Abstract
Cds1, a serine/threonine kinase, enforces the S-M checkpoint in the fission yeast Schizosaccharomyces pombe. Cds1 is required for survival of replicational stress caused by agents that stall replication forks, but how Cds1 performs these functions is largely unknown. Here we report that the forkhead-associated-1 (FHA1) protein-docking domain of Cds1 interacts with Mus81, an evolutionarily conserved damage tolerance protein. Mus81 has an endonuclease homology domain found in the XPF nucleotide excision repair protein. Inactivation of mus81 reveals a unique spectrum of phenotypes. Mus81 enables survival of deoxynucleotide triphosphate starvation, UV radiation, and DNA polymerase impairment. Mus81 is essential in the absence of Bloom's syndrome Rqh1 helicase and is required for productive meiosis. Genetic epistasis studies suggest that Mus81 works with recombination enzymes to properly replicate damaged DNA. Inactivation of Mus81 triggers a checkpoint-dependent delay of mitosis. We propose that Mus81 is involved in the recruitment of Cds1 to aberrant DNA structures where Cds1 modulates the activity of damage tolerance enzymes.
Genome integrity is vulnerable during DNA replication. The act of replication can convert a relatively benign single-strand DNA break to a cytotoxic double-strand break. A cyclobutane dimer, formed by UV irradiation, is sufficient to block the progression of DNA polymerases and to cause replication fork collapse (17). To cope with these problems, eukaryotic organisms have developed mechanisms for replicating DNA through and around damage in ways that cause minimal genome instability. Notable among the proteins involved in these processes are lesion bypass polymerases that can replicate through cyclobutane dimers (45). Recombination and double-strand break repair enzymes are also important, participating in the direct repair of DNA structure abnormalities that arise during DNA replication or permitting continued replication around sites of damage (17). These systems, and perhaps others that remain undiscovered, collectively form a genome defense system that allows tolerance of damage during DNA replication.
In the fission yeast Schizosaccharomyces pombe, the checkpoint kinase Cds1 is thought to regulate DNA damage tolerance systems (26, 28, 31). Cds1 is the presumptive homolog of Rad53 in the budding yeast Saccharomyces cerevisiae and Cds1 (also called Chk2) in humans (7, 9, 16, 27). One of its functions is to delay the onset of mitosis when genome replication encounters difficulties (8, 26, 47). Cds1 enforces this S-M checkpoint by regulating the Cdc25 and Mik1 mitotic control proteins (2, 8, 18, 47). In addition, Cds1 regulates the way in which DNA is replicated under conditions that cause replicational stress. For example, Cds1 is required to slow the replication of DNA that has been damaged by UV irradiation (26, 34). A similar function has been ascribed to Rad53 in budding yeast (32). Slowing of DNA replication partly involves the suppression of late-firing replication origins (35, 37). Activation of Cds1 or Rad53 has been linked to changes in the phosphorylation states of several different proteins (4, 11, 20, 21, 24, 33, 36, 38, 44), some of which are involved in DNA replication. Notably, human Cds1 has been implicated in the phosphorylation of BRCA1, a protein involved in DNA repair (24). BRCA1 does not exist in yeast; hence, evolutionarily conserved targets of Cds1 remain to be discovered. Indeed, the actual roles of Cds1 in DNA damage tolerance and of the proteins that it interacts with remain obscure.
Rad53 contains two forkhead-associated (FHA) domains (41). FHA1 is located near the amino terminus and is conserved in human Cds1 and fission yeast Cds1, while FHA2 is in a carboxyl-terminal extension that is unique to Rad53. FHA2 interacts with Rad9, a protein that is essential for the G2-M DNA damage checkpoint in budding yeast (41). We report here the initial result of a search for proteins that interact with the FHA1 domain of fission yeast Cds1. This search has uncovered a link between Cds1 and Mus81, a novel DNA damage tolerance protein that is highly conserved among eukaryotes.
MATERIALS AND METHODS
Yeast two-hybrid and UV assays.
The Cds1 FHA domain (residues 1 to 155) was amplified by PCR using primers AL56 (5′-CATATGGCCATGGCCATGGAAGAACCAGAAGAAGC-3′) and AL57 (5′-CGGGATCCTTAATCTCGTGAATGTTAAC-3′). The PCR product was digested with NcoI and BamHI and ligated into NcoI-BamHI-digested pAS2 to create pAL78. S. cerevisiae strain HF7c was transformed with pAL78 to generate strain AL319. The expression of the fusion protein gal4(1-147)HA-Cds1FHA was monitored in the strain AL319 by immunoblot analysis using antibodies against the hemagglutinin (HA) epitope. A library (a gift from S. Elledge, Baylor College) of S. pombe cDNAs expressed 3′ of the Gal4 activation domain in pACT was transformed into strain AL319. HIS3/3-AT selection gave more than 106 transformants that were subsequently screened for high lacZ expression. Mus81-myc was generated as described previously (3). Mus81-myc cells are indistinguishable from wild-type cells, including the response to UV and hydroxyurea (HU) treatment (our unpublished data). All UV sensitivity assays (see Fig. 1, 3, and 5) were performed two or more times, and representative results are shown.
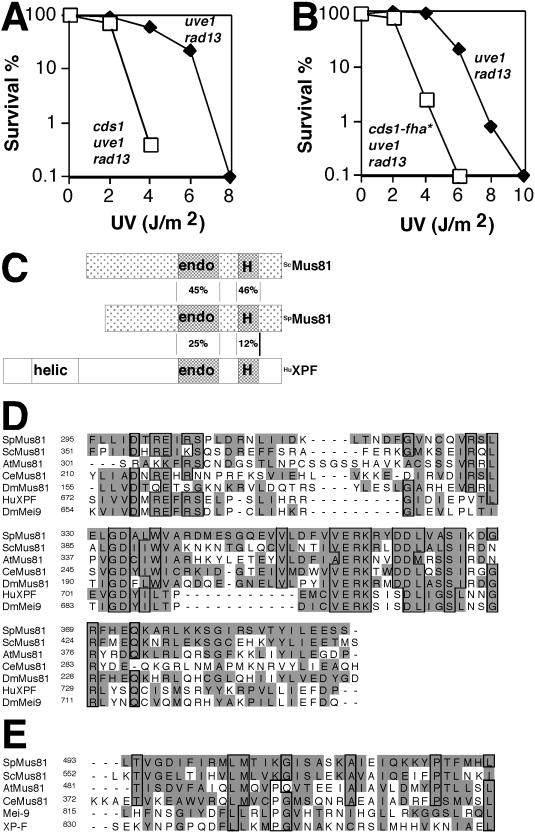
FIG. 1.
Identification of Mus81. (A) Cds1 is important for DNA damage tolerance. The uve1 rad13 and cds1 uve1 rad13 strains were assayed for UV survival (all results shown for UV sensitivity assays are representative examples of two or more experiments). (B) The cds1-fha* allele impairs DNA damage tolerance. uve1 rad13 and cds1-fha* uve1 rad13 cells were assayed for UV survival. (C) Schematic representation of Mus81 from S. pombe (SpMus81) and S. cerevisiae (ScMus81) and human XPF (HuXPF) (572, 632, and 905 amino acids, respectively). Percent identities between putative endonuclease (endo) and helix-hairpin-helix (H) domains are shown. The nonconserved helicase domain of XPF is shown (helic). Light shading depicts regions sharing more than 27% identity. (D) Alignment of endonuclease domains of Mus81 homologs and XPF family members. Sp, S. pombe; Sc, S. cerevisiae; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster, Hu, human. Shading highlights homologous residues, and the boxes show identities (PSI-BLAST results; alignment with ClustalW). (E) Alignment of the helix-hairpin-helix domains of Mus81 homologs and XPF family members.
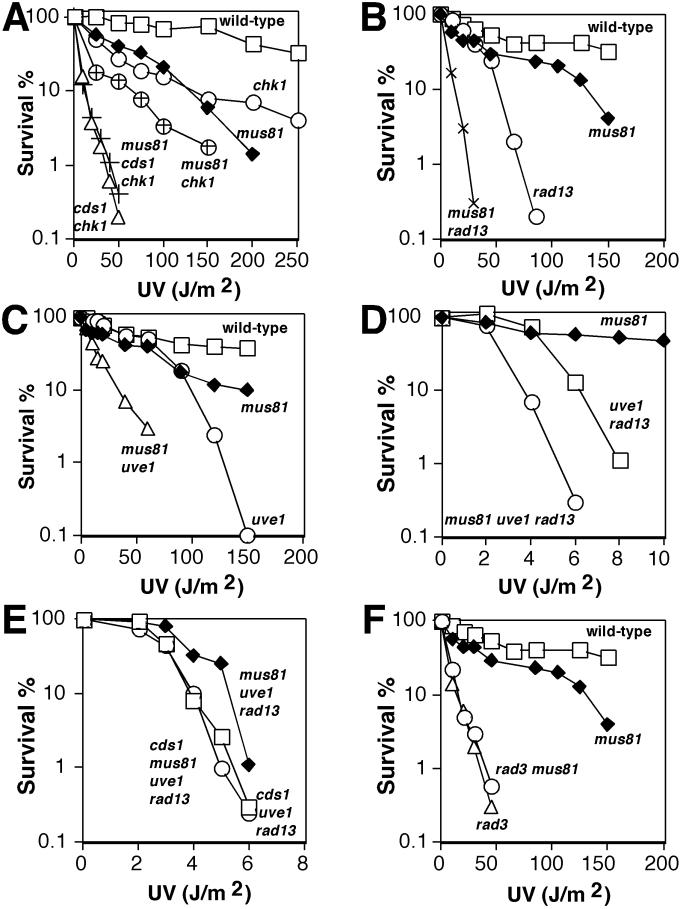
FIG. 3.
Mus81 is important for tolerance of UV damage. (A) UV survival is impaired in a mus81 mutant. UV survival rates of wild-type, chk1, mus81, cds1 chk1, mus81 chk1, and mus81 cds1 chk1 cells were measured. (B) The mus81 mutation diminishes UV survival in a NER-defective rad13 strain. Wild-type, mus81, rad13, and mus81 rad13 cells were tested for UV survival. (C) The mus81 mutation impairs UV survival in a UVER-defective uve1 strain. Wild-type, mus81, uve1, and mus81 uve1 cells were tested for UV survival. (D) Mus81 contributes to UV survival in the absence of NER and UVER. mus81, uve1 rad13, and mus81 uve1 rad13 cells were assayed for UV survival. (E) Mus81 appears to function in a Cds1-dependent UV tolerance pathway. cds1 uve1 rad13, mus81 uve1 rad13, and cds1 mus81 uve1 rad13 cells were assayed for UV survival. (F) Mus81 appears to function in a Rad3-dependent pathway for UV survival. Wild-type, mus81, rad3, and rad3 mus81 cells were assayed for UV survival. All results shown for UV sensitivity assays are representative of two or more experiments.
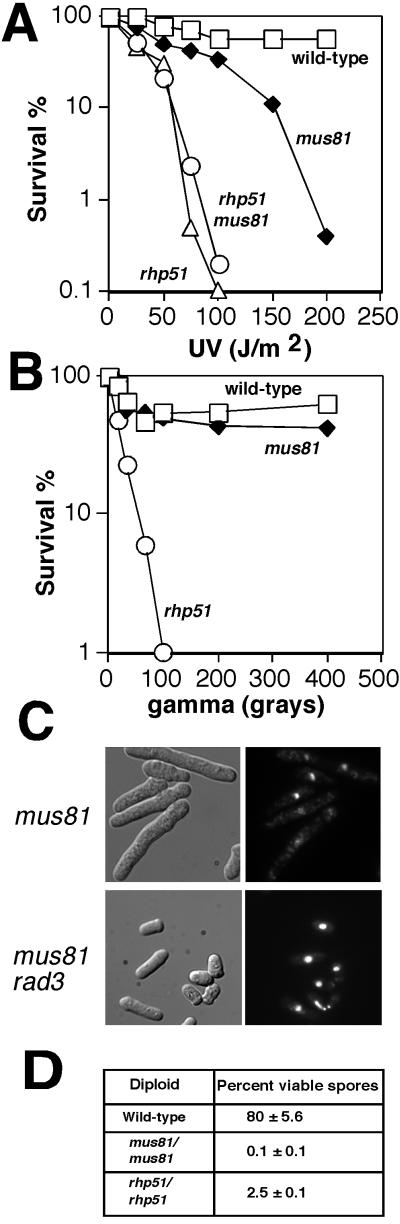
FIG. 5.
(A) Role of Mus81 in UV tolerance involves Rhp51-dependent recombination repair. Wild-type, mus81, rhp51, and rhp51 mus81 cells were tested for UV resistance. (B) Mus81 mutants are not significantly sensitive to ionizing radiation. Wild-type, mus81, and rhp51 cells were tested for resistance to ionizing radiation. All results shown for damage sensitivity assays are representative of two or more experiments. (C) mus81 cells are viable but display Rad3-dependent cell elongation. Mutant mus81 or mus81 rad3 cells were grown at 30°C in YES media, fixed in ethanol, and stained with DAPI to visualize DNA (right panels). Nomarski images are shown in the left panels. (D) Meiosis defect of Mus81. Wild-type diploids or diploids homozygous for mus81 or rhp51 were sporulated and plated on YES media to determine spore viability. Values are given as means ± standard deviations.
Protein and immunofluorescence techniques.
For all procedures cells were lysed in buffer A (50 mM Tris [pH 8], 150 mM NaCl, 2 mM EDTA, 10% glycerol, 0.2% Nonidet P-40, 5 μg of leupeptin, pepstatin, and aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride). Protein concentrations were normalized using optical density readings at a wavelength of 280 nm and resolved by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gels (acrylamide/bisacrylamide ratio, 200:1). Proteins were transferred to Immobilon membranes for Western blotting. Blots were probed with antibodies to the HA or myc epitopes following blocking of membranes in 5% milk in Tris-buffered saline and 0.3% Tween 20. The Cds1 kinase assay was performed as previously described (8). Phosphatase treatments were carried out with lambda phosphatase according to the guidelines in the New England Biolabs catalogue. The DNA stain DAPI (4′,6′-diamidino-2-phenylindole) was used to visualize nuclei. Cells were photographed using a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera. Images were acquired with IPlab Spectrum software (Signal Analytics Corporation).
Coimmunoprecipitations.
Cells were lysed in buffer A, and protein concentrations were normalized using optical density readings at a wavelength of 280 nm. A mixture of protein A-Sepharose and polyclonal anti-myc antiserum (Babco) was added to the lysates, followed by incubation at 4°C for 1.5 h. Complexes were collected by centrifugation and washed three times with lysis buffer before they were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Immunoblotting was performed as described above.
Strains.
Strains used in this study were as follows: PR109, wild type; NB2554, mus81::kanMx6; PS2345, rhp51::ura4+; PS2403, uve1::LEU2 rad13::ura4+; PS2402, uve1::LEU2; NR1587, rad13::ura4+; NB2573, cds1::ura4+ uve1::LEU2 rad13::ura4+; NB2574, cds1-fha* (a mutant allele; see below):2HA6His:ura4+:leu1+ uve1::LEU2 rad13::ura4+; NB2117, cds1::ura4+; NR1826, rad3::ura4+; NB2575, chk1::ura4+; BF2115, cds1::ura4+ chk1::ura4+; NB2566, mus81::kan chk1::ura4+; NB2555, mus81::kan rhp51::ura4+; NB2556, mus81::kan rad3::ura4+; NB2557, mus81::kan rad13::ura4+; NB2558, mus81::kan rad13::ura4+ uve1::LEU2; NB2559, mus81::kan uve1::LEU2; AL2228, pol1-1; AL2229, cdc6-23; NB2560, mus81::kan pol1-1; NB2561, mus81::kan cdc6-23; NB2562, cds1-fha*:2HA6His:ura4+:leu1+; NB2576, cds1:2HA6His:ura4+ mus81:13myc:kan; NB2564, cds1-kd:2HA6His:ura4+ mus81:13myc:kan; NB2577, cds1-fha*:2HA6His:ura4+:leu1+ mus81:13myc:kan; NB2565, cds1::ura4+ mus81:13myc:kan; NB2578, mus81:HA nmt1-GST:cds11-190; NB2579, mus81:HA nmt1-GST:cds1-fha*1-190; NB2580, cds1:2HA6His:ura4+ nmt1-GST:mus81-F; NB2581, cds1:2HA6His:ura4+ nmt1-GST:mus81-N; NB2582, cds1:2HA6His:ura4+ nmt1-GST:mus81-C; NB2583, cds1-kd:2HA6His:ura4+ nmt1-GST:mus81-F; NB2584, cds1-kd:2HA6His:ura4+ nmt1-GST:mus81-N; NB2585, cds1-kd:2HA6His:ura4+ nmt1-GST:mus81-C; NB2586, nmt1-GST:mus81-F; NB2587, nmt1-GST:mus81-N; NB2588, nmt1-GST:mus81-C; NB2589, mus81:13myc:kan; NB2590, mus81:13myc:kan cdc25-22.
RESULTS
Cds1 is important for tolerance of unrepaired UV lesions.
Replication of UV-damaged DNA is facilitated by lesion bypass polymerases and recombination enzymes. These enzymes do not excise UV photoproducts; hence, they function in what are termed damage tolerance mechanisms (17, 45). As defined in bacterial studies, damage tolerance proteins enhance the survival rates of mutant strains that cannot excise UV photoproducts. The survival of these mutants depends on the successful replication of the UV-damaged DNA, leading to a gradual dilution of the damage through cell division. It has been hypothesized that Cds1 is important for damage tolerance (31). To test this hypothesis, we determined whether Cds1 contributed to the survival of a mutant strain defective in nucleotide excision repair (NER) and UV damage excision repair (UVER). NER and UVER account for all detectable UV damage repair in fission yeast (46). NER was eliminated by the inactivation of rad13+, which encodes the homolog of S. cerevisiae Rad2p and human XPG. UVER was inactivated by a mutation of uve1+, which encodes the UV damage endonuclease. The cds1 mutation substantially enhanced the UV sensitivity of rad13 uve1 cells (Fig. 1A). These data support the proposition that Cds1 is important for UV damage tolerance. This finding correlates with the requirement for Cds1 in establishing an intra-S checkpoint (26, 34).
FHA1 domain is required for UV damage tolerance function of Cds1.
Cds1 contains an FHA1 domain hypothesized to mediate protein-protein interactions (15, 25, 41). To explore if the FHA1 domain is required for UV damage tolerance, cds1+ was replaced with cds1-fha*, an allele encoding mutations (S79A and H82A) at two highly conserved residues in the FHA domain. The cds1-fha* mutation substantially increased the UV sensitivity of rad13 uve1 cells (Fig. 1B). These data showed that the FHA1 domain is important for DNA damage tolerance.
FHA1 domain interacts with Mus81, a conserved DNA damage survival protein.
A yeast two-hybrid screen was performed with the region formed by amino acids 1 to 155 of Cds1, which contains the FHA1 domain. Three distinct but overlapping clones of a novel gene were identified. These clones failed to interact with a 1-to-155 construct that expressed mutant FHA (FHA*). Database searches revealed substantial identity to S. cerevisiae MUS81 (Fig. 1C). Budding yeast Mus81 interacts with the recombinational repair protein Rad54 in the two-hybrid system and leads to UV and methylmethanesulfonate sensitivity when inactivated (22). The yeast genes have uncharacterized sequence homologs in plants, nematodes, and fruit flies (Fig. 1D), as well as in humans (J. Vialard, personal communication).
Mus81 has two motifs found in the XPF family of NER endonucleases (1). Mutations of the human XPF gene ERRC1 cause a form of xeroderma pigmentosum (39). XPF homologs contain a highly conserved sequence, ERKX3D, which is proposed to form part of the endonuclease catalytic site (1). Mus81 contains this motif but lacks the amino-terminal helicase signature found in other members of the XPF family (Fig. 1D). Mus81 contains a predicted helix-hairpin-helix signature (amino acids 493 to 526) found in XPF and other proteins involved in DNA metabolism (Fig. 1E). A second helix-hairpin-helix signature has also been identified in the N terminus of Mus81 homologs (22). This domain is thought to allow nonspecific DNA binding via the phosphate backbone (14).
Coprecipitation of Cds1 and Mus81.
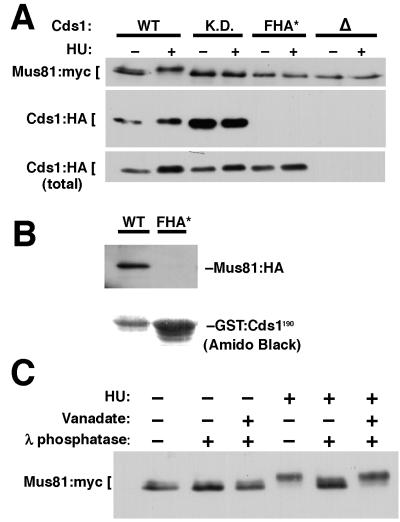
Immunoprecipitation studies established that Cds1 associates with Mus81 in vivo. Strains that expressed epitope-tagged Mus81:myc and Cds1:HA from genomic loci were used for these studies. Wild-type and kinase-dead Cds1:HA coprecipitated with Mus81:myc, whereas mutant Cds1-FHA*:HA did not (Fig. 2A). Cds1-FHA*:HA localization, stability, abundance, and solubility appeared to be equivalent to those of wild-type Cds1 (M. N. Boddy and P. Russell, unpublished data). Activation of Cds1:HA by HU treatment, which stalls replication forks by blocking deoxynucleotide triphosphate synthesis, did not significantly alter its interaction with Mus81:myc (Fig. 2A). For these studies we estimated that approximately 2 to 5% of the Cds1 was associated with Mus81 (M. N. Boddy and P. Russell, unpublished data). The association between Mus81 and Cds1 was confirmed in studies that expressed glutathione S-transferase (GST) fused to Mus81 or the Cds1 FHA domain in fission yeast. Mus81:HA coprecipitated with GST fused to the 1-to-190 region of Cds1 (GST-FHA1-190) but not with mutant GST-FHA*1-190 (Fig. 2B). A combination of two-hybrid and coimmunoprecipitation studies defined 141 amino acids in the N terminus of Mus81 (amino acids 175 to 314) capable of mediating the interaction with the Cds1 FHA domain (M. N. Boddy and P. Russell, unpublished data).
FIG. 2.
Mus81 and Cds1 associate in vivo. (A) Cells that expressed Mus81:myc and Cds1:HA from genomic loci were treated (+) or not treated (−) with HU. Immunoprecipitation with myc antibodies showed that Mus81:myc coprecipitated with Cds1:HA (WT) and Cds1 kinase dead (K.D.) but not with the Cds1 FHA mutant (FHA*). A Cds1 deletion strain (▵) served as a control. The bottom panel (total) is an immunoblot of Cds1:HA present in samples prior to immunoprecipitation. Note that lower-mobility forms of Mus81:myc were detected only in wild-type cells. (B) The 1-to-190 region of the Cds1 wild type (WT) or the FHA mutant (FHA*) was expressed from the nmt1 promoter as a GST fusion protein (GST:Cds1190) in a Mus81:HA strain. GST:Cds1190 proteins were purified and detected with amido black or immunoblotted with antibodies to HA. Mus81:HA coprecipitated with the wild-type but not with the mutant FHA domain. (C) Mus81 is a phosphoprotein. A Mus81:13myc strain was treated with HU (+) or not treated. (−) A Mus81:myc strain was immunoprecipitated and treated with λ phosphatase (+) or not treated, (−) either with (+) or without (−) the phosphatase inhibitor vanadate.
Cds1 regulates Mus81 phosphorylation in vivo.
Immunoblot analysis detected Mus81:myc as a closely migrating doublet (Fig. 2A). HU treatment caused Mus81:myc to migrate as a lower-mobility species, indicative of phosphorylation, as confirmed by phosphatase treatment (Fig. 2C). HU-induced phosphorylation of Mus81 was abolished in cds1 deletion, cds1-fha*, and cds1-kd (kinase-dead) strains (Fig. 2A). The lower-mobility form of Mus81:myc observed in asynchronous cells was also absent in the cds1 mutants. This species appears to be a phosphorylated form of Mus81 that is partially resistant to dephosphorylation by lambda phosphatase. A change in Mus81 mobility was not apparent for UV-irradiated cells (M. N. Boddy and P. Russell, unpublished data), but this result might be explained by the low activation rate of Cds1 in response to UV radiation compared to that in response to HU treatment (26).
Mus81 is important for DNA damage tolerance.
A mus81 deletion strain was viable but sensitive to UV irradiation (Fig. 3A). There are conflicting reports on the UV sensitivity of cds1 cells (26, 28). As previously observed (28), we have found that Cds1 mutant cells are not significantly sensitive to UV irradiation (M. N. Boddy and P. Russell, unpublished data). However, it is evident that Cds1 inactivation greatly enhances UV sensitivity in a chk1 mutant background (26) (Fig. 3A). Chk1 is essential for the G2-M DNA damage checkpoint (43); thus, the chk1 mutation highlights the importance of the DNA damage tolerance response during S phase (26). The mus81 and cds1 mutations reproducibly enhanced UV sensitivity in a chk1 background (Fig. 3A). Inactivation of mus81+ did not increase UV sensitivity in cds1 chk1 cells (Fig. 3A). The lack of synergy between mus81 and cds1 mutations indicated that Mus81 and Cds1 function in a related pathway. The enhanced UV sensitivity of cds1 chk1 cells relative to mus81 chk1 cells indicated that Mus81 is not the sole target of Cds1. The mus81 rad13 and mus81 uve1 mutants were more UV sensitive than any single mutant (Fig. 3B and C). Furthermore, the mus81 mutation enhanced the UV sensitivity of rad13 uve1 cells (Fig. 3D). These data implicated Mus81 in DNA damage tolerance. The mus81 mutation did not enhance the UV sensitivity in a cds1 rad13 uve1 background (Fig. 3E). As we observed in a chk1 background, the lack of synergy between mus81 and cds1 mutations in a rad13 uve1 background supported the conclusion that Cds1 and Mus81 function in a similar pathway. The Mus81 tolerance function appears to be checkpoint dependent, because mus81 did not exacerbate the UV sensitivity of a checkpoint-defective rad3 strain (Fig. 3F). Rad3 is required for Cds1 activity (8, 26).
Mus81 function was not restricted to UV tolerance. The mus81 mutant was sensitive to HU, although less sensitive than cds1 cells (Fig. 4A). A large fraction of the HU-treated mus81 cells were unable to exclude the vital stain phloxine B (M. N. Boddy and P. Russell, unpublished data), a result indicative of a role for Mus81 in HU survival. HU treatment activated Cds1 in mus81 cells (Fig. 4B), a finding which suggested that Mus81 acts downstream of Cds1. Thermosensitive alleles of DNA polymerase α or δ, but not ɛ, exhibited strong genetic interactions with mus81 (Fig. 4C). A similar genetic interaction was reported for cds1 and DNA polymerase α mutations (6). No interaction was observed between mus81 and cdc10-129 or cdc25-22, mutations that arrest cell cycle progression in G1 or G2, respectively.
FIG. 4.
Mus81 is important for survival under conditions that stall replication forks. (A) mus81 cells are sensitive to HU. Serial 10-fold dilutions (104 to 101) of cells were incubated on agar medium supplemented with no HU or 5 mM HU. (B) Cds1 is activated normally by HU treatment in mus81 cells. Wild-type (WT), mus81, and cds1 strains were incubated in the presence (+) or absence (−) of HU for 3 h. Cds1 activity was measured with the GST:Wee1152 substrate as previously described (6). SDS, sodium dodecyl sulfate. (C) The mus81 mutation lowers the restrictive temperature of thermosensitive DNA polymerase delta (polδts) (cdc6-23) and alpha (polαts) (pol1-1) alleles, shown at 28 and 33°C, respectively. The mus81 mutation does not lower the restrictive temperature of a polymerase epsilon (polɛts) (cdc20-m10) allele, shown at 33°C.
Many recombination proteins that are essential for the repair of double-strand breaks are also required for the repair of lesions that arise from replication of UV-damaged DNA. In fission yeast, the rhp51 mutation enhances the UV sensitivity of rad13 or uve1 cells (31). Moreover, we have confirmed that the rhp51 mutation also enhances the UV sensitivity of a rad13 uve1 double mutant which is unable to remove UV-induced lesions (M. N. Boddy and P. Russell, unpublished data). To evaluate if Mus81 and Rhp51 operate in the same pathway of UV damage tolerance, we measured the UV survival rate of a mus81 rhp51 double mutant. The mus81 mutation did not enhance the UV sensitivity of rhp51 cells (Fig. 5A). The mus81 single mutant exhibited intermediate sensitivity between that of wild-type and rhp51 cells. This result suggests that Mus81 functions in an Rhp51-dependent mechanism of UV damage tolerance.
UV radiation activates a Cds1-dependent intra-S-phase checkpoint that slows DNA synthesis (26, 34). Ionizing radiation, which causes DNA strand breaks, either does not activate this checkpoint or activates it very weakly (12, 34). We found that mus81 cells were insensitive to ionizing radiation (Fig. 5B). This phenotype contrasted with the profound sensitivity of rhp51 cells to ionizing radiation (Fig. 5B). These data suggest that Mus81, a presumptive endonuclease, is specifically required to cleave a class of DNA structures that form at stalled or collapsed replication forks. This activity is unnecessary for the repair of DNA breaks generated by ionizing radiation.
Mus81 is essential for viability in the absence of Rqh1.
The RecQ family of DNA helicases includes Bloom's syndrome protein in humans and Rqh1 in fission yeast. These proteins are important for maintaining genome integrity. Fission yeast rqh1 mutants have replication abnormalities that cause elevated recombination and sensitivity to HU (31, 40). It has been proposed that Rqh1 is also important for coping with stalled or collapsed replication forks (10). We found that rqh1 mus81 spores germinated but were inviable. These findings support the notion that Mus81 is important for correcting abnormal DNA structures that arise during replication. It is interesting that rqh1 rhp51 double mutants are viable (31). These results distinguish the functions of Mus81 and Rhp51 and suggest that the proteins may function both in partially dependent pathways, as in the case of UV damage tolerance, and independently, as indicated by their genetic interactions with rqh1 mutations.
Role of Mus81 in mitotic and meiotic divisions.
The mus81 deletion strain contained moderately elongated cells (Fig. 5C). This phenotype was reminiscent of rhp51, rhp54, and rhp55 recombination mutant cells that appear to trigger checkpoint arrest in the absence of extrinsic DNA-damaging agents (23, 29, 30). A mus81 rad3 culture had few elongated cells but frequent “cut” cells, in which DNA was unequally segregated to daughter cells, a phenotype that signals checkpoint failure (Fig. 5C). Hence, the mus81 mutation triggers a Rad3-dependent checkpoint delay of mitosis. It appears that Mus81, together with other recombinational repair enzymes, is important for the timely completion of DNA replication. Recombinational repair of collapsed replication forks that occur in the absence of DNA-damaging agents might explain why mus81 mutations trigger a Rad3-dependent checkpoint delay.
The role of Mus81 in meiosis was also investigated. In a mating of wild-type cells, approximately 80% of the resultant spores were viable (Fig. 5D). In contrast, approximately 0.1% of the spores from a mus81 × mus81 conjugation were viable (Fig. 5D). The effect of mus81 mutations on spore viability was even more severe than that observed in an rhp51 × rhp51 mating, in which approximately 2.5% of the spores germinated to produce colonies. The reason for the mus81 spore inviability has not been investigated, but it might arise from difficulties in either meiotic DNA replication or recombination.
DISCUSSION
Cds1 is best known and understood as a checkpoint kinase that delays mitosis when DNA synthesis is inhibited by HU (8, 26, 28). However, the S-M checkpoint activity of Cds1 is arguably not its most important function. This conclusion is based on the fact that cds1 cells exhibit very low viability when incubated in HU, even though mitosis is restrained by Chk1 (8, 26, 28). Thus, the recovery activity of Cds1 is centrally important for survival under replicational stress, but it is poorly understood. The function of Cds1 as a damage tolerance enzyme is even more mysterious. We undertook a screen to identify novel protein interactions involving Cds1, with the hope of ascertaining how Cds1 promotes damage tolerance and recovery from replicational stress. The central finding of this report is that Cds1 interacts physically with Mus81, a novel damage tolerance protein.
Cds1 is important for DNA damage tolerance.
DNA damage tolerance refers to mechanisms that facilitate successful replication of damaged DNA (17). If inactivation of a gene diminishes the UV survival of cells that are unable to repair UV lesions, then this gene can be considered to have importance in DNA damage tolerance. Cds1 was hypothesized to be involved in UV-induced DNA damage tolerance, but formal proof of this point, as defined above, was lacking (31). Thus, at the outset of these studies, it was essential to test whether cds1 mutations impair the UV survival of cells that cannot repair UV lesions. An affirmative answer was obtained in epistasis studies carried out with mutants defective for NER and UVER. Hence, we have provided formal evidence that Cds1 is important for DNA damage tolerance.
Using the same approach, we established that a functional FHA1 domain is required for the damage tolerance function of Cds1. This result provided the rationale for a yeast two-hybrid screen carried out with FHA1, which led to the identification of a protein that is highly related to budding yeast Mus81. The striking similarity to budding yeast Mus81 induced us to give the same name to the fission yeast protein. We have no convincing evidence that the two proteins are functionally analogous, but the possibility seems quite likely. Mus81 mutation in both yeasts results in sensitivity to UV but not ionizing radiation (this study and reference 22). Mus81 shares homology with the XPF family of endonucleases, suggesting a possible enzymatic activity for Mus81. Such an activity is speculative and remains to be established with biochemical assays. Mus81 is phosphorylated in a Rad3- and Cds1-dependent manner following exposure to HU. However, UV treatment does not result in a visible mobility change in Mus81 (Fig. 2C and data not shown). This may be due to the weaker activation of Cds1 by UV radiation than by HU treatment (26).
Although the phosphorylation state of Mus81 is dependent on Cds1, we have been unable to phosphorylate Mus81 with purified Cds1 in vitro (M. N. Boddy and P. Russell, unpublished data). This may be due to the absence of a cofactor or to the possibility that another Cds1-dependent kinase is responsible for the phosphorylation. At present, this precludes the mapping of phosphorylation sites on Mus81 and establishing a functional effect of such phosphorylation. It is interesting that the modification of Mus81 observed during an unperturbed cell cycle is S-phase specific and Cds1-Rad3 dependent (Fig. 2A and C) (M. N. Boddy and P. Russell, unpublished data). This strongly suggests that the replication checkpoint is activated by normal replication.
Mutant mus81 cells display phenotypes similar to and distinct from those of recombination repair mutants.
The role of recombination repair machinery in normal replication is becoming more apparent (19). In fission yeast, rhp54 (Rad54) cells exhibit a checkpoint-dependent delay in the cell cycle (30). This is true of other members of the Rad52 epistasis group of recombination repair proteins (42; M. N. Boddy and P. Russell, unpublished data). The defect in these cells appears to be manifested during replication. Mus81-defective cells, like recombination repair-defective mutants, show a checkpoint-dependent delay in the cell cycle. It is important to note that cds1 and rad3 cells, although slightly less viable than wild-type cells, show no profound cell cycle defect. This demonstrates that Mus81 has basal functions that are not dependent on the checkpoint proteins, including Cds1. Indeed, as also observed for recombination repair proteins, combining rad3 and mus81 mutations results in the accumulation of cut cells. This suggests that mus81 cells are defective in an aspect of DNA metabolism that, based on current data, is required for normal replication. Consistent with the similar phenotypes of Mus81 and recombination repair mutants, Mus81 appears to function in an Rhp51-dependent pathway for the tolerance of UV damage. The tolerance function may represent in part an augmentation of a mechanism that is required to repair stalled forks during normal replication. It is tempting to speculate that the Cds1-dependent phosphorylation of Mus81 stimulates or modulates this basal function.
A profound meiotic defect was also observed in both mus81 and rhp51 cells. Double-strand breaks are generated during meiosis, and recombination repair machinery is used to heal the breaks. This fact alone can explain the defect of rhp51 mutants in meiosis; however, a defect in meiotic replication cannot be excluded as a contributory factor. Interestingly, unlike recombination repair-defective rhp51 cells, mus81 cells are not defective in the repair of double-strand breaks induced by gamma irradiation. Therefore, Rhp51 and Mus81 appear to have both overlapping and distinct functions. These facts suggest that the meiotic defect of mus81 cells is in some way different from that of rhp51 cells. In fact, mus81 mutants show poorer spore viability than rhp51 mutants. Mus81 appears to be important for the mitotic S phase; therefore, the mus81 meiotic defect may be due to a problem in premeiotic replication. A role for Mus81 in resolving aberrant meiotic recombination structures cannot be excluded.
Genetic interactions with Rqh1 and DNA polymerase mutants.
The bacterial DNA helicase RecQ shares homology with a number of important helicases found in eukaryotes (40). This family includes the Bloom syndrome (BLM) and Werner syndrome (WRN) helicases that are mutated in human diseases that predispose patients to cancer. Fission yeast Rqh1 is closely related to BLM and shows similarly elevated levels of mitotic recombination (40). More recently, Rqh1 mutant cells have been suggested to accumulate the recombination intermediate termed X-DNA (Holliday junctions) (13). Rqh1 is proposed to catalyze reverse branch migration to prevent X-DNA accumulation in a nonrecombinogenic manner. In the absence of Rqh1, cells may resolve X-DNA structures by cleavage, resulting in recombinants. Interestingly, we observed that mus81 rqh1 double mutants are not viable. It is therefore possible that Mus81 is required for a step in the resolution of Holliday junctions (or other abnormal DNA structures) that accumulate in rqh1 cells. That Mus81 exhibits homology to a family of endonucleases may be relevant in this context. Further, we found that the deletion of Mus81 in cells containing thermosensitive alleles of DNA polymerases alpha and delta significantly reduced their restrictive temperatures (Fig. 4C). We found no such genetic interaction with a thermosensitive allele of DNA polymerase epsilon (Fig. 4C). Interestingly, X-DNA accumulates in S. cerevisiae mutants of DNA polymerases alpha and delta but not epsilon (48). Extrapolation of these results to fission yeast would provide support for the role of Mus81 in the resolution of X-DNA structures. Studies are under way to address these possibilities.
Conclusions.
We have uncovered a physical interaction between the evolutionarily conserved checkpoint kinase Cds1 and a novel damage tolerance protein, Mus81. Mus81 appears to function in a checkpoint- and recombination repair-dependent pathway for the tolerance of UV lesions. Mus81 is also required for normal cell cycle progression, potentially functioning during S phase to mitigate the recombinogenic properties of stalled replication forks. This basal role may be modified by interaction with Cds1 to promote the survival of lesions or conditions that impede the normal progression of replication forks, such as UV damage. It is noteworthy that human Cds1 is mutated in a subset of families that exhibit genetic inheritance of Li-Fraumeni cancer-prone syndrome (5). It will be important to determine if Cds1-defective cells derived from these patients exhibit the array of defects associated with inactivation of Cds1 in fission yeast and to consider the possibility that these defects might involve the human homolog of Mus81.
ACKNOWLEDGMENTS
We thank members of the Russell laboratory and the Scripps Cell Cycle Groups for their help and support. Strains were kindly provided by Antony Carr and A. Yasui, and the yeast two-hybrid library was kindly provided by S. Elledge.
M.N.B. was supported by a Special Fellowship from the Leukemia and Lymphoma Society. Work in W.-D. Heyer's laboratory was supported by the Swiss National Science Foundation. This work was funded by a National Institutes of Health grant awarded to P.R.
REFERENCES
- 1.Aravind L, Walker R W, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baber-Furnari B A, Rhind N, Boddy M N, Shanahan P, Lopez-Girona A, Russell P. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol Biol Cell. 2000;11:1–11. doi: 10.1091/mbc.11.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahler J, Wu J, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Phileppsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targetting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Bashkirov V I, King J S, Bashkirova E V, Schmuckli-Maurer J, Heyer W-D. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, Birch J M, Li F P, Garber J E, Haber D A. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 6.Bhaumik D, Wang T S F. Mutational effect of fission yeast polalpha on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. A human homolog of the checkpoint kinase Cds1 directly inhibits Cdc25. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 8.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 9.Brown A L, Lee C H, Schwarz J K, Mitiku N, Piwnica-Worms H, Chung J H. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc Natl Acad Sci USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraverty R K, Hickson I D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 11.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen P U, Bentley N J, Martinho R G, Nielsen O, Carr A M. Mik1 levels accumulate in S phase and may mediate an intrinsic link between S phase and mitosis. Proc Natl Acad Sci USA. 2000;97:2579–2584. doi: 10.1073/pnas.97.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claudette L D, Dixon J, Osman F, Whitby M C. Partial suppression of the fission yeast rqh1(−) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher D, Henckel J, Fersht A R, Jackson S P. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 16.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 18.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Cdc25 inhibited in vitro and in vivo by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber J E. DNA recombination: the replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 20.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 21.Huang M, Zhou Z, Elledge S J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 22.Interthal H, Heyer W-D. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 23.Khasanov F K, Savchenko G V, Bashkirova E V, Korolev V G, Heyer W-D, Bashkirov V I. A new recombinational DNA repair gene from Schizosaccharomyces pombe with homology to Escherichia coli RecA. Genetics. 1999;152:1557–1572. doi: 10.1093/genetics/152.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J S, Collins K M, Brown A L, Lee C H, Chung J H. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Smith G P, Walker J C. Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc Natl Acad Sci USA. 1999;96:7821–7826. doi: 10.1073/pnas.96.14.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay H, Griffiths D, Edwards R, Christensen P, Murray J, Osman F, Walworth N, Carr A. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka S, Huang M, Elledge S J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 28.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 29.Muris D F, Vreeken K, Carr A M, Broughton B C, Lehmann A R, Lohman P H, Pastink A. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muris D F, Vreeken K, Carr A M, Murray J M, Smit C, Lohman P H, Pastink A. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J Cell Sci. 1996;109:73–81. doi: 10.1242/jcs.109.1.73. [DOI] [PubMed] [Google Scholar]
- 31.Murray J M, Lindsay H D, Munday C A, Carr A M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 33.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Paolo Di Fiore P, Foiani M. Activation of rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santocanale C, Diffley J F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 36.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 37.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 38.Sidorova J M, Breeden L L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G, Hoeijmakers J H, Wood R D. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 40.Stewart E, Chapman C, Al-Khodairy F, Carr A, Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Hsiao J, Fay S F, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 42.Suto K, Nagata A, Murakami H, Okayama H. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol Biol Cell. 1999;10:3331–3343. doi: 10.1091/mbc.10.10.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 44.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 46.Yonemasu R, McCready S J, Murray J M, Osman F, Takao M, Yamamoto K, Lehmann A R, Yasui A. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:1553–1558. doi: 10.1093/nar/25.8.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 48.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]