00038.x [DOI] [Google Scholar]