Abstract
Background
Colorectal cancer (CRC) is one of the common gastrointestinal malignancies, tumor heterogeneity is the main cause of refractory CRC. Syndrome differentiation is the premise of individualized treatment of traditional Chinese medicine (TCM), but TCM syndrome lacks objective identification in CRC. This study is to investigate the correlation and significance of tumor heterogeneity and TCM syndromes classification in CRC.
Methods
In this study, we using scRNA-seq technology, investigate the significance of tumor heterogeneity in TCM syndromes classification on CRC.
Results
The results showed that 662 cells isolated from 11 primary CRC tumors are divided into 14 different cell clusters, and each cell subtype and its genes have different functions and signal transduction pathways, indicating significant heterogeneity. CRC tumor cell clusters have different proportions in Excess, Deficiency and Deficiency-Excess syndromes, and have their own characteristic genes, gene co-expression networks, gene functional interpretations as well as monocle functional evolution. Moreover, there were significant differences between the high expressions of MUC2, REG4, COL1A2, POSTN, SDPR, GPX1, ELF3, KRT8, KRT18, KRT19, FN1, SERPINE1, TCF4 and ZEB1 genes in Excess and Deficiency syndrome classification in CRC (P < 0.01).
Conclusions
The Excess and Deficiency syndromes classification may be related to tumor heterogeneity and its microenvironment in CRC.
Keywords: Excess and Deficiency syndromes classification, Colorectal cancer, Tumor heterogeneity, Tumor microcirculation, Single cell RNA sequencing
Background
Colorectal cancer (CRC) is one of the most common digestive tract cancers. It ranks third in the most common malignant tumors in the world. The incidence rate increased from 12 million cases in the early 70 s to 56 million cases now, with an annual growth rate of 4.2%, and mortality rate ranks second of the malignant tumor death spectrum [1, 2], seriously endangers human health. In addition to genetic and living habits, the heterogeneity of tumor and microenvironment may be the main internal cause in CRC [3, 4], it may be related to different cell subpopulations, gene status and phenotypes in tumors [5, 6].
High throughput cellular RNA sequencing technology has been widely used in transcriptome analysis to study transcriptional structure, splicing patterns, gene and transcriptional expression levels [7]. However, this sequencing method cannot be specific to a single cell, blurring the characteristics of different cell groups. Recently developed single cell RNA sequencing (scRNA-seq) technology is used to measure gene expression at the single cell level in cancer research [8]. It provides higher cell differential resolution than high-throughput RNA sequencing, and can analyze all cell types, gene expression profiles, characteristic genes and biological function evolution of all tumor cell types [9]. It has been applied to the study of tumor heterogeneity in some cancers, such as lung cancer [10], breast cancer [11], liver cancer [12] and CRC [13, 14].
Tumor heterogeneity is one of the characteristics of malignant tumors. In the process of tumor growth, after multiple divisions and proliferation of cells, its daughter cells show changes in molecular biology or genes, resulting in differences in tumor growth rate, invasion ability, sensitivity to drugs, prognosis, etc. [5, 6]. With the development of single-cell sequencing technology, it has been increasingly used for CRC tumor heterogeneity research. Recent researches have used single-cell RNA sequencing [14–20], single-cell DNA sequencing [21] and single-cell multi-omics [13, 22] technologies, from tumor cells [13–16], stem cells [17], circulating tumor cells (CTC) [18], granulocytes[13], T and B lymphocytes [19, 20], etc., explored their genetic heterogeneity, phenotypic heterogeneity, cell signaling pathway heterogeneity and single cell lineage development heterogeneity of tumor cells and their microenvironment in CRC. It improves our understanding of tumor heterogeneity in CRC, and may provide important information for clinical prognosis, identification of diagnostic markers, and personalized cancer treatment.
Treatment based on syndromes differentiation is a common method in the treatment of CRC in traditional Chinese medicine (TCM), the syndromes differentiation is the premise of individualized treatment in TCM, but the TCM syndrome, also called Zheng, is usually identified by the clinical manifestations of patients by the observation and the empirical judgment of TCM physicians, lack of objective detection and diagnosis [23]. Using single-cell sequencing technology to carry out biological detection can provide an objective data for TCM syndrome differentiation. This study used the second-generation scRNA-seq technology, Smart-seq2 to analyze the subgroup classification, characteristic genes and subgroup gene co-expression network, gene ontology (GO) and The Kyoto Encyclopedia of Genes and Genomes (KEGG) signal transduction pathways and changes in single-cell lineage development, explored the significance of tumor cells and their microcirculation heterogeneity in the TCM syndromes classification of CRC.
Materials and methods
Patients
In this study, 11 patients (7 males and 4 females) were enrolled at Shuguang hospital, Shanghai University of Traditional Chinese Medicine and Nanjing hospital of Traditional Chinese Medicine, Nanjing University of Traditional Chinese Medicine from March 2019 to February 2020. Among, the age ranged from 43 to 72 years, with a median of 64 years (Table 1). The diagnostic criteria of CRC follow the Diagnosis and Treatment Regulations on CRC (2010 Edition) issued by the Ministry of Health of the People’s Republic of China [24]. The TCM syndrome differentiation for CRC were according to the Guiding Principles for Clinical Research on New Drugs in TCM (3rd edition) [25] and TCM diagnostics (tenth edition). Every case was pattern identified by three TCM oncologists in associated chief position, and the collected data were assessed by Chi-square test for their consistency, and final judgment was made by a chief TCM oncologist. This research project was approved with the local ethics committee of Shuanghai University of TCM, and all patients were informed consent for this study.
Table 1.
Clinical characteristics and TCM syndrome types of CRC patients
| Patients (PT) |
Age | Sex | Histopathological diagnosis | Stages | TNM | Tumor sizes (CM) | TCM syndromes |
|---|---|---|---|---|---|---|---|
| PT1 | 69 | F | Rectal mushroom type | III b | T3N1bM0 | 4.0 × 3.5 | DES |
| PT2 | 43 | M | Transverse colon mushroom/ulcer type | II a | T3N0M0 | 4.7 × 3.5 | ES |
| PT3 | 49 | F | Ascending colon mushroom/ulcer type | II a | T3N0M0 | 9.0 × 8.5 × 4.0 | DS |
| PT4 | 58 | F | Rectal and sigmoid colon junction ulcer type | II a | T3N0M0 | 6.0 × 4.0 | ES |
| PT5 | 72 | F | Rectal ulcer type | III b | T3N2bM0 | 3.5 × 2.0 | DES |
| PT6 | 60 | M | Rectal ulcer type | II a | T3N0M0 | 7.5 × 6.0 × 0.8 | DS |
| PT7 | 64 | M | Rectal ulcer type | III b | T3N1M0 | 3.0 × 3.0 | ES |
| PT8 | 64 | M | Sigmoid colon mushroom/ulcer type | III b | T3N1bM0 | 6.5 × 4.0 | ES |
| PT9 | 68 | M | Rectal and sigmoid colon junction ulcer type | II a | T3N0M0 | 2.0 × 3.0 | ES |
| PT10 | 67 | M | Rectal ulcer type | II a | T3N0M0 | 2.5 × 2.5 | DS |
| PT11 | 51 | M | Rectal ulcer type | II a | T3N0M0 | 2.5 × 2.5 × 2.0 | DES |
Sample collection and preparation
The collected fresh tumor was from the above patients after surgical resection. Tumor tissue (TT) were transferred into pre-warmed DMEM medium containing 2 mg/ml collagenase P (Roche, USA) and 10 U/µl DNase I (Roche, USA), digested at 37 ℃, for 60 min, filter and centrifuge at 400 × g for 5 min, and then employed fluorescence activated cell sorting (FACS) to select live cells, After staining with 3 nM CFSE for 5 min, live single cells were manually selected.
ScRNA-seq library preparation and sequencing
ScRNA-seq was performed according to the manufacturer’s instructions of Smart-seq 2 (12). After lysis of single cells, reverse transcription was performed using Superscript reverse transcriptase (Takara, Japan) and locked TSO oligonucleotides, (Exiqon, Denmark). Full-length cDNA preamplification was conducted with 22 cycles of PCR amplification and HiFi-HotStart ReadyMix (KAPA Biosystems, USA). Subsequently, Ampure XP beads (Beckman, Brea, CA, USA.) were used for PCR purification. An Agilent high-sensitivity DNA chip was used to ensure the size and distribution of the cDNA library. Barcoded libraries were fragmented and tagged using a Nextera XT DNA sample preparation kit (Illumina). Then, we used reagents from the Nextera XT kit to amplify adapter-ligated fragments. Pooled libraries with unique N5-N7 barcodes were sequenced using a HiSeq 2500 instrument (Illumina) and single-end 50-bp read flow cells.
ScRNA-seq data pre-processing and quality control
Fastq reads were initially filtered using Trimmomatic. The remaining clean reads were aligned to UCSC human genome 19 (hg19) using Hisat 2. Next, we used Feature Counts software to quantify the expression of each gene, and counts were obtained for each sample. The expression level of each gene was converted to a transcript per million (TPM) value. Then, the expression values were log-normalized.
ScRNA-seq analysis
The ScRNA-seq analysis workflow included t-distribution stochastic neighbor embedding (tSNE) projection of each single cell, cluster analysis, identification of differentially expressed genes (DEGs) for each cell subpopulation, gene co-expression networks analysis, pathway enrichment analysis, and single cell trajectory analyses.
Unsupervised clustering and DEGs analyses
The expression tables of all cells were fed into an iteratively unsupervised clustering pipeline. We determined that the cells in the same cluster acted as the same subtype based on key genes mapping of different cell types using ‘Seurat’ package (V3.1.2). To assign gene markers for single cell clusters, DEGs were identified by calculating fold-change and p values between different groups using t-test method. We set a 1.5-fold cutoff of fold change and a false discovery rate (FDR) to P < 0.05, as the criteria for DEGs selection. This was determined using the ‘stats’ in R. DEGs heatmaps were generated with ‘Heatmap’ package (V1.0.12).
Gene co-expression network and pathway enrichment analyses
To construct the gene co-network, we constructed the network adjacency between genes, i and j, according to Pearson’s correlation between their expression profiles among cells. We then obtained the gene-to-gene co-expression adjacency matrix by computing the correlation co-efficient for these genes. Next, we selected the genes with high correlations (0.8 or greater) to draw a co-expression network graph using Cytoscape version 3.6.1. The pathway enrichment analysis was based on Gene Ontology Biological Processes (GOBP) andKEGG profiling by Metascape (http://metascape.org/) using P Value Cutoff 0.01. We selected a subgroup of representative genes from each cluster compared to the rest clusters for pathway enrichment and functional annotation.
Single cell trajectory analysis
To characterize the potential process of CRC cell functional changes and determine the potential lineage differentiation between diverse CRC cells, we applied Monocle 2 to perform a pseudo-time analysis of the evolution of CRC cells. Cells were chosen based on Seurat cluster identification results. All analyses were performed in R.
Statistical analysis
All data were expressed as means ± standard error of the mean, and the statistical analysis was performed using GraphPad Prism v8.0. Comparisons between groups were performed using a one-way ANOVA or Kruskal–Wallis or Wilcox test. Correlations between different gene expression profiles were examined by Pearson’s correlation analysis. Chi-square test was used to analyze the proportion of cell subpopulations. P < 0.05 was considered statistically significant.
Results
Clinical characteristics of patients
We generated scRNA-seq data from 662 cells isolated from tumor tissues of 11 patients with primary CRC, of which passed quality control. As shown in Table 1, CRCs were diagnosed pathologically, including 3 cases of colon cancer, 6 cases of rectal cancer, and 2 cases of borderline cancer of the rectum and colon. The clinical characteristics of these participants were recorded at the time of recruitment, including histopathological diagnosis, TNM stage, clinical stage, tumor size, and TCM syndrome type. According to TNM stage (AJCC 8 version), there were 7 cases in stage IIa and 4 cases in stage IIIb. All patients had a histopathological diagnosis of moderately differentiated adenocarcinoma. There were 5 cases in Excess syndrome (ES), 3 cases in Deficiency syndrome (DS), and 3 cases in Deficiency-Excess syndrome (DES).
Distribution of tumor cell subpopulations in CRC with TCM syndromes
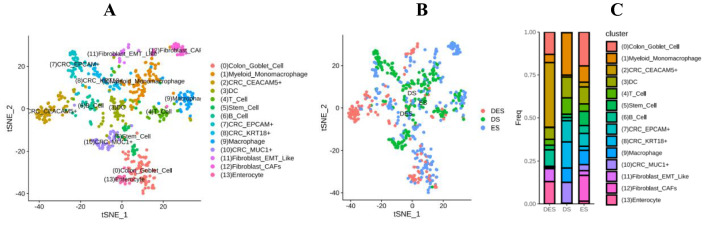
In order to clarify the relationship between CRC with TCM syndromes and the heterogeneity of intra-tumoral cells and their microenvironment, we analyzed the distribution of tumor single cell subpopulations of the three types of ES, DS and ESDS in CRC. The clustering analysis identified to partition the cells into 14 clusters. These clusters could be assigned to nine known cell lineages through marker genes, including marked with carcinoembryonic antigen-related cell adhesion molecules 5 (CEACAM5), epithelial cell adhesion molecules (EPCAM) +, Keratin 18 (KRT18) + and transmembrane glycoprotein Mucin 1 (MUC1) + 4 types of CRC cells, colon goblet cell, myeloid-derived monomacrophage, dendritic cells (DC), T lymphocytes, stem cells, B lymphocytes, macrophage, epithelial-mesenchymal transition (EMT) -like fibroblasts, EMT, cancer-related fibroblasts (CAFs) and enterocyte. The proportion of each cell lineage varies greatly among different samples (Fig. 1A and C), indicating there were the heterogeneity of single cells and their microenvironment in the tumor of CRC.
Fig. 1.
TSNE distribution of cell subpopulation and the proportion in CRC with Excess syndrome (ES), Deficiency syndrome (DS) and Excess-Deficiency syndrome (DES). A tSNE distribution of cell subpopulations; B the proportion of the three syndromes; C columnar graphic of cell subpopulations tSNE distribution and the three syndromes proportion
Moreover, we found that the percentages of cell subpopulations of CRC with ES, DS and DES were different (Table 2). The main distribution of cells in DES is CEACAM 5+ cells (37.67%). The main distribution in DS were myeloid-derived monomacrophages (24.71%), KRT18+ cells (15.21%), DC (12.17%) and MUC1+ cells (12.17%). Goblet cells (19.76%) and CAFs (15.02%) were the most distributed in ES (Fig. 1B and C).
Table 2.
Distribution of cell subpopulations in TCM syndrome types of CRC
| Cell types | Cell subpopulations | DES/Numbers (%) | DS/Numbers (%) | ES/Numbers (%) | P* |
|---|---|---|---|---|---|
| CRC cell | CEACAM5 + | 55 (37.67) | 3 (1.14) | 7 (19.76) | |
| EPCAM + | 0 (0.00) | 32 (12.17) | 19 (7.51) | ||
| KRT18 + | 1 (0.68) | 40 (15.21) | 6 (2.37) | ||
| MUC1 + | 1 (0.68) | 32 (12.17) | 9 (3.56) | ||
| Total | 57 (39.04) | 107 (40.68) | 41 (16.21) | < 0.001 | |
| Immune cell | Myeloid monomacrophage | 7 (4.79) | 65 (24.71) | 24 (9.49) | |
| DC | 10 (6.85) | 32 (12.17) | 25 (9.88) | ||
| T cell | 5 (3.42) | 25 (9.51) | 11 (4.35) | ||
| B cell | 14 (9.59) | 5 (1.90) | 11 (4.35) | ||
| Macrophage | 0 (0.00) | 22 (8.37) | 21 (8.30) | ||
| Total | 36 (24.66) | 149 (56.65) | 92 (36.36) | < 0.001 | |
| Stem cell | Stem cell | 4 (2.74) | 5 (1.90) | 21 (8.30) | < 0.005 |
| Fibroblast | EMT Like | 11 (7.53) | 0 (0.00) | 7 (2.77) | |
| CAFs | 0 (0.00) | 1 (0.38) | 38 (15.02) | ||
| Total | 11 (7.53) | 1 (0.38) | 45 (17.79) | < 0.001 | |
| Enterocyte | Enterocyte | 19 (13.01) | 0 (0.00) | 4 (1.58) | < 0.001 |
| Goblet cell | Goblet cell | 19 (13.01) | 1 (0.38) | 50 (19.76) | < 0.001 |
| Total | 14 | 146 | 263 | 253 |
*Chi-square test
In DES, the proportions of subpopulations of CRC cells, immune cells, stem cells, fibroblasts, colon epithelial cells and goblet cells accounted for 39.04, 24.66, 2.74, 7.53, 13.01, 13.01% respectively. In DS, that were 40.68, 56.65, 1.90, 0.38, 0 and 0.38%, respectively. In ES, the proportions were 16.21, 36.36, 8.30, 17.79, 1.58 and 19.76% respectively. Moreover, there were statistically significant differences (P < 0.005) (Table 2). It was indicated that the TCM syndrome classification in CRC may be related to the distributions of intra-tumor cell subpopulations and their heterogeneity.
Differential gene and functional annotation of tumor cell subpopulations in CRC with TCM syndromes
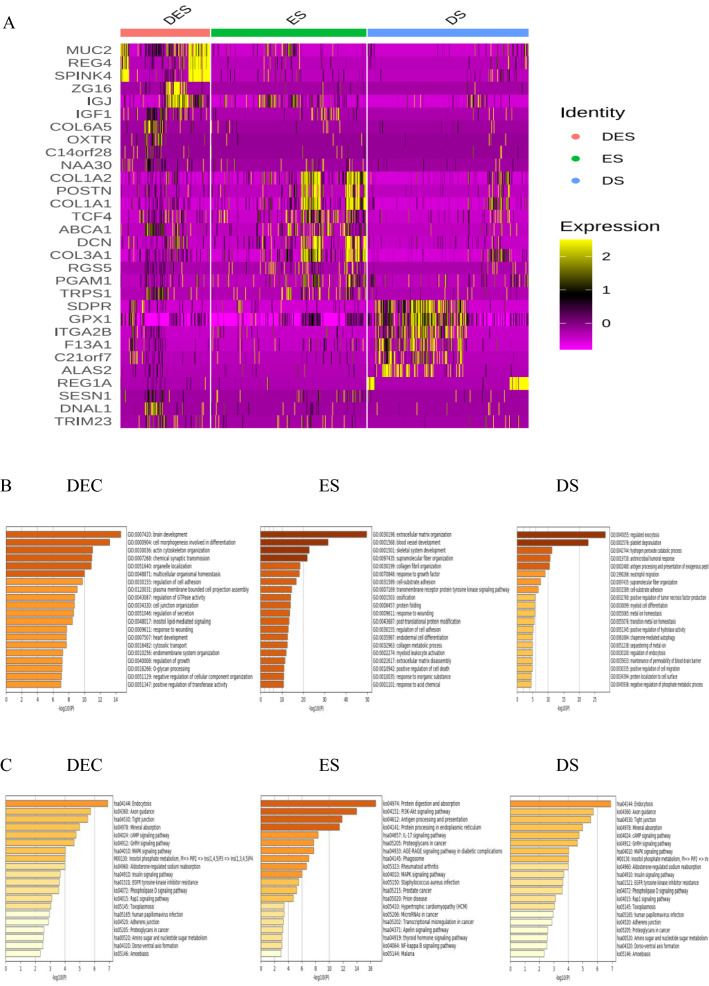
In order to clarify the relationship between TCM syndromes and cell phenotype in CRC, we analyzed theDEGs and functional annotations of tumor single cell subpopulations in CRC with TCM syndromes. We profiled the top 30 DEGs from distinct single cell subpopulations in CRC with the three types of syndromes using a heatmap (Fig. 2A). There were 686 DEGs (up-regulated 453, down-regulated 233) in ES, 1883 DEGs 1497 (up-regulated 1497, down-regulated 386) in DES and 2,295 DEGs (up-regulated 247, down-regulated 2048) in DS. As shown in Fig. 2, the high expressions of transmembrane glycoprotein Mucin 2 (MUC2) and regenerating islet-derived protein 4 (REG4) in DES were mainly related to brain development, cellogenesis, mitogen-activated protein kinase 1 (MAPK) and cyclic adenosine monophosphate (cAMP) signaling pathways. The high expressions of collagen type I alpha 2 (COL1A2) and periostin (POSTN) genes in ES were mainly related to vascular development and skeletal muscle development, and PI3k-Akt pathway. The high expressions of caveolae-associated protein 2 (CAVIN2 or SDPR) and glutathione peroxidase 1 (GPX1) genes in DS were mainly related to exocytosis, platelet decomposition and endocytosis (Fig. 2B and C), indicating that the TCM syndromes in CRC and may be related to the heterogeneity of genes and their function and signaling pathways in tumor cell subpopulations.
Fig. 2.
DEGs and functional annotations of tumor cells in CRC with Excess syndrome (ES), Deficiency syndrome (DS) and Excess-Deficiency syndrome (DES). A Top30 heat map of DEGs, in the expression level, yellower color was higher and redder color was lower; B GO/BP enrichment analysis; C KEGG signal pathway enrichment analysis
Co-expression network of high-expressed genes in tumor single cell of CRC with TCM syndromes
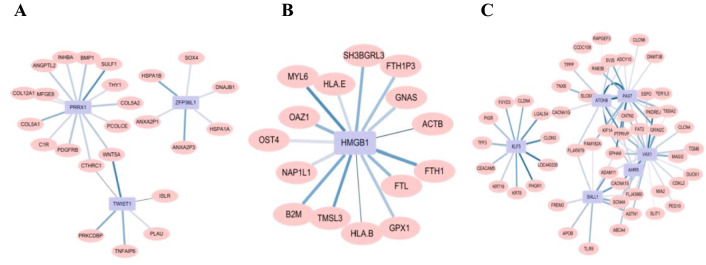
Further, we performed respectively the co-expression network analysis of high-expressed genes in ES, DS and DES with the transcription factor (TF) through spearman correlation, and screened them with a correlation coefficient greater than 0.5 or less than -0.5 and BH less than 0.05. The results were as shown in Fig. 3, the high expressed genes in the DES were mainly protein atonal homolog 8 (ATOH8), paired box protein Pax 7 (PAX7), ventral anterior homeobox 1 (VAX1), and their functions were mainly related to the reaction of purine-containing compounds, the secretion and decomposition of cortisol. The highly expressed gene in the DS was high mobility group protein B1 (HMGB1), and it related to iron ion transport and growth and development functions. The highly expressed genes in the ES were paired mesoderm homeobox protein 1 (PRRX1), mRNA decay activator protein ZFP36L (ZFP36L1) and twist-related protein 1 (TWIST1), and their functions were importantly related to embryogenesis and angiogenesis.
Fig. 3.
The high-expressed gene co-expression network in CRC with TCM syndromes. A Excess syndrome (ES); B Deficiency syndrome (DS); C Deficiency-Excess syndrome (DES). Pink ovals indicated highly expressed genes, and blue squares indicated TF. The lighter line color indicated lower correlation, and the thicker line indicated smaller p-value
Correlation of TCM syndromes and EMT-related DEGs of tumor single cells in CRC
In order to find potential molecular markers of TCM syndromes in CRC, we conducted Kruskal–Wallis test to screen keratin and EMT-related DEGs of tumor single cells in CRC with TCM syndromes. We found epithelium-specific ETS transcription factor 3 (ELF3), keratin 8, 18 and 19 (KRT8, 18 and 19), fibronectin 1 (FN1), serpin family Emember 1 (SERPINE1), transcription factor 4 (TCF4) and zinc finger E-box binding homeobox 1 (ZEB1) genes have significant differences in ES, DS and DES of CRC (P < 0.01) (Table 3).
Table 3.
Screening of EMT-related DEGs in tumor single cells in CRC
| Genes | P* | BH |
|---|---|---|
| ELF3 | 1.12E−07 | 6.32E−07 |
| KRT7 | 0.105616809 | 0.179548575 |
| KRT8 | 1.36E−08 | 1.15E−07 |
| KRT18 | 0.003262755 | 0.009244472 |
| KRT19 | 2.13E−10 | 3.63E−09 |
| CDH1 | 0.122199768 | 0.188854187 |
| GRHL1 | 0.247310338 | 0.28028505 |
| FN1 | 1.46E−05 | 6.19E−05 |
| SERPINE1 | 0.046275357 | 0.098335134 |
| TCF4 | 2.06E−05 | 7.01E−05 |
| VIM | 0.208640718 | 0.253349443 |
| TGFB2 | 0.173304473 | 0.226628926 |
| TGFB3 | 0.164412474 | 0.226628926 |
| SIX1 | 0.782746466 | 0.789963446 |
| NOTCH2 | 0.789963446 | 0.789963446 |
| ZEB1 | 0.006163609 | 0.014968766 |
| ZEB2 | 0.060271568 | 0.113846294 |
*Kruskal–Wallis test
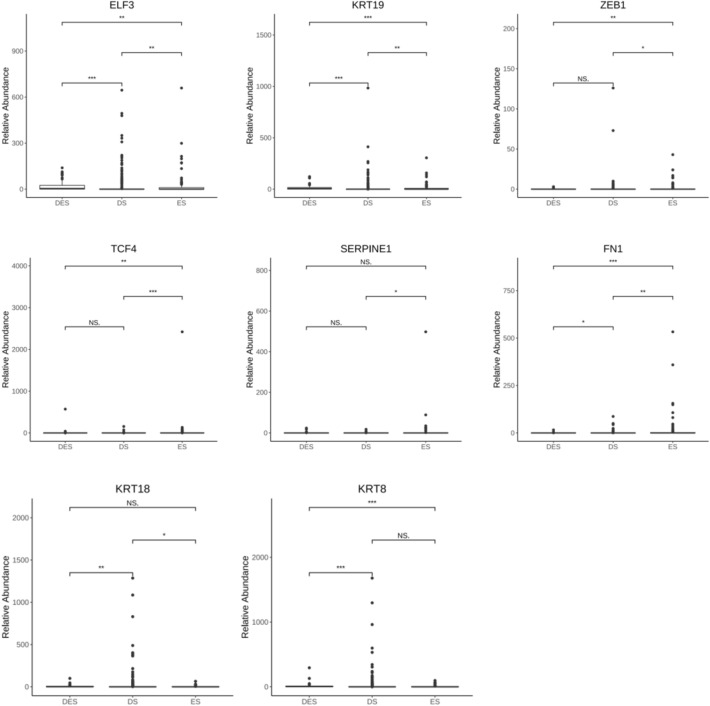
Furthermore, we used the Wilcox test to compare the roles of ELF3, KRT8, KRT18, KRT19, FN1, SERPINE1, TCF4 and ZEB1 genes in TCM syndrome differentiation of CRC with ES, DS and DES. The results were as shown in Fig. 4, ELF3, KRT18, KRT19 and FN1genes have significant difference between DES and DS or ES, ES and DS (P < 0.05 or P < 0.01 or P < 0.001); TCF4 and ZEB1 genes have significant difference between DES and ES or ES and DS (P < 0.05 or P < 0.01 or P < 0.001); KRT8 gene has significant differences between DES and ES or DS (P < 0.001); SERPINE1 gene has significant difference between ES and DS (P < 0.05), but there was no statistical difference between other TCM syndrome types (P > 0.05).
Fig. 4.
Correlation of ELF3, KRT8, KRT18, KRT19, FN1, SERPINE1, TCF4 and ZEB1 with Excess syndrome (ES), Deficiency syndrome (DS) and Excess-Deficiency syndrome (DES) in CRC. Analysis of variance, Wilcox test, *P < 0.05; **P < 0.01; ***P < 0.001; NS, no statistical significance
Differences in the evolution of tumor cell subpopulations in TCM syndromes of CRC
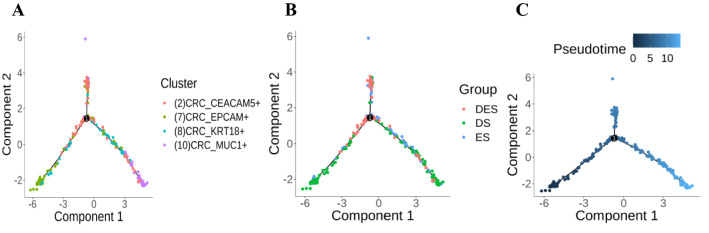
In order to clarify the relationship between TCM syndromes and the function evolution among tumor cell subpopulations in CRC, we used monocle (V2.16.0) to construct the cell chronological trajectory of the 4 subpopulations of tumor cells. The results were as shown in Fig. 5, the evolution trajectory of the subpopulation starts from the lower left corner to the lower right corner, from EPCAM+ cell subgroup, through CEACCAM5+ cell subgroup and KRT18+ cell subgroup, evolving MUC1+ cell subgroup. DES was mainly in the middle of the trajectory, ES was in the second half of the trajectory, and DS was all over the entire trajectory.
Fig. 5.
Monocle pseudotime analysis of tumor cell subpopulation evolution in CRC with TCM syndromes. A The evolutionary trajectory of CEACAM5 + , EPCAM + , KRT18 + and MUC1 + tumor cell subpopulations; B the distribution of Evidence (ES), Deficiency Syndrome (DS), and Deficiency and Excessive Syndrome (DES) on the evolutionary trajectory of tumor cell subpopulations; C the pseudotime trajectory of tumor cell subgroup evolution. The darker color is 'earlier', the pseudotime is 0; the lighter color is the more ‘late’, the pseudotime is greater
Discussion
CRC has been called “Intestinal accumulation”, “Accumulation” or “Intestinal mushroom” in TCM. Modern Chinese medicine physicians considered that the occurrence of CRC is due to the deficiency of Vital Qi, and the stagnation and accumulation of Evil Qi such as “Damp heat”, “Blood stasis” and “Poison stagnation” in the large intestine for long time, which resulted in the main pathogenesis in TCM of CRC is the “Deficiency of Vital Qi and Excess of Evil Qi” and “Combination of Deficiency and Excess” [26, 27]. Previous study on the distribution of clinical TCM syndromes in 760 cases of CRC has showed 565 in DS, 81 in ES, 52 in DES and 62 in no syndrome (NS), among DS is the most common syndrome type in CRC patients [27]. In this study, we used Smart-seq2 technology to explore the relationship between the heterogeneity of tumor cells and microcirculation and TCM syndromes in CRC through analyzing the classification of tumor cells and the proportion of each subgroup, the characteristic genes, gene co-expression network, the functional interpretation and the evolution of monocle functions.
Tumor heterogeneity is mainly divided into two types including inter-tumor heterogeneity and intra-tumor heterogeneity [28]. Almendro et al. [29] have classified intra-tumoral heterogeneity into cell, gene and functional heterogeneity, and temporal and spatial heterogeneity under the perspective of systems biology. Aaron S et al. [30] divided it into genetic and, phenotypic heterogeneity, cell signaling and pathway activity heterogeneity, and temporal and spatial heterogeneity. The manifestations of the tumor heterogeneity are complex and changeable. Different forms of heterogeneity may occur throughout the process of tumor formation and development. It needs to pay attention to is the relationship between the changes in molecular mechanisms and different functions in the process of the tumor heterogeneity. In this study, we found that the intra-tumor cells in CRC have 14 different cell subpopulations, 11 known cell lineages, and their genes have different functions and signal transduction pathways, as well as the development of different single cell lineages, showing obvious cells and genes and their phenotypic heterogeneity and cell signaling and pathway activity heterogeneity. Moreover, there were the different distributions of intra-tumor cell subpopulations in different TCM syndromes including ES, DS and DES, and the high expressions of MUC2 and REG4 in DES were mainly related to brain development, cytogenesis, MAPK and cAMP signaling pathways; the high expressions of COL1A2 and POSTN genes in ES were mainly related to vascular development, skeletal muscle development, and PI3k-Akt Pathway; SDPR and GPX1 genes were highly expressed in DS, mainly related to vomiting, platelet decomposition, and endocytosis. These results suggested the correlation of TCM syndromes classification and the heterogeneity of tumor cell subpopulations, and their characteristic genes, phenotypes and signaling pathways in CRC.
The tumor microenvironment includes all the components of non-cancerous solid tumors. It is a complex ecosystem composed of multiple cell types, which play different roles in tumor development, and is highly heterogeneous [31]. This study found 14 different cell subpopulations and 11 known cell lineages among 662 cells isolated from 11 primary CRC tumor tissues, including CRC cancer cells (CEACAM5 + , EPCAM + , KRT 18+ and MUC1+ cells), stem cells, immune cells (myeloid-monomacrophages, macrophages, DC, Goblet cells, T and B cells), fibroblast cells (ETM like and CAFs) and enterocyte. However, the proportion of cell subpopulations among ES, DS and DES in CRC is different. The main distribution of CEACAM5+ cells subpopulation was in DES; myeloid-monomacrophages, KRT 18+ cells, DC and MUC1+ cells subpopulations were the most distributed in DS; and goblet cells and fibroblasts subpopulations were the most distributed in ES. It was suggested that the TCM syndromes classification may be related to tumor cell subpopulations and the heterogeneity of tumor microenvironment in CRC.
Moreover, the pseudo-time analysis of monocle functional evolution showed that, 4-cell subpopulations of cancer cells in CRC with TCM syndromes appeared in different states and at different pseudo-times. Among, that of DES was mainly distributed in the middle part, ES was in the second part, and DS was all over the trajectory of pseudo-times, indicating that the DS run through the whole process of CRC tumor cell function evolution, the middle stage is the DES, and the later stage is mainly the ES. These results might provide scientific evidence to clarify the main pathogenesis of CRC in TCM, the "Deficiency of Vital Qi and Excess of Evil Qi" and "Combination of Deficiency and Excess" from the approach of tumor cell development.
CAFs are a kind of activated fibroblasts and are important stromal cells in the tumor microenvironment. In CRC, they interact with tumor cells to promote tumor occurrence, development and metastasis [32]. This study identified two cell subpopulations of fibroblasts including CAFs and EMT-like fibroblasts from CRC tumor tissues, which is consistent with the results of Li H et al. [16] in the previous research of tumor single cell in CRC. In this study, an EMT-related gene, fibronectin FN1 was highly expressed, and there was a significant difference between DES and ES, DES and DS or ES and DS in CRC. ZEB1 is a EMT transcription factor, it has been reported that the high expression of ZEB1 correlates with liver metastasis and poor prognosis in CRC [33, 34]. Moreover, TCF4, a transcription factor, participated in the regulation of Wnt/β-catenin signaling pathway in CRC cells [35, 36], and promotes adriamycin resistance and cell stemness by regulating the expression of EMT-related ZEB1 and ZEB2 [37]. This study found that the high expressions of TCF4 and ZEB1 genes have significant difference between DES and ES or ES and DS in CRC. It suggested that CAFs and EMT-related genes, the high expressions of FN1, ZEB1 and TCF4 in the tumor microenvironment may be involved in TCM syndromes classification in CRC.
Cytokeratin is the intermediate filament of the cell body, which can be divided into 20 different types according to its molecular weight and isoelectric point [38]. Among, KRT8, 18 and 19 have been reported to correlate with CRC [39], and their expression changes associate with progression towards neoplasia [40]. Moreover, KRT8 has been indicated epithelial to mesenchymal transition [41], and loss of K8 phosphorylation was also suggested to promote tumor migration and formation of metastasis [42]. KRT 18 and 19 increase in peripheral blood as soluble fragments in CRC [39]. In this study, Smart-seq2 analysis shows that, KRT8, 18 and 19 genes were enriched in the tumor tissues of CRC, and the high expressions of KRT18 and KRT19 genes were significant differences in DES and DS or ES, ES and DS classification, and the high expressions of KRT8 gene has significant differences between DES and ES or DES and DS in patients with CRC.
ELF3 is defined by their highly conserved ETS DNA binding domain and predominant epithelial-specific expression profile [43]. The ELF3 drives beta-catenin transactivation and associates with poor prognosis in CRC [44, 45]. We found that the high expression of ELF3 gene has a significant difference between DES and ES, DES and DS or ES and DS in CRC. SERPINE1, a clade E member of the serine protease inhibitor gene family and a prominent regulator of the pericellular proteolytic microenvironment [46]. In addition to play important roles in cell adhesion, migration and invasion, it has been reported to induce tumor vascularization and promote cell dissemination and tumor metastasis [47], and involved in the survival and prognosis of CRC [48, 49]. In this study, we found that, the high expressions of SERPINE1 has significant difference between ES and DS in CRC. The above results suggest that the high expressions of KRT19, KRT 18, KRT 8, ELF3 and SERPINE1 genes are potential candidates for identifying TCM syndrome types in CRC, which need to be confirmed by further researches.
The treatment based on TCM syndrome classification is the basic method and characteristic in the TCM treatment of CRC. Once understanding the tumor heterogeneity corresponding to different TCM syndromes, we will be able to utilize objective indicators related tumor heterogeneity to assist the diagnosis of TCM syndromes and treatments in CRC. Our results have shown that the different tumor heterogeneity may correlate with the different TCM syndromes in CRC, such as the distributions of intra-tumor cell subpopulations, genes and their function and signaling pathways, and gene co-expression network, DEGs and evolution of tumor cell subpopulations were also involved in TCM syndrome classification. Moreover, there were significant differences between the high expressions of MUC2, REG4, COL1A2, POSTN, SDPR, GPX1, ELF3, KRT8, KRT18, KRT19, FN1, SERPINE1, TCF4 and ZEB1 genes in Excess and Deficiency syndrome classification in CRC. These findings will be further validated by another approaches to finding a or a panel of potential biomarkers to assist the diagnosis of TCM syndromes, and also it may be helpful to further investigate the mechanism of occurrence and transformation of TCM syndromes, and finding therapeutic targets based on the TCM syndrome classification guiding treatments and/or assess the therapeutic effects and explore the mechanism of therapeutic effects in CRC.
Conclusions
Through the use of Smart-seq2 technology, this study found that CRC tumor cell subpopulations account for different proportions of ES, DS and DES, and have their characteristic genes, gene co-expression networks and their function and functional evolution. It suggested that the TCM syndrome classification in CRC may be related to the tumor heterogeneity and its microenvironment. In the future, the analyzed results of this study will be further validated, and single-cell multi-omics technology will be used to further explore the role of the tumor heterogeneity on the TCM syndromes classification and transformation in CRC.
Acknowledgements
Not applicable.
Abbreviations
- TCM
Traditional Chinese Medicine
- ES
Excessive syndrome
- DS
Deficiency syndrome
- DES
Deficiency and Excessive Syndrome
- NS
No syndrome
- CRC
Colorectal cancer
- TT
Tumor tissue
- DEGs
Differentially expressed genes
- scRNA-seq
Single cell RNA sequencing
- CTC
Circulating tumor cells
- FACS
Fluorescence activated cell sorting
- tSNE
T-distribution stochastic neighbor embedding
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GOBP
Ontology Biological Processes
- FDR
False discovery rate
- EMT
Epithelial–mesenchymal transition
- CAFs
Cancer-related fibroblasts
Authors’ contributions
YYL and SS drafted the manuscript. CZ, WWMZ, ML BZ and YL recruited patients, collected the clinical samples and information; YS and SZ carried out the detection; YYL, ZF and YL performed the data collection, analysis and interpretation. SS, SZ BJ and YL conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Key Program of National Natural Science Foundation of China (No.81330084), National Key Research and Development: Special Project for Research on the Modernization of Traditional Chinese Medicine (2018YFC1704204) and Shanghai Science and Technology Innovation Action Plan (20ZR1453700).
Availability of data and materials
All the data used to support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (Certificate No. 2014-345-41-01). All patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yiyu Lu, Chungen Zhou and Meidong Zhu contributed equally to this study
Contributor Information
Bin Jiang, Email: jbfirsth@aliyun.com.
Yunquan Luo, Email: yqluo09@sohu.com.
Shibing Su, Email: shibingsu07@163.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. 2017;23(28):5086–5096. doi: 10.3748/wjg.v23.i28.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusnir M, Cavalcante L. Inter-tumor heterogeneity. Hum Vaccines Immunother. 2012;8:1143–1145. doi: 10.4161/hv.21203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andor N, Trevor A, Graham TA, Marnix Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji CC. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22(1):105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao M, Zhong A, Patel N, Alur C, Vyas D. High throughput RNA sequencing utility for diagnosis and prognosis in colon diseases. World J Gastroenterol. 2017;23:2819–2825. doi: 10.3748/wjg.v23.i16.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang D, Peng M, Tang L, Ouyang J, Xiong F, Guo C, Tang Y, Zhou Y, Liao Q, Wu X, Wang H, Yu J, Li Y, Li X, Li G, Zeng Z, Tan Y, Xiong W. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res. 2021;40(1):81. doi: 10.1186/s13046-021-01874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, Liu S, Wu L, Jiang M, Hou Y. High throughput single cell RNA sequencing, bioinformatics analysis and applications. Adv Exp Med Biol. 2018;1068:33–43. doi: 10.1007/978-981-13-0502-3_4. [DOI] [PubMed] [Google Scholar]
- 10.Ma KY, Schonnesen AA, Brock A, Berg CVD, Eckhardt SG, Liu Z, Jiang N. Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight. 2019;4(4):e121387. doi: 10.1172/jci.insight.121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879–93.e13. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Xinglong WuX, Wen L, Tang F, Huang Y, Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26(3):304–19. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, Wang W, Yan J, Hu B, Guo H, Wang J, Gao S, Mao Y, Dong J, Zhu P, Xiu D, Yan L, Wen L, Qiao J, Tang F, Fu W. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362(6418):1060–3. doi: 10.1126/science.aao3791. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Song J, Zhao Z, Yang M, Chen M, Liu C, Ji J, Zhu D. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett. 2020;470:84–94. doi: 10.1016/j.canlet.2019.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, Wong M, Choi PJ, Wee LJK, Hillmer AM, Tan IB, Robson P, Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49(5):708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 16.Dai W, Zhou F, Tang D, Lin L, Zou C, Tan W, Dai Y. Single-cell transcriptional profiling reveals the heterogenicity in colorectal cancer. Medicine (Baltimore) 2019;98(34):e16916. doi: 10.1097/MD.0000000000016916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S. Data analysis in single-cell transcriptome sequencing. Methods Mol Biol. 2018;1754:311–326. doi: 10.1007/978-1-4939-7717-8_18. [DOI] [PubMed] [Google Scholar]
- 18.Tieng FYF, Baharudin R, Abu N, Mohd Yunos RI, Lee LH, Ab Mutalib NS. Single cell transcriptome in colorectal cancer-current updates on its application in metastasis, chemoresistance and the roles of circulating tumor cells. Front Pharmacol. 2020;27(11):135. doi: 10.3389/fphar.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Xu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, Konno H, Guo X, Ye Y, Gao S, Wang S, Hu X, Ren X, Shen Z, Ouyang W, Zhang Z. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Zhong Y, Zhuang Z, Xie J, Lu Y, Huang C, Sun Y, Wu L, Yin J, Yu H, Jiang Z, Wang S, Wang C, Zhang Y, Huang Y, Han C, Zhong Z, Hu J, Ouyang Y, Liu H, Yu M, Wei X, Chen D, Huang L, Hou Y, Lin Z, Liu S, Ling F, Yao X. Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer. Clin Transl Med. 2021;11(1):e253. doi: 10.1002/ctm2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, Navin NE. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017;27(8):1287–1299. doi: 10.1101/gr.209973.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA, Pronk A, Smakman N, Van Gorp J, Anderson E, Gamble SJ, Alder C, Van de Wetering M, Campbell PJ, Stratton MR, Clevers H. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556(7702):457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 23.Su SB, Lu A, Li S, Jia W. Evidence-based ZHENG: a traditional Chinese medicine syndrome. Evid Based Complement Alternat Med. 2012;2012:246538. doi: 10.1155/2012/246538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Department of Health, Ministry of Health Guidelines for diagnosis and treatment of colorectal cancer (2010 Edition) China Contin Med Educ. 2011;2011(49):97–104. [Google Scholar]
- 25.Ministry of Health of the People’s Republic of China. Guidelines for Clinical Research on New Chinese Medicine. Series 3, 1997.
- 26.Ji Q, Wang WH, Luo Y, Cai F, Lu Y, Deng W, Li Q, Su S. Characteristic proteins in the plasma of postoperative colorectal and liver cancer patients with Yin deficiency of liver-kidney syndrome. Oncotarget. 2017;8(61):103223–103235. doi: 10.18632/oncotarget.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang MD, Chen XL, Hu XQ, Xie XZ, Zhou CG, Jiang B, Qing Ji, Qi Li, Wang P, Meng ZQ, Wang WH, Su SB. TCM syndromes distribution in colorectal cancer and the correlation with western medicine treatment and clinical laboratory indicators. World J Tradit Chin Med. 2019;5(2):81–7. [Google Scholar]
- 28.Blanco-Calvo M, Concha Á, Figueroa A, Garrido F, Valladares-Ayerbes M. Colorectal cancer classification and cell heterogeneity: a systems oncology approach. Int J Mol Sci. 2015;16(6):13610–13632. doi: 10.3390/ijms160613610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 30.Meyer AS, Heiser LM. Systems biology approaches to measure and model phenotypic heterogeneity in cancer. Curr Opin Syst Biol. 2019;17:35–40. doi: 10.1016/j.coisb.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HO, Park WY. Single-cell RNA-Seq unveils tumor microenvironment. BMB Rep. 2017;50(6):283–284. doi: 10.5483/BMBRep.2017.50.6.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 33.Vu T, Datta PK. Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers (Basel) 2017;9(12):171. doi: 10.3390/cancers9120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-TCF signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 36.Goretsky T, Bradford EM, Ye Q, Lamping OF, Vanagunas T, Moyer MP, Keller PC, Sinh P, Llovet JM, Gao T, She QB, Li L, Barrett TA. Beta-catenin cleavage enhances transcriptional activation. Sci Rep. 2018;8(1):671. doi: 10.1038/s41598-017-18421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun S, Yang X, Qin X, Zhao Y. TCF4 promotes colorectal cancer drug resistance and stemness via regulating ZEB1/ZEB2 expression. Protoplasma. 2020;257(3):921–930. doi: 10.1007/s00709-020-01480-6. [DOI] [PubMed] [Google Scholar]
- 38.Nordgård O, Singh G, Solberg S, Jørgensen L, Halvorsen AR, Smaaland R, Brustugun OT, Helland Å. Novel molecular tumor cell markers in regional lymph nodes and blood samples from patients undergoing surgery for non-small cell lung cancer. PLoS ONE. 2013;8(5):e62153. doi: 10.1371/journal.pone.0062153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polari L, Alam CM, Nyström JH, Heikkilä T, Tayyab M, Baghestani S, Toivola DM. Keratin intermediate filaments in the colon: guardians of epithelial homeostasis. Int J Biochem Cell Biol. 2020;129:105878. doi: 10.1016/j.biocel.2020.105878. [DOI] [PubMed] [Google Scholar]
- 40.Evans CA, Rosser R, Waby JS, Noirel J, Lai D, Wright PC, Williams EA, Riley SA, Bury JP, Corfe BM. Reduced keratin expression in colorectal neoplasia and associated fields is reversible by diet and resection. BMJ Open Gastroenterol. 2015;2(1):e000022. doi: 10.1136/bmjgast-2014-000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knösel T, Emde V, Schlüns K, Schlag PM, Dietel M, Petersen I. Cytokeratin profiles identify diagnostic signatures in colorectal cancer using multiplex analysis of tissue microarrays. Cell Oncol. 2006;28:167–175. doi: 10.1155/2006/354295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuuchi E, Semba S, Kodama Y, Yokozaki H. Down-modulation of keratin 8 phosphorylation levels by PRL-3 contributes to colorectal carcinoma progression. Int J Cancer. 2009;124:1802–1810. doi: 10.1002/ijc.24111. [DOI] [PubMed] [Google Scholar]
- 43.Luk IY, Reehorst CM, Mariadason JM. ELF3, ELF5, EHF and SPDEF transcription factors in tissue homeostasis and cancer. Molecules. 2018;23(9):2191. doi: 10.3390/molecules23092191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang JL, Chen ZF, Chen HM, Wang MY, Kong X, Wang YC, Sun TT, Hong J, Zou W, Xu J, Fang JY. Elf3 drives beta-catenin transactivation and associates with poor prognosis in colorectal cancer. Cell Death Dis. 2014;5(5):e1263. doi: 10.1038/cddis.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakarai C, Osawa K, Matsubara N, Ikeuchi H, Yamano T, Okamura S, Kamoshida S, Tsutou A, Takahashi J, Ejiri K, Hirota S, Tomita N, Kido Y. (2012) Significance of ELF3 mRNA expression for detection of lymph node metastases of colorectal cancer. Anticancer Res. 2012;32(9):3753–8. [PubMed] [Google Scholar]
- 46.Longo CM, Higgins PJ. Molecular biomarkers of Graves’ ophthalmopathy. Exp Mol Pathol. 2019;106:1–6. doi: 10.1016/j.yexmp.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Wei X, He J, Tian X, Yuan S, Sun L. Plasminogen activator inhibitor-1 in cancer research. Biomed Pharmacother. 2018;105:83–94. doi: 10.1016/j.biopha.2018.05.119. [DOI] [PubMed] [Google Scholar]
- 48.Zeng C, Chen Y. HTR1D, TIMP1, SERPINE1, MMP3 and CNR2 affect the survival of patients with colon adenocarcinoma. Oncol Lett. 2019;18(3):2448–2454. doi: 10.3892/ol.2019.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Li C, Dong L, Chen X, Fan R. Identification and verification of three key genes associated with survival and prognosis of COAD patients via integrated bioinformatics analysis. Biosci Rep. 2020;40(9):BSR20200. doi: 10.1042/BSR20200141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data used to support the findings of this study are available from the corresponding author upon reasonable request.