Abstract
Background
The native potatoes (Solanum tuberosum subsp. tuberosum L.) grown in Chile (Chiloé) represent a new, unexplored source of endophytes to find potential biological control agents for the prevention of bacterial diseases, like blackleg and soft rot, in potato crops.
Result
The objective of this study was the selection of endophytic actinobacteria from native potatoes for antagonistic activity against Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum, and their potential to suppress tissue maceration symptoms in potato tubers. This potential was determined through the quorum quenching activity using a Chromobacterium violaceaum ATCC 12472 Wild type (WT) bioassay and its colonization behavior of the potato plant root system (S. tuberosum) by means of the Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) targeting technique. The results showed that although Streptomyces sp. TP199 and Streptomyces sp. A2R31 were able to inhibit the growth of the pathogens, only the Streptomyces sp. TP199 isolate inhibited Pectobacterium sp. growth and diminished tissue maceration in tubers (p ≤ 0.05). Streptomyces sp. TP199 had metal-dependent acyl homoserine lactones (AHL) quorum quenching activity in vitro and was able to colonize the root endosphere 10 days after inoculation.
Conclusions
We concluded that native potatoes from southern Chile possess endophyte actinobacteria that are potential agents for the disease management of soft rot and blackleg.
Keywords: Endophytic actinobacteria, Streptomyces sp., Quorum quenching, Confocal laser microscopy, Blackleg, Soft rot, Potato
Background
The world potato (S. tuberosum) production in 2018 was 368.2 million tons, with a yield of 209.4 hg ha- 1[1]. One of the greatest threats to productivity is loss caused by infectious bacterial diseases, which, once introduced into the crop, may persist and be disseminated in agricultural environments unnoticed or through latent (asymptomatic) infection of seed tubers [2, 3]. Research has estimated that 60% of these losses are due to rot, produced during the cultivation, transport, and storage of the tubers [4].
Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum are the etiological agents that cause blackleg [5] and soft rot (tubers) diseases on potato crops [6, 7]. They may enter the host through natural apertures (lenticels) or wounds and colonize the plant tissue without causing apparent signs or symptoms of the disease until the environmental conditions (free water, anaerobiosis, and temperature) are propitious for the disease development [4, 8]. The virulence of Pectobacterium sp. relies on plant cell wall-degrading enzymes (PCWDE), which disrupt the host cell integrity; however, they activate the synthesis of theses exoenzymes when reaching a high population density coordinated by quorum sensing [9] through a complex set of transcription factors and posttranscriptional regulators [10].
There is no efficient chemical control against these diseases, since the ability of bactericidal compounds to disinfect seed tubers is limited [3, 4]. The method to prevent latent infection in the mother tuber is to use seed tubers derived from material not contaminated by Pectobacterium sp., which can be obtained by cutting, rapid multiplication in vitro, or by botanical seeds [4, 7, 11, 12]. Biological control is proposed as an alternative and sustainable tool for preventing and controlling infectious diseases in plants [13, 14]. This involves studying the interactions of the controlling agent with the host and the pathogen to reduce the pathogen inoculum or to control the severity of disease symptoms by avoiding the expression of their virulence factors [14, 15].
In potato crops, the rhizospheric bacterial community appears to be dominated by alphaproteobacteria and actinobacteria [16], as does the endophytic bacterial community. However, this may vary according to management systems [17, 18]. On the other hand, different studies have found different isolates of actinobacteria with demonstrated antagonist activity against phytopathogens in vitro and in vivo [9, 19–23]. Facultative endophytes could be crucial to obtain plant probiotic agents because their adaptive behavior to colonize the root surface could be a decisive step for the expression of beneficial effects for the host plant [24–26].
The study of endophytes has emerged as an alternative method to control vascular wilting diseases [27]. Endophytes have been shown to stimulate plant growth [28–30] in addition to inducing biotic stress resistance [31], suppressing disease [32], and effectively competing for the space available for pathogens [13]. The potential of bacterial endophytes to inhibit different fungal and bacterial plant pathogens has been reported [33–35].
There is a unique germplasm of native potatoes in Chile on the island of Chiloé, which is considered a subcenter for the origin of potatoes grown worldwide. The island’s isolation has allowed the proliferation of a variety of native potatoes preserved in small fields, characterized by their shapes, sizes, colors, and phenological characteristics [36]. Currently there is no information on the endophytic microbiota associated with these native varieties; however, according to previous results in different varieties, endophytic actinobacteria is a promising bacterial group for the engineering approaches of the endospheric microbiome [37]. Therefore, we investigated the in vitro antagonist capacity of actinobacteria endophytes present in Chilean native potatoes against P. carotovorum subsp. carotovorum and P. atrosepticum to assess their potential as agents for the preventive disease management of blackleg and soft rot in potato crops.
Results
Characterization of endophytic actinobacteria
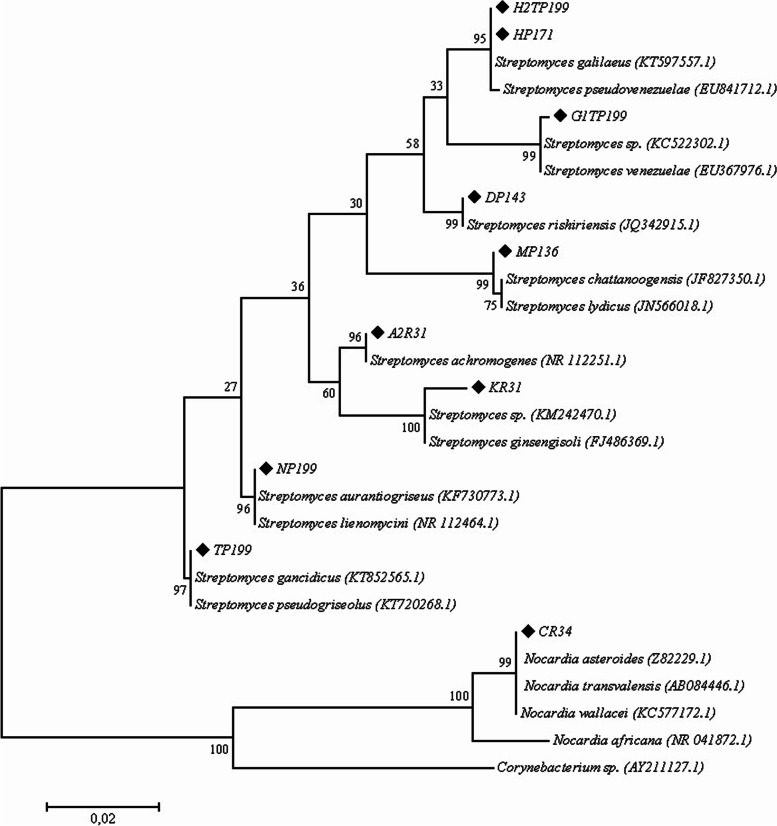
Ten isolates of putative endophyte actinobacteria were obtained from native Chiloé potatoes differentiated by their phenotypic characterization and 16S ribosomal gene analysis (Table 1, Fig. 3). Of these, nine isolates were related to the genus Streptomyces sp., and one isolate was related to the genus Nocardia sp. (Table 1, Fig. 3).
Table 1.
Macro- and microscopic phenotype descriptions of putative endophyte actinobacteria of native Chilean Solanum tuberosum subsp. tuberosum with their taxonomic identity according to 16S ribosomal gene analysis
| Isolate code | Origin of plant tissue | Culture media a | Aerial myceliab | Substrate myceliab | Soluble pigmentc | Filament diameter | Spore diameter | Spore disposition d | Identity by 16S ribosomal DNA partial gene | Closest taxa strain in NCBI | Nucleotide sequence | Similarity in Blast analysis | Accession in NCBI database | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISP2 | ISP3 | ISP4 | |||||||||||||
| H2TP199 | Leaf | B | MB | MB | gray | orange | + | 0.77 um | 0.62 um | Rectiflexibiles | Streptomyces sp. | Streptomyces galilaeus strain P-YIM40 (GenBank: MT533915.1) | 1051 bp | 100% | KY242591 |
| G1TP199 | Root | MB | MB | B | gray | orange | + | < 0.5 um | < 0.5 um | ND d | Streptomyces sp. | Streptomyces sp. strain GF22 (GenBank: MN826279.1) | 1036 bp | 99.2% | KY242592 |
| KR31 | Stem | B | B | B | white | brown | + | 0.72 um | < 0.5 um | Retinaculiaperti | Streptomyces sp. | Streptomyces sp. 7R006 (GeneBank: LC497916.1) | 803 bp | 98.9% | KY242593 |
| DP143 | Tubercle | MB | MB | MB | white/gray | brown | + | 0.67 um | < 0.5 um | Retinaculiaperti | Streptomyces sp. | Streptomyces sp. YRA147 (GenBank: JX430828.1) | 1104 bp | 99.8% | KY242596 |
| A2R31 | Stem | MB | EX | B | gray | brown | + | 0.77 um | 0.89 um | Rectiflexibiles | Streptomyces achromogenes | Streptomyces achromogenes strain NBRC 12735 | 969 bp | 100% | KY242590 |
| CR34 | Tubercle | MB | B | B | NDe | cream | – | < 0.5 um | < 0.5 um | ND e | Nocardia sp. | Nocardia sp. FGT33 (GenBank: KU382293.1) | 750 bp | 99.9% | KY296348 |
| HP171 | Root | MB | MB | EX | white | orange | – | 0.67 um | 0.66 um | Rectiflexibiles | Streptomyces sp. | Streptomyces galilaeus strain PSWC-10 (GenBank: MT275786.1) | 1027 bp | 99.9% | KY242594 |
| MP136 | Leaf | MB | MB | MB | black | yellow | – | 0.56 um | 0.66 um | Spira | Streptomyces sp. | Streptomyces sp. CH9-7 (GenBank: LC489240.1) | 1078 bp | 99.8% | KY242595 |
| TP199 | Tubercle | EX | B | MB | gray | yellow | – | 0.56 um | < 0.5 um | Straight to flexuous chains | Streptomyces sp. | Streptomyces sp. KK13-6 (GenBank: LC487830.1) | 1147 bp | 99.6% | KY228978 |
| NP199 | Tubercle | MB | MB | MB | gray | yellow | – | 0.77 um | 0.58 um | ND e | Streptomyces sp. | Streptomyces sp. HST09 (GenBank: KX130883.1) | 1114 bp | 99.3% | KY242597 |
aCulture media according to [38], ISP2 (yeast extract and– malt extract agar), ISP3 (oatmeal agar) and ISP4 (inorganic salts and– starch agar). Furthermore, excellent (EX, 76 to 100%), very good (MB, 51 to 75%), good (B, 26 to 50%), poor (P, 1 to 25%), and negative (N, 0%)
bDetermined on ISP2 medium according to [38]
cNegative: -, Positive: +. Determined in ISP1 medium according to [38]
dDescription according to [39] e ND Not determined
Fig. 3.
Phylogenetic tree based on an analysis of partial sequences of the 16S ribosomal gene. The maximum likelihood method shows relations between isolates H2T199, HP171, DP143, NP199, TP199, A2R31, KR31, MP136, CR34, and selected members of the phylum Actinobacteria from the NCBI database; Corynebacterium sp. (AY211127) was used as the tree root. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The bar indicates 0.02 substitutions per nucleotide
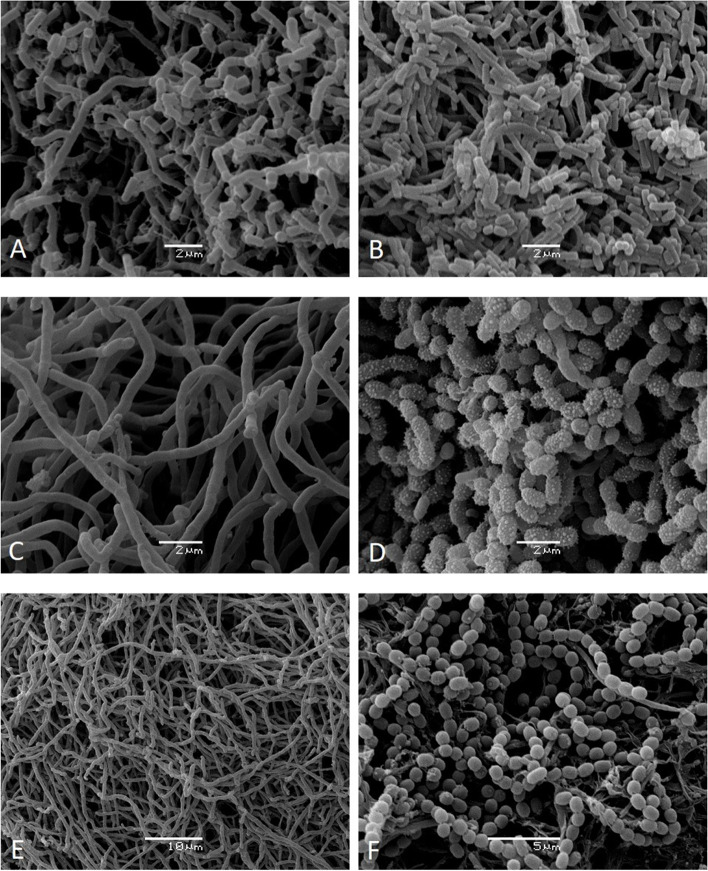
For the following ultramicroscopic visualization using scanning electron microscopy, three bacterial isolated were selected. Nocardia sp. CR34 was the only isolation obtained from this genus and was observed according the typical fragment mycelia in liquid (ISP1- tryptone-yeast extract broth) (Fig. 1A) and solid (ISP2- yeast extract and malt extract agar) culture (Fig. 1B).
Fig. 1.
Scanning electron microscopy. A Nocardia sp. CR34 growth in liquid ISP1 culture after 120 h of incubation at 120 rpm and 28 °C. B Nocardia sp. CR34 growth in ISP2-agar culture after 14 days of incubation at 28 °C. C Streptomyces sp. TP199 substrate mycelium growth in liquid ISP1 culture after 120 h of incubation at 120 rpm and 28 °C. D Streptomyces sp. TP199 aerial mycelium growth in ISP2-agar culture after 14 days of incubation at 28 °C. E Streptomyces sp. A2R31 substrate mycelium growth in liquid ISP1 culture after 120 h of incubation at 120 rpm and 28 °C. F Streptomyces sp. A2R31 aerial mycelium growth in ISP2-agar culture after 14 days of incubation at 28 °C. Scale bar: 5 μm
Then, Streptomyces sp. TP199 and Streptomyces sp. A2R31 were selected as they showed antimicrobial activity in vitro (Fig. 2), and their ultramicroscopic visualization showed the long chains of spore growth only in ISP2-agar typical of Streptomyces sp. (Fig. 1D and F).
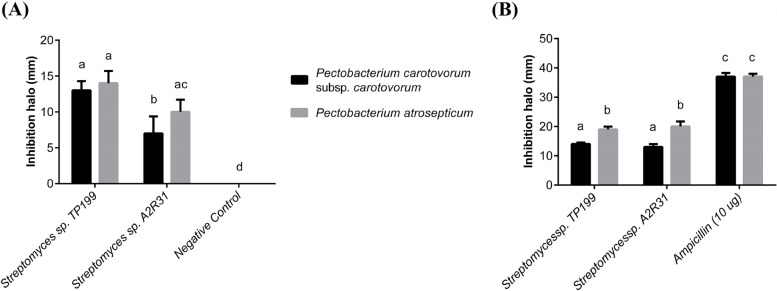
Fig. 2.
Assessment of the antagonistic activity of Streptomyces sp. TP199 and Streptomyces sp. A2R31. Isolates against P. carotovorum subsp. carotovorum and P. atrosepticum in vitro. A Inhibition growth of pathogens using the cross-streak method, negative control: pathogens without antagonistic; B agar plug method, positive control: ampicillin disc (10 μg). Plot mean with SD. Different letters on the top of error bars indicate significative statistical differences (Tukey-test, p ≤ 0,05), n = 4
For the Nocardia sp. CR34 isolate (Fig. 3), the morphology corresponded to characteristic pseudomycelial growth, both in ISP2-agar culture (Fig. 1B) and in liquid ISP1 culture (Fig. 1A). The other nine isolates were related to Streptomyces sp. (Fig. 3), showing mycelial morphology in liquid culture for isolates TP199 (Fig. 1C) and A2R31 (Fig. 1E). The spores could be distinguished on ISP2-agar culture, with a smooth texture in the A2R31 isolate (Fig. 1F) and a warty texture for the TP199 isolate (Fig. 1D).
Antagonism
Of the ten isolates obtained, only Streptomyces sp. TP199 and Streptomyces sp. A2R31 inhibited the growth of P. carotovorum subsp. carotovorum and P. atrosepticum in vitro. Streptomyces sp. TP199 generated inhibition halos of 13 ± 1.29 mm and 14 ± 1.71 mm against P. carotovorum subsp. carotovorum and P. atrosepticum, respectively, according to the cross-streak method, while Streptomyces sp. A2R31 generated inhibition halos of 7 ± 2.37 mm and 10 ± 1.71 mm against the same phytopathogens (Fig. 2A). Using the agar disc diffusion method, Streptomyces sp. TP199 generated inhibition halos of 14 ± 0.5 mm and 19 ± 0.96 mm against P. carotovorum subsp. carotovorum and P. atrosepticum, respectively, while the corresponding values for Streptomyces sp. A2R31 were 13 ± 0.96 mm and 20 ± 1.71 mm (Fig. 2B). Ampicillin, which was used as the positive control, generated inhibition halos of 37 ± 1.29 mm and 37 ± 0.96 mm against P. carotovorum subsp. carotovorum and P. atrosepticum, respectively (Fig. 2B).
The effects of Streptomyces sp. TP199 and Streptomyces sp. A2R31 to inhibit plant tissue maceration were screened by a potato tuber slice assay, which showed, for the control treatments: maceration halos of 4.29 ± 1.03 mm for the well with the sterile nutrient broth, 14.97 ± 0.49 mm for the well inoculated with P. atrosepticum, and 15.95 ± 2.83 mm for the well inoculated with the P. carotovorum subsp. carotovorum alone. On the other hand, when mixing pathogens and antagonists, the inocula resulted in macerated tissue halos of 11.87 ± 7.25 mm for the well inoculated with a mix of P. atrosepticum and Streptomyces sp. A2R31 in a 1:1 ratio, 5.16 ± 1.97 mm for the well inoculated with a mix of P. atrosepticum and Streptomyces sp. TP199 in a 1:1 ratio (Fig. 4), 11.03 ± 4.79 mm for the well inoculated with a mix of P. carotovorum subsp. carotovorum and Streptomyces sp. A2R31 in a 1:1 ratio, and 6.19 ± 3.25 mm for the well inoculated with a mix of P. carotovorum subsp. carotovorum and Streptomyces sp. TP199 in a 1:1 ratio (Figs. 4 and 5).
Fig. 4.
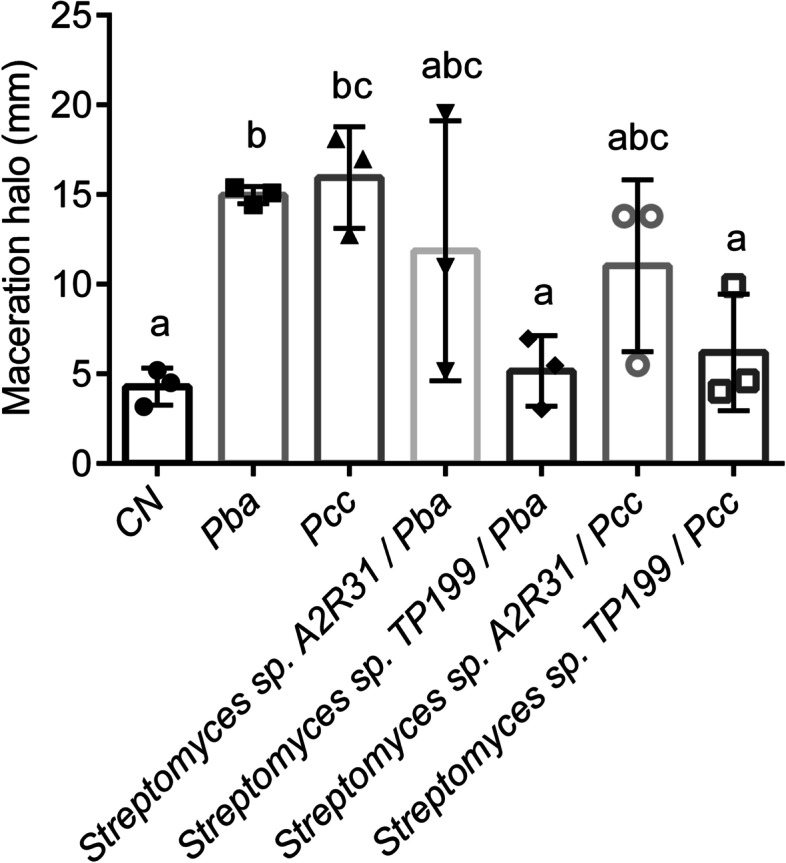
Maceration assay in tuber slices. Maceration tissue halos measured (mm) on tuber slices inoculated with Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum. These at a concentration of 108 cells/mL in combination with Streptomyces sp. A2R31 and Streptomyces sp. TP199 at a concentration of 108 spores/mL, incubated at 28 °C for 72 h; the treatments were: (i) P. carotovorum subsp. carotovorum, (ii) P. atrosepticum, (iii) Streptomyces sp. A2R31, (iv) Streptomyces sp. (v) P. carotovorum subsp. carotovorum + Streptomyces sp. A2R31 and (vi) P. carotovorum subsp. carotovorum + Streptomyces sp. TP199. Error bars indicate standard deviation of the media. CN: Sterile Nutrient Broth. Pba: P. atrosepticum. Pcc: P. carotovorum subsp. carotovorum. Plot mean with SD. Different letters on the top of error bars indicates statistical significative differences (Tukey-test, p ≤ 0,05), n = 3
Fig. 5.
Tuber slice assay. a. Wells inoculated with P. carotovorum subsp. carotovorum. b. Wells inoculated with Streptomyces sp. TP199. c. Wells inoculated with P. carotovorum subsp. carotovorum + Streptomyces sp. TP199. d. Wells inoculated with sterile nutrient broth. e. Uninoculated slice of the tuber
Quorum sensing inhibition through Chromobacterium violaceum ATCC 12472 wild type (WT) bioassay
To complement the inhibitory effect assay of Streptomyces sp. TP199 in the decreased tissue maceration in tubers caused by Pectobacterium sp., we assessed the interruption of communication signals that regulate the synthesis of pectinolytic enzymes at the transcription level in different strains of Pectobacterium sp. For the first experiment, the supernatant of Streptomyces sp. TP199 cultures showed no inhibition. Therefore, we proceeded in accordance with [40] and used an AHL (40 μM N-Hexanoyl-DL homoserine lactone (C6-AHL)) solution before performing the Chromobacterium violaceum ATCC 12472 (WT) bioassay. On the other hand, studies showed that certain metals are essential for cutting the ester bond in the lactone ring and for proper enzyme folding, and, in the following experiment, Streptomyces sp. TP199 was grown in ISP1 supplemented with solutions of three different metals (Mg, Zn, and Mn) at three different final concentrations (0.2, 1, and 2 mM, final). These 12 conditions, defined by the metallic nature and concentration, were tested for Streptomyces sp. TP199 from a sporulated culture in ISP2-agar. Streptomyces sp. Metal-free TP199, with acyl homoserine lactones (AHL) stimulation showed less inhibition around the control disk (Fig. 6). For each metal used at 1 and 2 mM, no colorless area appeared. All metal concentrations of 0.2 mM in Streptomyces sp. TP199 showed an area of inhibition (Fig. 6).
Fig. 6.
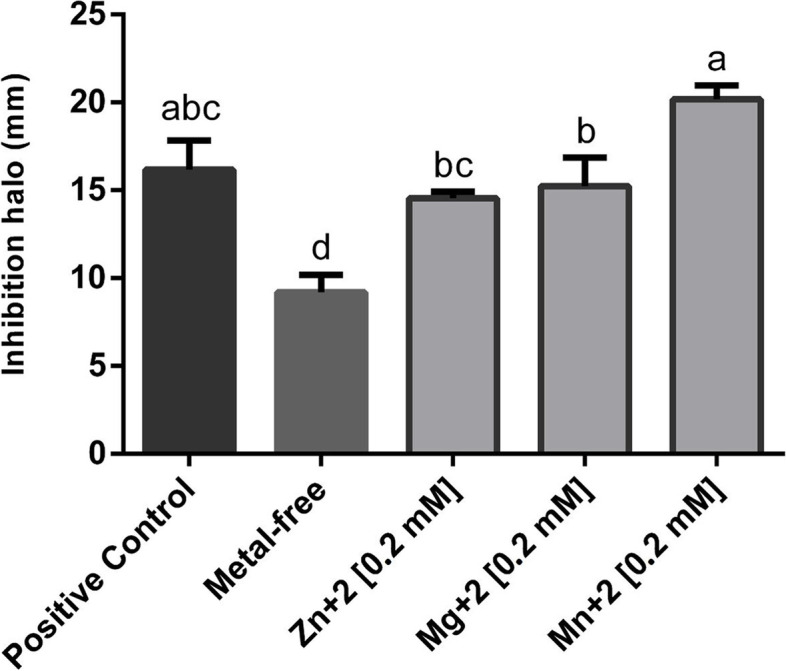
Inhibition of the violacein production of Chromobacterium violaceum ATCC 12472 (WT) by culture of Streptomyces sp. TP199. This according to the metal used. The bars represent the diameter sizes generated (inhibition halo in mm) of the colorless areas. Plot mean with SD. Different letters on the top of error bars indicate statistical significative differences (Tukey-test, p ≤ 0,05), n = 3
Endosphere colonization of Streptomyces sp. TP199 in roots potato plants
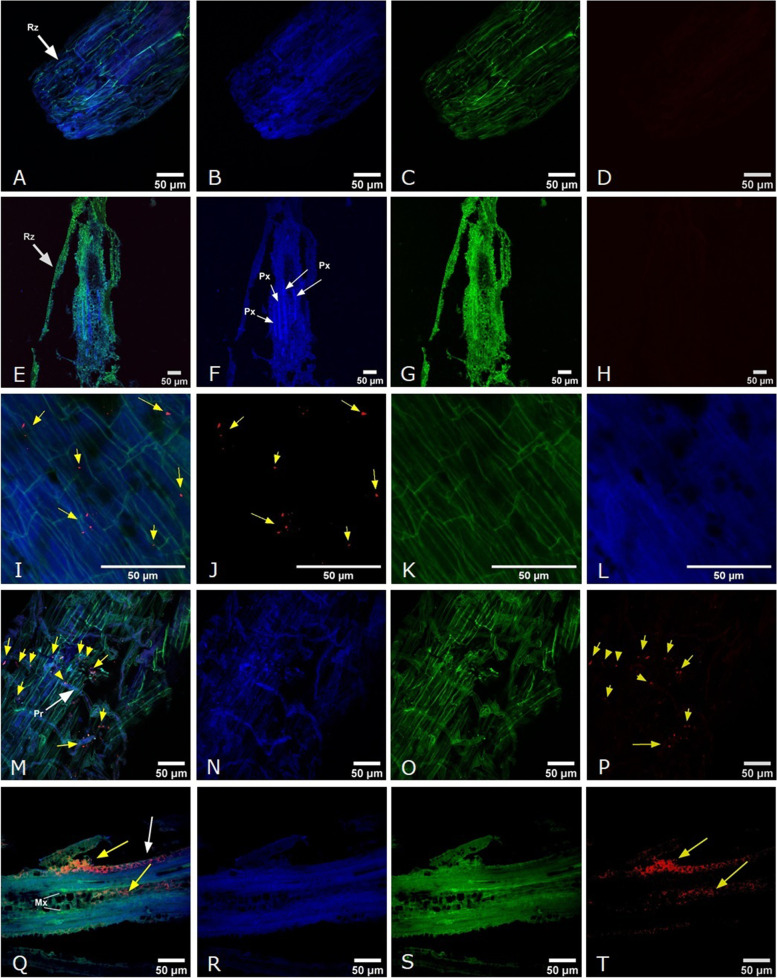
Two-photon laser scanning confocal microscopy was used to obtain insights into the Streptomyces sp. TP199 root colonization pattern, which was targeted by 23S rRNA with a Cy5 dye double-labeled HGC69a probe (Fig. 7D, H, J, P, T). The identification of the colonization zones in the plant was possible by the autofluorescence of the plant tissue, which is visualized in blue (Fig. 7B, F, L, N, R) and green (Fig. 7C, G, K, O, S). Therefore, in the merged image, the signal from Streptomyces sp. TP199 (visualized in red) is contrasted with the autofluorescence of plant tissue (Fig. 7A, E, I, M, Q). The first step of colonization was visualized on a rhizodermal (Fig. 7I) and in the root piliferous zone cells 3 days after inoculation (Fig. 7M). In the case of the histological sections, which correspond to 10 days after inoculation, the greater intensity in the fluorescence corresponding to the HGC69a probe was observed in the area of root hair growth and the metaxylem (Fig. 7Q). The zoom of the vascular bundle’s plant cells allowed us to visualize that the points of greater intensity of the HGC69a probe’s fluorescence were presented in the cell walls and intercellular spaces of eukaryotic cells.
Fig. 7.
Streptomyces sp. TPP199 root colonization patterns targeted by 23S rRNA with Cy5 dye double-labeled HGC69a. Probe visualized by two-photon laser scanning microscopy where autofluorescence of the plant tissue is shown in blue (B, F, L, N, R) and green (C, G, K, O, S) separately, while the probe is in red separately (D, H, J, P, T) and in a merged image (A, E, I, M, Q). A-D Apical root from a not-inoculated plant, scale bar of 50 μm (day 3 of the assay). E-H Longitudinal histological section root from a not-inoculated plant, scale bar of 90 μm (day 10 of the assay). I-L Rhizodermis from a 3 day post-inoculated plant, scale bar of 20 μm. M-P Root hair growth area from a 3 day post-inoculated plant, scale bar of 60 μm. Q-T Longitudinal histological section root from a 10 day post-inoculated plant, scale bar of 60 μm. A, E, I, M, and Q merged image. B, F, J, N, and R blue (excitation-emission: 405/410–480 nm). C, G, K, O, and S green (excitation-emission: 488/490–560 nm) separately, while the 23S rRNA with Cy5 dye double-labeled HGC69a probe is in D, H, L, P, and T red separately (excitation-emission: 633/638–747 nm). Rz: rhizodermis, Pr: root hair. Mx: metaxylem. Yellow arrows indicate Streptomyces sp. TP199 targeted with Cy5 dye double-labeled HGC69a probe fluorescence, and white arrows indicate the plant morphology
Discussion
Actinobacteria is a bacterial phylum differentiated by the high content of G + C in its DNA, recognized as one of the main producers of natural bioactive compounds of microbial origin [41] and its biotechnological application prospects [42].
In this study, the presence of facultative endophytic actinobacteria was determined for the first time in native Chilean potatoes by the selective humic acid medium (HV) culture, most of them from the Streptomyces genus, which is consistent with the results from cultivation and cultivation-independent population analysis of bacterial endophytes in potato (S. tuberosum) [17, 34, 43–47]. According to our results, the morphological and molecular characterization of the isolates obtained was consistent with the genera Streptomyces, and not related to pathogenic species or Nocardia [39, 48].
Ten isolates of actinobacteria were selected by considering that the disinfection controls presented no growth when cultured. However, this validation is subject to the restrictions governing microorganism culturability [49], as they might be epiphytes resistant to the disinfection process [50, 51]. Thus, the isolates reported here should be considered facultative endophytes, with the exception of Streptomyces sp. TP199, which was validated as an endophyte by microscopy [30].
The in vitro results for antagonism (cross streak and diffusion with agar discs) resulted in the selection of Streptomyces sp. A2R31 and Streptomyces sp. TP199 as antagonists of Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum. We observed that only the Streptomyces sp. TP199 isolate produced a significant reduction in the tuber tissue maceration assay relative to inoculation with the two phytopathogens alone. This phenomenon may, therefore, be linked to the relative inoculation times of the antagonist and the pathogen since according to [52, 53], a Streptomyces sp. isolate was able to reduce the severity of rot symptoms in plants and tubers by 65 to 95% when it was inoculated 24 h before the pathogens. Moreover, the nine facultative actinobacteria endophytes reported in this study without antimicrobial activity could be studied for another plant–microbial interaction.
Isolates of Streptomyces were previously reported as antagonists of Pectobacterium through an in vitro assessment of anti-microbial activity; for example, the authors in [53] reported four isolates of Streptomyces that inhibited the growth of six different strains of Pectobacterium (strains B21, K6, M2, Kh6, CFBP5890, and CFBP5889) with inhibition halos in the range of 11 to 27.33 mm, comparable to the results obtained in this work. A further 13 isolates of actinobacteria with antagonistic activity against Erwinia chrysanthemi 3937VIII, a phytopathogen that causes soft rot, were obtained; the study also presented notable results from an isolate of Streptomyces cinereoruber evaluated as a biocontroller [54].
The synthesis of antimicrobial compounds produced by Streptomyces sp. TP199 and Streptomyces sp. A2R31 could be induced by the presence of the pathogen in the medium, and this could be the reason why no antimicrobial activity against P. carotovorum subsp. carotovorum and Pectobacterium atrosepticum was observed in the supernatants of the actinobacterial cultures, since the induction of secondary metabolites was reported under the conditions of microbial co-cultures [55, 56]. To test this hypothesis and distinguish a bactericidal effect, Streptomyces sp. TP199 and P. carotovorum subsp. carotovorum was co-cultivated, and no inhibition was observed in the P. carotovorum subsp. carotovorum growth, according to the live and dead cells count recording and Gompertz model analysis.
Then, to explain the lack of bactericidal effect on P. carotovorum subsp. carotovorum growth and an effective maceration tissue control in the potato slices assay, we considered a direct effect on the virulence of the pathogen, as shown by [40, 57–60], who assessed the interruption of communication signals that regulated the synthesis of pectinolytic enzymes at the transcription level in different strains of Pectobacterium sp. Consequently, the inhibition of violacein synthesis through the Chromobacterium violaceum ATCC 12472 (WT) bioassay plates were evaluated to test the AHL inhibition mechanism in Streptomyces sp. TP199. First, no inhibition was observed when the supernatants were taken; every 12 h of Streptomyces sp. TP199 growth was tested; however, using the research of [61], we reviewed that some AHL-lactonases need a metal in order to degrade the AHL in an optimum way. Different metals were tested, in this experiment, in addition to AHL including: manganese, calcium, magnesium, and zinc. The choice of these metals was made according to their frequency in different strains, in the AHL degradation [61]. Through the results in C. violaceum plates, violacein inhibition appeared only for 0.2 mM for each metal, which is a promising result for the inhibition quorum sensing mechanism possibly related to an AHL degrading enzyme metal-dependent and not currently defined for Streptomyces sp. species [59, 60].
Finally, this taxa has been described as part of the plant microbiota, colonizing different compartments, such as the endosphere, phyllosphere, and rhizosphere [28, 62]. To determine the colonization capacity of Streptomyces sp. TP199 in a commercial potato crop, like an endophyte, it was inoculated in the root system of potato cv. Pukará-INIA plants, which were obtained by micropropagation to obtain plant material free of pathogens. Bearing in mind that the combination of the use of DOPE-FISH with the use of confocal laser microscopy (CLSM) facilitated the exploration of micro-habitats and allowed the observation of microorganisms associated with their host in situ with excellent results, these techniques were used and resulted in the visualization of Streptomyces targeted with a double-labeled Cyt5 dye HGC69a probe in the piliferous zone and into the xylem vessels 10 days post-inoculation, similar to the results of [63]; however, it is necessary to make a distinction regarding the passive or active mechanism and to determine if this could confer protection to the plants against Pectobacterium sp. infection [62, 64].
To understand this behavior, future studies must determine the chemical nature, concentrations, and diffusion properties of the assemblage of active molecules secreted by the antagonists, interpret more precisely the potential of Streptomyces sp. TP199 against P. carotovorum subsp. carotovorum and P. atrosepticum, and validate the mechanism related to blocking the virulence expression of Pectobacterium sp. inside the plant and in the tubers [14, 59].
Conclusions
In conclusion, two isolates of endophytic actinobacteria from native Chilean potatoes (Solanum tuberosum L.), Streptomyces sp. TP911 and Streptomyces sp. A2R31, were found to possess antagonistic activity in vitro against Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum; however, only Streptomyces sp. TP199 decreased the tuber tissue maceration (approximately 38 and 34%, respectively), and responded to the presence of antimicrobial compounds and AHL signal interference with metal-dependent enzymatic action. Streptomyces sp. TP199 also acted as a vegetable probiotic by being able to colonize the root endosphere of potato plants of cv. Pukará-INIA. This suggests that the native Chilean potato possesses endophytic actinobacteria that could be potential agents for the management of the diseases of blackleg and soft rot in potato crops.
Methods
Isolation of endophyte actinobacteria
Tubers of native potato (S. tuberosum subsp. tuberosum) obtained from Chiloé (Southern Chile) were made available by the National Agricultural Research Institute (INIA) Remehue, Osorno (accession number Ch. P11). This institution made the formal identification of the plant material used. A voucher specimen of this material has been deposited in a publicly available herbarium of INIA Remehue (Deposition number not available). They were sown in pots containing sterile trumao soil (Soil derived from frequent volcanic ash in southern Chile) and kept in a greenhouse for 5 months at INIA Quilamapu, Chillán (VIII Region of Chile) to collect plant tissue. Leaves, stems, roots, and tubers were collected from plants that presented no visible disease symptoms in the greenhouse. After collection, the tissues were washed in a soapy solution, and then 2 to 3 g of each tissue was resuspended in phosphate-buffered saline (PBS) solution with Tween-20 (0.01%) and sonicated at 60 Hz for 3 min [65, 66]. The tissues were then immersed in ethanol 70% for 1 min, sodium hypochlorite (NaOCl) at 3% for 4 min, washed three times in sterile water, and finally dried on sterile absorbent paper under a laminar airflow hood. To obtain only endophytes from the phylum Actinobacteria, slices of approx. Four square millimeter were placed on humic acid medium (HV) [67] and supplemented with nystatin (0.05 g/mL) and cycloheximide (0.05 g/mL), designed for the specific growth actinobacteria. They were incubated at 30 °C for 30 days [66, 68]. The surface disinfection of the tissues was validated in HV agar incubated at 28 °C for at least 30 days [66, 68].
Characterization of the actinobacteria
Macroscopic morphological characterization was conducted out according to the guidelines of [38] in three different culture media: ISP2 (yeast extract and malt extract agar), ISP3 (oatmeal agar) and ISP4 (inorganic salts and starch agar). Growth was categorized by the growth area on a Petri dish as a function of the total area and expressed as a percentage in the following categories: excellent (EX, 76 to 100%), very good (MB, 51 to 75%), good (B, 26 to 50%), poor (P, 1 to 25%), and negative (N, 0%). Measurements were taken using the ImageJ program [69]. The isolates were incubated at 28 °C and observed after 21 days. The production of soluble pigments, the color of the aerial mycelium, and the substrate mycelium color were recorded [39, 48]. Cultures were obtained using the slide technique and incubated for 14 days with ISP2 as the growth medium, then observed under the microscope.
The coverslips with growth were removed and placed on another slide fixed with absolute methanol for 15 min and stained with crystal violet for 1 min. After rinsing and drying, the coverslip was examined under an optical microscope. In each case, the filaments and spore morphology was observed, and the diameter was measured [39, 48]. Isolates representing different taxonomic genera were selected for the examination of cell morphology by scanning electron microscopy [39], both as solid cultures on ISP2 after 14 days of incubation at 28 °C and as liquid cultures in ISP1 (tryptone-yeast extract broth) after 120 h of incubation with shaking at 120 rpm and 28 °C. The products were preserved in 4% glutaraldehyde at 5 °C and processed with scanning electron microscopy at the Microscopy Centre of the University of Concepción, Concepción, Chile.
The taxonomy was determined via 16S ribosomal genes by cultivating the actinobacterial isolates in ISP1 for 120 h at 28 °C and inoculating them to a final concentration of 2.5 × 108 spores/mL. The samples were centrifuged at 12,000 rpm and 4 °C, and the DNA was extracted from the pellets using the ZR Soil Microbe DNA MiniPrepTM kit following the manufacturer’s instructions. For isolates that did not form spores, DNA was extracted from an even covering on ISP2 medium [38]. The 16S ribosomal gene was amplified with the universal primers for bacteria: 9-27F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1541R (5′-AAGGAGGTGATCCAACC-3′) [70]. The PCR product was viewed by electrophoresis in a 1.2% (w/v) agarose gel with ethidium bromide (10 mg/mL) in a transilluminator with UV light [71]. Sequencing services were provided by Macrogen Korea (http://www.macrogen.com/). The rDNA 16S sequences were analyzed using the BLASTn tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences produced by Blast were selected with > 98% identity.
MEGA7 software was used to construct the phylogenetic tree [72] using the maximum likelihood method based on the model of [73]. The tree was validated statistically by bootstrapping with 1000 iterations [74], and the initial tree for the heuristic search was obtained by applying the Neighbor Joining method to an estimated distance matrix using the compound maximum probability approach.
Antibacterial activity
The strains of Pectobacterium carotovorum subsp. carotovorum and Pectobacterium atrosepticum were supplied by the Phytopathology Laboratory of INIA CRI Remehue, Osorno, Chile. The antagonistic activity of the actinobacterial isolates was evaluated using the cross streak agar diffusion technique and agar discs [53, 54]. The cross streak agar procedure is a simple way to test antagonistic properties of actinobacteria through an easy and a rapid semi-quantitative screening, where the inhibition halos could be measure, then let establish a spectrum of inhibiting properties of actinobacteria [75]. The Pectobacterium strains were used in the exponential phase (108 cell/mL) cultured in nutrient broth under shaking at 110 rpm and 28 °C for 18 h. For cross streak evaluation, the actinobacterial isolates were sown in a strip across the diameter of the dish on nutrient agar and incubated at 28 °C until sporulation. Then, the Pectobacterium was sown in a strip perpendicular to the actinobacteria and incubated for 48 h at 28 °C to observe the formation of the inhibition halos. For evaluation on agar discs, Pectobacterium was sown evenly on nutrient agar and then placed on an agar disc (6 mm) with sporulated actinobacteria on ISP2, as well as an agar disc with sterile ISP2 as a blank and a disc with ampicillin (10 μg) as a positive control. These were incubated at 28 °C for 48 h to observe the formation of inhibition halos. Both assays were performed in triplicate for each isolate, and the measured halos were analyzed by means of Tukey HSD test [76].
Maceration assay in tuber slices
Tubers of Pukará-INIA potatoes were washed with a soapy solution and resuspended in PBS with Tween-20 [0.01%], then sonicated at 60 Hz for 10 min. The tubers were disinfected in sequence in 3% sodium hypochlorite (NaOCl) for 4 min and 70% ethanol for 1 min and flamed. Then, the slices were placed in a laminar flow chamber (for drying). Slices 7 to 8 mm thick were cut and placed in Petri dishes with sterile absorbent paper, and three wells (6 mm) were made in each slice [77–79]. A pathogenic suspension of each strain of Pectobacterium (108 cells/mL) was mixed with a suspension of actinobacteria; spores (108 spores/mL) at a 1:1 ratio. The control suspensions were prepared in the same way but with only the pathogen or only the antagonist, mixing the strains with sterile PBS at a 1:1 ratio. A volume of 30 μL of each inoculum was added to each well, and the absorbent paper under the tuber slices was moistened with sterile water. The sterile nutrient broth was used as a negative control. The dishes were then incubated at 28 °C for 72 h, and the macerated tissue diameter around the wells was measured. Each assay was performed in triplicate [77–79], and the mean diameter per treatment was compared using Tukey HSD test (p ≤ 0.05 [76];).
Metals tests and AHL- inhibition assays
Bioassay plates were made with LB-Agar 1,5% medium, covered by a solution of Chromobacterium violaceum ATCC 12472 (WT) at 107 cells/mL from the inoculum, in Soft Luria Bertani (LB)-Agar 0,7% [80]. In the first experiment, filter discs were filled with 20 μL of each sampling time supernatant (every 12 h of Streptomyces sp. TP199 for 7 days) previously passed through 0,22 μm filters to eliminate the possible presence of bacteria. Two serial dilutions at ½ and ¼ were made from these supernatants [57, 58]. In the second experiments, Streptomyces sp. TP199 were grown on ISP2 agar culture for 3 days at 28 °C and then subcultured in ISP1 medium for 1 day. An inoculum of 108 cells/mL of this strain was made. One hundred fifty microliter of both cultures were further incubated in two 96 well microplates at 28 °C with 120 rpm shaking for 3 days.
The metals solutions were adjusted at pH = 7 (MnCl2, Zn2+, and MgSO4) and added at final concentrations of 0,2, 0,5, 1, and 2 mM, according to the experiment. One triplicate of wells did not receive a metal solution. Negative controls were made with filling two triplicates of wells, respectively, with metal solutions only and ISP1 medium. Forty microliters of each well were mixed with an equal volume of a 40 μM N-Hexanoyl-DL homoserine lactone (C6-AHL) [40]. The mixture was incubated at 28 °C with a 120 rpm shaking during 2 h and the reaction was stopped by heating at 95 °C during 10 min. Negative controls were made with the LB liquid medium and positive controls with vanillin (0,01 g/mL), previously proven to have AHL-inhibition in the laboratory. All plates were incubated 24 h at 30 °C to allow the purple development. An inhibition halo of the purple pigment was observed when AHLs production by C. violaceum ATCC 12472 (WT) was inhibited. We measured and recorded the results. The mean diameter per treatment was compared using Tukey HSD test (p ≤ 0.05 [76];).
Plant inoculation
Twenty plants were obtained by micropropagation and acclimated in a sterile sand substrate under controlled environmental conditions. The photoperiod was maintained with 16 h light and 8 h dark, 70% relative humidity (R.H.), and 20 ± 2 °C for approximately 3 weeks, until vigorous root growth was observed [81]. Then, 10 plants were selected to control treatment, and 10 plants were inoculated with 108 spores/mL of Streptomyces sp. TP199. The root system was inoculated by the dipping method for 10 min, keeping the spores’ movements softly. Then, they were transplanted to independent pots in the sterile substrate, and the conditions of controlled growth were maintained.
Root tissue samples were collected from the control and inoculated plants on days 3 and 10 post-inoculation and were fixed in 4% paraformaldehyde for 24 h and subsequently stored in PBS/EtOH 95.5° (1: 1) at − 20 °C until processing. The HM525 cryostat was used to obtain 30 μm thick cross-sections, which were placed on microscope slides that were previously treated with polylysine (1 mg/mL).
Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH)
The probe HGC69a (5′-TATAGTTACCACCGCCGT-3′ )[82] was used for the 16S rRNA gene whose specificity was verified in the Probe Check database (http://microbial-ecology.net/) and was labeled with fluorophore Cy5 at the 5 ‘and 3’ ends of the nucleotide sequence (emission and excitation outside the autofluorescence range of the plant tissue )[83]. Whole root samples and histological root sections on microscope slides (the edge was drawn with hydrophobic pencil) were processed and were pre-treated with achromopeptidase (1 mg/mL) at 37 °C for 15 min and then serially dehydrated with ethanol (50–99.9%, 30 min each stage). A first hybridization step was performed (0.02 mM Tris-HCl, 0.01% SDS, 0.9 M NaCl, and 25% formamide), the probe was added to a final concentration of 15 ng/μL in the dark, and then the samples were incubated at 48 ± 1 °C for 3 h. Subsequently, the samples were drained in a washing solution (0.02 mM Tris-HCl, 0.01% SDS, and 5 M NaCl), preheated to 51 °C, and incubated in the same solution at 51 °C for 20 min. Finally, the samples were dried in the dark in the laminar flow chamber, the immersion solution was added to maintain fluorescence (Prolong, Sigma), and the coverslip was placed.
Visualization by two-photon confocal laser microscopy
A two-photon confocal laser microscope LSM780 NL0 Zeiss (Advanced Microscopy Center, CMA BIO-BIO, CONICYT PIA ECM-12 Project) linked to the ZEN blue software was used for the visualization and image capture from the treated root samples. Different lasers were used for different ranges of excitation and emission, corresponding to the excitation/emission spectra: 488/490–560 nm, 405/410–480 nm (UV light), and 633/638–747 nm for the probe (far-red light). The Image J program [69] was used to process the images.
Acknowledgements
Not Applicable.
Abbreviations
- ATCC
American Type Culture Collection
- WT
Wild type
- DOPE-FISH
Double labeling of oligonucleotide probes for fluorescence in situ hybridization
- AHL
Acyl homoserine lactones
- PCWDE
Pectobacteria relies on plant cell wall-degrading enzymes
- ISP1
Tryptone-yeast extract broth
- ISP2
Yeast extract and– malt extract agar
- ISP3
Oatmeal agar
- ISP4
Inorganic salts and– starch agar
- EX
Excellent
- MB
Very good
- B
Good
- P
Poor
- N
Negative
- NCBI
National Center for Biotechnology Information
- ND
Not determined
- Pcc
P. carotovorum subsp. carotovorum
- Pba
P. atrosepticum
- 23S rRNA
23S ribosomal RNA
- HV
Humic acid medium
- INIA
National Agricultural Research Institute
- PBS
Phosphate-buffered saline
- HSD
Honestly significant difference
- RH
Relative humidity
- SDS
Sodium dodecyl sulfate
- UV
Ultraviolet
Authors’ contributions
Conceptualization N.P., S.M. A.F., I.A., and H.U.; methodology, N.P., P.L., A.M., and C.T.; software, N.P. and A.M.; validation, N.P., P.L., A.M., C.T., and H.U.; formal analysis, N.P., C.T., P.R., and H.U.; investigation, N.P., P.L., and A.M.; resources, H.U., S.M., A.F., and I.A.; data curation, N.P. and H.U.; writing—original draft preparation, N.P. and H.U.; writing—review and editing, N.P., P.R., and H.U.; visualization, N.P. and P.R.; supervision, H.U. and S.M..; funding acquisition, H.U., S.M., A.F., and I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundacion para la Innovacion Agraria (FIA), grant number PYT20150093 and by the Agencia Nacional de Investigación y Desarrollo (ANID) Of Chile through the Fondef ID N° 19i10382.
Availability of data and materials
The sequencing data generated in this study are submitted to the Gene-Bank Database (https://www.ncbi.nlm.nih.gov/nuccore, accessions numbers: KY242591, KY242592, KY242593, KY242596, KY242590, KY296348, KY242594, KY242595, KY228978 and KY242597).
Solanum tuberosum Pukará-INIA is available according to the number 1-20-09-2016-G0-128 of the Registry of Varieties Suitable for Certification of Chile, (https://www.sag.gob.cl/).
Solanum tuberosum (native from the Chiloe region, Chile) was obtained from the Chilean germplasm network belonging to the National Institute of Agricultural Research (https://web.inia.cl/blog/tag/red-de-bancos-de-germoplasma/). Accession number Ch. P11_9.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.FAO. Food and Agriculture Organization of the United Nations . Faostat statistical database, Potatoes Crop Production, and Yield. Rome: FAO; 2020. [Google Scholar]
- 2.Pérombelon M. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol J. 2002;51(1):1–12. doi: 10.1046/j.0032-0862.2001.Shorttitle.doc.x. [DOI] [Google Scholar]
- 3.Vreugdenhil D, Bradshaw J, Gebhardt C, Govers F, Taylor MA, MacKerron DK, et al. Potato biology and biotechnology: advances and perspectives. Wageningen: Elsevier; 2011.
- 4.Mantsebo CC, Mazarura U, Goss M, Ngadze E. The epidemiology of Pectobacterium and Dickeya species and the role of calcium in postharvest soft rot infection of potato (Solanum tuberosum) caused by the pathogens: a review. Afr J Agric Res. 2014;9(19):1509–1515. doi: 10.5897/AJAR2013.8558. [DOI] [Google Scholar]
- 5.Plant KP, LaPatra SE. Advances in fish vaccine delivery. Dev Comp Immunol. 2011;35(12):1256–1262. doi: 10.1016/j.dci.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Hauben L, Moore ER, Vauterin L, Steenackers M, Mergaert J, Verdonck L, et al. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst Appl Microbiol. 1998;21(3):384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- 7.Charkowski AO. Biology and control of Pectobacterium in potato. Am J Potato Res. 2015;92(2):223–229. doi: 10.1007/s12230-015-9447-7. [DOI] [Google Scholar]
- 8.Toth IK, Bell KS, Holeva MC, Birch PR. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol. 2003;4(1):17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 9.Khaskheli MA, Wu L, Chen G, Chen L, Hussain S, Song D, et al. Isolation and characterization of root-associated bacterial endophytes and their biocontrol potential against major fungal phytopathogens of rice (Oryza sativa L.) Pathogens. 2020;9(3):172. doi: 10.3390/pathogens9030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Põllumaa L, Alamäe T, Mäe A. Quorum sensing and expression of virulence in Pectobacteria. Sensors. 2012;12(3):3327–3349. doi: 10.3390/s120303327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czajkowski R, Perombelon MC, van Veen JA, van der Wolf JM. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review. Plant Pathol J. 2011;60(6):999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. [DOI] [Google Scholar]
- 12.Vreugdenhil D. The canon of potato science: 39. Dormancy Potato Res. 2007;50(3-4):371. doi: 10.1007/s11540-008-9068-3. [DOI] [Google Scholar]
- 13.Compant S, Duffy B, Nowak J, Clément C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71(9):4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez-Pacheco MM, Bernal-Mercado AT, Vázquez-Armenta FJ, González-Aguilar G, Lizardi-Mendoza J, Madera-Santana T, et al. Quorum sensing interruption as a tool to control virulence of plant pathogenic bacteria. Physiol Mol Plant Pathol. 2019;106:281–291. doi: 10.1016/j.pmpp.2019.04.002. [DOI] [Google Scholar]
- 15.Campbell R, Campbell R. Biological control of microbial plant pathogens. Cambridge: Cambridge University Press; 1989.
- 16.İnceoğlu Ö, Al-Soud WA, Salles JF, Semenov AV, van Elsas JD. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One. 2011;6(8):e23321. doi: 10.1371/journal.pone.0023321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pageni BB, Lupwayi NZ, Larney FJ, Kawchuk LM, Gan Y. Populations, diversity and identities of bacterial endophytes in potato (Solanum tuberosum L.) cropping systems. Can J Plant Sci. 2013;93(6):1125–1142. doi: 10.4141/cjps2013-166. [DOI] [Google Scholar]
- 18.Bulgarelli D, Schlaeppi K, Spaepen S, Van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 19.Dutta J, Thakur D. Evaluation of antagonistic and plant growth promoting potential of Streptomyces sp. TT3 isolated from tea (Camellia sinensis) rhizosphere soil. Curr Microbiol. 2020;1:–10. 10.1007/s00284-020-02002-6. [DOI] [PubMed]
- 20.Errakhi R, Lebrihi A, Barakate M. In vitro and in vivo antagonism of actinomycetes isolated from Moroccan rhizospherical soils against Sclerotium rolfsii: a causal agent of root rot on sugar beet (Beta vulgaris L.) J Appl Microbiol. 2009;107(2):672–681. doi: 10.1111/j.1365-2672.2009.04232.x. [DOI] [PubMed] [Google Scholar]
- 21.Bubici G, Marsico AD, D’Amico M, Amenduni M, Cirulli M. Evaluation of Streptomyces sp. for the biological control of corky root of tomato and Verticillium wilt of eggplant. Appl Soil Ecol. 2013;72:128–134. doi: 10.1016/j.apsoil.2013.07.001. [DOI] [Google Scholar]
- 22.Evangelista-Martínez Z. Isolation and characterization of soil Streptomyces species as potential biological control agents against fungal plant pathogens. World J Microbiol Biotechnol. 2014;30(5):1639–1647. doi: 10.1007/s11274-013-1568-x. [DOI] [PubMed] [Google Scholar]
- 23.Alper D, Güven K. Şahi̇n N. isolation, plant growth-promoting traits, antagonistic effects on clinical and plant pathogenic organisms and identification of actinomycetes from olive rhizosphere. Microb Pathog. 2020;104134. 10.1016/j.micpath.2020.104134. [DOI] [PubMed]
- 24.Dubey A, Malla MA, Kumar A, Dayanandan S, Khan ML. Plants endophytes: unveiling hidden agenda for bioprospecting toward sustainable agriculture. Crit Rev Biotechnol. 2020:1–22. 10.1080/07388551.2020.1808584. [DOI] [PubMed]
- 25.Pang Y, Liu X, Ma Y, Chernin L, Berg G, Gao K. Induction of systemic resistance, root colonisation and biocontrol activities of the rhizospheric strain of Serratia plymuthica are dependent on N-acyl homoserine lactones. Eur J Plant Pathol. 2009;124(2):261–268. doi: 10.1007/s10658-008-9411-1. [DOI] [Google Scholar]
- 26.Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ Microbiol. 2005;7(11):1809–1817. doi: 10.1111/j.1462-2920.2005.00889.x. [DOI] [PubMed] [Google Scholar]
- 27.Eljounaidi K, Lee SK, Bae H. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases–review and future prospects. Biol Control. 2016;103:62–68. doi: 10.1016/j.biocontrol.2016.07.013. [DOI] [Google Scholar]
- 28.Berg G, Grube M, Schloter M, Smalla K. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol. 2014;5:148. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Cocq K, Gurr SJ, Hirsch PR, Mauchline TH. Exploitation of endophytes for sustainable agricultural intensification. Mol Plant Pathol. 2017;18(3):469–473. doi: 10.1111/mpp.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fadiji A, Babalola O. Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi J Biol Sci. 2020. 10.1016/j.sjbs.2020.08.002. [DOI] [PMC free article] [PubMed]
- 31.Martínez-Hidalgo P, García JM, Pozo MJ. Induced systemic resistance against Botrytis cinerea by Micromonospora strains isolated from root nodules. Front Microbiol. 2015;6:922. doi: 10.3389/fmicb.2015.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marian M, Ohno T, Suzuki H, Kitamura H, Kuroda K, Shimizu M. A novel strain of endophytic Streptomyces for the biocontrol of strawberry anthracnose caused by Glomerella cingulata. Microbiol Res. 2020;234:126428. doi: 10.1016/j.micres.2020.126428. [DOI] [PubMed] [Google Scholar]
- 33.Reiter B, Pfeifer U, Schwab H, Sessitsch A. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 2002;68(5):2261–2268. doi: 10.1128/AEM.68.5.2261-2268.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sessitsch A, Reiter B, Berg G. Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can J Microbiol. 2004;50(4):239–249. doi: 10.1139/w03-118. [DOI] [PubMed] [Google Scholar]
- 35.Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol Ecol. 2005;51(2):215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Solano Solis J, Morales Ulloa D, Anabalón RL. Molecular description and similarity relationships among native germplasm potatoes (Solanum tuberosum ssp. tuberosum L.) using morphological data and AFLP markers. Electron J Biotechnol. 2007;10(3):436–443. doi: 10.2225/vol10-issue3-fulltext-14. [DOI] [Google Scholar]
- 37.Quiza L, St-Arnaud M, Yergeau E. Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front Plant Sci. 2015;6:507. doi: 10.3389/fpls.2015.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirling ET, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16(3):313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 39.Li Q, Chen X, Jiang Y, Jiang C. Morphological identification of actinobacteria. Actinobacteria-basics and biotechnological applications. Rijeka: InTech; 2016. pp. 59–86. [Google Scholar]
- 40.Chankhamhaengdecha S, Hongvijit S, Srichaisupakit A, Charnchai P, Panbangred W. Endophytic actinomycetes: a novel source of potential acyl homoserine lactone degrading enzymes. Biomed Res Int. 2013;2013. 10.1155/2013/782847. [DOI] [PMC free article] [PubMed]
- 41.Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 42.Singh R, Dubey AK. Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front Microbiol. 2018;9:1767. doi: 10.3389/fmicb.2018.01767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sessitsch A, Reiter B, Pfeifer U, Wilhelm E. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol Ecol. 2002;39(1):23–32. doi: 10.1111/j.1574-6941.2002.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 44.Barnett BA, Holm DG, Koym JW, Wilson RG, Manter DK. Site and clone effects on the potato root-associated core microbiome and its relationship to tuber yield and nutrients. Am J Potato Res. 2015;92(1):1–9. doi: 10.1007/s12230-014-9405-9. [DOI] [Google Scholar]
- 45.Pageni BB, Lupwayi NZ, Akter Z, Larney FJ, Kawchuk LM, Gan Y. Plant growth-promoting and phytopathogen-antagonistic properties of bacterial endophytes from potato (Solanum tuberosum L.) cropping systems. Can J Plant Sci. 2014;94(5):835–844. doi: 10.4141/cjps2013-356. [DOI] [Google Scholar]
- 46.Kõiv V, Roosaare M, Vedler E, Kivistik PA, Toppi K, Schryer DW, et al. Microbial population dynamics in response to Pectobacterium atrosepticum infection in potato tubers. Sci Rep. 2015;5:11606. doi: 10.1038/srep11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Someya N, Kobayashi YO, Tsuda S, Ikeda S. Molecular characterization of the bacterial community in a potato phytosphere. Microbes Environ. 2013;28(3):295–305. doi: 10.1264/jsme2.ME13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki KI, Ludwig W, et al. Bergey's manual® of systematic bacteriology: volume five the actinobacteria, part A and B. New York: Springer; 2012.
- 49.Alain K, Querellou J. Cultivating the uncultured: limits, advances and future challenges. Extremophiles. 2009;13(4):583–594. doi: 10.1007/s00792-009-0261-3. [DOI] [PubMed] [Google Scholar]
- 50.Castiblanco LF, Sundin GW. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J Integr Plant Biol. 2016;58(4):362–372. doi: 10.1111/jipb.12428. [DOI] [PubMed] [Google Scholar]
- 51.Richter-Heitmann T, Eickhorst T, Knauth S, Friedrich MW, Schmidt H. Evaluation of strategies to separate root-associated microbial communities: a crucial choice in rhizobiome research. Front Microbiol. 2016;7:773. doi: 10.3389/fmicb.2016.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baz M, Tran D, Kettani-Halabi M, Samri S, Jamjari A, Biligui B, et al. Calcium-and ROS-mediated defence responses in BY2 tobacco cells BY nonpathogenic Streptomyces sp. J Appl Microbiol. 2012;112(4):782–792. doi: 10.1111/j.1365-2672.2012.05248.x. [DOI] [PubMed] [Google Scholar]
- 53.Baz M, Lahbabi D, Samri S, Val F, Hamelin G, Madore I, et al. Control of potato soft rot caused by Pectobacterium carotovorum and Pectobacterium atrosepticum by Moroccan actinobacteria isolates. World J Microbiol Biotechnol. 2012;28(1):303–311. doi: 10.1007/s11274-011-0820-5. [DOI] [PubMed] [Google Scholar]
- 54.El Karkouri A, El Hassani FZ, El Mzibri M, Benlemlih M, El Hassouni M. Isolation and identification of an actinomycete strain with a biocontrol effect on the phytopathogenic Erwinia chrysanthemi 3937VIII responsible for soft rot disease. Ann Microbiol. 2010;60(2):263–268. doi: 10.1007/s13213-010-0036-1. [DOI] [Google Scholar]
- 55.Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender J-L. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv. 2014;32(6):1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Wu C, Zacchetti B, Ram AF, Van Wezel GP, Claessen D, Choi YH. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci Rep. 2015;5:10868. doi: 10.1038/srep10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong Y-H, Zhang X-F, Xu J-L, Zhang L-H. Insecticidal bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl Environ Microbiol. 2004;70(2):954–960. doi: 10.1128/AEM.70.2.954-960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong Y-H, Xu J-L, Li X-Z, Zhang L-H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci U S A. 2000;97(7):3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maisuria VB, Nerurkar AS. Interference of quorum sensing by Delftia sp. VM4 depends on the activity of a Novel N-Acylhomoserine lactone-acylase. PLoS One. 2015;10(9):e0138034. doi: 10.1371/journal.pone.0138034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velasco-Bucheli R, Hormigo D, Fernández-Lucas J, Torres-Ayuso P, Alfaro-Ureña Y, Saborido AI, et al. Penicillin acylase from Streptomyces lavendulae and aculeacin a acylase from Actinoplanes utahensis: two versatile enzymes as useful tools for quorum quenching processes. Catalysts. 2020;10(7):730. doi: 10.3390/catal10070730. [DOI] [Google Scholar]
- 61.Hong K-W, Koh C-L, Sam C-K, Yin W-F, Chan K-G. Quorum quenching revisited—from signal decays to signalling confusion. Sensors. 2012;12(4):4661–4696. doi: 10.3390/s120404661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toumatia O, Compant S, Yekkour A, Goudjal Y, Sabaou N, Mathieu F, et al. Biocontrol and plant growth promoting properties of Streptomyces mutabilis strain IA1 isolated from a Saharan soil on wheat seedlings and visualization of its niches of colonization. S Afr J Bot. 2016;105:234–239. doi: 10.1016/j.sajb.2016.03.020. [DOI] [Google Scholar]
- 64.Kubheka GC, Coutinho TA, Moleleki N, Moleleki LN. Colonization patterns of an mCherry-tagged Pectobacterium carotovorum subsp. brasiliense strain in potato plants. Phytopathology. 2013;103(12):1268–1279. doi: 10.1094/PHYTO-02-13-0049-R. [DOI] [PubMed] [Google Scholar]
- 65.Coombs JT, Franco CM. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69(9):5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaewkla O, Franco CM. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb Ecol. 2013;65(2):384–393. doi: 10.1007/s00248-012-0113-z. [DOI] [PubMed] [Google Scholar]
- 67.Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol. 1987;65(5):501–509. doi: 10.1016/0385-6380(87)90108-7. [DOI] [Google Scholar]
- 68.Shimizu M. Endophytic actinomycetes: biocontrol agents and growth promoters. In: Bacteria in agrobiology: Plant growth responses. Berlin: Springer; 2011. p. 201–20.
- 69.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7-8):518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar V, Bisht GS, Institu S. An improved method for isolation of genomic DNA from filamentous actinomycetes. Int J Eng Technol Manag Appl Sci. 2010;2:2. [Google Scholar]
- 72.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 74.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. doi: 10.1086/286013. [DOI] [PubMed] [Google Scholar]
- 75.Sugathan S, Manilal A, Selvin J, Idhayadhulla A, Kumar RS, Panikkar M. Evaluating the antagonistic potential of seaweed-associated marine bacteria collected from the southwest coast of India. Asian J Anim Vet Adv. 2012;7:578–587. doi: 10.3923/ajava.2012.578.587. [DOI] [Google Scholar]
- 76.Lee S, Lee DK. What is the proper way to apply the multiple comparison test? Korean J Anesthesiol. 2018;71(5):353. doi: 10.4097/kja.d.18.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jafra S, Przysowa J, Czajkowski R, Michta A, Garbeva P, Van der Wolf J. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can J Microbiol. 2006;52(10):1006–1015. doi: 10.1139/w06-062. [DOI] [PubMed] [Google Scholar]
- 78.Krzyzanowska D, Potrykus M, Golanowska M, Polonis K, Gwizdek-Wisniewska A, Lojkowska E, et al. Rhizosphere bacteria as potential biocontrol agents against soft rot caused by various Pectobacterium and Dickeya spp. strains. J Plant Pathol. 2012;94(2):367–378. [Google Scholar]
- 79.Zamani M, Behboudi K, Ahmadzadeh M. Quorum quenching by Bacillus cereus U92: a double-edged sword in biological control of plant diseases. Biocontrol Sci Tech. 2013;23(5):555–573. doi: 10.1080/09583157.2013.787046. [DOI] [Google Scholar]
- 80.Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T. N-acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol Lett. 2008;279(1):124–130. doi: 10.1111/j.1574-6968.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- 81.Volkova L, Urmantseva V, Burgutin A. Stress-protective effect of phenylpropanoid complex on potato plants in vitro. Russ J Plant Physiol. 2014;61(2):255–261. doi: 10.1134/S1021443714010166. [DOI] [Google Scholar]
- 82.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Compant S, Muzammil S, Lebrihi A, Mathieu F. Visualization of grapevine root colonization by the Saharan soil isolate Saccharothrix algeriensis NRRL B-24137 using DOPE-FISH microscopy. Plant Soil. 2013;370(1-2):583–591. doi: 10.1007/s11104-013-1648-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data generated in this study are submitted to the Gene-Bank Database (https://www.ncbi.nlm.nih.gov/nuccore, accessions numbers: KY242591, KY242592, KY242593, KY242596, KY242590, KY296348, KY242594, KY242595, KY228978 and KY242597).
Solanum tuberosum Pukará-INIA is available according to the number 1-20-09-2016-G0-128 of the Registry of Varieties Suitable for Certification of Chile, (https://www.sag.gob.cl/).
Solanum tuberosum (native from the Chiloe region, Chile) was obtained from the Chilean germplasm network belonging to the National Institute of Agricultural Research (https://web.inia.cl/blog/tag/red-de-bancos-de-germoplasma/). Accession number Ch. P11_9.