Abstract
We have performed the fluorescently labeled amplified-fragment length polymorphism (FAFLP) method on 97 strains of the food-borne pathogen Salmonella enterica subsp. enterica comprising seven different serovars using the restriction enzymes EcoRI and MseI. From the total FAFLP fingerprinted strains, 81 were compared with pulsed-field gel electrophoresis (PFGE) typing of the same strains. The FAFLP method showed a discriminatory power equal to that of PFGE. We report a fast, robust, and high-resolution adaptation of the AFLP assay for fingerprinting S. enterica subsp. enterica serovars with capillary electrophoresis that can be scaled to high throughput on automated analysis instruments.
Salmonella enterica subsp. enterica is one of the main causes of human food-borne enteric infection in the world (4, 12). It is of great public health significance that strains of Salmonella can be rapidly identified and distinguished even within the same serovars. In this study, the amplified-fragment length polymorphism (AFLP) technique has been applied to the generation of genomic DNA fingerprints from Salmonella serovars. AFLP is a DNA fingerprinting method based on restriction cutting of DNA and stringent PCR amplification of the resulting fragments (14). The AFLP method generates fingerprints from DNA of both eukaryotic and prokaryotic origins without any prior knowledge of the sequence. The method involves the following steps: (i) digestion of DNA with a rare- and a frequent-cutter restriction enzyme and (ii) subsequent ligation with specific adapters to all restriction fragments. The resulting adapter-ligated fragments are templates for selective amplification with PCR primers directed at restriction-enzyme-specific target sequences on the adapters. The PCR primers can be labeled radioactively, and the fragments can be separated on sequencing gels (5, 14). We have used an approach where the primers were labeled with fluorescent dye (2). We then separated the dye-labeled fragments by capillary electrophoresis, which is currently one of the most suitable methods for fragment analysis and is now often used for typing small alterations at microsatellite loci (6, 7). The use of capillary electrophoresis with the addition of an internal standard enables us to size the fluorescent-AFLP (FAFLP) fragments with a resolution of ±1 bp. The internal standard also allows us to compare fingerprint patterns directly from different runs and to compare new patterns with previously stored fingerprints. Pulsed-field gel electrophoresis (PFGE) is currently the method of choice for typing Salmonella strains. PFGE has good discriminatory power (9) and has proven highly useful and reliable in outbreak situations (10, 11, 13). The PFGE method is, however, more labor-intensive than AFLP and more difficult to adapt for automation. We have chosen FAFLP as a new rapid genotyping method and combined it with the high resolution of capillary electrophoresis to obtain accurately sized fragments, which can be stored in a database for later comparisons and epidemiological analysis. To assess the performance of the FAFLP technique, we compared it with PFGE typing of the same Salmonella strains used for FAFLP.
Bacterial strains.
In all, 97 strains from seven different serovars of S. enterica subsp. enterica were used in this study. Strains from a major serovar Typhimurium outbreak were included. All strains used were obtained from the strain collection at the National Reference Laboratory for Enteropathogenic Bacteria at the National Institute of Public Health, Oslo, Norway, which serotypes bacterial enteropathogens isolated by microbiological laboratories throughout Norway.
FAFLP and PFGE analysis.
Genomic DNA was extracted using a commercial kit (Easy-DNA; Invitrogen BV, Leek, The Netherlands). We used a modification of the AFLP protocol first described by Vos et al. (14). The choice of the EcoRI-MseI enzymes was based on previous studies (1, 14). The combination which gave the fingerprint patterns reported here was the EcoRI(0)-MseI(C) primer combination. This resulted in fingerprints with bands up to about 500 bp in length. Several PCR protocols were tested with annealing temperatures from 56 to 65°C, and the effects of PCR preamplification steps were examined. The EcoRI and MseI adapters and primers were as previously published (14). The EcoRI PCR primer was 5′ labeled with the dye FAM (5-carboxyfluorescein). The XbaI adapters were 5′-CTA GCG TAC GCA GTC-3′ and 5′-CTC GTA GAC TGC GTA CG-3′. The XbaI PCR primer sequence was 5′-GAC TGC GTA CGC TAG A-3′ with 5′ FAM label. For the restriction cutting and ligation, 500 ng of genomic DNA was incubated at 37°C for 2 h in a 40-μl solution containing 1× One Phor All buffer (Pharmacia, Uppsala, Sweden) with 4 U of rare-cutting enzyme (EcoRI or XbaI), 4 U of frequent-cutting enzyme (MseI) (New England Biolabs, Beverly, Mass.), and 50 ng of bovine serum albumin per μl. After 2 h, a 10-μl ligation mix was added containing 5 pmol of EcoRI or XbaI adapters, 50 pmol of MseI adapter, 1 mM ATP, 1 U of T4 DNA ligase (New England Biolabs), and 50 ng of bovine serum albumin per μl in 1× One Phor All buffer (Pharmacia). Incubation was continued for 3 h at 37°C. After 3 h, 50 μl of Tris-EDTA buffer was added, to make the PCR template solution. One microliter of the PCR template solution was used in a 20-μl PCR mix containing 10 pmol of primer for rare-cutting enzyme (EcoRI or XbaI), 10 pmol of primer for frequent-cutting enzyme (MseI), 2 mM (each) deoxynucleoside triphosphate, and 0.4 U of Taq polymerase (Sigma, St. Louis, Mo.) in 1× Taq buffer supplied with enzyme. The PCR was carried out on a Perkin-Elmer GeneAmp PCR system 9700 (Perkin-Elmer Inc., Norwalk, Conn.). We used a high annealing temperature for the first 10 cycles (“touchdown” PCR) to ensure specific primer matches and to reduce PCR artifacts (3). The temperature profile was as follows (EcoRI-MseI): 95°C denaturation for 5 min followed by 10 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 45 s; then 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and finally a 5-min extension step at 72°C. The same profile was run for the XbaI-MseI combination with the annealing temperature set at 60°C for the first 10 cycles and 56°C for the next 30 cycles. The PCR product was diluted 1:2, and 1 μl was taken out for capillary electrophoresis on an ABI-310 Genetic Analyzer (Perkin-Elmer) with POP4-polymer and Genescan TAMRA-500 as internal standard in each sample (Perkin-Elmer).
A standard XbaI macrorestriction was subjected to PFGE (8). The DNA fragments were separated in 1% SeaKem GTG agarose (FMC Bioproducts, Rockland, Maine) with 0.25× modified Tris-borate-EDTA buffer for 22 h at 350 V and 12°C, with pulse times from 5 to 40 s, using a Beckman Gene Line II (Beckman, Fullerton, Calif.). Both the FAFLP and the PFGE fingerprints were analyzed visually. The FAFLP fingerprints were superimposed and visually compared with the GeneScan software, and the threshold for assigning a peak was set to 200 relative fluorescence (Perkin-Elmer).
Results and discussion.
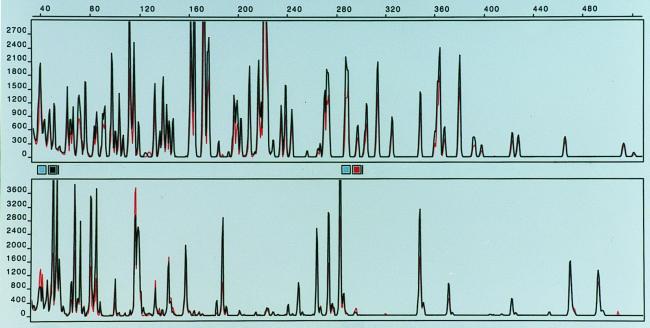
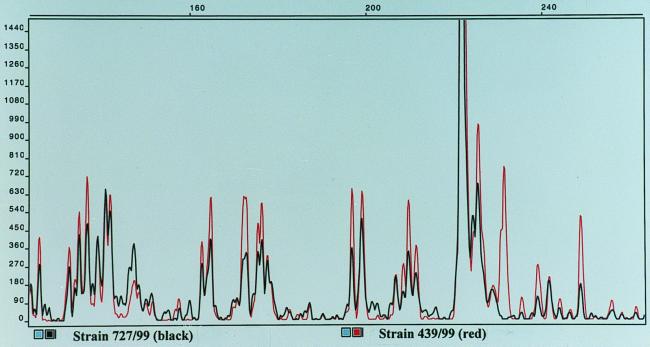
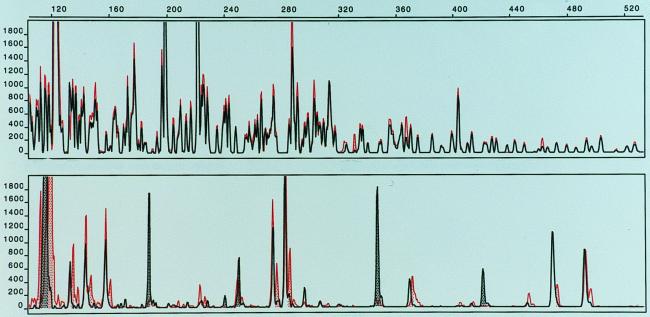
The FAFLP fingerprinting allowed 48 distinct patterns to be distinguished among the 97 Salmonella strains examined. The FAFLP fingerprints were clearly distinguishable between different serovars of Salmonella, and band variation within strains of the same serovar was used to identify specific genotypes. We report that the FAFLP method, using the EcoRI and MseI enzymes, generated about the same number of distinct fingerprint profiles as did PFGE for Salmonella. When PFGE analysis was performed on a selection of 81 strains comprising all 48 FAFLP patterns, we found 43 different profiles. The FAFLP and PFGE methods both resolved 74 of 81 (91.4%) strains into distinct and comparable profile groups in which the same strains were grouped together by both methods (Table 1). The methods differed only in five strains of serovar Dublin, one strain of serovar Enteritidis, and one strain of serovar Hindmarsh (Table 1). In Fig. 1, the FAFLP fingerprints from an isolate from a patient infected with serovar Typhimurium in an outbreak in western Norway were compared with those of a strain from the suspected source of infection. The patient strain displayed here, as well as the majority of isolated strains from the outbreak, showed perfect identity with the suspected source. Small variations in band intensities could sometimes be seen, as measured by the height of the peaks. Such variations were sometimes observed when DNA from the same strain was subject to separate FAFLP analyses. This could probably be the result of slightly different effectiveness of the cutting-ligation reaction or in the PCR amplification step; however, the band pattern, measured in sizes (base pairs) of the fragments, remained constant when DNA from the same strain was independently extracted and subjected to separate FAFLP reactions. Using PFGE, we could not discriminate between five unrelated strains of serovar Dublin, but these strains gave somewhat different FAFLP fingerprints (Table 1). In Fig. 2, we show the FAFLP fingerprint patterns for the serovar Dublin strains 439/99 and 727/99 superimposed. These strains gave identical PFGE profiles (data not shown). The FAFLP patterns differ by several low-intensity bands (below 200 relative fluorescence) and one major band (the area is enlarged for better view). The remaining serovar Dublin profiles were also separated by the position of one major band; thus, the serovar Dublin strains appeared very homogeneous, and the designation of six profiles based on one major band can be questioned, but is still reported because of the identical matches seen between the outbreak strains (Table 1 and Fig. 1). The serovar Enteritidis strain 978/98 was separated into a distinct pattern by PFGE, which was not seen with FAFLP. To further investigate if the serovar Enteritidis PFGE profile 15 could be discriminated from PFGE profile 10 (Table 1) by FAFLP, we used an additional rare-cutter restriction enzyme for the FAFLP analysis. The XbaI restriction enzyme was chosen because it was used for macrorestriction of DNA for PFGE which was able to discriminate these profiles (10 and 15). Figure 3 shows the fingerprint patterns of strain 355/98 (profile 10) and strain 978/98 (profile 15) superimposed. Our difficulty in typing serovar Enteritidis by AFLP is in agreement with a previous study on Salmonella in which strains of serovar Enteritidis with identical phage type specificities were indistinguishable by AFLP (1). The use of fluorescent dyes completely removes the need to work with radioactive isotopes, and there is no need for a gel fixation step and development of film. This combined with one PCR amplification step and a 5-h cutting-ligation reaction makes this a rapid fingerprinting method. In addition, there is no need for gel casting and waiting for polymerization with the ABI-310 apparatus, which further reduces time and labor.
TABLE 1.
Salmonella FAFLP and PFGE fingerprint profiles
| Serovar and strain no. | FAFLP profile
|
PFGE profile | Serovar and strain no. | FAFLP profile
|
PFGE profile | |||
|---|---|---|---|---|---|---|---|---|
| MseI(C)-EcoRI(0) | MseI(0)-XbaI(0) | MseI(C)- EcoRI(0) | MseI(0)- XbaI(0) | |||||
| Salmonella serovar Bovismorbificans | ||||||||
| 93/96 | E1 | NDa | 1 | |||||
| 583/96 | E2 | ND | 2 | |||||
| 2411/96 | E3 | ND | 3 | |||||
| 1425/97 | E4 | ND | 4 | |||||
| 45/98 | E5 | ND | 5 | |||||
| 807/98 | E6 | ND | 6 | |||||
| 2162/98 | E7 | ND | 7 | |||||
| 2514/98 | E8 | ND | 8 | |||||
| Salmonella serovar Dublin | ||||||||
| 727/99 | E9 | X1 | 9 | |||||
| 775/97 | E10 | X1 | 9 | |||||
| 846/97 | E11 | X1 | 9 | |||||
| 2873/98 | E12 | X1 | 9 | |||||
| 2987/98 | E13 | X1 | 9 | |||||
| 439/99 | E14 | X1 | 9 | |||||
| 491/99 | E14 | X1 | 9 | |||||
| Salmonella serovar Enteritidis | ||||||||
| 1000/98 | E15 | ND | 10 | |||||
| 978/98 | E15 | X6 | 15 | |||||
| 355/98 | E15 | X2 | 10 | |||||
| 1016/98 | E16 | ND | 11 | |||||
| 977/98 | E17 | X3 | 12 | |||||
| 962/98 | E18 | X4 | 13 | |||||
| 973/98 | E19 | X5 | 14 | |||||
| Salmonella serovar Hindmarsh | ||||||||
| 2161/98 | E20 | ND | 7 | |||||
| 3808/92 | E21 | ND | 16 | |||||
| 2178/94 | E22 | ND | 17 | |||||
| Salmonella serovar Minnesota | ||||||||
| St. 99 | E23 | ND | 18 | |||||
| St. 218 | E24 | ND | 19 | |||||
| 2797/85 | E25 | ND | 20 | |||||
| 1118/97 | E26 | ND | 21 | |||||
| E1699/87 | E27 | ND | 22 | |||||
| E1700/87 | E28 | ND | 23 | |||||
| BE837/97 | E29 | ND | 24 | |||||
| BE1006/97 | E29 | ND | 24 | |||||
| BE1037/97 | E29 | ND | 24 | |||||
| BE1053/97 | E29 | ND | 24 | |||||
| Salmonella serovar Oranienburg | ||||||||
| 2742/97 | E30 | ND | 25 | |||||
| 2654/97 | E30 | ND | 25 | |||||
| 1273/97 | E31 | ND | 26 | |||||
| 1592/97 | E31 | ND | 26 | |||||
| 1840/97 | E32 | ND | 27 | |||||
| 2262/97 | E33 | ND | 28 | |||||
| 310/97 | E34 | ND | 29 | |||||
| 479/97 | E35 | ND | 30 | |||||
| Salmonella serovar Typhimurium | ||||||||
| 2125/71 | E36 | ND | 31 | |||||
| 6313/76 | E36 | ND | 31 | |||||
| 374/82 | E36 | ND | 31 | |||||
| 659/73 | E37 | ND | 32 | |||||
| 177/81 | E37 | ND | 32 | |||||
| 178/81 | E37 | ND | 32 | |||||
| 652/82 | E37 | ND | 32 | |||||
| 124/83 | E37 | ND | 32 | |||||
| 717/83 | E37 | ND | 32 | |||||
| 177/84 | E37 | ND | 32 | |||||
| 94/85 | E37 | ND | 32 | |||||
| 289/86 | E37 | ND | 32 | |||||
| 2010/76 | E38 | ND | 33 | |||||
| 399/84 | E38 | ND | 33 | |||||
| 230/99b | E39 | ND | 34 | |||||
| 232/99b | E39 | ND | 34 | |||||
| 233/99b | E39 | ND | 34 | |||||
| 243/99b | E39 | ND | ND | |||||
| 244/99b | E39 | ND | 34 | |||||
| 245/99b | E39 | ND | 34 | |||||
| 247/99b | E39 | ND | ND | |||||
| 249/99b | E39 | ND | ND | |||||
| 250/99b | E39 | ND | ND | |||||
| 286/99b | E39 | ND | ND | |||||
| 288/99b | E39 | ND | ND | |||||
| 251/99b | E39 | X9 | ND | |||||
| 254/99b | E39 | ND | 34 | |||||
| 255/99b | E39 | X9 | 34 | |||||
| 256/99b | E39 | ND | ND | |||||
| 289/99b | E39 | ND | ND | |||||
| 290/99b | E39 | X9 | ND | |||||
| 291/99b | E39 | ND | ND | |||||
| 304/99b | E39 | X9 | ND | |||||
| 193/99b | E39 | X9 | 34 | |||||
| 549/99b | E39 | X9 | 34 | |||||
| 1964/96 | E39 | X9 | ND | |||||
| 1862/98 | E39 | ND | ND | |||||
| 3446/98 | E39 | ND | ND | |||||
| 246/99b | E40 | X7 | 35 | |||||
| 3019/98 | E40 | ND | 35 | |||||
| 287/99b | E41 | X8 | 36 | |||||
| 1990/96 | E42 | ND | 37 | |||||
| 217/96 | E42 | ND | 38 | |||||
| 1965/96 | E43 | ND | ND | |||||
| 435/99 | E43 | ND | 38 | |||||
| 436/99 | E43 | ND | 38 | |||||
| 437/99 | E43 | ND | 38 | |||||
| 1574/96 | E44 | ND | 39 | |||||
| 1805/98 | E45 | ND | 40 | |||||
| 2819/98 | E45 | ND | 40 | |||||
| 1609/96 | E46 | X11 | 41 | |||||
| 3635/93 | E46 | ND | 41 | |||||
| 2193/97 | E47 | ND | 42 | |||||
| 2232/97 | E48 | X10 | 43 | |||||
ND, not done.
Strains from a serovar Typhimurium outbreak in Norway in 1999.
FIG. 1.
FAFLP fingerprints of Salmonella serovar Typhimurium strains. (Top) EcoRI(0)-MseI(C) fingerprints of strains 549/99 (red) and 290/99 (black) superimposed. (Bottom) XbaI(0)-MseI(0) fingerprints of strains 549/99 (red) and 290/99 (black) superimposed.
FIG. 2.
Superimposed FAFLP fingerprints of Salmonella serovar Dublin strains.
FIG. 3.
FAFLP fingerprints of Salmonella serovar Enteritidis strains. (Top) EcoRI(0)-MseI(C) fingerprints of strains 978/98 (red) and 355/98 (black) superimposed. (Bottom) XbaI(0)-MseI(0) fingerprints of strains 978/98 (red) and 355/98 (black) superimposed.
In conclusion, we present a rapid adaptation of the FAFLP protocol coupled with capillary electrophoresis fragment separation. We show that the FAFLP method is discriminative at the level of PFGE with the possible exception of serovar Enteritidis intraserovar separation. The discriminatory power of FAFLP was increased when a second rare cutter (XbaI) was added. The ABI-310 apparatus allows for multiple dyes in the same run; thus, the XbaI-MseI and EcoRI-MseI reactions (with separate dyes) can be mixed before the electrophoresis and run simultaneously. A ±1-bp resolution in runs of about 30 min is achieved. With the introduction of new multicapillary instruments, this FAFLP method can be scaled to very high throughput.
REFERENCES
- 1.Aarts H J, van-Lith L A, Keijer J. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett Appl Microbiol. 1998;26:131–135. doi: 10.1046/j.1472-765x.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- 2.Desai M, Tanna A, Wall R, Efstratiou A, George R, Stanley J. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J Clin Microbiol. 1998;36:3133–3137. doi: 10.1128/jcm.36.11.3133-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez T M, Motarjemi Y, Miyagawa S, Kaferstein F K, Stohr K. Foodborne salmonellosis. World Health Stat Q. 1997;50:81–89. [PubMed] [Google Scholar]
- 5.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 6.Kleparnik K, Mala Z, Havac Z, Blazkova M, Holla L, Bocek P. Fast detection of a (CA)18 microsatellite repeat in the IgE receptor gene by capillary electrophoresis with laser-induced fluorescence detection. Electrophoresis. 1998;19:249–255. doi: 10.1002/elps.1150190218. [DOI] [PubMed] [Google Scholar]
- 7.Le H, Fung D C, Yu B, Trent R J. Capillary electrophoresis: new technology for DNA diagnostics. Pathology. 1998;30:304–308. doi: 10.1080/00313029800169496. [DOI] [PubMed] [Google Scholar]
- 8.Liu S L, Sanderson K E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992;174:1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–162. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 10.Ridley A M, Threlfall E J, Rowe B. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J Clin Microbiol. 1998;36:2314–2321. doi: 10.1128/jcm.36.8.2314-2321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor J P, Barnett B J, del-Rosario L, Williams K, Barth S S. Prospective investigation of cryptic outbreaks of Salmonella agona salmonellosis. J Clin Microbiol. 1998;36:2861–2864. doi: 10.1128/jcm.36.10.2861-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd E C. Epidemiology of foodborne diseases: a worldwide review. World Health Stat Q. 1997;50:30–50. [PubMed] [Google Scholar]
- 13.Van-Beneden C A, Keene W E, Strang R A, Werker D H, King A S, Mahon B, Hedberg K, Bell A, Kelly M T, Balan V K, Mac K W, Fleming D. Multinational outbreak of Salmonella enterica serotype Newport infections due to contaminated alfalfa sprouts. JAMA. 1999;281:158–162. doi: 10.1001/jama.281.2.158. [DOI] [PubMed] [Google Scholar]
- 14.Vos P, Hogers R, Bleeker M, Reijans M, van-de-Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]