Abstract
A sight threatening, pterygium is a common ocular surface disorders identified by fibrovascular growth of the cornea and induced by variety of stress factors, like ultraviolet (UV) exposure. However, the genes involved in the etiopathogenesis of this disease is not well studied. Herein, we identified the gene expression pattern of pterygium and examined the expression of pterygium-related genes in UV-B-induced human primary cultured corneal epithelial cells (HCEpCs), telomerase immortalized human corneal epithelial (hTCEpi), primary conjunctival fibroblast (HConFs) and primary pterygium fibroblast cells (HPFCs). A careful analysis revealed that the expression of 10 genes was significantly modulated (by > 10-fold). Keratin 24 (KRT24) and matrix metalloproteinase 9 (MMP-9) were dramatically upregulated by 49.446- and 24.214-fold, respectively. Intriguingly, UV-B exposure (50 J/m2) induced the upregulation of the expressions of MMP-9 in corneal epithelial cells such as HCEpCs and hTCEpi. Furthermore, UV-B exposure (100 and/or 200 J/m2) induced the upregulation of the expressions of MMP-9 in fibroblast such as HConFs and HPFCs. The exposure of HCEpCs to 100 and 200 J/m2 UV-B induced significant expressions of KRT24 mRNA. Nevertheless, no expression of KRT24 mRNA was detected in HConFs and HPFCs. The findings provide evidence that the progression of pterygium may involve the modulation of extracellular matrix-related genes and vasculature development and the up-regulation of KRT24 and MMP-9 by UV stress. UV radiation may promote the modulation of these pterygium-related genes and induce the initiation and progression of human pterygium.
Keywords: Pterygium, MMP-9, KRT24, Corneal epithelial cells, Conjunctival fibroblast, UV-B
1. Introduction
Eye is maximally exposed to environmental stressors like ultraviolet (UV) radiation, which eventually affects cellular integrity of cells by aberrantly regulating gene expression and function. Recent compelling evidence revealed that UV radiation is clinical risk factor for progression of pterygium. A pterygium is an epithelial and fibrovascular proliferation of the conjunctiva (Cameron, 1983; Coroneo et al., 1999). If not retarded, an advancing pterygium generates significant changes in the refractive index and corneal curvature (Bedrossian, 1960; Tomidokoro et al., 2000) and its progression would subsequently invade the cornea, forming a wing-like shape and causing visual loss. It is estimated that more than 200million people is affected due to pterygium (Lucas et al., 2008). Nonetheless, effective treatment for pterygium has not yet been identified, and surgical removal is the only way to remove a pterygium at present (Gris et al., 2000). Hence, it is highly imperative to identify the factors (genes) contributing to pterygium etiology and thereby develop gene targeted therapy.
Several studies have shown that pterygium formation is stimulated by various factors, including UV radiation (Zhou et al., 2016). UV exposure of pterygium epithelial cells generates proinflammatory cytokines, such as IL6 and IL8 (Di Girolamo et al., 2006). In addition, UV-B-driven oxidative stress can induce other inflammatory factors, like IL1α and TNFα that can lead other delirious signaling by activating MMPs Also, the progression of a pterygium may be the result of excessive wound healing, cell proliferation, and a limbal stem-cell deficiency triggered by oxidative stress due to UV exposure, and/or an inflammatory mediator, immunologic mechanisms, growth factors, or viruses on the ocular surface (Coroneo et al., 1999; Di Girolamo et al., 2004; Jaworski et al., 2009; Threlfall and English, 1999). The parts of pterygium are head (apical part present in cornea), neck (limbal part) and body (scleral part). There is evidence that UV-mediated limbal damage accelerates pterygium pathogenesis, and this has been reviewed extensively by Coroneo and colleagues (Coroneo, 1993; Coroneo et al., 1999; Di Girolamo et al., 2004). Furthermore, UV radiation is known to cause oxidative DNA damage, and proposed to be a major event in the pathogenesis of pterygium. From the above reports, it appears that pterygium formation is multifactorial, however, the mechanisms underlying the development of pterygium are not fully known (Coroneo, 1993; Threlfall and English, 1999).
In the context of primary pterygium, there are various reports on oxidative stress and the antioxidant defense balance (Balci et al., 2011; Kormanovski et al., 2014). Enhanced levels of nitric oxide and reduced levels antioxidants, such as superoxide dismutase and catalase have been shown in primary pterygium (Balci et al., 2011; Kormanovski et al., 2014). The marker 8-hydroxydeoxyguanosine, a sensitive and stable marker commonly used to identify oxidative damage to DNA, has been observed in pterygium in some studies (Kau et al., 2006; Maxia et al., 2008; Tsai et al., 2005), dictating the a major role of oxidative stress in pterygium pathobiology.
Moreover, the progression of a pterygium is signalized by the degradation of the corneal basement membrane and extracellular matrix (ECM) and a proliferation of fibrovascular tissue (Cameron, 1983; Coroneo et al., 1999; Di Girolamo et al., 2004). It has been observed that fibroblasts from pterygium have characteristics of myofibroblasts (Dushku and Reid, 1994; Kato et al., 2007). Also, the expression of several types of matrix metalloproteinases (MMPs) (MMP-1, 2, 3, 7, and 9) increases in pterygium (Chao et al., 2011; John-Aryankalayil et al., 2006; Yang et al., 2009). It has been demonstrated that MMP-2 and -9 are overexpressed in pterygium tissue and fibroblasts isolated from pterygium (Yang et al., 2009), suggesting that the levels of these MMPs in pterygium and its fibroblasts may be linked to the progression of pterygium.
Herein, by employing more extensive, microarray study, we identified relative gene expression profiles that are modulated in apical portion of pterygium compared to normal uninvolved conjunctival tissues. In addition, we compared the gene expressions between apical and basal portion of pterygium. Specifically, we also investigated the effect of UV-B exposure on the expression of MMP-9 proposed to play role in pterygium progression, using human pterygium tissues. The outcomes of the study can pave the way to develop therapeutic agents for the treatment/prevention of pterygium based upon target gene(s).
2. Materials and methods
2.1. Human pterygium sampling
This study was approved by the institutional review board of Kanazawa Medical University (Approval code: 78) with the appropriate informed consent obtained from all patients. Fifteen pterygium specimens were collected from 12 patients aged 70.539 ± 11.163 (mean ± standard deviation: SD) years and as controls, conjunctival tissues from conjunctivochalasis (Con) were obtained from four patients aged 79.750 ± 3.775 years at Kanazawa Medical University Hospital. There was no significant difference in age between the two groups. Three samples taken from the right eye of three patients were used for the microarray, and three samples taken from the left eye were used for the reverse transcribed quantitive real-time polymerase chain reaction (RT-qPCR) method. In addition, 9 samples collected from 9 patients were used for the RT-qPCR method in addition to the above 3 samples. As the normal controls, it was impossible to collect the normal conjunctiva near the limbus from the patient because the consent of the patient could not be obtained. Further, maintaining the normal conjunctiva is important in the treatment of other eye diseases in the future, and therefore excision of the normal conjunctiva from human patients is difficult. Due to these reasons, control conjunctival samples were obtained from conjunctivochalasis specimens which were excised by making a crescentic excision of the loose inferior bulbar conjunctiva starting with a peritomy approximately 3 mm posterior to the limbus including the conjunctival laxative part exposed from the eyelid margin.
Excised pterygium tissues were divided into apical portion including head (apical part present in cornea) and neck (limbal part) (PA) and basal portions (PB). PA and PB tissues were further divided laterally into two identical and symmetric pieces (Fig. 1). One piece was stored in RNAlater solution (Ambion®, ThermoFisher Scientific Japan, Tokyo) to stabilize the RNA until processing for a microarray or RT-qPCR. The other piece was frozen and used immediately for the isolation of protein as described as below or fixed with 10% Formalin Neutral Buffer Solution (Wako, Osaka, Japan) for immunohistochemistry.
Fig. 1.
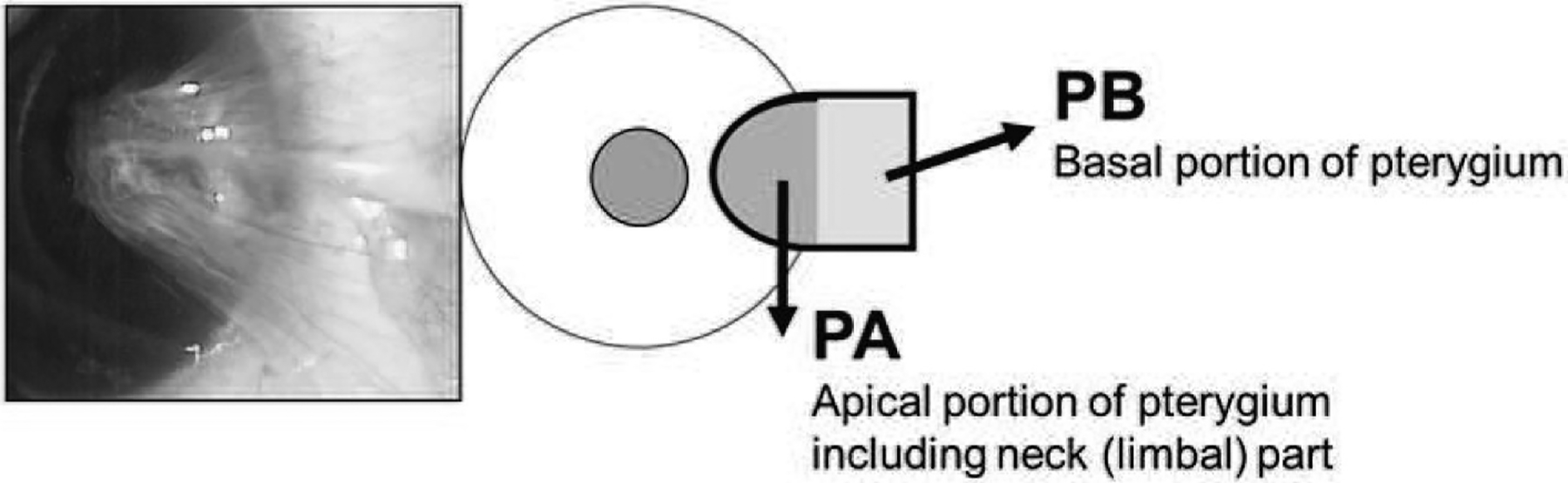
Method of pterygium sample collection. Human pterygium tissues were surgically exciced and divided in apical portion including head and neck (limbal part) (PA) and basal portion (PB).
2.2. Cell culture and UV-B treatment
Primary human corneal epithelial cells (HCEpCs) were obtained from Cell Applications (San Diego, CA, USA). Human telomerase-immortalized corneal epithelial cell line (hTCEpi) was obtained from Evercyte GmbH (Vienna, Austria). HCEpCs and hTCEpi were cultured with KGM™-2 Keratinocyte Growth Medium-2 (Lonza Japan Ltd, Osaka, Japan) at 37 °C in an air-CO2 (19:1) atmosphere. Primary human conjunctival fibroblasts (HConFs) were obtained from ScienCell Research Laboratories (Carlsbad, CA) and cultured with Fibroblast Medium (ScienCell) at 37 °C in an air-CO2 (19:1) atmosphere. Primary pterygium fibroblast cells (HPFCs) were established by ourselves from human pterygium specimens by using a technique previously reported (Li et al., 2001). HPFCs were subsequently spread in 75-cm2 culture flasks (Nunc, Roskilde, Denmark) in with Fibroblast Medium (ScienCell) and 100 U/ml penicillin and 100 μg/mL streptomycin (ThermoFisher) at 37 °C in an air-CO2 (19:1) atmosphere. First, 1 × 105 of HCEpCs, HConFs, hTCEpis and HPFCs were seeded on 35-mm culture dishes at 37 °C in an air-CO2 (19:1) atmosphere for 24 h.
For the UV-B irradiation assays (Fig. 2A), the HConFs, HCEpCs and HPFs were placed in 1 mL of 1X phosphate-buffered saline (PBS), and irradiated with 0, 50, 100, or 200 J/m2 UV-B once per day for 2 and/or 4 days,. The medium was then changed to fresh medium. Ultraviolet-B light was generated by a 15-W UV-B light source (312 nm), with its intensity standardized using a UV light meter (UVP, Upland, CA). hTCEpis were placed in 1 mL of 1X PBS, and irradiated with 0, 50 or 100 J/m2 UV-B once per day for 2 days (Fig. 2B). Experiments were repeated three times for each cell type.
Fig. 2.
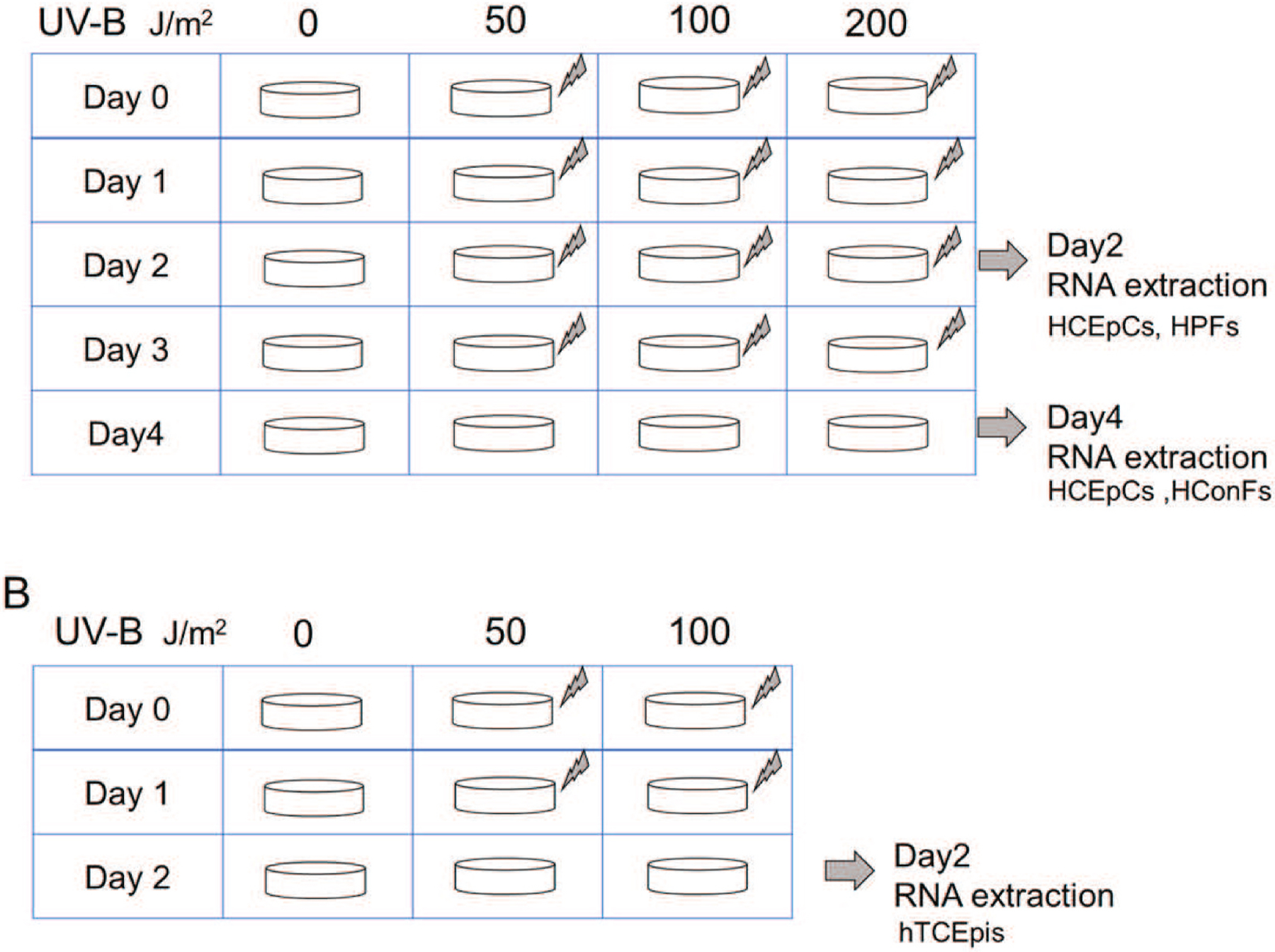
Illustration of method for culture experiments. UV-B was irradiated with varying intensity (A: 0, 50, 100, 200 J/m2) or (B: 0, 50, 100 J/m2) every day, and RNA extraction was performed on the second and/or fourth day.
2.3. RNA extraction
Total RNA from each human sample and cultured HConFs, HCEpCs, HPFC and hTCEpi cells was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) by following the manufacturer’s protocol. Samples of RNA were set aside for a RT-qPCR to verify the results obtained from the microarray analysis.
2.4. Microarray analysis and gene ontology analysis
Pterygium-related genes were screened by microarray analysis using pterygium samples as follows. Three apical portions of pterygium specimens from primary patients (PA-1, PA-2, PA-3) and one control conjunctival tissue (Con-1) were used for the microarray analysis, and all four samples were processed for the microarray analysis as follows. We used the Agilent Low Input Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA) for RNA labeling. Briefly, for each labeling, 100 ng of total RNA was reverse-transcribed in the presence of CDS primer mix and MMLV reverse transcriptase. Cyanine 3-CTP was coupled to the first-strand cDNA. Labeled cDNA was hybridized to SurePrint G3 Human 8 × 60K microarray kit v2 (Agilent) using the Agilent® Gene Expression Hybridization Kit according to the manufacturer’s protocol. Air-dried slides were scanned with an Agilent Microarray Scanner. Scanned images were analyzed with Feature Extraction Software (v. 10.7.3.1) (Agilent).
Per chip normalization was done by dividing each gene’s measurement by the specific control measurements or by the average intensity in the single array. Normalized data were exported for the subsequent analysis. Genes with a normalized ratio >2.0-fold or <0.5-fold between three samples were selected as significant genes for the progression of pterygium.
2.5. RT-qPCR
To measure the expression of mouse and human MMP-9 and KRT24 mRNAs, we conducted a relative quantification of mRNA using a Prism7300 (Applied Biosystems, ThermoFisher Scientific Japan). The comparative Ct method was used for the relative quantification of miRNA expression. The PCR amplification was performed using TaqMan Universal Master Mix and pre-developed human MMP-9 and KRT24 probe mix (Applied Biosystems). The relative quantity of each mRNA was determined using the comparative Ct method and then normalized using a pre-developed TaqMan ribosomal RNA control reagent VIC probe as an endogenous control (Applied Biosystems).
2.6. Statistical analysis
For all quantitative data collected, the statistical analysis was conducted with Turkey test or Student’s t-test when appropriate. The data are presented as the mean ± SD of the indicated number of experiments. A significant difference between the control and treatment group was defined as a p-value <0.05 for two or more independent experiments.
3. Results
3.1. Gene expression profiling and analysis of pterygium
As described above, three apical portions of pterygium specimens from primary patients (PA-1, PA-2, PA-3) and one control conjunctival tissue (Cont-1) were used for the microarray analysis. The data for the microarray analysis was deposited to Gene expression omnibus (GEO) database (Accession number: GSE151872). There were 957 genes in the PA-1 group, 1110 genes in the PA-2 group, and 1194 genes in the PA-3 group that showed significant changes of greater than 2.0-fold. Table 1 provides the list of the 8 top-ranked genes that showed significant changes (>2.0-fold) in each three pterygium patients (PA-1-3). Table 2 provides the list of the 30 top-ranked genes that showed significant changes (>2.0-fold) that were common to all three pterygium patients (PA-1-3).
Table 1.
Top 10 genes ranked by the greatest differences in log2 ratio between control and PA1–3.
| PA1 | PA2 | PA3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Probe Name | Fold Chane Log2 ratio | Gene Symbol | Probe Name | Fold Change Log2 ratio | Gene Symbol | Probe Name | Fold Change Log2 ratio | Gene Symbol |
| A 23 P4387 | 6.864 | KRT24 | A_23_P4387 | 4.877 | KRT24 | A_23_P40174 | 5.586 | MMP9 |
| A 33 P3311503 | 4.935 | SMR3B | A_23_P93141 | 4.686 | GSTA5 | A_23_P7313 | 4.962 | SPP1 |
| A 23 P29965 | 4.632 | SMR3B | A_33_P3273885 | 4.429 | A_23_P18452 | 4.420 | CXCL9 | |
| A 24 P870620 | 4.431 | PTN | A_23_P40174 | 4.395 | MMP9 | A_24_P131589 | 4.162 | CD86 |
| A 23 P214144 | 4.250 | COL10A1 | A_32_P60065 | 4.255 | F2RL2 | A_33_P3329078 | 4.088 | HBG1 |
| A 23 P132760 | 3.977 | TRH | A_23_P214300 | 4.146 | GSTA2 | A_21_P007321 | 4.007 | |
| A 33 P3241269 | 3.837 | CES1 | A_23_P69030 | 3.689 | COL8A1 | A_32_P157927 | 3.880 | |
| A 24 P626951 | 3.783 | A_23_P7313 | 3.629 | SPP1 | A_21_P007446 | 3.816 | ||
| A 33 P3304668 | 3.780 | COL1A1 | A_33_P3364240 | 3.615 | PAEP | A_19_P00809119 | 3.800 | CASC15 |
| A_33_P3281435 | 3.533 | A_33_P233040 | 3.493 | SERPINB11 | A_24_P70183 | 3.453 | MYH11 | |
Table 2.
Top 30 genes that showed significant changes of greater than 5.0-fold in common to 3 pterygium samples.
| Gene Symbol | GenBank Accession | Gene Name | Ave(n = 3) | SD |
|---|---|---|---|---|
| KRT24 | NM_019016 | keratin 24 | 49.446 | 21.618 |
| MMP9 | NM_004994 | matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | 24.214 | 10.393 |
| PTN | NM_002825 | pleiotrophin | 12.844 | 7.892 |
| GSTA5 | NM_153699 | glutathione S-transferase alpha 5 | 12.809 | 10.397 |
| SMR3B | NM_006685 | submaxillary gland androgen regulated protein 3B | 12.722 | 15.471 |
| CXCL9 | NM_002416 | chemokine (C-X-C motif) ligand 9 | 12.220 | 7.955 |
| GSTA2 | NM_000846 | glutathione S-transferase alpha 2 | 11.350 | 5.891 |
| COL8A1 | NM_001850 | collagen, type VIII, alpha 1 | 10.628 | 2.661 |
| CES1 | NM_001025195 | carboxylesterase 1 | 10.570 | 3.885 |
| COL10A1 | NM_000493 | collagen, type X, alpha 1 | 10.316 | 8.039 |
| GZMA | NM_006144 | granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 8.177 | 1.592 |
| UCHL1 | NM_004181 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 7.635 | 1.961 |
| TMEM119 | NM_181724 | transmembrane protein 119 | 7.213 | 1.765 |
| PTGFR | NM_001039585 | prostaglandin F receptor (FP) | 7.174 | 2.224 |
| NFATC4 | NM_001136022 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | 7.068 | 1.801 |
| PI16 | NM_153370 | peptidase inhibitor 16 | 6.765 | 1.703 |
| COMP | NM_000095 | cartilage oligomeric matrix protein | 6.725 | 1.665 |
| GDF10 | NM_004962 | growth differentiation factor 10 | 6.567 | 3.089 |
| EPS15 | AK129853 | epidermal growth factor receptor pathway substrate 15 | 6.510 | 1.071 |
| CES1 | NM_001266 | carboxylesterase 1 | 6.493 | 4.419 |
| FAP | NM_004460 | fibroblast activation protein, alpha | 6.441 | 1.111 |
| MYH11 | NM_001040113 | myosin, heavy chain 11, smooth muscle | 6.330 | 1.886 |
| SEMA3D | NM_152754 | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D | 5.949 | 2.084 |
| ITGBL1 | NM_004791 | integrin, beta-like 1 (with EGF-like repeat domains) | 5.906 | 1.925 |
| SERPINB11 | NM_080475 | serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene) | 5.882 | 4.718 |
| APOC1 | NM_001645 | apolipoprotein C-I | 5.679 | 3.761 |
| ARHGAP44 | NM_014859 | Rho GTPase activating protein 44 | 5.641 | 0.694 |
| CYS1 | NM_001037160 | cystin 1 | 5.592 | 1.349 |
| GJC1 | NM_005497 | gap junction protein, gamma 1, 45 kDa | 5.567 | 1.120 |
| HBA221:31 | NM_000517 | hemoglobin, alpha 2 | 5.496 | 3.325 |
The most highly up-regulated gene family that was detected in all three samples was KRT24, the superfamily of intermediate filament proteins, which showed a >49.446-fold higher expression compared to the control. MMP-9, which belongs to a class of enzymes that is involved in the degradation of ECM, was up-regulated by more than 24.214-fold. Our gene ontology analyses revealed an upregulation of a set of genes classified in ECM, extracellular regions, regulation of cell migration, and vasculature development in all three pterygium samples (Table 3).
Table 3.
Gene ontology analysis that showed significant changes of greater than 5.0-fold in common to 3 pterygium samples.
| GO ACCESSION | GO Term | p-value |
|---|---|---|
| GO:0031012 | extracellular matrix | 2.99E-33 |
| GO:0044421 | extracellular region part | 9.94E-31 |
| GO:0005576 | extracellular region | 2.85E-29 |
| GO:0005578 | proteinaceous extracellular matrix | 2.35E-25 |
| GO:0030198 | extracellular matrix organization | 7.38E-24 |
| GO:0043062 | extracellular structure organization | 8.80E-24 |
| GO:0022617 | extracellular matrix disassembly | 8.47E-17 |
| GO:0005615 | extracellular space | 2.93E-16 |
| GO:0001944 | vasculature development | 4.80E-16 |
| GO:0001568 | blood vessel development | 5.50E-15 |
| GO:0044420 | extracellular matrix part | 1.66E-14 |
| GO:0030334 | regulation of cell migration | 9.65E-14 |
| GO:0048731 | system development | 5.69E-13 |
| GO:2000145 | regulation of cell motility | 8.80E-13 |
| GO:0044259 | multicellular organismal macromolecule metabolic process | 9.34E-13 |
| GO:0030574 | collagen catabolic process | 8.77E-13 |
| GO:0051270 | regulation of cellular component movement | 1.51E-12 |
| GO:0032963 | collagen metabolic process | 1.64E-12 |
| GO:0072358 | cardiovascular system development | 1.87E-12 |
| GO:0032501|GO:0050874 | multicellular organismal process | 1.69E-12 |
Significant changes that were <0.5-fold were detected in 897 genes in the PA-1 specimens, 1516 genes in the PA-2 specimens, and 1785 genes in the PA-3 specimens. Table 4 is the list of the 30 top-ranked genes that showed significant changes <0.5-fold in common to all groups. Gene ontology analyses revealed a down-regulation of genes related to the defense response to viruses, the cellular response to type I interferon, the type I interferon signaling pathway, and the response to type I interferon in two of the three pterygium patients’ samples (Table 5).
Table 4.
Top 30 genes (a) and analysis of gene ontology features (b) that showed significant changes of smaller than 0.5-fold in common to 3 pterygium samples.
| Gene Symbol | GenBank Accession | Gene Name | Ave(n = 3) | SD |
|---|---|---|---|---|
| DEFB4A | NM_004942 | defensin, beta 4A | 0.109 | 0.065 |
| CEACAM5 | NR_038428 | chromosome X open reading frame 28 | 0.135 | 0.084 |
| KRT32 | NM_002278 | keratin 32 | 0.159 | 0.082 |
| RHCG | NM_016321 | Rh family, C glycoprotein | 0.163 | 0.060 |
| NXPH4 | NM_007224 | neurexophilin 4 | 0.170 | 0.068 |
| FOSB | NM_006732 | FBJ murine osteosarcoma viral oncogene homolog B | 0.173 | 0.080 |
| RNF151 | NM_174903 | ring finger protein 151 | 0.189 | 0.083 |
| TUBBP5 | NR_027156 | tubulin, beta pseudogene 5 | 0.195 | 0.204 |
| C10orf99 | NM_207373 | chromosome 10 open reading frame 99 | 0.200 | 0.028 |
| GDF15 | NM_004864 | growth differentiation factor 15 | 0.207 | 0.049 |
| NWD1 | NM_001007525 | NACHT and WD repeat domain containing 1 | 0.214 | 0.114 |
| CRNN | NM_016190 | cornulin | 0.215 | 0.205 |
| SPINK2 | NM_021114 | serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) | 0.224 | 0.080 |
| TRNP1 | NM_001013642 | TMF1-regulated nuclear protein 1 | 0.233 | 0.071 |
| CCDC60 | NM_178499 | coiled-coil domain containing 60 | 0.234 | 0.006 |
| MUC20 | NM_001098516 | mucin 20, cell surface associated | 0.239 | 0.083 |
| UCA1 | NR_015379 | urothelial cancer associated 1 (non-protein coding) | 0.242 | 0.118 |
| ATF3 | NM_001040619 | activating transcription factor 3 | 0.249 | 0.101 |
| KRT34 | NM_021013 | keratin 34 | 0.251 | 0.113 |
| ATF3 | NM_001674 | activating transcription factor 3 | 0.251 | 0.138 |
| LOC100506810 | NR_038856 | uncharacterized LOC100506810 | 0.252 | 0.105 |
| ZNF812 | NM_001199814 | zinc finger protein 812 | 0.255 | 0.053 |
| FOS | NM_005252 | FBJ murine osteosarcoma viral oncogene homolog | 0.256 | 0.056 |
| OAS1 | NM_002534 | 2′–5′-oligoadenylate synthetase 1, 40/46 kDa | 0.257 | 0.005 |
| LINC00265 | NR_026999 | long intergenic non-protein coding RNA 265 | 0.261 | 0.014 |
| CRYM | NM_001888 | crystallin, mu | 0.262 | 0.049 |
| PADI1 | NM_013358 | peptidyl arginine deiminase, type I | 0.262 | 0.129 |
| ABO | NM_020469 | ABO blood group (transferase A, alpha 1–3-N-acetylgalactosaminyltransferase; transferase B, alpha 1–3-galactosyltransferase) | 0.265 | 0.049 |
| SNORA54 | NR_002982 | small nucleolar RNA, H/ACA box 54 | 0.267 | 0.087 |
| TNFSF15 | NM_005118 | tumor necrosis factor (ligand) superfamily, member 15 | 0.268 | 0.096 |
Table 5.
Gene ontology analysis that showed significant changes of smaller than 0.5-fold in common to 3 pterygium samples.
| GO ACCESSION | GO Term | p-value |
|---|---|---|
| GO:0051607 | defense response to virus | 3.65E-06 |
| GO:0071357 | cellular response to type I interferon | 3.94E-05 |
| GO:0034340 | response to type I interferon | 4.53E-05 |
| GO:0060337 | type I interferon signaling pathway | 3.41E-05 |
| GO:0009615 | response to virus | 4.10E-04 |
| GO:0016266 | O-glycan processing | 4.32E-04 |
3.2. Validation of the expression of KRT24 and MMP-9 mRNA modulated in microarray
The results of the microarray analysis presented above showed the modulation in the expression levels of several genes during the progression of pterygium, indicating the importance of these genes’ regulatory roles in the gene expression during pterygium development. However, we observed that genes belonging to the same class/family had similar expression profiles, suggesting that they may not be differentiated in a microarray analysis due to the possibility of cross-hybridization. To avoid this, we selected KRT24 and MMP-9, whose expression was modulated in our microarray data, and we validated their expression by real-time RT-qPCR (Fig. 3).
Fig. 3.
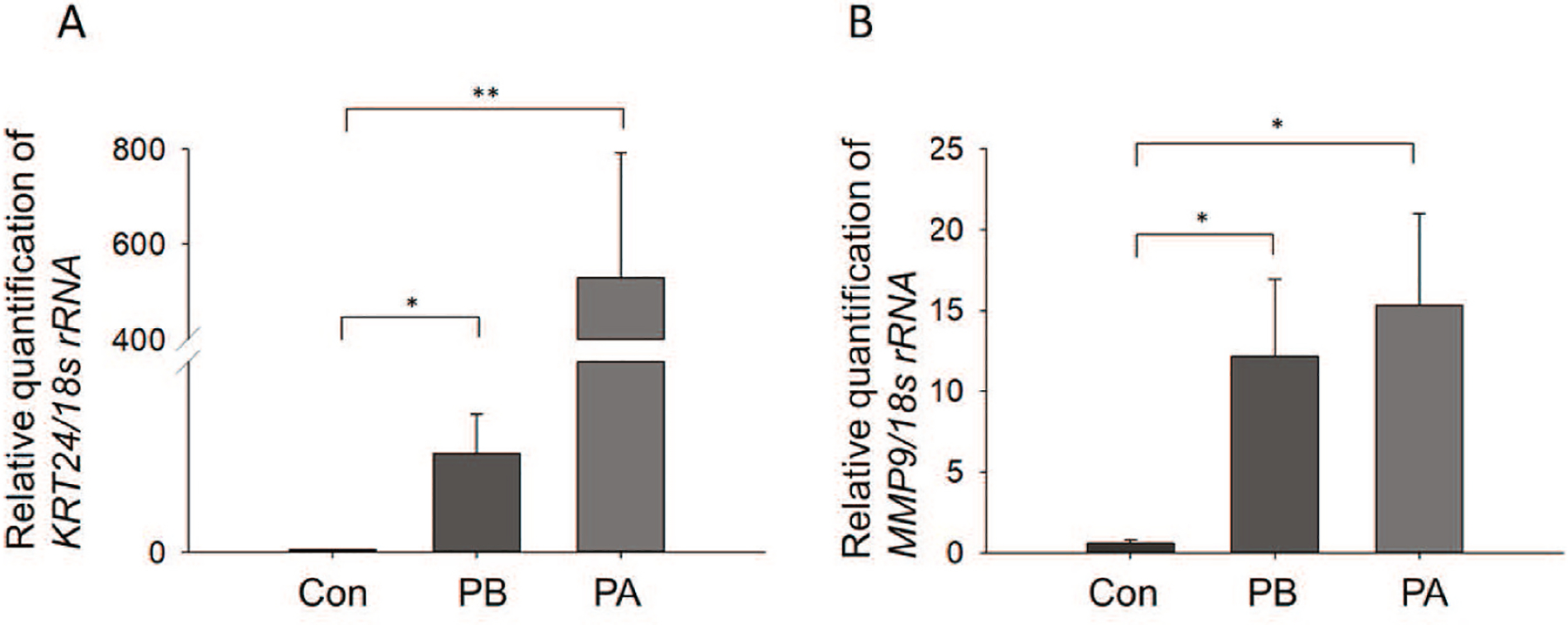
Expression of MMP-9 and KRT24 mRNAs in human pterygium samples. (A) Comparison of MMP-9 mRNA levels between three control conjunctiva tissues and 12 pterygium tissues at the basal and apical portion of each. *p < 0.05 and **p < 0.025; between control conjunctiva, basal and apical portion of pterygium. (B) Comparison of KRT24 mRNA levels between three control conjunctiva tissues and 12 pterygium tissues at the basal and apical portion. *p < 0.025 between control conjunctiva, basal and apical portion of pterygium. Data are mean ± SD. Con: Control; PA: Apical portion of pterygium; PB: Basal portion of pterygium.
We analyzed samples of three normal conjunctiva of conjunctivochalasis as control samples (Con) and 12 pterygium samples for the real time RT-qPCR. KRT24 (A) and MMP-9 (B) mRNA were dramatically altered in both the apical and basal portions of pterygium (Fig. 3A and B). The expression of KRT24 mRNA in the basal (PB) and apical portions (PA) of pterygium was significantly up-regulated in comparison to the expression in the control samples (Con) (Fig. 3A, *p < 0.05 and **p < 0.025 vs. control). The expression of MMP-9 mRNA in the basal and apical portions of the pterygium samples was significantly up-regulated compared to the control samples (Fig. 3B, *p < 0.025vs. control).
3.3. UV-B-induced expression of KRT24 and/or MMP-9 mRNA in HCEpC, HConF, hTCEpi and HPFC
To determine whether the expressions of KRT24 and/or MMP-9 mRNA are up-regulated in cells exposed to UV-B which is proposed to be a causative factor of pterygium, we exposed HCEpC and HConF cells exposed to 0, 50, 100, or 200 J/m2 UV-B once a day for 4 days, and total RNA extracts were prepared on Day 2 and/or Day 4 from each. We then performed an RT-qPCR (n = 3 in each group). The exposure of HCEpC cells to 100 and 200 J/m2 UV-B also induced significant expressions of KRT24 mRNA on Day 4 (Fig. 4A; *p < 0.02, 0 vs 200 J/m2; **p < 0.04, 50 vs 200 J/m2). The exposure of HCEpC cells to 50 J/m2 UV-B significantly induced the expression of MMP-9 on Day 2 and 4 (Fig. 4B; *p < 0.007, ***p < 0.001 vs. 0 J/m2). On Day2 and 4, the expression of MMP-9 was significantly decreased after the exposure of 100 and 200 J/m2 (Fig. 4B; *p < 0.007, **p < 0.05, ***p < 0.001 vs. 50 J/m2). UV-B In addition, the exposure of HConF cells to 50 or 200 J/m2 UV-B induced a significant expression of MMP-9 mRNA on Day 4 (*p < 0.001, **p < 0.03 vs. 0 J/m2) (Fig. 5A). In compared toHConF cells after exposure of UV-B for 4 days, the exposure of HConF cells to 100 J/m2 UV-B induced a significant up-regulation of MMP-9 mRNA (***p < 0.005) (Fig. 5A). The exposure of HPF cells to 100 and 200 J/m2 UV-B significantly induced the expression of MMP-9 on Day2 (Fig. 5B; *p < 0.05 vs. 200 J/m2). The exposure of hTCEpi cells to 50 J/m2 UV-B significantly induced the expression of MMP-9 on Day 2 (Fig. 5C; *p < 0.025 vs. 0 and 100 J/m2). No expression of KRT24 mRNA was detected in HConF cells and HPF cells. There was no significant change in expression of KRT24 after UV-B exposure to hTCEpi cells, because expression of KRT24 was very low level in hTCEpi cells (Data not shown). Our analyses demonstrated that KRT24 is specifically expressed in HCEpCs.
Fig. 4.
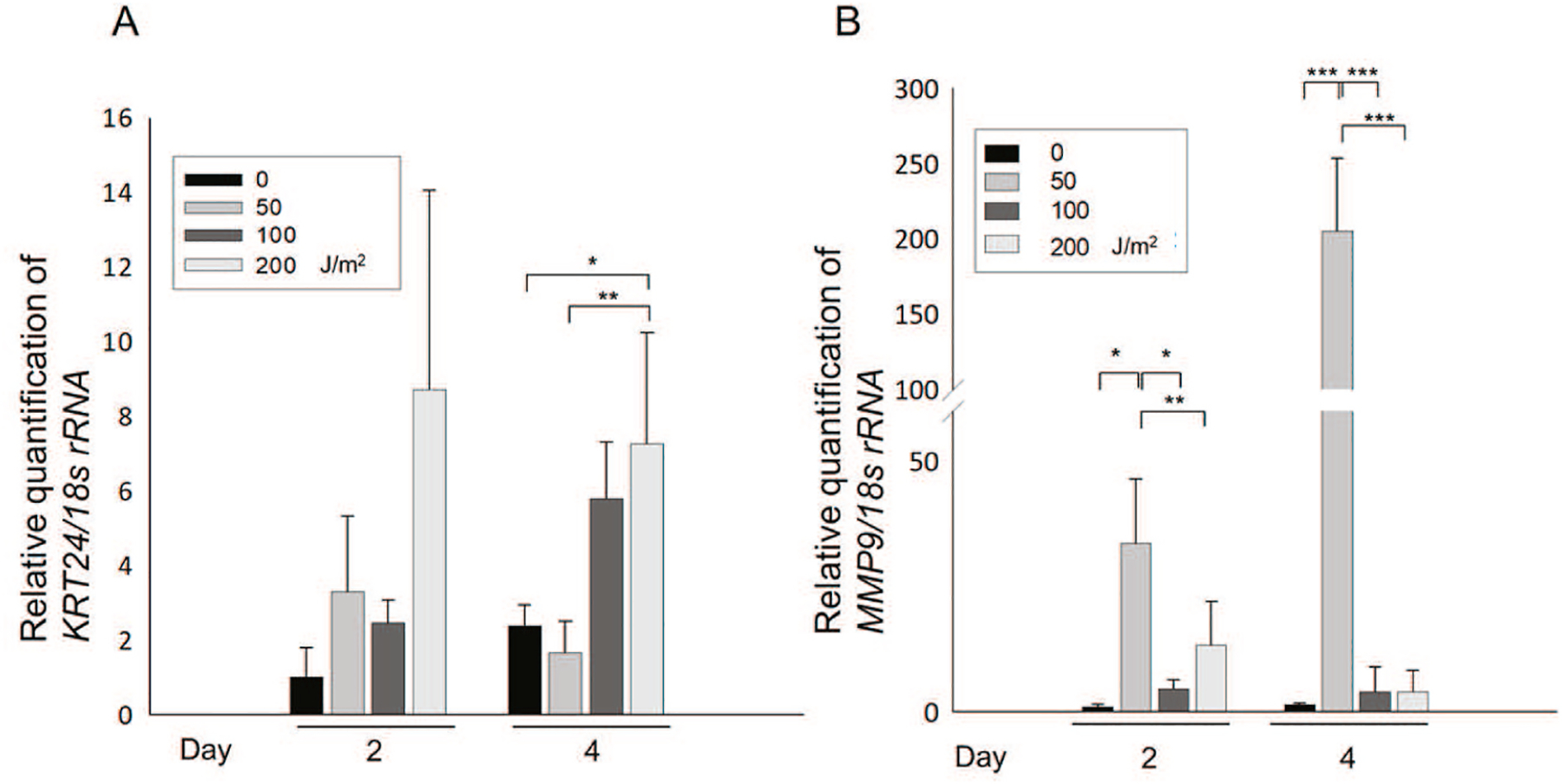
Expression of MMP-9 (A) and KRT24 (B) mRNA in HCEpC cells after UV-B exposure. As shown in Fig. 2A, after culturing for 2 and 4 days, the cells were collected and studied by a real-time RT-qPCR. (A) KRT24 mRNA levels in HCEpCs exposed to 0–200 J/m2 UV-B. *p < 0.02, **p < 0.04 between 0 and 200 J/m2 or 50 and 200 J/m2 UV-B exposure on day 4. Data are mean ± SD (n = 6). (B) MMP-9 mRNA levels in HCEpCs exposed to 0–200 J/m2 UV-B. *p < 0.007, **p < 0.05, ***p < 0.001 between 0 and 50 J/m2, or 50 and 100 or 200 J/m2 UV-B exposure on Day2 and 4. Data are mean ± SD (n = 6).
Fig. 5.
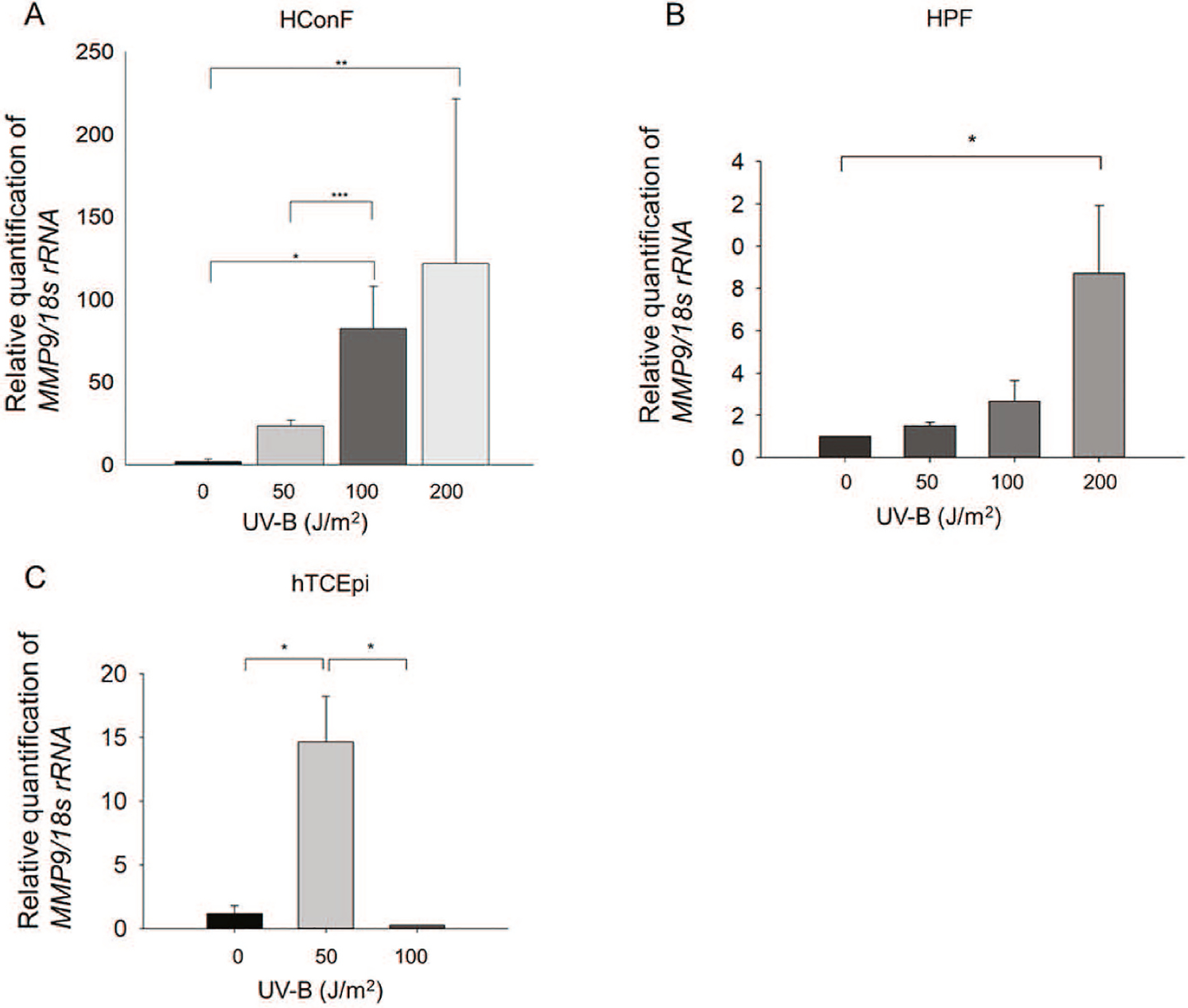
Expression of MMP-9 mRNA in HConFC, HPF and hTCEpi cells after UV-B exposure. (A) As shown in Fig. 2A, after culturing for 4 days, the cells were collected and studied by real-time RT-qPCR. MMP-9 mRNA levels in HConFCs exposed to 0–200 J/m2 UV-B. *p < 0.001, **p < 0.03, ***p < 0.005 between 0 and 50 or 100 J/m2 UV-B exposure. Data are mean ± SD (n = 6). (B) As shown in Fig. 2A, after culturing for 2 days, the cells were collected and studied by RT-qPCR. MMP-9 mRNA levels in HPFs exposed to 0–200 J/m2 UV-B. *p < 0.05. Data are mean ± SD (n = 3). (C) As shown in Fig. 2B, after culturing for 2 days, the hTCEpis were collected and studied by RT-qPCR. MMP-9 mRNA levels in hTCEpis exposed to 0–100 J/m2 UV-B. *p < 0.025. Data are mean ± SD (n = 3).
4. Discussion
A gene expression analysis of pterygium demonstrated that several pathways were significantly affected. The analysis revealed that the tissue-specific markers, KRT24 and MMP-9 were the most abundant in the apical portion of the pterygium. KRT24 is a member of the type I (acidic) keratin family, which belongs to the superfamily of intermediate filament proteins (Schweizer et al., 2006) that was identified in Naegeli-Franceschetti-Jadassohn syndrome and dermatopathia pigmentosa reticularis (Sprecher et al., 2002). KRT24 influences the cellular response to proapoptotic signals and the routing of membrane proteins in polarized epithelial cells (Hong et al., 2007). Further, KRT24 was consistently up-regulated in the mucosa of colorectal cancer patients (Hong et al., 2007). KRT24 has also been reported as a terminally differentiated gene for corneal stromal cells (Hashmani et al., 2013).
In our present study, KRT24 was highly up-regulated in the apical portion of pterygium specimens in both the microarray analysis and RT-q PCR compared to the conjunctival control tissues and basal portion of pterygium tissue. In compared to control conjunctiva, the expression of KRT24 was also significantly increased in the basal portion of the pterygium, which may have not included corneal epithelial cells. Recently, it was found that KRT24 was up-regulated in pterygium using microarray analysis compared to its expression in tissues from conjunctiva, but it could not confirm by RT-qPCR analysis (Zhang and Liu, 2019). Furthermore, Keratin K24, which is encoded by the KRT24 gene that is located at one end of the type I keratin gene cluster, is highly expressed in the superficial layer of the corneal epithelium point to a predominant role of human K24 in the cornea (Ehrlich et al., 2019). The expression of KRT24 was not detected in the HConF cells, and its expression in the control conjunctival tissues was low-level. We speculate that KRT24 is related to the transformation of limbal tissues to pterygium and that KRT24 may be a marker of pterygium.
Moreover, based upon accumulating evidence, we think that UV-B induces the expressions of genes that may contribute to the progression of pterygium. Toward this, cells were exposed to UV-B as described in ‘Materials and Methods’ section. In cultured HCEpC cells, we found, for the first time that expression of KRT24 was induced after UV-B exposure in HCEpC cells (Fig. 4A). Pterygium is a condition of the ocular surface characterized by squamous cell metaplasia and goblet cell hyperplasia. We believe that because UV-B is one of the causative factors of pterygium, UV-B-driven aberrant KRT24 expression could induce the epithelial transformation of limbal stem cells, resulting in the pathogenesis of pterygium.
Our findings regarding the upregulation of MMP-9 are consistent with those of previous studies. We also observed that MMP-9 mRNA was highly upregulated in both apical and basal portions of pterygium. MMP-9 (gelatinase B, 92-kDa gelatinase, 92-kDa type IV collagenase) is a member of the MMP family, which plays a role in the proteolysis of ECM (Kahari and Saarialho-Kere, 1999; Yang et al., 2009). Gelatinases are able to perform the final degradation of fibrillar collagens after their first cleavage by collagenases (Kahari and Saarialho-Kere, 1999). The expression of various MMPs is increased in epithelial cells and fibroblasts in pterygium (Dushku et al., 2001; Seet et al., 2012; Yang et al., 2009). An increased expression of MMPs dissolves Bowman’s layer in the cornea and induces angiogenesis and the migration and proliferation of pterygium onto the cornea (Yang et al., 2009). In addition, MMP-9 can degrade cell matrix proteins such as fibronectin, laminin, elastin, and various collagens (types I, II, and V) (Kahari and Saarialho-Kere, 1999; Okada et al., 1995).
Furthermore, our work revealed that UV-B increased the expression of MMP-9 mRNA in HCEpC, hTCEpi, HConF and HPFCs. In hCEpCs and hTCEpi, the expression of MMP9 was increased only by UV-B irradiation at 50 J/m2. In corneal epithelial cells, MMP9 may be induced only by mild UV-B irradiation and may be involved in the early pterygium development and degradation of corneal epithelium. Moreover, when the intensity of UV-B irradiation is increased (>100 J/m2), the expression of MMP9 in fibroblasts such as HConFs and HPFs was increased. In conjunctival fibroblast, stronger intensity of UV-B irradiation (>100 J/m2) may be needed for MMP9 induction to induce the pathogenesis of pterygium. In an earlier study, it was observed that the expression of MMP-1 was induced by UV-B exposure (Di Girolamo et al., 2003). Collectively, the significant inductions of MMP-9 and KRT24 mRNAs after UV-B exposure suggest a plausible role of UV-B in the pathophysiology of pterygia. Our gene ontology analysis revealed the genes that may be related to the progression of pterygium; for example, ECM-related genes and genes related to vasculature development and the regulation of cell migration were significantly increased. Pterygia are highly vascularized, proliferative, degenerative and invasive ocular surface lesions that originate at the corneal limbus (Di Girolamo et al., 2003; Dushku and Reid, 1994). The genes that we observed in the microarray analysis may be involved in the progression of pterygia and provide a clue to develop specific transcription based therapy for treating pterygium, in future.
Moreover, several research groups have shown that altered corneal limbal epithelial cells undergo epithelial-mesenchymal transition (EMT) influenced by the Wnt/β-catenin pathway and microRNA (miRNA)-200 (Ando et al., 2011; Kato et al., 2007; Kim et al., 2016). In addition, the fibrovascular change is more severe in recurrent pterygia than at the initial site of presentation (Tan et al., 1997; Touhami et al., 2005). It was reported that myofibroblasts with tumor-inducing phenotypes express α smooth muscle actin (αSMA) and that αSMA was also expressed in pterygia (Kato et al., 2007; Touhami et al., 2005). Pterygium has also been described as a benign neoplastic lesion (Dushku et al., 2001; Tan et al., 2000; Weinstein et al., 2002). Thus, pterygia may have tumor-like characteristics. We surmised that since myofibroblastic changes in pterygia are observed in the basal portion, the EMT may be induced in the basal portion of conjunctival tissues, leading to the progression of pterygia.
Our analyses also demonstrated that defensins were highly down-regulated in the pterygium tissues. The gene ontology analysis showed the genes that function in the defense response to viruses and the response to type I interferon. Defensins are cationic antimicrobial peptides characterized by the presence of six cysteine residues linked to form three disulfide bridges. Two forms of human defensin, α and β, are recognized, depending on the location and connectivity of the cysteines. β-defensins are expressed by many epithelia including the cornea (Ganz and Lehrer, 1995; Haynes et al., 1999; McDermott et al., 2003). Six human β-defensins (hBD-1 through –6) have been identified to date (Bensch et al., 1995; Garcia et al., 2001a, 2001b; McDermott et al., 2003; Tomita et al., 2002; Yamaguchi et al., 2002). hBD-1 is constitutively expressed, whereas hBD-2 and -3 are inducible by cytokines and bacterial products. hBD-4 appears to have a more limited distribution than hBD-1,- 2, or -3 (Garcia et al., 2001a, 2001b).
Defensins have a broad spectrum of antimicrobial activity, being effective against many Gram-positive and -negative bacteria, some fungi, and enveloped viruses (Ganz and Lehrer, 1995). Human papillomavirus (HPV) infection has been reported as a possible inducing factor of pterygium. A closer look of the literature indicated that the prevalence of HPV in ocular surface diseases varies over a wide range (0%–100%) (Di Girolamo, 2012; Woods et al., 2013). This variance may be linked to the many different types of assays applied to detect the virus, as well as geography and genetic susceptibility (Di Girolamo, 2012; Woods et al., 2013). The existing literature indicated that approx. 60% of the published studies identified HPV in pterygia tissues, with an overall prevalence of 19% (Di Girolamo, 2012; Woods et al., 2013). The decreased defense response against a virus in limbal tissues may allow the infection of HPV or other viruses or bacteria and may induce the progression of a pterygium.
5. Conclusion
In conclusion, our study provides the evidence the involvement of UV-B and UV-B-driven aberrant expression of MMPs, and KRT24 in the development of pterygia. The finding may help in the identification of new therapeutic target and the design of new approaches for the treatment and prevention of pterygia.
Funding
This work was supported by grants from Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP 17K16991 (to NK), a Grant for Promoted Research from Kanazawa Medical University [S2016-9] (to NK) and National EYE Institute, National Institute of Health (NIH) (EY024589) to (DPS).
Abbreviations:
- UV
ultraviolet
- HCEpCs
human primary cultured corneal epithelial cells
- hTCEpi
telomerase immortalized human corneal epithelial
- HConFs
primary conjunctival fibroblast
- HPFCs
primary pterygium fibroblast cells
- KRT24
Keratin 24
- MMP-9
matrix metalloproteinase 9
- ROS
reactive oxygen species
- ECM
extracellular matrix
- SD
standard deviation
- Con
conjunctivochalasis
- PA
apical portion
- PB
basal portions
- RT-qPCR
reverse transcribed quantitive real-time polymerase chain reaction
- PBS
phosphate-buffered saline
- ANOVA
one-way analysis of variance
- EMT
epithelial-mesenchymal transition
- miRNA
micro RNA
- αSMA
α smooth muscle actin
- hBD
β-defensins
- HPV
human papillomavirus
Footnotes
Declaration of competing interest
All authors have no conflicts of interest to disclose relating to this work.
References
- Ando R, Kase S, Ohashi T, Dong Z, Fukuhara J, Kanda A, Murata M, Noda K, Kitaichi N, Ishida S, 2011. Tissue factor expression in human pterygium. Mol. Vis 17, 63–69. [PMC free article] [PubMed] [Google Scholar]
- Balci M, Sahin S, Mutlu FM, Yagci R, Karanci P, Yildiz M, 2011. Investigation of oxidative stress in pterygium tissue. Mol. Vis 17, 443–447. [PMC free article] [PubMed] [Google Scholar]
- Bedrossian RH, 1960. The effects of pterygium surgery on refraction and corneal curvature. Arch. Ophthalmol 64, 553–557. [DOI] [PubMed] [Google Scholar]
- Bensch KW, Raida M, Magert HJ, Schulz-Knappe P, Forssmann WG, 1995. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 368, 331–335. [DOI] [PubMed] [Google Scholar]
- Cameron ME, 1983. Histology of pterygium: an electron microscopic study. Br. J. Ophthalmol 67, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SC, Hu DN, Yang PY, Lin CY, Yang SF, 2011. Overexpression of urokinase-type plasminogen activator in pterygia and pterygium fibroblasts. Mol. Vis 17, 23–31. [PMC free article] [PubMed] [Google Scholar]
- Coroneo MT, 1993. Pterygium as an early indicator of ultraviolet insolation: a hypothesis. Br. J. Ophthalmol 77, 734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroneo MT, Di Girolamo N, Wakefield D, 1999. The pathogenesis of pterygia. Curr. Opin. Ophthalmol 10, 282–288. [DOI] [PubMed] [Google Scholar]
- Di Girolamo N, 2012. Association of human papilloma virus with pterygia and ocular-surface squamous neoplasia. Eye 26, 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo N, Chui J, Coroneo MT, Wakefield D, 2004. Pathogenesis of pterygia: role of cytokines, growth factors, and matrix metalloproteinases. Prog. Retin. Eye Res 23, 195–228. [DOI] [PubMed] [Google Scholar]
- Di Girolamo N, Coroneo MT, Wakefield D, 2003. UVB-elicited induction of MMP-1 expression in human ocular surface epithelial cells is mediated through the ERK1/2 MAPK-dependent pathway. Invest. Ophthalmol. Vis. Sci 44, 4705–4714. [DOI] [PubMed] [Google Scholar]
- Di Girolamo N, Wakefield D, Coroneo MT, 2006. UVB-mediated induction of cytokines and growth factors in pterygium epithelial cells involves cell surface receptors and intracellular signaling. Invest. Ophthalmol. Vis. Sci 47, 2430–2437. [DOI] [PubMed] [Google Scholar]
- Dushku N, John MK, Schultz GS, Reid TW, 2001. Pterygia pathogenesis: corneal invasion by matrix metalloproteinase expressing altered limbal epithelial basal cells. Arch. Ophthalmol 119, 695–706. [DOI] [PubMed] [Google Scholar]
- Dushku N, Reid TW, 1994. Immunohistochemical evidence that human pterygia originate from an invasion of vimentin-expressing altered limbal epithelial basal cells. Curr. Eye Res 13, 473–481. [DOI] [PubMed] [Google Scholar]
- Ehrlich F, Laggner M, Langbein L, Burger P, Pollreisz A, Tschachler E, Eckhart L, 2019. Comparative genomics suggests loss of keratin K24 in three evolutionary lineages of mammals. Sci. Rep 9, 10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T, Lehrer RI, 1995. Defensins. Pharmacol. Ther 66, 191–205. [DOI] [PubMed] [Google Scholar]
- Garcia JR, Jaumann F, Schulz S, Krause A, Rodriguez-Jimenez J, Forssmann U, Adermann K, Kluver E, Vogelmeier C, Becker D, Hedrich R, Forssmann WG, Bals R, 2001a. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 306, 257–264. [DOI] [PubMed] [Google Scholar]
- Garcia JR, Krause A, Schulz S, Rodriguez-Jimenez FJ, Kluver E, Adermann K, Forssmann U, Frimpong-Boateng A, Bals R, Forssmann WG, 2001b. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. Faseb. J 15, 1819–1821. [PubMed] [Google Scholar]
- Gris O, Guell JL, del Campo Z, 2000. Limbal-conjunctival autograft transplantation for the treatment of recurrent pterygium. Ophthalmology 107, 270–273. [DOI] [PubMed] [Google Scholar]
- Hashmani K, Branch MJ, Sidney LE, Dhillon PS, Verma M, McIntosh OD, Hopkinson A, Dua HS, 2013. Characterization of corneal stromal stem cells with the potential for epithelial transdifferentiation. Stem Cell Res. Ther 4, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes RJ, Tighe PJ, Dua HS, 1999. Antimicrobial defensin peptides of the human ocular surface. Br. J. Ophthalmol 83, 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Ho KS, Eu KW, Cheah PY, 2007. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin. Canc. Res 13, 1107–1114. [DOI] [PubMed] [Google Scholar]
- Jaworski CJ, Aryankalayil-John M, Campos MM, Fariss RN, Rowsey J, Agarwalla N, Reid TW, Dushku N, Cox CA, Carper D, Wistow G, 2009. Expression analysis of human pterygium shows a predominance of conjunctival and limbal markers and genes associated with cell migration. Mol. Vis 15, 2421–2434. [PMC free article] [PubMed] [Google Scholar]
- John-Aryankalayil M, Dushku N, Jaworski CJ, Cox CA, Schultz G, Smith JA, Ramsey KE, Stephan DA, Freedman KA, Reid TW, Carper DA, 2006. Microarray and protein analysis of human pterygium. Mol. Vis 12, 55–64. [PubMed] [Google Scholar]
- Kahari VM, Saarialho-Kere U, 1999. Matrix metalloproteinases and their inhibitors in tumour growth and invasion. Ann. Med 31, 34–45. [DOI] [PubMed] [Google Scholar]
- Kato N, Shimmura S, Kawakita T, Miyashita H, Ogawa Y, Yoshida S, Higa K, Okano H, Tsubota K, 2007. Beta-catenin activation and epithelial-mesenchymal transition in the pathogenesis of pterygium. Invest. Ophthalmol. Vis. Sci 48, 1511–1517. [DOI] [PubMed] [Google Scholar]
- Kau HC, Tsai CC, Lee CF, Kao SC, Hsu WM, Liu JH, Wei YH, 2006. Increased oxidative DNA damage, 8-hydroxydeoxy- guanosine, in human pterygium. Eye 20, 826–831. [DOI] [PubMed] [Google Scholar]
- Kim KW, Park SH, Kim JC, 2016. Fibroblast biology in pterygia. Exp. Eye Res 142, 32–39. [DOI] [PubMed] [Google Scholar]
- Kormanovski A, Parra F, Jarillo-Luna A, Lara-Padilla E, Pacheco-Yepez J, Campos-Rodriguez R, 2014. Oxidant/antioxidant state in tissue of primary and recurrent pterygium. BMC Ophthalmol. 14, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DQ, Lee SB, Gunja-Smith Z, Liu Y, Solomon A, Meller D, Tseng SC, 2001. Overexpression of collagenase (MMP-1) and stromelysin (MMP-3) by pterygium head fibroblasts. Arch. Ophthalmol 119, 71–80. [PubMed] [Google Scholar]
- Lucas RM, McMichael AJ, Armstrong BK, Smith WT, 2008. Estimating the global disease burden due to ultraviolet radiation exposure. Int. J. Epidemiol 37, 654–667. [DOI] [PubMed] [Google Scholar]
- Maxia C, Perra MT, Demurtas P, Minerba L, Murtas D, Piras F, Corbu A, Gotuzzo DC, Cabrera RG, Ribatti D, Sirigu P, 2008. Expression of survivin protein in pterygium and relationship with oxidative DNA damage. J. Cell Mol. Med 12, 2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott AM, Redfern RL, Zhang B, Pei Y, Huang L, Proske RJ, 2003. Defensin expression by the cornea: multiple signalling pathways mediate IL-1beta stimulation of hBD-2 expression by human corneal epithelial cells. Invest. Ophthalmol. Vis. Sci 44, 1859–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Naka K, Kawamura K, Matsumoto T, Nakanishi I, Fujimoto N, Sato H, Seiki M, 1995. Localization of matrix metalloproteinase 9 (92-kilodalton gelatinase/type IV collagenase = gelatinase B) in osteoclasts: implications for bone resorption. Lab. Invest 72, 311–322. [PubMed] [Google Scholar]
- Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA, Rogers MA, Wright MW, 2006. New consensus nomenclature for mammalian keratins. J. Cell Biol 174, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet LF, Tong L, Su R, Wong TT, 2012. Involvement of SPARC and MMP-3 in the pathogenesis of human pterygium. Invest. Ophthalmol. Vis. Sci 53, 587–595. [DOI] [PubMed] [Google Scholar]
- Sprecher E, Itin P, Whittock NV, McGrath JA, Meyer R, DiGiovanna JJ, Bale SJ, Uitto J, Richard G, 2002. Refined mapping of Naegeli-Franceschetti- Jadassohn syndrome to a 6 cM interval on chromosome 17q11.2-q21 and investigation of candidate genes. J. Invest. Dermatol 119, 692–698. [DOI] [PubMed] [Google Scholar]
- Tan DT, Chee SP, Dear KB, Lim AS, 1997. Effect of pterygium morphology on pterygium recurrence in a controlled trial comparing conjunctival autografting with bare sclera excision. Arch. Ophthalmol 115, 1235–1240. [DOI] [PubMed] [Google Scholar]
- Tan DT, Tang WY, Liu YP, Goh HS, Smith DR, 2000. Apoptosis and apoptosis related gene expression in normal conjunctiva and pterygium. Br. J. Ophthalmol 84, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall TJ, English DR, 1999. Sun exposure and pterygium of the eye: a dose-response curve. Am. J. Ophthalmol 128, 280–287. [DOI] [PubMed] [Google Scholar]
- Tomidokoro A, Miyata K, Sakaguchi Y, Samejima T, Tokunaga T, Oshika T, 2000. Effects of pterygium on corneal spherical power and astigmatism. Ophthalmology 107, 1568–1571. [DOI] [PubMed] [Google Scholar]
- Tomita T, Nagase T, Ohga E, Yamaguchi Y, Yoshizumi M, Ouchi Y, 2002. Molecular mechanisms underlying human beta-defensin-2 gene expression in a human airway cell line (LC2/ad). Respirology 7, 305–310. [DOI] [PubMed] [Google Scholar]
- Touhami A, Di Pascuale MA, Kawatika T, Del Valle M, Rosa RH Jr., Dubovy S, Tseng SC, 2005. Characterisation of myofibroblasts in fibrovascular tissues of primary and recurrent pterygia. Br. J. Ophthalmol 89, 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YY, Cheng YW, Lee H, Tsai FJ, Tseng SH, Lin CL, Chang KC, 2005. Oxidative DNA damage in pterygium. Mol. Vis 11, 71–75. [PubMed] [Google Scholar]
- Weinstein O, Rosenthal G, Zirkin H, Monos T, Lifshitz T, Argov S, 2002. Overexpression of p53 tumor suppressor gene in pterygia. Eye 16, 619–621. [DOI] [PubMed] [Google Scholar]
- Woods M, Chow S, Heng B, Glenn W, Whitaker N, Waring D, Iwasenko J, Rawlinson W, Coroneo MT, Wakefield D, Di Girolamo N, 2013. Detecting human papillomavirus in ocular surface diseases. Invest. Ophthalmol. Vis. Sci 54, 8069–8078. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Nagase T, Makita R, Fukuhara S, Tomita T, Tominaga T, Kurihara H, Ouchi Y, 2002. Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J. Immunol 169, 2516–2523. [DOI] [PubMed] [Google Scholar]
- Yang SF, Lin CY, Yang PY, Chao SC, Ye YZ, Hu DN, 2009. Increased expression of gelatinase (MMP-2 and MMP-9) in pterygia and pterygium fibroblasts with disease progression and activation of protein kinase C. Invest. Ophthalmol. Vis. Sci 50, 4588–4596. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu F, 2019. Elevation of S100 calcium-binding protein A7 in recurrent pterygium. Exp Ther Med 18, 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WP, Zhu YF, Zhang B, Qiu WY, Yao YF, 2016. The role of ultraviolet radiation in the pathogenesis of pterygia (Review). Mol. Med. Rep 14, 3–15. [DOI] [PubMed] [Google Scholar]


